Background: Mechanisms whereby local bone-derived factors regulate FGF-23 are unclear.
Results: Low and high molecular weight FGF-2 stimulated FGF-23 promoter activity in osteoblasts through membrane FGFR-mediated PLCγ and MAPK activation of NFAT and Ets1 and integrative nuclear FGFR1 signaling involving cAMP/CBP/CREB signaling pathways, respectively.
Conclusion: Membrane FGFR and intranucelar FGFR/CBP pathways regulate FGF-23 transcription.
Significance: Paracrine/autocrine FGFs control hormonal FGF-23.
Keywords: Calcineurin, Cyclic AMP (cAMP), Fibroblast Growth Factor (FGF), Fibroblast Growth Factor Receptor (FGFR), Osteoblast
Abstract
Fibroblastic growth factor receptor 1 (FGFR1) signaling pathways are implicated in the regulation of FGF-23 gene transcription, but the molecular pathways remain poorly defined. We used low molecular weight (LMW, 18 kDa) FGF-2 and high molecular weight (HMW) FGF-2 isoforms, which, respectively, activate cell surface FGF receptors and intranuclear FGFR1, to determine the roles of membrane FGFRs and integrative nuclear FGFR1 signaling (INFS) in the regulation of FGF-23 gene transcription in osteoblasts. We found that LMW-FGF-2 induced NFAT and Ets1 binding to conserved cis-elements in the proximal FGF-23 promoter and stimulated FGF-23 promoter activity through PLCγ/calcineurin/NFAT and MAPK pathways in SaOS-2 and MC3T3-E1 osteoblasts. In contrast, HMW-FGF-2 stimulated FGF-23 promoter activity in osteoblasts through a cAMP-dependent binding of FGFR1 and cAMP-response element-binding protein (CREB) to a conserved cAMP response element (CRE) contiguous with the NFAT binding site in the FGF-23 promoter. Mutagenesis of the NFAT and CRE binding sites, respectively, inhibited the effects of LMW-FGF-2 and HMW-FGF-23 to stimulate FGF-23 promoter activity. FGF-2 activation of both membrane FGFRs and INFS-dependent FGFR1 pathways may provide a means to integrate systemic and local regulation of FGF-23 transcription under diverse physiological and pathological conditions.
Introduction
FGF-23 is a member of the hormone-like FGF gene family that also includes FGF-19 and FGF-21 (1). FGF-23 is predominately secreted by osteoblasts and osteocytes in bone (2). Hormone-like FGF-23 evolved from paracrine/autocrine FGFs through the emergence of a novel C terminus that permits diffusion and heparin-independent binding to a binary membrane receptor complex consisting of FGFR and α-Klotho, a type I membrane β-glycosidase (4–9). FGF-23 activation of FGFR/α-Klotho complexes in the kidney leads to reductions in Npt2a co-transporters and inhibition of renal tubular phosphate reabsorption and reductions in circulating 1,25(OH)2D levels through inhibition of Cyp27b1 and stimulation of Cyp24 enzymes regulating vitamin D metabolism (3–9).
Pathological increments in circulating FGF-23 concentrations underlie acquired and hereditary forms of hypophosphatemic rickets, whereas decrements in FGF-23 cause hereditary tumoral calcinosis (10). Physiologically, FGF-23 participates in systemic and local regulatory networks that control serum phosphate and 1,25(OH)2D levels. As a counter-regulatory hormone for 1,25(OH)2D, elevations of FGF-23, which is induced by 1,25-(OH)2D, parathyroid hormone (PTH), or calcium (11), results in reductions in serum phosphorus levels and suppression of 1,25(OH)2D production (14, 15). FGF-23 also coordinates bone mineralization with renal handling of phosphate through poorly defined local processes that involve classical paracrine FGFR1 activation (1, 11–22).
Emerging data suggests that the earlier evolved paracrine/autocrine FGFR signaling pathways remain linked to the more recent hormonal FGF-23 (1, 10, 23). First, FGF-23 is increased in osteoglophonic dysplasia, which is caused by activating mutations in FGFR1 (24, 25). Second, ligands for FGFR1, including FGF-1, LMW-FGF-2, and FGF-7, as well as HMW-FGF-2, are significantly increased in the Hyp and/or Dmp1 knock-out mouse models of FGF-23 excess (10, 26–28). Third, pharmacological inhibition of FGFR1 blocks FGF-23 transcription in bone both in vitro and ex vivo (10, 29, 30). Fourth, the administration of monoclonal FGFR1 activating antibodies stimulates FGF-23 production and induces hypophosphatemia (31). Finally, and most specifically, conditional deletion of FGFR1 in osteocytes of the Hyp mouse model reduces FGF-23 expression in bone (32).
The FGF-2 gene produces 18-kDa low molecular weight (LMW)2 FGF-2 and 22–34-kDa high molecular weight FGF-2 isoforms created by alternative initiation codons (33). Membrane signaling involves extracellular LMW-FGF-2 formation of ternary complexes with cell surface FGF receptors and heparin-sulfate proteoglycans (34). Cell surface FGFRs are principally coupled to PI3K/Akt, RAS/MAPK, and PLCγ intracellular signaling pathways (35). In contrast, HMW-FGF-2 isoforms have an N-terminal nuclear localization sequence (NLS) that leads to nuclear localization and activation of intracellular FGFR1/CBP/CREB signaling pathways (also called integrative FGFR1 nuclear pathway or INFS) (36). INFS appears to be the earliest evolved FGFR signaling mechanism (37). To understand the roles of membrane FGFR1 and INFS in regulating FGF-23 transcription, we examined the effects LMW-FGF-2 and HMW-FGF-2 on FGF-23 transcription in osteoblasts cell lines.
