Background: Bacterial virulence-associated type III secretion system (T3SS) assembly requires a dedicated enzyme to penetrate peptidoglycan (PG).
Results: We structurally characterized a T3SS PG-lytic enzyme, identified catalytically important residues, and characterized its activity.
Conclusion: The active site is similar to lysozymes and lytic transglycosylases and interaction with the T3SS enhances activity.
Significance: Structural information is critical for development of drugs targeting T3SS PG-lytic enzymes.
Keywords: Bacterial Pathogenesis, Cell Wall, Peptidoglycan, Type III Secretion System (T3SS), X-ray Crystallography
Abstract
The Gram-negative bacterium enteropathogenic Escherichia coli uses a syringe-like type III secretion system (T3SS) to inject virulence or “effector” proteins into the cytoplasm of host intestinal epithelial cells. To assemble, the T3SS must traverse both bacterial membranes, as well as the peptidoglycan layer. Peptidoglycan is made of repeating N-acetylmuramic acid and N-acetylglucosamine disaccharides cross-linked by pentapeptides to form a tight mesh barrier. Assembly of many macromolecular machines requires a dedicated peptidoglycan lytic enzyme (PG-lytic enzyme) to locally clear peptidoglycan. Here we have solved the first structure of a T3SS-associated PG-lytic enzyme, EtgA from enteropathogenic E. coli. Unexpectedly, the active site of EtgA has features in common with both lytic transglycosylases and hen egg white lysozyme. Most notably, the β-hairpin region resembles that of lysozyme and contains an aspartate that aligns with lysozyme Asp-52 (a residue critical for catalysis), a conservation not observed in other previously characterized lytic transglycosylase families to which the conserved T3SS enzymes had been presumed to belong. Mutation of the EtgA catalytic glutamate, Glu-42, conserved across lytic transglycosylases and hen egg white lysozyme, and this differentiating aspartate diminishes type III secretion in vivo, supporting its essential role in clearing the peptidoglycan for T3SS assembly. Finally, we show that EtgA forms a 1:1 complex with the building block of the polymerized T3SS inner rod component, EscI, and that this interaction enhances PG-lytic activity of EtgA in vitro, collectively providing the necessary strict localization and regulation of the lytic activity to prevent overall cell lysis.
Introduction
Diarrheal diseases account for ∼18% of deaths of children under the age of 5 years (1). In 2011, one of the most prevalent diarrhea inducing pathogens, enteropathogenic Escherichia coli (EPEC),4 caused an estimated 79,000 deaths in this age category of children (2). Following ingestion, EPEC adheres intimately to the epithelial cells of the small intestine and causes effacement of microvilli and formation of actin pedestals beneath the bacterium (3–6). Once adhered to the intestinal epithelium, EPEC assembles a virulence-specific protein transport system (referred to as the type III secretion system or T3SS). The syringe-like T3SS injects more than 20 different virulence proteins from the EPEC cytoplasm directly into the host cytoplasm. Critical for pathogenesis, delivered effectors manipulate host cell processes such as cytoskeleton dynamics, inflammatory signaling pathways, cell cycle progression, and apoptosis (7). The T3SS machinery is also conserved among other Gram-negative pathogens such as Shigella, Salmonella, Yersinia, and Pseudomonas, underscoring the importance of understanding such a key virulence factor at a molecular level.
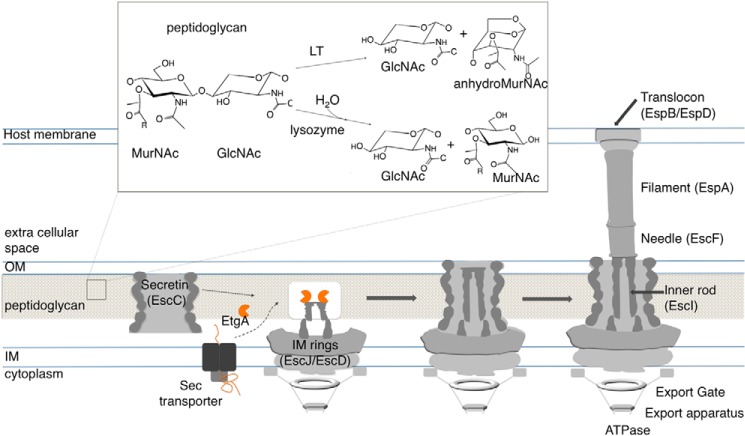
The EPEC T3SS (as well as effector proteins and their chaperones) is encoded in a pathogenicity island called LEE (locus of enterocyte effacement) that is arranged in five polycistronic operons (LEE1–LEE5). The T3SS is composed of more than 20 proteins that oligomerize in a highly regulated and hierarchical fashion to form a contiguous channel through both bacterial membranes and the host membrane. The first components to assemble are the cytoplasmic export apparatus, inner membrane basal body rings, and outer membrane embedded secretin (Fig. 1). The export apparatus, which includes an ATPase, export gate, and autoprotease, shuttles T3SS substrates from the bacterial cytoplasm into the secretion channel. The basal body, which spans the bacterial membranes, is formed by concentric inner membrane rings composed of EscJ and EscD, and an outer membrane-embedded secretin, EscC (8–10). The inner rod component (EscI), which shares sequence homology with the well characterized filamentous extracellular needle (EscF), is presumed to create a channel through the basal body inner and outer membrane rings (11, 12), leading to the subsequent needle appendage that continues the channel through the extracellular space. In EPEC, the needle in turn is capped by a filamentous extension composed of EspA (13–17) that facilitates the span of the T3SS across the microvilliated surface of the infected host gut epithelial cells. In the final step of assembly, the translocon (EspB and EspD) assembles at the tip of the EspA filament and inserts into the host membrane to form a pore (18) for direct delivery of virulence effectors into the host cell.
FIGURE 1.
Schematic overview of T3SS apparatus components and role of EtgA during assembly. EtgA is transported to the periplasm by the Sec secretion system. It interacts with the inner rod, EscI, in the bacterial periplasm and locally clears peptidoglycan during assembly. After assembly of the inner rod and needle, the EspA filament and translocon form. Inset, peptidoglycan is composed of repeating MurNAc-GlcNAc disaccharide. Both lytic transglycosylases and lysozymes cleave the β-1,4 glycosidic linkage, but lysozyme uses a hydrolysis mechanism to produce MurNAc and GlcNAc, whereas lytic transglycosylases produce 1,6-anhydromuramoyl product along with GlcNAc. OM, outer membrane layer; IM, inner membrane layer.
Assembly of molecular transport systems (such as the T3SS) that span both bacterial membranes requires local rearrangements in the peptidoglycan layer (19–22). Peptidoglycan is composed of glycan strands of β-1,4-glycosidic linked GlcNAc and N-acetylmuramic acid (MurNAc) disaccharide, which are cross-linked by short 4–5-residue peptides to create a mesh-like layer with an average pore size between 2 and 7 nm (23, 24). Proteins smaller than ∼50 kDa can diffuse freely through the peptidoglycan, but the peptidoglycan layer acts as a barrier to larger proteins and protein complexes (23). The periplasmic region of the T3SS is ∼17 nm in diameter at the widest point, and most species encode a T3SS-specific “specialized” PG-lytic enzyme. PG-lytic enzymes such as lytic transglycosylases degrade peptidoglycan by cleaving the β-1,4 glycosidic linkage between GlcNAc and MurNAc disaccharide, releasing a 1,6-anhydromuramoyl product (Fig. 1, inset). The 1,6-anhydromuramoyl product released by lytic transglycosylases is recycled for production of new peptidoglycan (25). Lysozymes have the same substrate specificity as lytic transglycosylases; however, the reaction catalyzed by lysozyme uses a water molecule to hydrolyze the glycosidic linkage between GlcNAc and MurNAc disaccharide, releasing GlcNAc and MurNAc products (Fig. 1, inset).
The EPEC LEE pathogenicity island encodes a T3SS-specialized PG-lytic enzyme, EtgA, located between the LEE1 operon and the grlR (global regulator of LEE repressor) and grlA (global regulator of LEE activator) operon (11). EtgA was classified as a putative family 1A lytic transglycosylase because of the presence of conserved sequence motifs (11). The N terminus of EtgA has a signal peptide that targets it for transport to the periplasm by the Sec-dependent general secretory pathway (26). An early study that used a yeast two-hybrid assay to survey interactions between LEE encoded proteins suggested that EtgA binds the inner rod subunit, EscI (27). Peptidoglycan degrading activity of EtgA, as well as T3SS PG-lytic enzyme homologs from Salmonella and Shigella (IagB and IpgF, 41 and 35% sequence identity, respectively), was shown by zymogram, and mutation of a conserved glutamate (Glu-42 in EPEC) abrogates activity (26, 28). Deletion of etgA from the EPEC homolog Citrobacter rodentium, a mouse bacterial pathogen similar to EPEC, impedes T3S and attenuates virulence (29). Accordingly, deletion of etgA from EPEC decreases T3SS assembly and reduces T3SS-dependent hemolysis of erythrocytes (26). However, it has been reported that disruption of the Salmonella ipgF gene had no effect on HeLa cell invasion, although this may not be reflective of the situation in the naturally targeted epithelial cells (30).
E. coli has at least seven known lytic transglycosylases (Slt70, MltA, MltB, MltC, MltD, MltE, and MltF) that play a role in peptidoglycan remodeling during cell growth and division (31). EtgA shares sequence similarity with consensus motifs I, II, and III of the catalytic domain of family 1A lytic transglycosylase Slt70, which has been well characterized at the molecular level (32–34). Despite low sequence homology, the catalytic domain of Slt70 is structurally similar to goose-type lysozyme (32, 33). Both lytic transglycosylases (such as Slt70) and goose-type lysozyme lack the catalytic aspartate (Asp-52) that is generally found in lysozymes, such as the prototypical hen egg white lysozyme. In the proposed lytic transglycosylase mechanism, the catalytic Glu acts as a general acid to donate a proton to the glycosydic oxygen, resulting in bond cleavage and formation of an oxocarbenium ion transition state, stabilized by the formation of an oxozolinium intermediate. The next step of the lytic transglycosylase mechanism involves an intramolecular nucleophilic attack by the C6 hydroxyl of the MurNAc (probably aided by abstraction of a proton from the hydroxyl group by the oxyanionic form of the catalytic Glu) on the C1 carbon of the oxozolinium intermediate, resulting in formation of the 1,6-anhydro product (21, 32–34).
Because activity of PG-lytic enzymes can be fatal to the bacterium, their expression, localization, and activity must be tightly regulated. Expression of EtgA in EHEC is negatively regulated by the presence of GrlA, an activator of T3SS gene expression (35). Presumably, this allows for synthesis of T3SS components prior to transport of EtgA to the periplasm (35). Additionally, activity of specialized PG-lytic enzymes may be spatially regulated by physical interaction with other components of the molecular transport system. For instance, VirB1, a PG-lytic enzyme associated with the type IV secretion system (involved in conjugation, DNA uptake, and effector translocation), interacts with several apparatus components (36). Likewise, the interaction of EtgA with the T3SS inner rod, EscI, may spatially restrict the activity of EtgA. Interestingly, the flagellar secretion system, which is evolutionarily related to the T3SS, has a modular PG-lytic enzyme called FlgJ (Salmonella), that has PG-lytic activity in its C-terminal domain, whereas its N-terminal domain forms the rod cap itself, ensuring that peptidoglycan is cleared as the rod assembles (37, 38). Furthermore, lytic transglycosylases involved in cell wall remodeling during growth and division are lipoproteins that are spatially restricted by association with the periplasmic leaflet of the outer membrane (21, 39).
In this study, we obtained the first known structure of a T3SS-specific PG-lytic enzyme (conserved with type II and IV secretion system and type IV pili-associated PG-lytic enzymes), encompassing the catalytic core of EPEC EtgA. Based on structural similarity to other lytic transglycosylases and lysozymes, we mutated putative catalytic residues in the EtgA active site and tested the effect of each mutant on type III secretion. Additionally, we expressed and co-purified recombinant EtgA with its binding partner, the inner rod protein EscI, and determined the stoichiometry of the EscI·EtgA complex. Finally, we show that EscI stabilizes EtgA and enhances its peptidoglycan degrading activity. Secretion system-associated PG-lytic enzymes such as EtgA are attractive drug targets, because they represent one of the few enzymatic components of the T3SS. Drugs that target a component of the pathogen required for virulence, but not for replication per se, may lessen selective pressure and emergence of resistance factors (40). Structural information for the specialized virulence-associated T3SS PG-lytic enzymes will be critical for developing drugs that specifically target these enzymes.
EXPERIMENTAL PROCEDURES
Cloning of Expression Vectors for Crystallography
The coding region of EtgA residues 19–152 was amplified from EPEC genomic DNA and cloned into pET 21a vector (untagged), as well as pET28a vector with an N-terminal His10 tag by restriction-free cloning. EtgA mutants were generated by QuikChange PCR. The coding region of EscI residues 24–137 was amplified from EPEC genomic DNA and cloned by restriction-free PCR into pET 28a vector with an N-terminal His10 tag.
Protein Expression and Purification
For protein production for crystallization trials, EtgA (19–152) D60N-pET21 was co-transformed with N-terminally His10-tagged EscI (24–137)-pET28 in E. coli BL21 (λDE3). To produce selenomethionine-labeled protein, cultures were grown in M9 medium (supplemented with 1 mm MgSO4, 0.1 mm CaCl2, 0.01 mm FeCl3, 1 mg of thiamine, and 1% glucose) with 50 μg/ml kanamycin and 100 μg/ml ampicillin at 37 °C to an A600 of 0.7, and then 0.05 g of selenomethionine was added per liter of culture. After 30 min of growth with selenomethionine, the culture was induced with 1 mm IPTG for 18 h at 20 °C. Cells were harvested by centrifugation, and cell pellets were resuspended in lysis buffer containing 20 mm Hepes, pH 7.5, 500 mm NaCl, 50 mm imidazole, 1 mm BME, and one complete protease inhibitor mixture tablet (Roche) and lysed by French press. Cell lysate was centrifuged at 45,000 rpm for 1 h at 4 °C. The supernatant was loaded on a 1-ml nickel-nitrilotriacetic acid column, which was pre-equilibrated with wash buffer (20 mm Hepes, pH 7.5, 500 mm NaCl and 50 mm imidazole, 1 mm BME). EtgA (19–152) D60N and EscI (24–137) complex was eluted with elution buffer (20 mm Hepes, pH 7.5, 500 mm NaCl, 500 mm imidazole, 1 mm BME). Elution fractions were pooled and concentrated and loaded onto a Superdex 75 10/30 column equilibrated with buffer (Hepes, pH 7.5, 500 mm NaCl, 5 mm BME). EtgA (19–152) D60N and EscI (24–137) complex eluted as a single peak and protein-containing fractions were pooled and concentrated to 20 mg/ml for crystallization trials.
To express EtgA·EscI complex for the in vitro activity assay, WT EtgA (19–152)-pET21 (or the catalytic mutants E42A or D60N) was co-transformed with N-terminally His10-tagged EscI (24–137)-pET28 in E. coli BL21 (λDE3). Cultures were grown in 2 liters of LB medium with 50 μg/ml kanamycin and 100 μg/ml ampicillin and induced with 1 mm IPTG at an A600 of 0.6 at 20 °C for 20 h. Cells were harvested and lysed, and protein was purified as described above, but without BME. To produce EtgA for activity assays, WT EtgA (19–152)-pET28 (or the catalytic mutants E42A or D60N) was transformed into E. coli BL21 (λDE3) and expressed as above. Cells were harvested and lysed, and EtgA was purified on a 1-ml nickel-nitrilotriacetic acid column, as described above. EtgA was prepared fresh, immediately before the activity assay.
Crystallization of EtgA
EtgA (19–152) D60N·EscI (24–137) complex was set up extensively for crystallization but was recalcitrant to crystallization. Limited in situ proteolysis (41) of the EtgA (19–152) D60N·EscI (24–137) complex using a 1:1000 molar ratio of chymotrypsin:EtgA (19–152) D60N·EscI (24–137) complex was set up in sitting drop vapor diffusion and produced crystals in multiple conditions containing 2-propanol as the precipitant. Crystals typically appeared after 3 days in the optimized condition (0.1 m imidazole, pH 7.3–7.5, with 20–23% 2-propanol) at room temperature. Crystals were cryoprotected with a solution of mother liquor with 25% glycerol and flash frozen in liquid nitrogen.
Data Collection, Structure Determination, and Refinement
Crystals were screened, and multiwavelength anomalous dispersion data were collected at the Lawrence Berkeley National Laboratory Advanced Light Source on Beamline 8.2.1 at a peak wavelength of 0.9785 Å, an inflection wavelength of 0.9797 Å, and a high energy remote wavelength of 0.9611 Å. Data were processed with xia2 using XDS to index all frames, XSCALE to scale, and Aimless to merge (42). Phases were obtained using autoSHARP, which located one selenomethionine (43). The structure was built, refined, and validated using autoSHARP, Coot, Refmac, and the PDB_REDO server (43–47). The figures were made using the UCSF Chimera package (48).
Scanning Electron Microscopy of BL21 E. coli Expressing EtgA
BL21 (DE3) E. coli was transformed with WT EtgA (19–152)-pET27, with a PelB signal sequence for export to the periplasm. Cells were grown in LB medium until an A600 of 0.6, and then induced with 0.1 mm IPTG. Cells were imaged by scanning electron microscopy both with IPTG, 1 h after induction, and without IPTG at the same time point. Samples were prepared by filtering onto a 0.4-μm nucleopore filter in a Swinex holder, in fixative (4% EM grade formaldehyde, 2.5% glut), followed by microwave processing. The samples were removed from the filter holder, processed in an inverted position through postfixation in buffered osmium tetroxide (1%) washes, and alcohol dehydration prior to drying. Following drying, the samples were mounted on aluminum scanning electron microscopy stubs using a conductive (silver) paste, dried, and sputter-coated with gold. Samples were imaged with a Hitachi S-4700 field emission scanning electron microscope.
Size Exclusion Chromatography-Multiangle Light Scattering
Purified EtgA (19–152)·EscI (10–137) complex at a concentration of 2 mg/ml was injected over a Superdex 75 HR 10/30 column (GE Healthcare) pre-equilibrated in buffer (20 mm Hepes, pH 7.5, 500 mm NaCl) and analyzed with a DAWN® HELEOS-II® 18-angle light-scattering detector and Optilab® T-rEXTM differential refractometer (Wyatt Technology). All detectors were normalized using a 2 mg/ml monomeric bovine serum albumin standard. Data analysis was performed using ASTRA 6 software (Wyatt Technology).
Cloning, Expression, and Purification of EHEC Inner Rod Protein EprJ
The coding region of EprJ was generated by PCR from EHEC genomic DNA and inserted into pMAL-c2x vector by restriction-free cloning. pMAL-c2X-EprJ plasmid was transformed into BL21 (DE3) cells and expressed as described above for EtgA. Cells were resuspended in buffer (20 mm Hepes, pH 7.5, 500 mm NaCl), lysed by French press, and centrifuged at 45,000 rpm for 45 min. Lysate was passed over 3 ml of amylose resin and then resin was washed with 15 ml of wash buffer (20 mm Hepes, pH 7.5, 500 mm NaCl). MBP-EprJ fusion protein was eluted with 5 ml of elution buffer (20 mm Hepes, pH 7.5, 500 mm NaCl, 250 mm maltose). The MBP tag was cleaved overnight with Factor Xa protease. EprJ was purified over a Superose 6 column in buffer (20 mm Hepes, pH 7.5, 150 mm NaCl) and eluted in the void volume of the column.