EXPERIMENTAL PROCEDURES
Cell Culture and Promoter Analysis
MC3T3-E1 osteoblast precursor cells and SaOS-2 osteoblast cells were cultured according to American Type Culture Collection guidelines. Briefly, 3–5 × 104 cells were seeded in 6-cm diameter tissue culture plates in α-MEM (Life Technologies, Grand Island, NY) with 10% fetal calf serum at 37 °C in the presence of 5% CO2 in a humidified incubator. Cells were plated 18 h before transfection and fed fresh medium 4 h before transfection. FGF-23 promoters (mFGF-23 and hFGF-23) DNA were constructed into a pGL3 basic reporter gene (Promega, Madison, WI). To create mutations of NFAT, CREB, or both NFAT/CREB sites in the FGF-23 promoter, a GENEART Site-directed Mutagenesis System (Life Technologies) was used by following the manufacturer's instructions. All FGF-23 reporter plasmid DNAs were introduced into MC3T3-E1 or SaOS-2 cells using cationic liposomes (LipofectAMINE2000, Life Technologies). Co-transfections (0.25 μg of FGF-23 promoter plasmid DNAs with FGFR1, FGFR2, FGFR3, FGFR4, HMW-FGF-2, or FGFR1(TK−), FGFR1(SP−/NLS), FGFR1(TK−/SP−/NLS) plasmid DNAs) was carried out for 16–18 h, and then cells were washed twice with phosphate-buffered saline and incubated in fresh medium containing 10% fetal calf serum for 38 h. LMW-FGF-2, Cyclosporine A (Sigma), or U0126 (Cell Signaling Technology) was added to the cell cultures 24 h before the cells were harvested. To standardize the transfection efficiency, 0.1 μg of pRL-CMV vector (pRL Renilla reniformis luciferase control reporter vector; Promega) was cotransfected in all experiments. Cells were harvested 72 h after transfection and lysed in 50 μl of reporter lysis buffer (Promega). A luciferase assay (20 μl of cell lysed) was performed using a dual luciferase assay kit (Promega), and activity was measured with an Optocomp 1 luminometer (MGM Instruments, Inc., Hamden, CT). Promoter activity (mean ± S.D. of triplicate samples in relative fold-changes) is represented by the reporter activity normalized to the pRL-CMV control. Graphs represent typical results of 3–4 separate experiments. To induce osteogenic differentiation for MC3T3 and SaOS-2 cells, cells were first cultured in the α-MEM containing 10% FBS for 3 days to grow cells to about 80% confluence. Then, these cells were cultured in serum-free medium supplemented with 100 μg/ml of ascorbic acid, 10 mm β-glycerophosphate for another 18 days. Supernatants from day 21 cells cultured in the conditional medium were collected and concentrated using Amicon Ultra-4 Centrifugal Filters (10K) (Millipore, Temecula, CA). Full-length FGF-23 levels were measured using the FGF-23 ELISA kit (Kainos Laboratories, Tokyo, Japan) following the manufacturer's recommendations.
Chromatin Immunoprecipitation (ChIP) Assays
ChIP assays were performed with a kit from Cell Signaling Technology (Danvers, MA) according to the manufacturer's instructions with the following modifications. Approximately 4 × 107 MC3T3-E1 or SaOS-2 cells in 100-cm2 culture dishes were cross-linked in 1% formaldehyde solution (Fisher Scientific, Pittsburgh, PA) for 10 min at room temperature, the cross-linking was stopped by adding 1 m glycine for 5 min at room temperature. Cells were washed twice with ice-cold 1× phosphate-buffered saline solution (PBS) (Life Technologies) and harvested in 1-ml of 1× PBS + protease inhibitor mixture (PIC) + phenylmethylsulfonyl fluoride (PMSF) (Sigma). Cells were then pelleted by centrifugation at 1500 × g for 5 min at 4 °C and the pellet was resuspended in 10 ml of ice-cold buffer A with DTT, PIC, and PMSF on ice for 10 min. Cell nuclei were pelleted by centrifugation at 3000 × g for 5 min at 4 °C. The nuclei pellet was then washed in 10 ml of ice-cold buffer B with DTT, and resuspended in 1.0 ml of buffer B with DTT. Micrococcal nuclease was added to the nuclei and incubated for 20 min at 37 °C. Digestion was stopped by adding 100 μl of 0.5 m EDTA, and the nuclei were pelleted by centrifugation at 13,000 × g for 1 min at 4 °C. The nuclear pellet was resuspended in 0.5 ml of 1× ChIP buffer with PIC and PMSF for sonication using the VirSonic Ultrasonic cell disrupter 100 (VirTis, Gardiner, NY) to shear the DNA to an average length of 300 to 500 base pairs (six, 15-s bursts on ice). Samples were stored at −80 °C before use. For chromatin immunoprecipitation, 200 μl of the cross-linked chromatin preparation were added to 800 μl of ChIP buffer with PIC. Immunoprecipitations were carried out with 2 μg of antibodies (CREB, Est-1, or NFATc1 from Santa Cruz Biotechnology, Santa Cruz, CA, and FGFR1 from Cell Signaling) and 0.2 μg of p-CREB (Santa Cruz). Histone H3 rabbit mAb (2 μg) and normal rabbit IgG (1 μg) were used as positive or negative controls, respectively. After overnight immunoprecipitation on a rotator at 4 °C, ChIP samples were then washed and eluted and DNA was uncross-linked with NaCl and subsequently treated with proteinase K, Tris-HCl, and EDTA. DNA was purified and subjected to PCR amplification of the FGF-23 promoter DNA containing putative binding sites for CREB and NFAT transcription factors using specific primers (mouse: forward primer, 5′-tgacccagggtcacagata-3′, reverse primer, 5′-gaactgaggtggggatcctg-3′; human: forward primer, 5′-cgcctccggggtctttgca-3′, reverse primer, 5′-gtggctggtgctgagatttga-3′).
RT-PCR
Total RNA was extracted from SaOS-2 and MC3T3-E1 osteoblasts using an RNeasy Mini Kit (Qiagen) according to the manufacturer's instructions. cDNAs were synthesized by using a iScriptTM cDNA Synthesis Kit (Bio-Rad). RT-PCR was performed to examine the FGF-23 mRNA, FGFR1, FGFR2, FGFR3, and FGFR4 in the SaOS-2 and MC3T3-E1 osteoblasts using a pair of primers as follows: human FGF-23, 5′-cagcatgagcgtcctcagag-3′ (forward) and 5′-gccagcatcctctgatctgatc-3′ (reverse); mouse FGF-23, 5′-caactggggaagcctgacc-3′ (forward) and 5′-ccttcgagtcatggctcctg-3′ (reverse); human FGFR1, 5′-aaggacaaacccaaccgttgacc-3′ (forward) and 5′-cccaaagtctgctatcttcatcac-3′ (reverse); human FGFR2, 5′-ctgtgccgaatgaagaacacgacc-3′ (forward) and 5′-cccaaagtctgctatcttatcac-3′ (reverse); human FGFR3, 5′-tgaggacacaggtgtggacacagg-3′ (forward) and 5′-tctgccggatgctgccaaacttgt-3′ (reverse); human FGFR4, 5′-ctcgccggcctcgtgagtctagat-3′ (forward) and 5′-cccaaagtctgctatcttcatcac-3′ (reverse); mouse FGFR1, 5′-aacctctaaccgcagaac-3′ (forward) and 5′-gagactccacttccacag-3′ (reverse); mouse FGFR2, 5′-atcatcgcctgctccatc-3′ (forward) and 5′-gctgttgttactgctgttcc-3′ (reverse); mouse FGFR3, 5′-cttcagtgtgcgtgtaac-3′ (forward) and 5′-ttctttgccattcttcagc-3′ (reverse); mouse FGFR4, 5′-atgaccgtcgtacacaatcttac-3′ (forward) and 5′-tgtccagtagggtgcttgc-3′ (reverse) and mouse GAPDH, 5′-tatgtcgtggagtactgg-3′ (forward) and 5′-agtgatggcatggactgtgg-3′ (reverse).