Imaging of EprJ by Negative Stain Electron Microscopy
Purified EprJ (1 μl of 0.3 mg/ml) was applied to a glow-discharged carbon coated transmission electron microscopy grid (Ted Pella, Inc.). After drying, the grid was stained with 1 μl of Nano-W® (methylamine tungstate; Nanoprobes) for 30 s. Excess stain was removed by blotting with a Whatman paper. Grids were imaged with a FEI Tecnai G2 200-kV transmission electron microscope.
Generation of C. rodentium and EPEC etgA Deletion Mutants for T3SS Secretion Assay
An in-frame deletion mutant of etgA, formerly known as rorf3, was generated in C. rodentium strain DBS100 and characterized as previously described (29). An etgA deletion mutant was also generated in the streptomycin-resistant derivative of EPEC O127:H6 strain E2348/69 using the sacB gene-based allelic exchange method and the suicide vector pCVD442 (49). A 4044-bp DNA fragment containing the EPEC etgA gene, as well as ∼1.8 kb of flanking sequences on both sides, was amplified by PCR using primers EPescT-F (5′-ATGAATGAGATAATGACGGTCATAGTATC-3′) and EPcesD-1 (5′-CTCAATGACCTTCATTCTTATGCC-3′). The PCR product was cloned into pCRII-TOPO (Invitrogen), and the resultant plasmid was used as template for inverse PCR using primers EPetgA-RD (5′-GCTAGCTCAGAAGGCAATACGCAATG-3′, NheI) and EPetgA-DF (5′-GCTAGCTGAAATGAGAATGATACTCAG-3′, NheI) to create an internal deletion in the etgA gene. The inverse PCR product was digested with NheI, gel-purified, treated with T4 DNA ligase, and transformed into E. coli strain DH10B. The DNA fragment containing the etgA gene with the internal deletion and its flanking regions was then subcloned as a SacI/XbaI fragment into the suicide vector pCVD442 to generate pCVD-ΔEPetgA. The etgA gene in the suicide vector has an internal deletion from nucleotides 37 to 384 (∼76% of the coding region), and an NheI site was introduced at the deletion site. Plasmid pCVD-ΔEPetgA was transformed into E. coli strain SM10λpir by electroporation and introduced into EPEC strain E2348/69 by conjugation. After sucrose selection as previously described (49), EPEC colonies resistant to sucrose and streptomycin but sensitive to ampicillin, indicative of allelic exchange and loss of the suicide vector, were screened by colony PCR for deletion of etgA. The obtained EPEC etgA mutant was further verified by PCR.
Complementation Constructs for EPEC and C. rodentium etgA Deletion Mutants
Constructs expressing EPEC or C. rodentium etgA in the plasmid pACYC184 (New England Biolabs), which has a moderate copy number of 30–50/cell in bacteria, were toxic to EPEC and C. rodentium when grown under type III secretion inducing conditions, and caused partial bacterial lysis, probably because of the peptidoglycan-hydrolyzing activities of EtgA. We next generated EtgA complementation constructs in the vector pZS*24MCS (EXPRESSYS), which has a much lower copy number of three to five per cell than pACYC184. The coding region of EPEC etgA, as well as its 159-bp upstream promoter region, was amplified by PCR using primers EtgAcom-1 (5′-GGATCCATTTGTTCTATCCATAAGC-3′, BamHI) and EPetgAcom-2 (5′-GTCGACGATTCGTATTGCGATAGACCTTG-3′, SalI). Likewise, the C. rodentium etgA gene and its 149-bp promoter region were amplified using primers EtgAcom-1 (5′-GGATCCATTTGTTCTATCCATAAGC-3′, BamHI) and CRetgAcom-2 (5′-GTCGACGATCCGTATTGCAATGGATATTG-3′, SalI). These PCR products were digested with BamHI and SalI, gel-purified, and ligated into BamHI/SalI-treated pZS*24MCS to generate pZS*-EPetgA and pZS*-CRetgA, respectively. The constructs were confirmed by DNA sequencing and transformed via electroporation into EPEC and/or C. rodentium etgA deletion mutants for complementation. The EPEC and Citrobacter ΔetgA mutants can be complemented by either pZS*-EPetgA or pZS*-CRetgA, indicating that EPEC etgA and Citrobacter etgA are functionally exchangeable. Based on this observation, we were able to test variants of EPEC etgA generated by site-directed mutagenesis in the Citrobacter ΔetgA mutant, which has a much more pronounced type III secretion defect than the EPEC ΔetgA mutant.
Type III Secretion Assay for EPEC and C. rodentium
EPEC and C. rodentium strains were grown overnight in LB broth containing appropriate antibiotics at 37 °C in a shaker at 225 rpm. The cultures were then diluted 1:40 into 3 ml of prewarmed DMEM (HyClone) supplemented with 4500 mg/liter glucose, 4 mm l-glutamine, and 110 mg/liter sodium pyruvate without any antibiotics in a 6-well tissue culture plate (Corning Inc.) and grown statically at 37 °C for 6 h in a tissue culture incubator containing 5% CO2 (v/v) to induce type III secretion. The cultures were centrifuged at 16,100 × g for 10 min to pellet the bacteria, and the bacterial pellet was resuspended in SDS-PAGE sample buffer to generate whole cell lysates. The bacterial growth medium supernatant was collected and passed through a Millex-GV 0.22-μm filter unit (Millipore) to remove any remaining bacteria, and the proteins in the supernatant were precipitated with 10% (v/v) TCA. After centrifugation at 16,100 × g for 30 min, the protein pellet was dried in air and dissolved in SDS-PAGE sample buffer, with the residual TCA neutralized with 0.5 μl of saturated Tris. The amount of the sample buffer used to resuspend the bacterial pellet or dissolve the precipitated proteins was normalized according to the A600 values of the cultures to ensure equal loading of the samples.
Thermal Stability Assay
EtgA (19–152) or EtgA (19–152)·EscI (24–137) complex thermostability was measured as a function of its temperature-dependent aggregation by differential static light scattering (StarGazer-2; Harbinger Biotechnology and Engineering Corporation) according to the method of Vedadi et al. (50). Briefly, 10 μl of 0.4 mg/ml protein in 100 mm Hepes, pH 7.5, 300 mm NaCl was heated from 25 to 85 °C at a rate of 1 °C/min in individual wells of a clear-bottomed 384-well plate (Corning 3540, Rochester, NY). To test the importance of the EtgA disulfide bond for protein stability, 0.4 mg/ml EtgA (19–152)·EscI (24–137) complex (in 100 mm Hepes, pH 7.5, 300 mm NaCl) was incubated with a serial dilution of reducing agent (either DTT or TCEP) ranging in concentration from 1 μm to 10 mm. Protein aggregation, as a measure of the intensity of scattered light, was scanned every 30 s with a CCD camera. The integrated intensities were plotted against temperature, where the inflection point of each fitted curve, using a Boltzmann regression, was defined as the aggregation temperature, Tagg. The Kagg for DTT and TCEP was determined by a four-parameter logistic curve using SigmaPlot software (Systat Software Inc.).
EtgA Activity Assay
EtgA activity assays were performed using an Enzchek® lysozyme assay kit. In summary, 450 μg/ml of peptidoglycan labeled with fluorescein (Enzchek®) was serially diluted in half with 50 mm Hepes pH 7 buffer to create a series of 12 reaction mixtures. A final concentration of 1 μm of protein (WT EtgA·EscI, EtgA E42A·EscI, EtgA D60N·EscI, WT EtgA, EtgA E42A, or EtgA D60N) was added to each reaction. As a separate negative control, an equal volume of buffer was added in place of protein to a peptidoglycan dilution series. All reactions were done in triplicate with a final reaction volume of 10 μl and transferred to a nonbinding surface 384-well low volume plate (Corning® 3820). Fluorescence was measured at 37 °C with an excitation/emission wavelength of 485/530 nm using a Bio-Tek® Synergy H4 microplate reader. Reactions were read every 30 s for 1 h, and data were analyzed using SigmaPlot software.
RESULTS
Structural Characterization of the Catalytic Core of EtgA
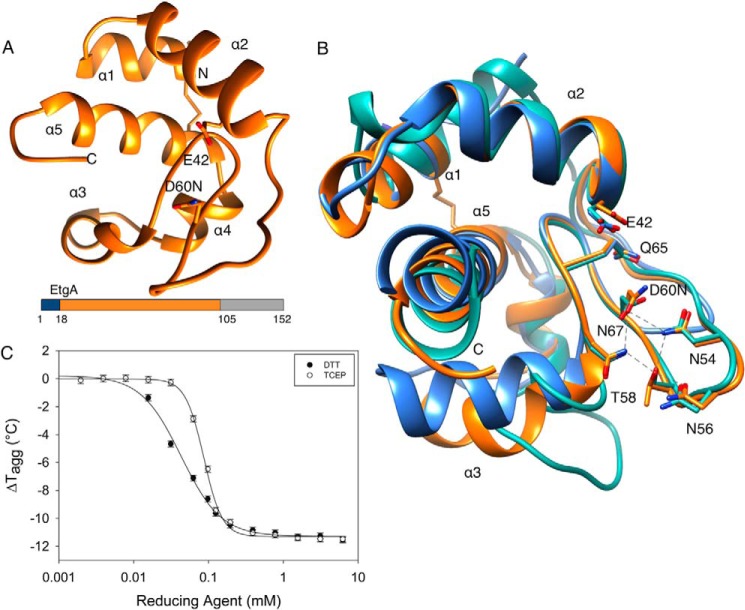
To gain insight into the activity EtgA, we have solved the x-ray crystal structure of its catalytic core. Initially, a complex of the inner rod EscI (24–137) and EtgA (19–152) D60N (necessary for stabilization because in isolation EtgA is highly unstable and prone to rapid precipitation) was set up for crystallization trials but was recalcitrant to crystallization. In situ limited proteolysis of the EscI (24–137)·EtgA (19–152) D60N complex with chymotrypsin produced crystals that diffracted to 2.0 Å resolution (Table 1). The structure of the crystallized proteolytic product was solved using multiwavelength anomalous dispersion phasing of selenomethionine derivative crystals and was revealed to encompass the catalytic core of EtgA, residues 19–105 (Table 1 and Fig. 2A). The catalytic core of EtgA has a globular, mainly α-helical fold (helices denoted as α1–5) with a β-strand region (residues 47–67) closely resembling the catalytic domain of E. coli lytic transglycosylase Slt70 (PDB code 1QTE; residues 451–618). Accordingly, a DALI search revealed Slt70 (along with another E. coli family 1 lytic transglycosylase, MltE) to be among the top structural homologs, with a z score of 11.2 and a root mean square deviation of 2.1 Å over 79 aligned Cα atoms. C-lysozyme (PDB code 2FBD) also had a high z score (9.0) with an root mean square deviation of 2.2 Å over 80 aligned Cα atoms. Alignment of EtgA with the catalytic region of Slt70 (PDB code 1QTE) and the top lysozyme DALI hit (PDB code 2FBD) revealed that the core helices of EtgA (α2 and α5) closely align with both lysozyme and Slt70, corresponding to a common “scaffold” present in many lytic transglycosylases and lysozymes (Fig. 2B). Interestingly, EtgA helices α1 and α5 are joined by a disulfide bond formed between Cys-20 and Cys-89, residues that are conserved in T3SS PG-lytic enzymes, as well as PG-lytic enzymes associated with the type IV pilus, bundle-forming pilus, and the type II secretion system (Figs. 2A and 3). This conserved disulfide bond stabilizes the fold of the protein, because incubation with increasing concentration of reducing agent (DTT or TCEP) causes a maximum change in aggregation temperature of 11.5 °C (Fig. 2C). Notably, a disulfide is absent from the same region in lysozyme and Slt70 (Fig. 2), suggesting that the extra stabilization provided by the disulfide is only present in T3SS PG-lytic enzymes and their macromolecular complex-associated homologs.