Western Blot Analysis
SaOS-2 cells (1 × 106) were cultured in 10-cm dishes overnight as described above and transfected with human FGFR1 plasmid DNA (1 μg) with or without co-transfection of HMW-FGF-2 plasmid DNA. Transfections were carried out for 16–18 h, and then cells were washed twice with phosphate-buffered saline and incubated in fresh medium containing 10% fetal calf serum for 38 h. Forskolin (10 μm) was added to the cell cultures 24 h before cells were harvested. Nuclear and cytoplasmic proteins were isolated using a NE-PER Nuclear and Cytoplasmic Extraction kit (Thermo Scientific, Rockford, IL). Protein contents in the samples were quantified and stored at −80 °C until use. We loaded 150 μg of cell lysates to each well of SDS-PAGE gel. For electrophoresis, samples were prepared by mixing 3× SDS loading buffer (Cell Signaling) with 1× DTT. About 50 μg of protein were loaded onto NuPAGE 4–12% BisTris gels (Invitrogen). Proteins were separated at 150 V for 60 min and transferred to nitrocellulose membrane (Invitrogen). Membranes were blocked with Superblock blocking buffer in TBST (Thermo Scientific, Rockford, IL) for 30 min and then incubated with primary antibody (FGFR1, FGFR3, FGFR4, 1:1000, Cell Signaling Technology. FGFR2, 1:1000, Abcam, Cambridge, MA) with gentle agitation overnight at 4 °C. After 3 washes with TBST (15 min once and 2× 5 min), the membrane was incubated with secondary antibody in a Superblock blocking buffer at room temperature for 1 h. Membrane was then washed 4 times (15 min and 3× 5 min) and subjected to ECL (Thermo Scientific) and analyzed with the FOTO/Analyst Luminary/FX imaging work station (FOTODYNE INCORPORATED, Hartland, WI). Western blot using β-actin antibody or Lamin B antibody (Santa Cruz Biotechnology) was used as an internal control of protein loadings of cytoplasmic or nuclear proteins, respectively.
Statistics
We evaluated differences between two groups by unpaired t test and multiple groups by one-way analysis of variance. All values are expressed as mean ± S.D. All computations were performed using GraphPad Prism 5 (GraphPad Software Inc., La Jolla, CA).
RESULTS
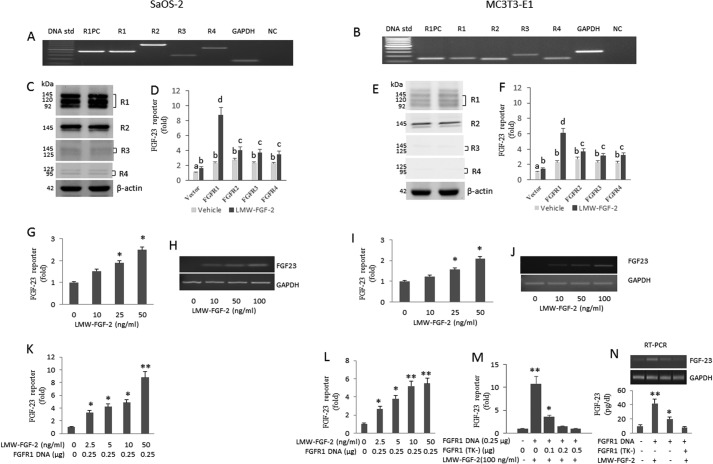
We investigated membrane and INFS signaling, respectively, by assessing the response to treatment with LMW-FGF-2 (also known as basic FGF) and by transfection of HMW-FGF-2 cDNA. We evaluated the response in both human SaOS-2 osteoblasts and mouse-derived MC3T3-E1 osteoblasts, which are known to express FGF-23 (32, 38). In SaOS-2 osteoblasts, we detected all four FGFR transcripts by RT-PCR (Fig. 1A). The apparent expression of FGFR1 and FGFR2 was greater than FGFR4 transcripts; FGFR3 expression was minimally detected in SaOS-2 by RT-PCR (Fig. 1A). We found that MC3T3-E1 osteoblasts express FGFR1, -2, and -3 transcripts. We also identified FGFR-4 transcripts in MC3T3-E1 osteoblasts (Fig. 1B), which were previously reported to be absent in these cells (39).
FIGURE 1.
LMW-FGF-2 up-regulates FGF-23 gene transcription through FGFR1 signaling. A and B, expression of FGFR1, -2, -3, and -4 in SaOS2 and MC3T3-E1 osteoblast cells. R1PC, FGFR1 positive control; R1, FGFR1; R2, FGFR2; R3, FGFR3; and R4, FGFR4; NC, negative control. C and E, Western blot analysis of R1, R2, R3, and R4 in SaOS-2 or MC3T3-E1 cells, respectively. D and F, effect of co-transfection of R1, R2, R3, or R4 on FGF-23 promoter activity with or without LMW-FGF-2 (50 ng/ml) treatment in SaOS-2 or MC3T3-E1 cells, respectively. G and I, dose-dependent stimulation of human and mouse promoter activity by LMW-FGF-2 (10–50 ng/ml). H and J, RT-PCR analysis of FGF-23 gene expression simulated by LMW-FGF-2 in SaOS-2 and MC3T3-E1 osteoblasts cultured in a conditional medium containing 50 μg/ml of ascorbic acid and 5 mm β-glycerophosphate for 3 weeks. LMW-FGF-2 (0–100 ng/ml) was added to the medium at day 20 of culture and cells were cultured for another 24 h before harvesting for RNA isolation. K and L, co-transfection of FGFR1 enhanced the effect of LMW-FGF-2 on FGF-23 promoter activity in both SaOS2 and MC3T3-E1 osteoblasts. M, dominate-negative FGFR1(TK−) construct blocked the stimulatory effect of LMW-FGF-2 on mFGF-23 promoter activity in a dose-dependent manner. N, LMW-FGF-2 (100 ng/ml) stimulated FGF-23 gene expression in FGFR1 overexpression MC3T3-E1 cells. Dominate-negative FGFR1(TK−) construct (1.0 μg) blocked the stimulatory effect of LMW-FGF-2 (100 ng/ml) on FGF-23 gene expression in FGFR1 overexpression MC3T3 cells cultured in a conditional medium for 3 weeks. RT-PCR of FGF-23 mRNA (top) and FGF-23 protein (bottom) are shown. Data are expressed as the mean ± S.D. from three independent experiments. Values sharing the same superscript in different groups are not significantly different. *, p < 0.05; **, p < 0.01 versus control vector group, respectively.
We also evaluated FGFR1, FGFR2, FGFR3, and FGFR4 protein expression by Western blot analysis using specific monoclonal antibodies for each FGFR in both cell lines. Consistent with the results from RT-PCR analysis, we detected the expected size bands corresponding to all four FGFR proteins in SaOS-2 osteoblasts. The expression of FGFR1 and FGFR2 was greater than FGFR3 and FGFR4 proteins in SaOS-2 osteoblasts (Fig. 1C). We also found that MC3T3-E1 osteoblasts expressed FGFR1 and FGFR2 (Fig. 1E), but FGFR3 and FGFR4 were minimally detected in these cells (Fig. 1E).
Membrane FGFR Regulation of FGF-23
LMW-FGF-2 Stimulates FGF-23 Promoter Activity Principally through FGFR1
To study the role of specific FGFRs in FGF-23 gene regulation, human p700-FGF-23 proximal promoter and mouse p600-FGF-23 proximal promoter-luciferase constructs were, respectively, co-transfected with vector DNA, FGFR1, FGFR2, FGFR3, or FGFR4 in SaOS-2 and MC3T3-E1 cells and stimulated with LMW-FGF-2 (50 ng/ml). We found that LMW-FGF-2 resulted in a minimal 2-fold increase in promoter activity in control (vector-transfected) SaOS-2 cells, reflecting endogenous FGFR activation (Fig. 1D). Co-transfection of FGFR1, FGFR2, FGFR3, or FGFR4 in the absence of LMW-FGF-2, also resulted in a minimal 2–3-fold increase in FGF-23 promoter activity as compared with vector control in SaOS-2 cells. LMW-FGF-2 in FGFR1-cotransfected SaOS-2 cells resulted in a marked 9-fold increase of FGF-23 promoter activity (Fig. 1D). In contrast, LMW-FGF-2 resulted in only a 3–4-fold increase in FGFR2-, FGFR3-, or FGFR4-cotransfected SaOS-2 cells (Fig. 1D). Similar results were found in MC3T3-E1 cells (Fig. 1F).
We found that LMW-FGF-2 resulted in a dose-dependent increase in promoter activity, reaching an approximate 2–3-fold increase in both cell types at LMW-FGF-2 concentrations of 50 ng/ml (Fig. 1, G and I). LMW-FGF-2 also stimulated increments in endogenous FGF-23 transcripts in SaOS-2 and MC3T3-E1 osteoblasts as assessed by RT-PCR (Fig. 1, H and J).
To further investigate the specific role of FGFR1, we tested the effects of LMW-FGF-2 to activate the FGF-23 promoter in the presence of a co-transfected wild-type FGFR1 or FGFR1(TK−) mutant cDNA construct that lacks the tyrosine kinase domain and forms non-functional dimers with endogenous FGFR1 receptors. Co-transfection of FGFR1 cDNA resulted in a 6–10-fold stimulation in FGF-23 promoter activity for SaOS-2 and MC3T3-E1 osteoblasts (Fig. 1, K and L). This response was enhanced compared with the 2–3-fold induction of FGF-23 by endogenous FGFRs. In contrast, overexpression of the dominant-negative FGFR1(TK−) mutant resulted in inhibition of LMW-FGF-2-mediated FGFR1 stimulation of the FGF-23 promoter in MC3T3-E1 osteoblasts in a dose-dependent manner (Fig. 1M). LMW-FGF-2 also stimulated FGFR1-mediated FGF-23 gene expression in MC3T3-E1, which was inhibited by co-transfection of the dominant-negative FGFR1(TK−) construct (Fig. 1N), as determined by RT-PCR of FGF-23 mRNA (top) and FGF-23 protein (bottom).
To assess the effects of LMW-FGF-2 on FGF-23 protein expression in cultured osteoblasts, we compared FGF-23 secretion in the medium of MC3T3-E1 osteoblasts after transfection with wild-type FGFR1 or FGFR1(TK−) mutant constructs. FGF-23 levels in conditioned medium were ∼10 pg/dl before LMW-FGF-2 stimulation and increased to ∼40 pg/dl after administration of LMW-FGF-2 to cells transfected with FGFR1. In contrast, FGFR1(TK−)-transfected osteoblasts did not increase the FGF-23 levels in response to LMW-FGF-2 stimulation (Fig. 1J).
FGFR-dependent Activation of the FGF-23 Promoter through PLCγ and Calcineurin-NFAT Signaling Cascade
Cell surface FGFR1 is coupled to at least three major signaling pathways, including PI3K/AKT, Ras/MAPK, in which ERK plays a central role, and PLCγ pathways (40, 41). We have previously shown that ERK inhibitors (PD98059, which inhibits MAPK kinase kinase and U0126, which inhibits MEK1 and MEK2) inhibit LMW-FGF-2-mediated FGFR1 activation of the FGF-23 promoter in MC3T3-E1 cells (32). In contrast, the phosphoinositide 3-kinase inhibitor wortmannin had no effect of LMW-FGF-2 stimulation of FGF-23 promoter activity in MC3T3-E1 osteoblasts (32).
Because the calcineurin-NFAT cascade is an important downstream target of PLCγ (42) and Ets-1 is an key cis-acting element in the MAPK/ERK signaling cascade (43), we examined whether the proximal FGF-23 promoter possessed NFAT and Ets-1 binding sites. We found a putative consensus Ets-1 site that overlaps an NFAT site adjacent to a CREB binding site at positions −182 and −165 bp in the mouse FGF-23 promoter (44). These sites were conserved in the human FGF-23 promoters.