TABLE 1.
Data collection and refinement statistics
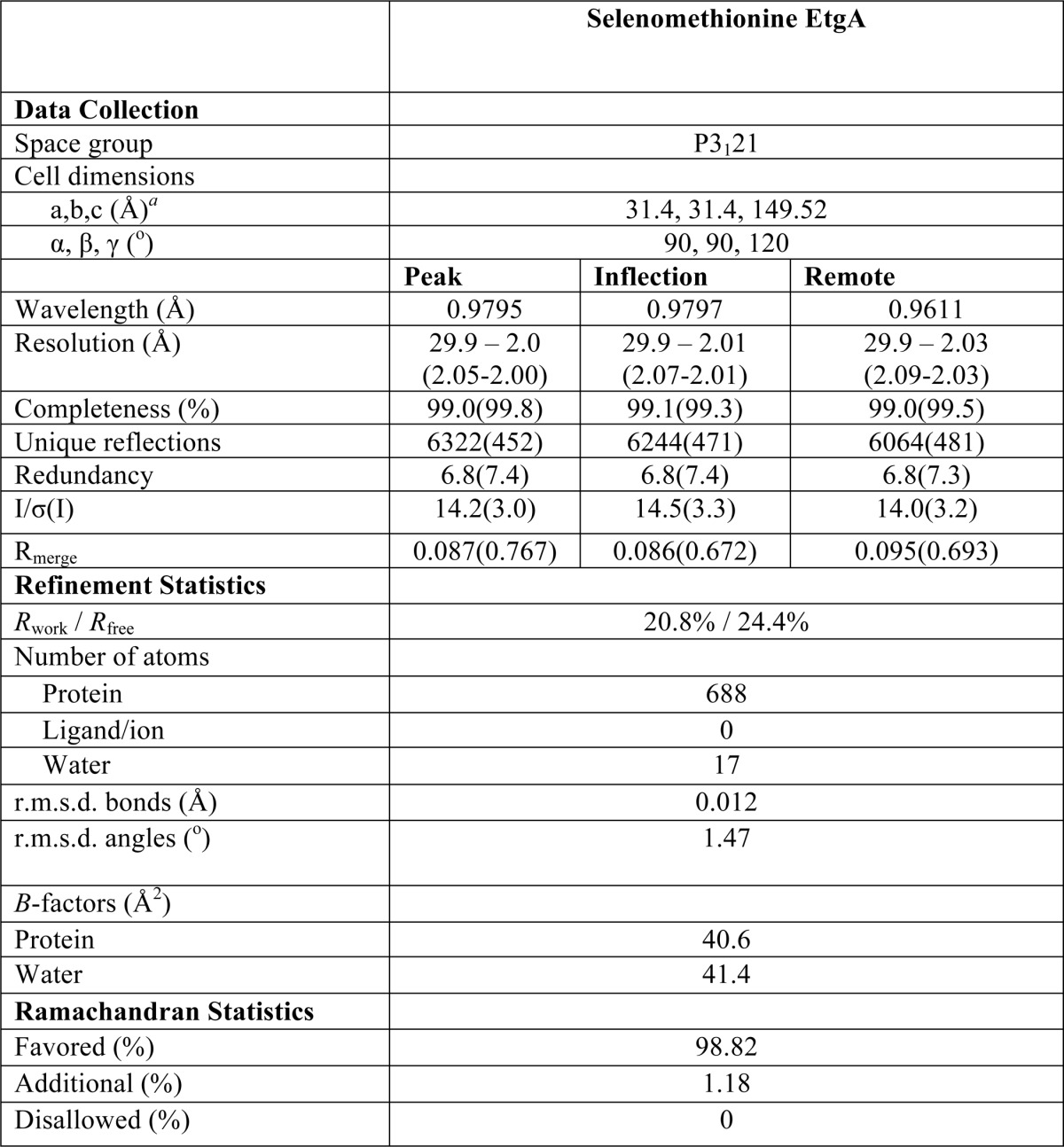
a Values in parentheses represent the highest resolution shell.
FIGURE 2.
Structure of the catalytic core of EtgA. A, the crystallized chymotrypsin proteolyzed fragment of EtgA corresponds to the catalytic core, encompassing residues 19–105, as shown in the bottom schematic (orange corresponds to the region in the crystal structure, blue corresponds to the Sec secretion signal, and gray is the region of EtgA absent from the structure). The catalytic glutamate (Glu-42) and a conserved aspartate (D60N) are shown as sticks. B, EtgA active site shares similarity with both lysozyme (PDB code 2FBD) and Slt70 (PDB code 1QTE), as shown by alignment of EtgA (orange) with Slt70 (blue) and lysozyme (cyan). The catalytic glutamate (Glu-42) has a similar position in all three structures. The lysozyme catalytic aspartate aligns with EtgA D60N. Other active site residues present in lysozyme and EtgA but absent from Slt70 include Asn-54, Asn-56, Thr-58, and Asn-67, which form a hydrogen-bonding network with D60N (residues shown as sticks, and hydrogen bonds shown as dashed lines). C, to test the importance of the EtgA disulfide bond for stability, differential static light scattering was measured to monitor aggregation of EtgA·EscI complex with increasing concentration of reducing agent (DTT or TCEP). The change in aggregation temperature (Tagg) was plotted as a function of reducing agent concentration.
FIGURE 3.
Multiple sequence alignment of T3SS specialized PG-lytic enzyme EtgA with other macromolecular machine-associated peptidoglycan-cleaving enzymes, lytic transglycosylase Slt70 and C-lysozyme. Alignment of EtgA (E. coli, C7BUG6), IagB (Salmonella enterica serovar Typhi, P43018), IpgF (Shigella flexneri, Q07568), and YsaH (Yersinia enterocolitica, Q9KKJ1) with type II secretion system PG-lytic enzyme OrfC (Burkholderia pseudomallei, Q9ZF87), bundle-forming pilus PG-lytic enzyme Bfp H (E. coli 0127:H6, Q47073), type IVB pilus PG-lytic enzyme PilT (S. enterica serovar Typhi, Q9ZIU8), Slt70 lytic transglycosylase (E. coli, POAGC3), and lysozyme (Musca domestica, Q7YT16) is shown. The macromolecular machine-associated PG-lytic enzymes contain the glutamate general acid (denoted by *) present in both Slt70 and lysozyme, as well as an aspartate (denoted by *), which is present and required for catalysis in lysozyme but absent from Slt70. EtgA residues conserved with lysozyme that form a hydrogen-bonding network with the conserved Asp are boxed and labeled as Lysozyme-like residues. Cysteines that form a disulfide bond in EtgA and are conserved with other T3SS, type IV pili, and bundle-forming pili, and the type II secretion system PG-lytic enzymes are boxed and labeled. A motif conserved between Slt70 and T3SS-associated PG-lytic enzymes is also boxed and labeled. Sequence alignment was done using clustal omega and the image was generated using Chimera. Swiss-Prot/TrEMBL accession numbers for each sequence are shown above in brackets.