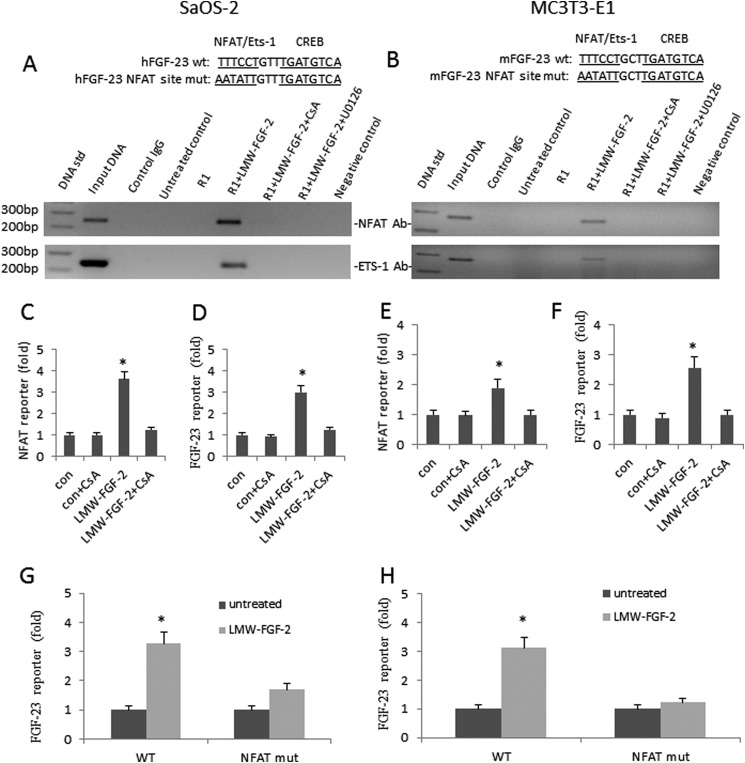
To establish that LMW-FGF-2 stimulates NFAT or Ets-1 binding to the consensus NFAT or Ets-1 binding site in the FGF-23 promoter, we performed ChIP assays using an NFAT or Ets-1 antibody and RT-PCR on the immunoprecipitates with primers to amplify the region spanning the putative NFAT or Ets-1 binding sites. We found that stimulation of either SaOS-2 or MC3T3-E1 osteoblasts with LMW-FGF-2 resulted in binding of NFAT and Ets-1 to the endogenous FGF-23 proximal promoter region (Fig. 2, A and B). This binding was inhibited by treatment of osteoblasts with the calcineurin inhibitor CsA (Fig. 2, A and B) or with the MEK1 and MEK2 inhibitor U0126 (Fig. 2, A and B). Interestingly, inhibition of calcineurin by CsA or ERK1/2 by U0126 also blocks Ets-1 and NFAT binding to the FGF-23 promoter in both osteoblast cell lines (Fig. 2, A and B).
FIGURE 2.
LMW-FGF-2 up-regulates FGF-23 gene transcription via NFAT and MAPK pathways. A and B, LMW-FGF-2 enhanced NFAT and Ets-1 binding to the endogenous FGF-23 promoter as determined by CHIP assay using NFAT or Ets-1 antibody. First lane, 100-bp DNA standard; second lane, input ChIP DNA; third lane, nonspecific IgG; fourth lane, untreated control cells; fifth lane, FGFR1 co-transfected cells; sixth lane, FGFR1 co-transfected cells treated with LMW-FGF-2 (50 ng/ml) for 24 h; seventh lane, FGFR1 co-transfected cells treated with LMW-FGF-2 (50 ng/ml) in the presence of CsA (1 μm) for 24 h; eighth lane, FGFR1 co-transfected cells treated with LMW-FGF-2 (50 ng/ml) in the presence of U0126 (10 μm) for 24 h; and ninth lane, negative control. C and E, LMW-FGF-2 stimulates NFAT activity in both SaOS-2 and MC3T3-E1 osteoblasts. Cyclosporine A (1 μm) blocks the effect of LMW-FGF-2 on NFAT reporter gene expression. D and F, LMW-FGF-2 stimulates FGF-23 promoter activity in both SaOS-2 and MC3T3-E1 osteoblasts. Cyclosporine A (1 μm) blocks the effect of LMW-FGF-2 on FGF-23 reporter gene expression. G and H, LMW-FGF-2 stimulates both human and mouse FGF-23 promoter activities. NFAT site mutation blocks LMW-FGF-2 effect. Data are expressed as the mean ± S.D. from three independent experiments. *, p < 0.05 versus control vector group.
To evaluate the role of phospholipase Cγ-induced NFAT signaling, we used a NFAT reporter gene (Qiagen, Valencia, CA) to test the ability of LMW-FGF-2 to stimulate NFAT promoter activity in SaOS-2 and MC3T3-E1 osteoblasts. Incubation with LMW-FGF-2 stimulated NFAT activity by ∼3-fold in both SaOS-2 and MC3T3-E1 osteoblasts (Fig. 2, C and E). To test the ability of the calcineurin inhibitor CsA to block LMW-FGF-2 stimulation of NFAT and FGF-23 promoter activity, we treated the cells with CsA. Incubation with CsA resulted in inhibition of FGF-2 stimulation of NFAT and FGF-23 reporter activity in both SaOS-2 and MC3T3-E1 osteoblasts, achieving an almost complete inhibition at concentrations of 1.0 μm (Fig. 2, C–F).
Finally, to confirm the role of NFAT signaling through the identified cis-acting element, we mutated the NFAT site in both the human and mouse FGF-23 promoter (Fig. 2, G and H). LWM-FGF-2 effects to significantly stimulate the wild-type FGF-23 promoter activity in SaOS-2 and MC3T3-E1 osteoblasts was lost when the transfected NFAT mutant FGF-23 promoter was stimulated with LMW-FGF-2 (Fig. 2, G and H). The identification of NFAT protein binding to and activation of NFAT sites following LMW-FGF-2 stimulation is consistent with a role for calcineurin pathways mediating the effects of membrane FGFR1 to activate the FGF-23 promoter.
Integrative Nuclear FGFR1 Signaling
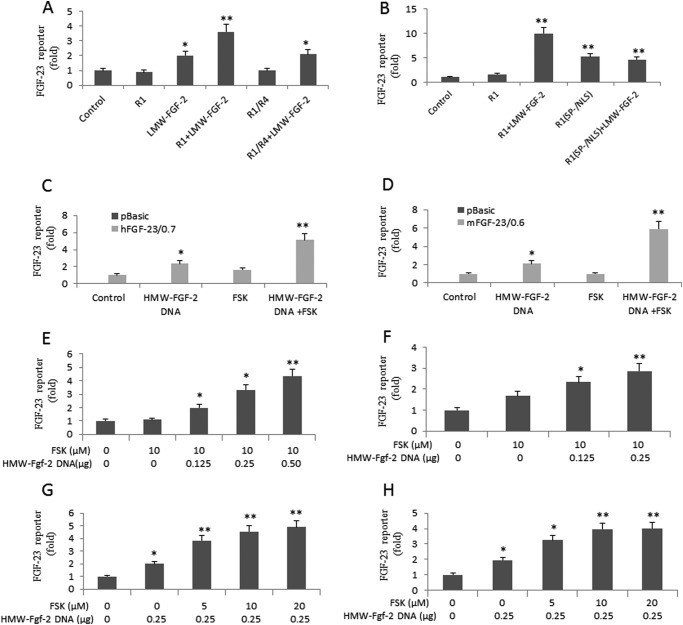
To explore the possibility for a role of INSF signaling, we initially tested “membrane arrested” FGFR1–4, a chimeric receptor in which the transmembrane domain of FGFR1 is replaced with the transmembrane domain of FGFR4 to limit receptor internalization, and the FGFR1(SP−/NLS)-mutated receptor, in which the secretory peptide (SP) required for plasma membrane insertion has been replaced with a NLS to promote nuclear uptake. Co-transfection of wild-type FGFR1 had no effect on FGF-23 promoter activity in the absence of ligand. However, addition of LMW-FGF-2 resulted in a 4-fold enhancement of FGF-23 promoter activity in FGFR1-transfected cells compared with control cells. Cells stimulated with LMW-FGF-2 alone only resulted in a 2-fold enhancement of FGF-23 promoter activity, reflecting the function of endogenous FGFRs. Co-transfection of the chimeric FGFR1/FGFR4 construct did not result in LWM-FGF-2 stimulation of FGF-23 promoter activity above that of endogenous FGFR activation in MC3T3-E1 osteoblasts (Fig. 3A). In contrast, the FGFR1 (SP−/NLS) mutant, which preferentially translocates to the nucleus, stimulated a 5-fold increase of FGF-23 promoter activity in the absence of the exogenous ligand (Fig. 3B). The addition of LMW-FGF-2 failed to further stimulate FGF-23 promoter-luciferase activity in cells transfected with the FGFR1(SP−/NLS) (Fig. 3B). These findings are consistent with the presence of INSF signaling in osteoblasts.
FIGURE 3.
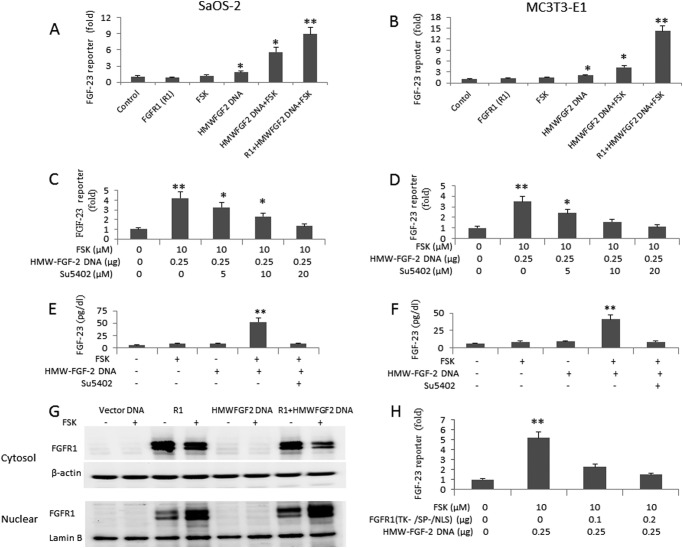
HMW-FGF-2 and cAMP activate integrative nuclear FGFR1 signaling pathway. A, effect of the chimeric FGFR1/FGFR4 construct on LWM-FGF-2 (25 ng/ml) stimulation of FGF-23 promoter activity in MC3T3-E1 osteoblasts. B, effect of the FGFR1 (SP−/NLS) mutant (0.25 μg) on LWM-FGF-2 (100 ng/ml) stimulation of FGF-23 promoter activity in MC3T3-E1 osteoblasts. C and D, co-transfection of HMW-FGF-2 (0.25 μg) stimulates FGF-23 promoter activity in both human and mouse osteoblast cells, whereas FSK (10 μm) alone shows little effect. Combination of HMW-FGF-2 and FSK enhances FGF-23 promoter activity by more than 5-fold as compared with controls. E and F, dose-dependent up-regulation of FGF-23 promoter activity by HMW-FGF-2 with fixed FSK (10 μm). G and H, dose-dependent up-regulation of FGF-23 promoter activity by FSK with fixed HMW-FGF-2 (0.25 μg). Data are expressed as the mean ± S.D. from three independent experiments. Values sharing the same superscript in different groups are not significantly different. *, p < 0.05; **, p < 0.01 versus control vector group, respectively.
To directly test the role of INSF signaling in the regulation of FGF-23 promoter activity, we transfected a HMW-FGF-2 cDNA that encodes the intracellular ligand for intranuclear FGFR1 (36) into osteoblasts. We also tested the co-dependence of cAMP on HMW-FGF-2 regulation of FGF-23 promoter activity, because FGFR1 and HMW-FGF-2 co-localize in the nuclear matrix to activate gene transcription through interactions with the CBP (36, 45, 46).
In both SaOS-2 and MC3T3-E1 osteoblasts, transfection of HMW-FGF-2 (0.25 μg) alone resulted in an approximate 2-fold stimulation of FGF-23 promoter activity (Fig. 3, C and D). Surprisingly, addition of only forskolin (FSK, 10 μm) to the medium had no significant effects to stimulate the FGF-23 promoter activity in either SaOS-2 or MC3T3-E1 osteoblasts. However, in the presence of fixed amounts of FSK (10 μm), we observed a marked accentuation of the ability of HMW-FGF-2 to stimulate FGF-23 promoter activity. The combined effect of HWM-FGF-2 and FSK resulted in a 5–6-fold increase in FGF-23 promoter activity (Fig. 3, C and D). In the presence of FSK, a dose-dependent effect of HMW-FGF-2 to increase the FGF-23 promoter activity in both SaOS-2 and MC3TE-E1 osteoblasts was observed (Fig. 3, E and F). Conversely, in cells transfected with fixed amounts of transfected HMW-FGF-2 (0.25 μg), FSK also show a dose-dependent stimulation of FGF-23 promoter activity (Fig. 3, G and H). Thus, consistent with current models of INSF signaling, HMW-FGF-2 and cAMP-dependent pathways act in a cooperative fashion.
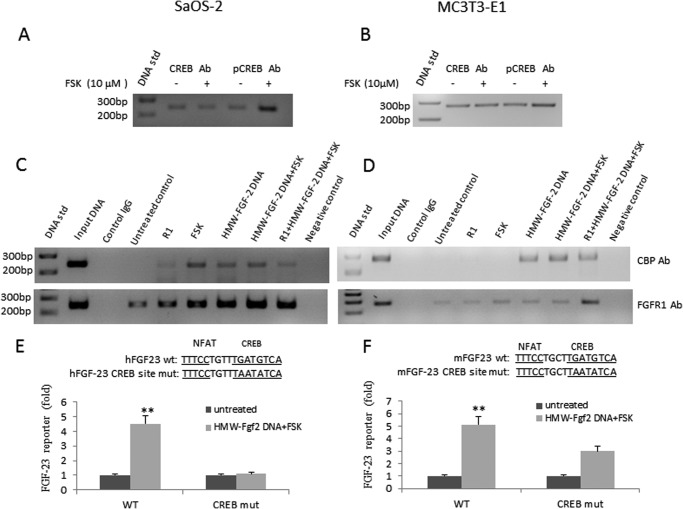
Next, we examined if CREB binds to the CRE site of the FGF-23 promoter located adjacent to the NFAT cis-element. We performed ChIP assays using CREB and p-CREB antibodies. We found low levels of CREB binding to the CRE site in the FGF-23 promoter under unstimulated conditions in both SaOS-2 and MC3T3-E1 osteoblasts (Fig. 4, A and B). FSK treatment increased pCREB binding to the FGF-23 promoter.