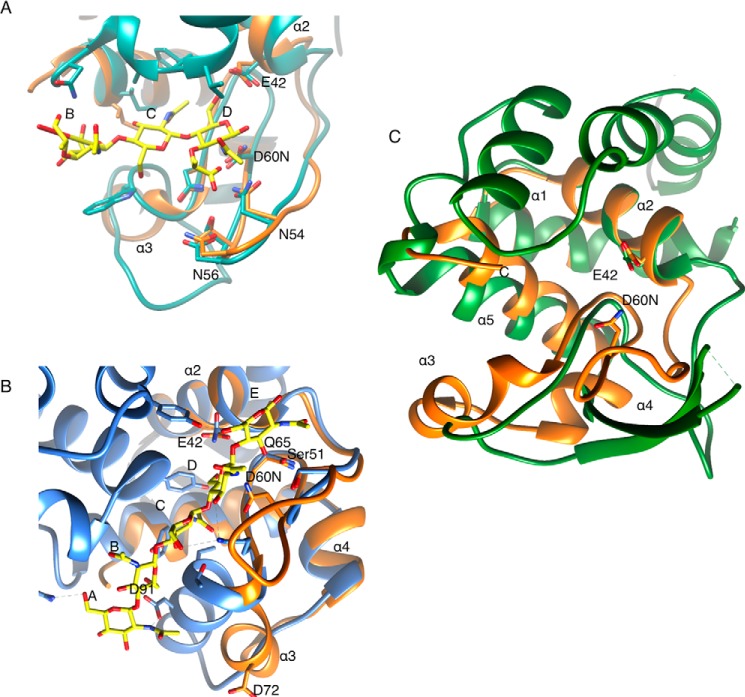
Strikingly, EtgA contains an extension of the second turn of its β-hairpin (residues 53–60), which aligns with the corresponding region in lysozyme, but is absent from Slt70 and other structurally characterized lytic transglycosylases (Fig. 2B). The presence of this region in EtgA is particularly interesting, because it contains an aspartate (Asp-60) in the same position as the lysozyme catalytic Asp-52 (in our structure this aspartate has been mutated to an asparagine because it conferred greater protein yields and stability for crystallography). This residue (D60N) forms a hydrogen-bonding network with the side chains of Asn-54, Asn-56, Thr-58, and Asn-67 (Fig. 2B). This hydrogen-bonding network was first described in HEW lysozyme and forms a platform against which the GlcNAc sugar packs in the D (−1) site (51). Indeed, this aspartate and the residues forming the hydrogen-bonding network are also conserved in Shigella and Salmonella EtgA homologs, IpgF and IagB, as well as homologs associated with bundle-forming pilus and type II secretion system assembly (Fig. 3). Based on observations using 2-fluorochitobiosyl, lysozyme Asp-52 has been proposed to act as a nucleophile, generating the glycosyl-enzyme intermediate, followed by protonation of the leaving group oxygen (4-OH) of GlcNAc by the general/acid base Glu-35 and attack by water (52). Earlier proposals had suggested alternatively, that lysozyme Asp-52, restrained by the conserved hydrogen-bonding network described above, would provide charge stabilization of the developed oxocarbenium in an X displacement reaction rather than act as a nucleophile per se. Conversely, lytic transglycosylases studied to date lack a catalytic aspartate. An overlay of EtgA with lysozyme bound to MurNAc-GlcNAc-MurNAc trisaccharide reveals that EtgA Asn-54, Asp-60, and Glu-42 align with the corresponding lysozyme residues to contact substrate in the D (−1) site (Fig. 4A). Comparison of MltE in complex with chitopentaose (i.e. (GlcNAc)5) to EtgA reveals that several residues of MltE that make hydrogen bonds or van der Waals contacts with chitopentaose are conserved and overlay with EtgA residues (Fig. 4B). Of note, EtgA Gln-65 and Ser-51 align with the corresponding residues in MltE that contact chitopentaose in the E (+1) and D (−1) positions.
FIGURE 4.
Comparison of EtgA active site with Slt70 and lysozyme active site bound to peptidoglycan-like fragments and flagellar PG-lytic enzyme FlgJ. A, overlay of EtgA (orange) with HEW lysozyme (PDB code 9LYZ, cyan) bound to MurNAc-GlcNAc-MurNAc trisaccharide (yellow sticks), centered on subsite D. B, overlay of EtgA (orange) with E. coli lytic transglycosylase MltE (PDB code 4HJZ, blue) bound to chitopentaose (yellow sticks). C, structural comparison of EtgA with the flagellar N-acetylglucosaminidase FlgJ reveals that the core scaffold of EtgA (α1, α2, and α5) aligns with the flagellar-associated peptidoglycan cleaving enzyme, FlgJ (PDB code 2ZYC); however, the β-turn and α3 and α4 of EtgA share little structural similarity to FlgJ. EtgA is shown in orange, and labels refer to EtgA residues and helices. FlgJ is shown in green.
Surprisingly, given the evolutionary relationship between the flagella and T3SS (53), structural comparison of the flagella PG-lytic enzyme, FlgJ, to EtgA reveals structural homology to the scaffold helices (α1, α2, and α5 in EtgA) but markedly different structure of the β-hairpin and α3 and α4 regions of EtgA (Fig. 4C) (54). These differences are reflected in a relatively weak (compared with lysozyme and Slt70) DALI z score of 4.3 and root mean square deviation of 3.4 Å over 70 aligned Cα atoms. Additionally, structure-based sequence alignment of FlgJ (PDB code 2ZYC) with EtgA reveals only 9% sequence identity. The β-hairpin of FlgJ extends further (and is also partially disordered in this structure) than that of EtgA and protrudes from the globular fold of the structure. Although the EtgA general acid Glu-42 aligns with FlgJ Glu-185, FlgJ lacks a catalytic aspartate corresponding to EtgA Asp-60.
EtgA Requires a Catalytic Glutamate and Aspartate for Activity in Vivo
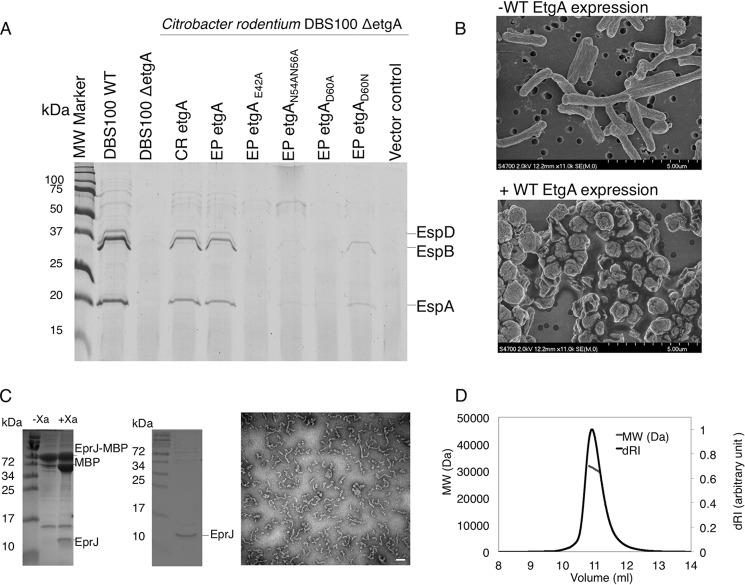
Given the unexpected and unprecedented (for lytic transglycosylase enzymes) structural alignment of EtgA Asp-60 and β-hairpin region with lysozyme catalytic Asp-52 and β-hairpin, and their conservation in T3SS encoding species, we mutated the conserved residues and tested the effect on type III secretion. The ΔetgA mutant of both C. rodentium and EPEC displayed a reduced type III secretion phenotype. The Citrobacter ΔetgA mutant exhibited a more than 90% reduction in type III secretion when compared with the wild-type strain (29), whereas the EPEC ΔetgA mutant only had 50% attenuation of type III secretion, possibly because of functional redundancy with a housekeeping PG-lytic enzyme (data not shown; (26)). Because greater reduction in type III secretion was useful for testing the effect of EtgA catalytic mutants, the Citrobacter ΔetgA mutant was used for the following experiments. Citrobacter ΔetgA could be complemented by both EPEC and Citrobacter etgA (91% sequence identity) expressed on a low copy number plasmid (3–5 copies/cell). The low copy number plasmid was critical for successful complementation, because complementation of etgA with a moderate copy number plasmid (30–50 copies/cell) was toxic to EPEC and Citrobacter when grown under type III secretion inducing conditions and caused partial bacterial lysis (data not shown), probably because of the peptidoglycan-hydrolyzing activities of EtgA. As expected, mutation of the catalytic glutamate (E42A) abrogated type III secretion (Fig. 5A). Complementation with etgA N54A N56A (two asparagines located in the β-hairpin that are conserved in lysozyme and form a hydrogen-bonding network with Asp-60) severely decreased type III secretion (Fig. 5A). Mutation of EtgA Asp-60 (conserved with lysozyme catalytic Asp-52) to alanine also caused a severe decrease in type III secretion, indicating an important role in peptidoglycan cleavage.
FIGURE 5.
Analysis of EtgA activity in vivo and interaction with the inner rod. A, type III secretion assay of C. rodentium etgA mutant complemented with EPEC etgA and its site-directed mutants. C. rodentium DBS100 ΔetgA was complemented with EPEC etgA with mutations of key conserved active site residues. T3SS secreted proteins (translocon components EspB and EspD, as well as EspA filament) from C. rodentium grown in DMEM were analyzed by 16% SDS-PAGE and stained by Coomassie G250. B, scanning electron microscopy images of BL21 (DE3) E. coli without expression of EtgA (top panel) and after induction of EtgA expression (bottom panel). EtgA was fused to a PelB signal peptide for export to the periplasm. D, stoichiometry of EtgA and inner rod (EscI) complex by size exclusion chromatography-multiangle light scattering. EscI (10–137) and EtgA (19–152) were co-purified, injected over a Superdex 75 10/300 column, and analyzed by multiangle light scattering. EscI (10–137) and EtgA (19–152) were shown to form a monodisperse complex of 31,400 Da (± 0.4%), corresponding to a 1:1 ratio. C, purification of MBP tagged EprJ (EscI homolog in EHEC) by amylose affinity chromatography (left gel) followed by cleavage of the MBP tag and size exclusion chromatography (right gel). Purified EprJ was imaged by negative stain electron microscopy. The white scale bar corresponds to 100 nm. MW, molecular mass.
EtgA Associates with the T3SS through Interaction with the Inner Rod, EscI
T3SS-associated PG-lytic enzymes may be spatially regulated through interaction with secretion system components to prevent undesired peptidoglycan cleavage and cell lysis. Indeed, we show that expression of WT EtgA (19–152) fused to a PelB signal sequence for periplasmic localization in BL21 (DE3) E. coli (a nonpathogenic expression strain lacking a T3SS) caused severe cell lysis, as shown by scanning electron microscopy images comparing morphology of BL21 (DE3) E. coli without induction of EtgA expression (Fig. 5B, top panel) and with induction of EtgA expression (Fig. 5B, bottom panel). Because unregulated expression and export of EtgA to the periplasm is autolytic, a targeting mechanism must be in place to ensure that periplasmic EtgA localizes to the nascent T3S apparatus to clear peptidoglycan. To better understand the interaction between the inner rod and EtgA and how it may serve as a tethering mechanism, we sought to co-express and co-purify the two proteins to characterize the interaction in vitro. Although full-length EscI (residues 1–142) was insoluble, as was also observed for expression of the Salmonella homolog PrgJ (55), several N- and C-terminally truncated polyhistidine-tagged constructs of EscI successfully co-expressed and co-purified with untagged EtgA (19–152). The minimal region of EscI required for interaction with EtgA encompassed residues 50–137, whereas the longest EscI construct that could be co-expressed encompassed residues 10–137. Size exclusion chromatography-multiangle light scattering was used to characterize the stoichiometry between EscI (10–137) and EtgA (19–152) and showed that the two proteins interact in a 1:1 ratio to form a complex of 31,400 Da ± 0.4% (Fig. 5D). Based on sequence similarity to the T3SS needle component, EscI is predicted to polymerize into a filamentous rod-like structure (11, 12) that presumably lies within and connects the inner and outer membrane ring structures of the T3SS basal body. Although we were unable to express and purify a soluble filamentous form of EscI, we could express and purify the EHEC homolog (EprJ, 26% identity) by fusion to an N-terminal MBP solubilization tag. Following cleavage of the MBP tag, EprJ formed heterogenous, filamentous structures, as observed by negative stain electron microscopy (Fig. 5C), showing directly for the first time that full-length EprJ is capable of filamentous polymerization. Although it is unknown precisely how EtgA would localize along a filamentous form of the inner rod, our data nevertheless shows that in this interaction, each EscI component binds only one EtgA protein via residues encompassing the central region of EscI.
Interaction with the Inner Rod EscI Enhances the Activity of EtgA in Vitro
To determine whether the inner rod affects the PG-lytic activity of EtgA, we compared activity of EtgA versus EtgA·EscI complex in vitro using a fluorescently labeled peptidoglycan substrate. An initial buffer screen showed the optimal pH for EtgA activity is between 6 and 7.5, so subsequent activity assays were done at pH 7. EtgA had an apparent Vmax of 95 ± 5 RFU/min, whereas EtgA·EscI complex had nearly an 8-fold higher apparent Vmax of 720 ± 15 RFU/min (Fig. 6A and Table 2). Both samples had a similar apparent Km, suggesting that the inner rod does not affect binding of peptidoglycan to EtgA (Table 2). Mutation of the catalytic glutamate (E42A) and aspartate (D60N) rendered EtgA inactive (both alone and in complex with the inner rod), underscoring the importance of both residues for catalysis (Fig. 6A). The observation that EtgA catalytic mutants in complex with EscI were inactive shows that EscI does not have any peptidoglycan cleaving activity itself but acts to enhance the activity of EtgA. A thermal aggregation assay was used to compare the stability of EtgA to the stability EtgA in complex with EscI. EtgA had an aggregation temperature (Tagg) of 40 °C, whereas EtgA·EscI had a Tagg of 48 °C (Fig. 6B), suggesting that EscI stabilizes EtgA. The stabilizing effect of the inner rod on EtgA may contribute toward the increase in EtgA activity observed in the presence of the inner rod.
FIGURE 6.
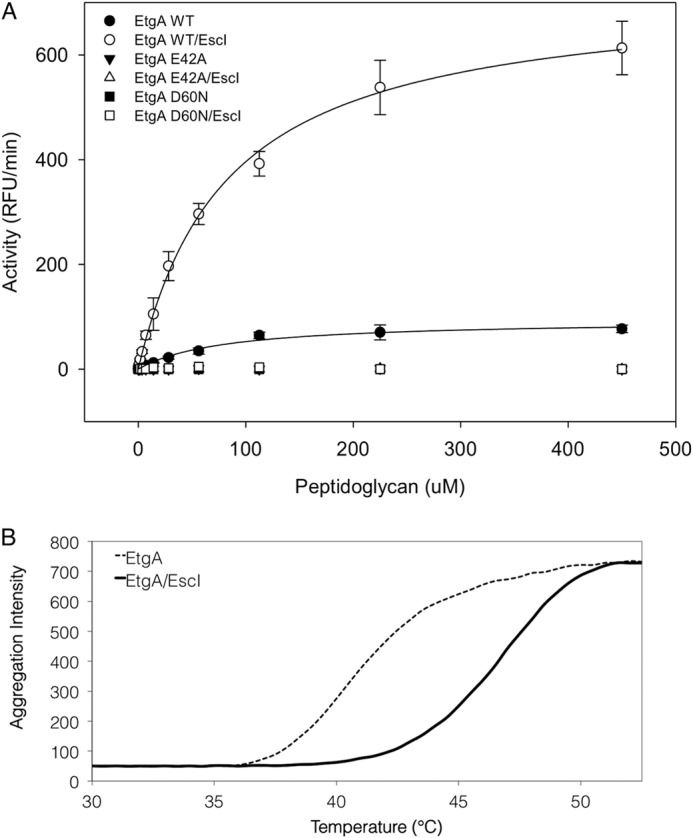
The inner rod enhances EtgA peptidoglycan cleaving activity. A, EtgA (19–152) and EtgA (19–152)·EscI (24–137) complex was incubated with fluorescein-labeled peptidoglycan, and the activity was monitored by measuring fluorescence with an excitation/emission wavelength of 485/530 nm over 1 h. Conserved EtgA active site residues Glu-42 and Asp-60 were also mutated and assayed for activity, both with and without the inner rod EscI. B, thermal aggregation of EtgA (19–152) or EtgA (19–152)·EscI (24–137) complex was monitored over increasing temperature by differential static light scattering (aggregation intensity).
TABLE 2.
EtgA peptidoglycan cleavage parameters
| Apparent Vmax | Apparent Km | |
|---|---|---|
| RFU/min/μm EtgA | μm | |
| EtgA (19-152) WT | 95 ± 5 | 80 ± 15 |
| WT EtgA(19-152)/ EscI (24-137) | 720 ± 15 | 80 ± 5 |
DISCUSSION
Clusters of genes encoding type II, III, and IV secretion systems (as well as type IV pili) often encode a PG-lytic enzyme. Based on sequence homology with family 1A lytic transglycosylases, these specialized secretion system PG-lytic enzymes were presumed to function as a lytic transglycosylase with creation of N-acetylglucosamine and 1,6-anhydromuramic acid products (20). In our crystal structure of T3SS-specific PG-lytic enzyme EtgA, we see features that are conserved with both lytic transglycosylases and HEW lysozyme. The most surprising feature of the EtgA structure is a β-hairpin loop and aspartate that aligns remarkably well with the lysozyme β-hairpin loop and catalytic aspartate. This conserved aspartate previously went unnoticed in the literature, because sequence alignments were done with Slt70, which, like all other characterized lytic transglycosylases, lacks a catalytic aspartate. Accordingly, we have shown mutation of the aligned aspartate decreased type III secretion and abrogated peptidoglycan cleaving activity, underscoring its importance for catalysis in these virulence systems.
Currently, little is known about the mechanistic details of how specialized PG-lytic enzymes facilitate assembly of macromolecular complexes. Recently, a PG-lytic enzyme associated with the flagella, FlgJ, was shown to cleave peptidoglycan between GlcNAc and MurNAc saccharides, acting as an endo-specific N-acetylglucosaminidase (56). Another study showed that Helicobacter pylori and Salmonella typhimurium required activity of housekeeping lytic transglycosylases for functionality of the flagella (i.e. motility), but not flagella assembly (57). This work proposed a model in which the N-actelyglucosaminidase activity of FlgJ clears peptidoglycan to facilitate flagella assembly, and subsequently housekeeping (involved in cell growth and cell wall maintenance) lytic transglycosylases remodel the opening in the peptidoglycan, producing 1,6-anhydromuramoyl peptidoglycan ends that interact with MotB, the flagellar motor protein (56, 57). Does the difference in structure (and likely activity) between type III-associated PG-lytic enzyme EtgA and the flagellar FlgJ somehow play a nuanced role in assembly of each system, or does it reflect a different evolutionary acquirement of a PG-lytic enzyme? The cell wall products produced by N-acetylglucosaminidase, lysozyme, and lytic transglycosylase would differ, creating a distinct chemical environment around the secretion system. Multiple components of the type III secretion apparatus interact with peptidoglycan (58) and may selectively bind particular peptidoglycan moieties.
Supporting the idea that PG-lytic enzymes are intricately involved in macromolecular complex assembly, the tomato plant pathogen Pseudomonas syringae pv. tomato DC300 encodes three PG-lytic enzymes (HopP1, HrpH, and HopAJ1) that are up-regulated during type III assembly (59). HopP1 has homology to EtgA (26% sequence identity), and we observe that it contains both a conserved catalytic glutamate and aspartate, whereas HrpH and HopAJ1 do not share sequence homology with EtgA and have only a catalytic glutamate. A combination of deletions of HopP1, HprH, and HopAJ1 causes a decrease in virulence and effector translocation, but not a decrease in effector secretion (59). Indeed, many type II, III, and IV secretion system gene clusters encode a specific PG-lytic enzyme, yet the deletion phenotype is often only attenuated for virulence. The variability in deletion phenotype, as also observed in our analysis of C. rodentium and EPEC ΔetgA mutants here, can then perhaps be attributed to functional redundancy with other PG-lytic enzymes or the ability of the secretion apparatus to insert in naturally occurring holes in the peptidoglycan sacculus. It is possible that specialized PG-lytic enzymes play a multifaceted role in assembly, modifying local peptidoglycan in such a way that it is a recruitment signal for peptidoglycan-interacting components of the secretion system.