FIGURE 4.
INFS up-regulates FGF-23 gene transcription through the CREB pathway. A and B, ChIP assay shows CREB binds to endogenous FGF-23 promoter in both SaOS-2 and MC3T3-E1 osteoblasts. Simulation of cells with forskolin (10 μm) increases pCREB binding. C and D, CBP and FGFR1 bind to the endogenous FGF-23 promoter and such binding is further enhanced after activation of INFS by a combination of FSK and HMW-FGF-2. First lane, 100-bp DNA ladder; second lane 2, input ChIP DNA; third lane, nonspecific IgG; fourth lane, untreated control; fifth lane, co-transfection of FGFR1; sixth lane, FSK-treated cells; seventh lane, co-transfection of HMW-FGF-2; eighth lane, co-transfection of HMW-FGF-2 plus FSK; ninth lane, Co-transfection of FGFR1 and HMW-FGF-2 plus FSK; tenth lane, negative control. E and F, mutation of CREB site blocks the effect of HMW-Fgf-2 (0.25 μg) and FSK (10 μm) on FGF-23 promoter activity. Data are expressed as the mean ± S.D. from three independent experiments. **, p < 0.01 versus control vector group, respectively.
To investigate if HMW-FGF-2 and FGFR1 form a nuclear signaling complex that binds to the CREB binding site in the FGF-23 promoter, we next performed ChIP assays using CBP or FGFR1 antibodies. In SaOS-2 osteoblasts, we detected FGFR1 but not CBP binding to the CREB site in the FGF-23 promoter under unstimulated conditions (Fig. 4C). Transfection of FGFR1 or treatment with FSK resulted in an increase in both FGFR1 and CBP binding. Transfection of HMW-FGF-2 alone and in combination with FSK treatment and FSK treatment plus FGFR1 co-transfection, maximally increased binding of FGFR1 to the endogenous FGF-23 promoter (Fig. 4C). Similar increments in FGFR1 and CBP binding to CREB were observed in MC3T3-E1 transfected with HMW-FGF-2 and treated with FSK, and co-transfected with FGFR1 (Fig. 4D). MC3T3-E1 osteoblasts differed from SaOS-2 in the lack of CBP binding to CREB after FGFR1 transfection or FSK treatment alone (Fig. 4D).
To investigate the functional relevance of the ChIP assay results, we performed mutagenesis of the CREB binding site. Mutation of CREB completely blocked the ability of combined HMW-FGF-2 and FSK to stimulate FGF-23 promoter activity in SaOS-2 (Fig. 4E). In MC3T3-E1 osteoblasts the response to HMW-FGF-2 and FSK stimulation by FGF-23 promoter activity was also inhibited (Fig. 4F).
To further explore the role of FGFR1, we examined the effect of co-transfection of FGFR1 on combined HMW-FGF-2 and FSK-stimulated FGF-23 promoter activity. Transfection of FGFR1 resulted in a 10–15-fold increase in FGF-23 promoter activity in SaOS-2 and MC3T3-E1 osteoblasts following co-transfection of HMW-FGF-2 and FSK treatments (Fig. 5, A and B). We also examined the effects of the FGFR-specific chemical inhibitor SU5042 on the ability of HMW-FGF-2/FSK to activate FGF-23 promoter activity and production of FGF-23 protein in SaOS-2 and MC3T3-E1 osteoblasts. We found that treatment with SU5042 resulted in a dose-dependent inhibition of FGF-23 promoter activity (Fig. 5, C and D) and FGF-23 protein levels in conditioned media (Fig. 5, E and F) in both cell lines. We further demonstrated that HMW-FGF-2/cAMP-induced INSF signaling involves the translocation of FGFR1 from cytoplasmic into nuclear (Fig. 5G). We showed that osteoblasts expressed a low level of endogenous FGFR1 as determined by Western blot. Then, we overexpressed a human FGFR1 cDNA by transient transfection into SaOS-2 cells and studied the integrative nuclear FGFR1 translocation stimulated by HMW-FGF-2 with or without forskolin treatment. We found that cAMP increased nuclear FGFR1 accumulation in osteoblasts. Co-transfection of HMW-FGF-2 also increased the nuclear FGFR1 level. Moreover, a combination of forskolin and HMW-FGF-2 further increased FGFR1 translocation from cytosolic to nuclear (Fig. 5G). Dominant-negative FGFR1 (TK−/SP−/NLS) mutant blocks HMW-FGF-2/cAMP-induced INSF signaling (Fig. 5H).
FIGURE 5.
Up-regulation of FGF-23 by HMW-FGF-2 and FSK requires functional FGFR1. A and B, effect of co-transfection of FGFR1 on FGF-23 promoter activity stimulated by INFS signaling. C and D, the FGFR1 inhibitor Su5402 blocks the effect of HMW-FGF-2 and FSK on FGF-23 promoter activity in a dose-dependent manner in both human and mouse osteoblasts. E and F, a combination of HMW-FGF-2 and FSK stimulates FGF-23 secretion by SaOS-2 and MC3T3 osteoblast cells cultured in a conditioned medium containing 50 μg/ml of ascorbic acid and 5 mm β-glycerophosphate for 3 weeks. Treatment of these cells with Su5402 (10 μm) during the last 24 h of culture blocks FGF-23 expression. G, Western blot analysis of FGFR1 cytoplasmic and unclear allocations stimulated by forskolin with or without HMW-FGF-2 co-transfections. β-Actin or Lamin B blot was used for internal controls. H, the dominant-negative FGFR1 (TK−/SP−/NLS) mutant blocked the effect of HMW-GFG-2/cAMP on FGF-23 promoter activity in a dose-dependent manner. Data are expressed as the mean ± S.D. from three independent experiments. Values sharing the same superscript in different groups are not significantly different. *, p < 0.05; **, p < 0.01 versus control vector group, respectively.