Activities of PG-lytic enzymes are tightly regulated temporally and spatially to prevent erroneous peptidoglycan cleavage and autolysis. Expression of EtgA is negatively regulated by GrlA (an activator of T3SS gene expression), presumably to allow expression of T3SS components before expression and export of EtgA to the periplasm (35). Once in the periplasm, EtgA interacts with the T3SS inner rod component EscI, which likely polymerizes into a filament. Our in vitro peptidoglycan cleavage assay shows that EtgA is nearly eight times more active in the presence of the inner rod, suggesting that the inner rod not only spatially restricts the activity of EtgA but may also enhance its activity. Once transported to the periplasm by the Sec secretion system, EtgA would be marginally active until it binds EscI (which is secreted through the T3SS), preventing destructive uncontrolled cell lysis. This may be a common theme for secretion system associated PG-lytic enzymes, because others such as VirB1 from the type IV secretion system have been shown to interact with components of the secretion apparatus (36).
In conclusion, the structure of EtgA reveals that, despite sequence similarity with family 1A lytic transglycosylases, EtgA possesses a catalytic glutamate and aspartate and a β-hairpin region that are remarkably similar to lysozyme. Although EtgA and lysozyme share a conserved sequence in this region, the homology was previously unnoticed, and EtgA (as well as other homologs associated with the type II, III, and IV secretion systems) was presumed to act as a lytic transglycosylase. Additionally, we show that the peptidoglycan cleaving activity of EtgA is enhanced in the presence of the inner rod EscI. The low level of activity detected for EtgA in the absence of EscI may serve as a mechanism to prevent uncontrolled lysis prior to interaction with EscI. Future work will focus on characterization of the mechanism and reaction products of EtgA to definitively classify it as a lysozyme or lytic transglycosylase.
Acknowledgments
We thank the beamline staff at Lawrence Berkeley National Laboratory Advanced Light Source on Beamline 8.2.1 for assistance with data collection and Derrick Horne, Elaine Humphries, Bradford Ross and the University of British Columbia Bioimaging facility staff for assistance with electron microscopy. We thank D. King and J. Bergeron for insightful discussion on muramidases and T3SS assembly.
This work was funded by CIHR operating grants to NCJS and BBF and operating funding from the HHMI International Scholar program to NCJS.
- EPEC
- enteropathogenic E. coli
- T3SS
- type III secretion system
- PG
- peptidoglycan
- MurNAc
- N-acetylmuramic acid
- IPTG
- isopropyl β-d-thiogalactopyranoside
- PDB
- Protein Data Bank
- MBP
- maltose-binding protein
- BME
- β-mercaptoethanol
- RFU
- relative fluorescence units
- TCEP
- tris(2-carboxyethyl)phosphine.
REFERENCES
- 1. Bryce J., Boschi-Pinto C., Shibuya K., Black R. E., and WHO Child Health Epidemiology Reference Group. (2005) WHO estimates of the causes of death in children. Lancet 365, 1147–1152 [DOI] [PubMed] [Google Scholar]
- 2. Lanata C. F., Fischer-Walker C. L., Olascoaga A. C., Torres C. X., Aryee M. J., Black R. E., Child Health Epidemiology Reference Group of the World Health Organization, and UNICEF (2013) Global causes of diarrheal disease mortality in children <5 years of age: a systematic review. PloS one 8, e72788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Jerse A. E., Yu J., Tall B. D., Kaper J. B. (1990) A genetic locus of enteropathogenic Escherichia coli necessary for the production of attaching and effacing lesions on tissue culture cells. Proc. Natl. Acad. Sci. U.S.A. 87, 7839–7843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Moon H. W., Whipp S. C., Argenzio R. A., Levine M. M., Giannella R. A. (1983) Attaching and effacing activities of rabbit and human enteropathogenic Escherichia coli in pig and rabbit intestines. Infect. Immun. 41, 1340–1351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Rosenshine I., Ruschkowski S., Stein M., Reinscheid D. J., Mills S. D., Finlay B. B. (1996) A pathogenic bacterium triggers epithelial signals to form a functional bacterial receptor that mediates actin pseudopod formation. EMBO J. 15, 2613–2624 [PMC free article] [PubMed] [Google Scholar]
- 6. Taylor C. J., Hart A., Batt R. M., McDougall C., McLean L. (1986) Ultrastructural and biochemical changes in human jejunal mucosa associated with enteropathogenic Escherichia coli (0111) infection. J. Pediatr. Gastroenterol. Nutr. 5, 70–73 [DOI] [PubMed] [Google Scholar]
- 7. Wong A. R., Pearson J. S., Bright M. D., Munera D., Robinson K. S., Lee S. F., Frankel G., Hartland E. L. (2011) Enteropathogenic and enterohaemorrhagic Escherichia coli: even more subversive elements. Mol. Microbiol. 80, 1420–1438 [DOI] [PubMed] [Google Scholar]
- 8. Schraidt O., Marlovits T. C. (2011) Three-dimensional model of Salmonella's needle complex at subnanometer resolution. Science 331, 1192–1195 [DOI] [PubMed] [Google Scholar]
- 9. Spreter T., Yip C. K., Sanowar S., André I., Kimbrough T. G., Vuckovic M., Pfuetzner R. A., Deng W., Yu A. C., Finlay B. B., Baker D., Miller S. I., Strynadka N. C. (2009) A conserved structural motif mediates formation of the periplasmic rings in the type III secretion system. Nat. Struct. Mol. Biol. 16, 468–476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Yip C. K., Kimbrough T. G., Felise H. B., Vuckovic M., Thomas N. A., Pfuetzner R. A., Frey E. A., Finlay B. B., Miller S. I., Strynadka N. C. (2005) Structural characterization of the molecular platform for type III secretion system assembly. Nature 435, 702–707 [DOI] [PubMed] [Google Scholar]
- 11. Pallen M. J., Beatson S. A., Bailey C. M. (2005) Bioinformatics analysis of the locus for enterocyte effacement provides novel insights into type-III secretion. BMC Microbiol. 5, 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Sal-Man N., Deng W., Finlay B. B. (2012) EscI: a crucial component of the type III secretion system forms the inner rod structure in enteropathogenic Escherichia coli. Biochem. J. 442, 119–125 [DOI] [PubMed] [Google Scholar]
- 13. Daniell S. J., Takahashi N., Wilson R., Friedberg D., Rosenshine I., Booy F. P., Shaw R. K., Knutton S., Frankel G., Aizawa S. (2001) The filamentous type III secretion translocon of enteropathogenic Escherichia coli. Cell. Microbiol. 3, 865–871 [DOI] [PubMed] [Google Scholar]
- 14. Knutton S., Rosenshine I., Pallen M. J., Nisan I., Neves B. C., Bain C., Wolff C., Dougan G., Frankel G. (1998) A novel EspA-associated surface organelle of enteropathogenic Escherichia coli involved in protein translocation into epithelial cells. EMBO J. 17, 2166–2176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Sekiya K., Ohishi M., Ogino T., Tamano K., Sasakawa C., Abe A. (2001) Supermolecular structure of the enteropathogenic Escherichia coli type III secretion system and its direct interaction with the EspA-sheath-like structure. Proc. Natl. Acad. Sci. U.S.A. 98, 11638–11643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Wang Y. A., Yu X., Yip C., Strynadka N. C., Egelman E. H. (2006) Structural polymorphism in bacterial EspA filaments revealed by cryo-EM and an improved approach to helical reconstruction. Structure 14, 1189–1196 [DOI] [PubMed] [Google Scholar]
- 17. Yip C. K., Finlay B. B., Strynadka N. C. (2005) Structural characterization of a type III secretion system filament protein in complex with its chaperone. Nat. Struct. Mol. Biol. 12, 75–81 [DOI] [PubMed] [Google Scholar]
- 18. Ide T., Laarmann S., Greune L., Schillers H., Oberleithner H., Schmidt M. A. (2001) Characterization of translocation pores inserted into plasma membranes by type III-secreted Esp proteins of enteropathogenic Escherichia coli. Cell. Microbiol. 3, 669–679 [DOI] [PubMed] [Google Scholar]
- 19. Dijkstra A. J., Keck W. (1996) Peptidoglycan as a barrier to transenvelope transport. J. Bacteriol. 178, 5555–5562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Koraimann G. (2003) Lytic transglycosylases in macromolecular transport systems of Gram-negative bacteria. Cell Mol. Life Sci. 60, 2371–2388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Scheurwater E., Reid C. W., Clarke A. J. (2008) Lytic transglycosylases: bacterial space-making autolysins. Int. J. Biochem. Cell Biol. 40, 586–591 [DOI] [PubMed] [Google Scholar]
- 22. Scheurwater E. M., Burrows L. L. (2011) Maintaining network security: how macromolecular structures cross the peptidoglycan layer. FEMS Microbiol. Lett. 318, 1–9 [DOI] [PubMed] [Google Scholar]
- 23. Demchick P., Koch A. L. (1996) The permeability of the wall fabric of Escherichia coli and Bacillus subtilis. J. Bacteriol. 178, 768–773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Meroueh S. O., Bencze K. Z., Hesek D., Lee M., Fisher J. F., Stemmler T. L., Mobashery S. (2006) Three-dimensional structure of the bacterial cell wall peptidoglycan. Proc. Natl. Acad. Sci. U.S.A. 103, 4404–4409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Uehara T., Suefuji K., Valbuena N., Meehan B., Donegan M., Park J. T. (2005) Recycling of the anhydro-N-acetylmuramic acid derived from cell wall murein involves a two-step conversion to N-acetylglucosamine-phosphate. J. Bacteriol. 187, 3643–3649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. García-Gómez E., Espinosa N., de la Mora J., Dreyfus G., González-Pedrajo B. (2011) The muramidase EtgA from enteropathogenic Escherichia coli is required for efficient type III secretion. Microbiology 157, 1145–1160 [DOI] [PubMed] [Google Scholar]
- 27. Creasey E. A., Delahay R. M., Daniell S. J., Frankel G. (2003) Yeast two-hybrid system survey of interactions between LEE-encoded proteins of enteropathogenic Escherichia coli. Microbiology 149, 2093–2106 [DOI] [PubMed] [Google Scholar]
- 28. Zahrl D., Wagner M., Bischof K., Bayer M., Zavecz B., Beranek A., Ruckenstuhl C., Zarfel G. E., Koraimann G. (2005) Peptidoglycan degradation by specialized lytic transglycosylases associated with type III and type IV secretion systems. Microbiology 151, 3455–3467 [DOI] [PubMed] [Google Scholar]
- 29. Deng W., Puente J. L., Gruenheid S., Li Y., Vallance B. A., Vázquez A., Barba J., Ibarra J. A., O'Donnell P., Metalnikov P., Ashman K., Lee S., Goode D., Pawson T., Finlay B. B. (2004) Dissecting virulence: systematic and functional analyses of a pathogenicity island. Proc. Natl. Acad. Sci. U.S.A. 101, 3597–3602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Allaoui A., Ménard R., Sansonetti P. J., Parsot C. (1993) Characterization of the Shigella flexneri ipgD and ipgF genes, which are located in the proximal part of the mxi locus. Infect. Immun. 61, 1707–1714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Vollmer W., Joris B., Charlier P., Foster S. (2008) Bacterial peptidoglycan (murein) hydrolases. FEMS Microbiol. Rev. 32, 259–286 [DOI] [PubMed] [Google Scholar]
- 32. Thunnissen A. M., Dijkstra A. J., Kalk K. H., Rozeboom H. J., Engel H., Keck W., Dijkstra B. W. (1994) Doughnut-shaped structure of a bacterial muramidase revealed by x-ray crystallography. Nature 367, 750–753 [DOI] [PubMed] [Google Scholar]
- 33. Thunnissen A. M., Rozeboom H. J., Kalk K. H., Dijkstra B. W. (1995) Structure of the 70-kDa soluble lytic transglycosylase complexed with bulgecin A: implications for the enzymatic mechanism. Biochemistry 34, 12729–12737 [DOI] [PubMed] [Google Scholar]
- 34. van Asselt E. J., Thunnissen A. M., Dijkstra B. W. (1999) High resolution crystal structures of the Escherichia coli lytic transglycosylase Slt70 and its complex with a peptidoglycan fragment. J. Mol. Biol. 291, 877–898 [DOI] [PubMed] [Google Scholar]
- 35. Yu Y. C., Lin C. N., Wang S. H., Ng S. C., Hu W. S., Syu W. J. (2010) A putative lytic transglycosylase tightly regulated and critical for the EHEC type three secretion. J. Biomed. Sci. 17, 52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Höppner C., Carle A., Sivanesan D., Hoeppner S., Baron C. (2005) The putative lytic transglycosylase VirB1 from Brucella suis interacts with the type IV secretion system core components VirB8, VirB9 and VirB11. Microbiology 151, 3469–3482 [DOI] [PubMed] [Google Scholar]
- 37. Hirano T., Minamino T., Macnab R. M. (2001) The role in flagellar rod assembly of the N-terminal domain of Salmonella FlgJ, a flagellum-specific muramidase. J. Mol. Biol. 312, 359–369 [DOI] [PubMed] [Google Scholar]
- 38. Nambu T., Minamino T., Macnab R. M., Kutsukake K. (1999) Peptidoglycan-hydrolyzing activity of the FlgJ protein, essential for flagellar rod formation in Salmonella typhimurium. J. Bacteriol. 181, 1555–1561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Blackburn N. T., Clarke A. J. (2002) Characterization of soluble and membrane-bound family 3 lytic transglycosylases from Pseudomonas aeruginosa. Biochemistry 41, 1001–1013 [DOI] [PubMed] [Google Scholar]
- 40. Baron C., Coombes B. (2007) Targeting bacterial secretion systems: benefits of disarmament in the microcosm. Infect. Disord. Drug Targets 7, 19–27 [DOI] [PubMed] [Google Scholar]
- 41. Dong A., Xu X., Edwards A. M., Chang C., Chruszcz M., Cuff M., Cymborowski M., Di Leo R., Egorova O., Evdokimova E., Filippova E., Gu J., Guthrie J., Ignatchenko A., Joachimiak A., Klostermann N., Kim Y., Korniyenko Y., Minor W., Que Q., Savchenko A., Skarina T., Tan K., Yakunin A., Yee A., Yim V., Zhang R., Zheng H., Akutsu M., Arrowsmith C., Avvakumov G. V., Bochkarev A., Dahlgren L. G., Dhe-Paganon S., Dimov S., Dombrovski L., Finerty P., Jr., Flodin S., Flores A., Gräslund S., Hammerström M., Herman M. D., Hong B. S., Hui R., Johansson I., Liu Y., Nilsson M., Nedyalkova L., Nordlund P., Nyman T., Min J., Ouyang H., Park H. W., Qi C., Rabeh W., Shen L., Shen Y., Sukumard D., Tempel W., Tong Y., Tresagues L., Vedadi M., Walker J. R., Weigelt J., Welin M., Wu H., Xiao T., Zeng H., Zhu H., Midwest Center for Structural Genomics, and Structural Genomics Consortium (2007) In situ proteolysis for protein crystallization and structure determination. Nat. Methods 4, 1019–1021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Winter G., Lobley C. M., Prince S. M. (2013) Decision making in xia2. Acta Crystallogr. D Biol. Crystallogr. 69, 1260–1273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Vonrhein C., Blanc E., Roversi P., Bricogne G. (2007) Automated structure solution with autoSHARP. Methods Mol. Biol. 364, 215–230 [DOI] [PubMed] [Google Scholar]
- 44. Joosten R. P., Long F., Murshudov G. N., Perrakis A. (2014) The PDB_REDO server for macromolecular structure model optimization. IUCrJ 1, 213–220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Emsley P., Lohkamp B., Scott W. G., Cowtan K. (2010) Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 66, 486–501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Winn M. D., Ballard C. C., Cowtan K. D., Dodson E. J., Emsley P., Evans P. R., Keegan R. M., Krissinel E. B., Leslie A. G., McCoy A., McNicholas S. J., Murshudov G. N., Pannu N. S., Potterton E. A., Powell H. R., Read R. J., Vagin A., Wilson K. S. (2011) Overview of the CCP4 suite and current developments. Acta Crystallogr. D Biol. Crystallogr. 67, 235–242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Murshudov G. N., Vagin A. A., Dodson E. J. (1997) Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. D Biol. Crystallogr. 53, 240–255 [DOI] [PubMed] [Google Scholar]
- 48. Pettersen E. F., Goddard T. D., Huang C. C., Couch G. S., Greenblatt D. M., Meng E. C., Ferrin T. E. (2004) UCSF Chimera-a visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612 [DOI] [PubMed] [Google Scholar]
- 49. Donnenberg M. S., Kaper J. B. (1991) Construction of an eae deletion mutant of enteropathogenic Escherichia coli by using a positive-selection suicide vector. Infect. Immun. 59, 4310–4317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Vedadi M., Niesen F. H., Allali-Hassani A., Fedorov O. Y., Finerty P. J., Jr., Wasney G. A., Yeung R., Arrowsmith C., Ball L. J., Berglund H., Hui R., Marsden B. D., Nordlund P., Sundstrom M., Weigelt J., Edwards A. M. (2006) Chemical screening methods to identify ligands that promote protein stability, protein crystallization, and structure determination. Proc. Natl. Acad. Sci. U.S.A. 103, 15835–15840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Strynadka N. C., James M. N. (1991) Lysozyme revisited: crystallographic evidence for distortion of an N-acetylmuramic acid residue bound in site D. J. Mol. Biol. 220, 401–424 [DOI] [PubMed] [Google Scholar]
- 52. Vocadlo D. J., Davies G. J., Laine R., Withers S. G. (2001) Catalysis by hen egg-white lysozyme proceeds via a covalent intermediate. Nature 412, 835–838 [DOI] [PubMed] [Google Scholar]
- 53. Abby S. S., Rocha E. P. (2012) The non-flagellar type III secretion system evolved from the bacterial flagellum and diversified into host-cell adapted systems. PLoS Genet. 8, e1002983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Hashimoto W., Ochiai A., Momma K., Itoh T., Mikami B., Maruyama Y., Murata K. (2009) Crystal structure of the glycosidase family 73 peptidoglycan hydrolase FlgJ. Biochem. Biophys. Res. Commun. 381, 16–21 [DOI] [PubMed] [Google Scholar]
- 55. Zhong D., Lefebre M., Kaur K., McDowell M. A., Gdowski C., Jo S., Wang Y., Benedict S. H., Lea S. M., Galan J. E., De Guzman R. N. (2012) The Salmonella type III secretion system inner rod protein PrgJ is partially folded. J. Biol. Chem. 287, 25303–25311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Herlihey F. A., Moynihan P. J., Clarke A. J. (2014) The essential protein for bacterial flagella formation FlgJ functions as a β-N-acetylglucosaminidase. J. Biol. Chem. 289, 31029–31042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Roure S., Bonis M., Chaput C., Ecobichon C., Mattox A., Barrière C., Geldmacher N., Guadagnini S., Schmitt C., Prévost M. C., Labigne A., Backert S., Ferrero R. L., Boneca I. G. (2012) Peptidoglycan maturation enzymes affect flagellar functionality in bacteria. Mol. Microbiol. 86, 845–856 [DOI] [PubMed] [Google Scholar]
- 58. Pucciarelli M. G., García-del Portillo F. (2003) Protein-peptidoglycan interactions modulate the assembly of the needle complex in the Salmonella invasion-associated type III secretion system. Mol. Microbiol. 48, 573–585 [DOI] [PubMed] [Google Scholar]
- 59. Oh H. S., Kvitko B. H., Morello J. E., Collmer A. (2007) Pseudomonas syringae lytic transglycosylases coregulated with the type III secretion system contribute to the translocation of effector proteins into plant cells. J. Bacteriol. 189, 8277–8289 [DOI] [PMC free article] [PubMed] [Google Scholar]