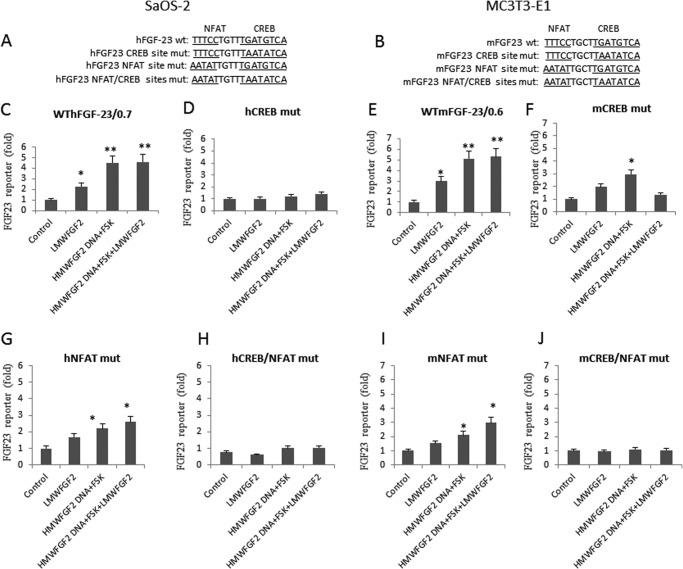
Finally, we examined the effects of inactivation of the CREB and NFAT binding sites for the FGF-23 promoter by LMW-FGF-2 and HMW-FGF-2 stimulation of FGF-23 transcription (Fig. 6, A and B). LMW-FGF-2 stimulated FGF-23 promoter activity 2–3-fold, whereas HMW-FGF-2 plus FSK stimulated FGF-23 promoter activity ∼5-fold in the wild-type promoter. Addition of LMW-FGF-2 to the combined HMW-FGF-2 plus FSK did not stimulate the FGF-23 promoter activity further (Fig. 6, C and E, 4th column). Mutation of the CREB binding site completely eliminated effects of both LMW-FGF-2, and combined HMW-FGF-2 and FSK to stimulate FGF-23 promoter activity in SaOS-2 osteoblasts (Fig. 6D). Mutation of the CREB binding site also inhibited LMW-FGF-2 and HMW-FGF-2 plus FSK stimulation of the FGF-23 promoter in MC3T3-E1 osteoblasts (albeit less completely than in SaOS-2 cells) (Fig. 6F). Thus, CREB binding to CRE is important for both LMW- and HMW-FGF-2 signaling in osteoblasts. Mutation of the NFAT binding site prevented the effects of LMW-FGF-2 and HMW-FGF-2 plus FSK significantly to the stimulated FGF-23 promoter activity in both SaOS-2 and MC3T3-E1 osteoblasts (Fig. 6, G and I), although the magnitude of the response was less (compare Fig. 6, G and I with C and E). Mutation of both CREB and NFAT binding sites resulted in loss of LMW-FGF-2 and HMW-FGF-2 plus FSK effects in both SaSO-2 and MC3T3-E1 osteoblasts (Fig. 6, H and J).
FIGURE 6.
Membrane and INFS FGFR1 signaling regulates FGF-23 in a cooperative fashion. A and B, human and mouse FGF-23 promoter DNA sequence containing wild-type NFAT and CREB sites, and NFAT site mutation, CREB mutation, or both NFAT and CREB site mutations. Effect of LMW-FGF-2 (50 ng/ml), HMW-FGF-2 and FSK, or HMW-Fgf-2 and FSK plus LMW-FGF-2 on the promoter activity of the wild-type FGF-23 reporter (C and E), CREB mutant (D and F), NFAT mutant (G and I), or CREB/NFAT mutant (H and J), respectively. Data are expressed as the mean ± S.D. from three independent experiments. Values sharing the same superscript in different groups are not significantly different. *, p < 0.05; **, p < 0.01 versus control vector group, respectively.
DISCUSSION
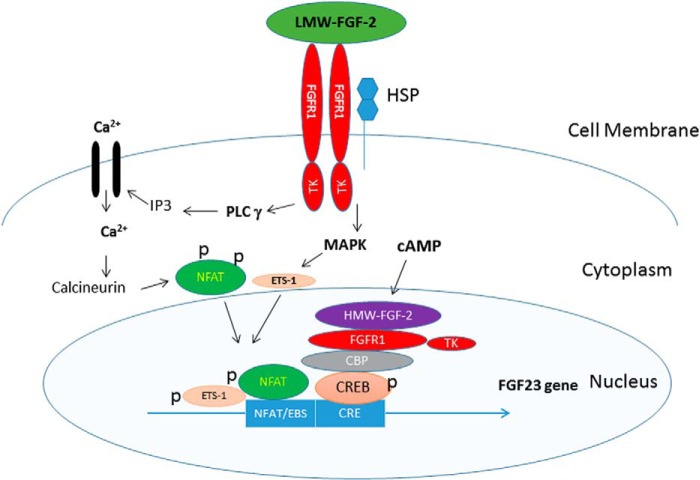
Our studies show that LMW-FGF-2 and HMW-FGF-2 activate the FGF-23 promoter in osteoblasts through the respective FGFR membrane and INFS pathways (Fig. 7). LWM-FGF-2 activation of membrane FGFRs is coupled to FGF-23 gene transcription through activation of PLCγ-dependent NFAT and MAPK-dependent signaling leading to binding of phosphorylated NFAT and ETS-1 to binding sites in the proximal FGF-23 promoter. In contrast, HMW-FGF-2 acts in concert with cAMP-dependent pathways to stimulate FGF-23 promoter activity through a FGFR/HMW-FGF-2/CBP/CREB complex that binds to the CRE in the proximal FGF-23 promoter. To our knowledge this is the first report demonstrating a role for the intracrine FGFR1 and CREB nuclear regulator complex regulating FGF-23 hormone transcription in osteoblasts (47).
FIGURE 7.
Schematic representation of FGFR1 signaling in FGF-23 gene transcription.
The close proximity between NFAT and CRE binding sites and the effects of mutations of either NFAT or CRE to attenuate HMW-FGF-2/FSK stimulation of the FGF-23 promoter suggest that the NFAT and CRE sites may be functionally coupled. Cross-talk between these signaling pathways could occur at multiple levels, including MAPK activation of both ETS-1 and NFAT, and LMW-FGF-2 and HMW-FGF-2 activating cAMP-dependent CREB/INFS pathways. However, HMW-FGF-2 in the presence of FSK is more potent than LMW-FGF-2 in activating the FGF-23 promoter in both human-derived SaOS-2 and mouse-derived MC3T3-E1 osteoblasts.
These findings are important for several reasons. First, we establish the importance of HMW-FGF-2 regulation of the FGF-23 promoter activity in vitro and establish the involvement of the INFS pathway in regulating FGF-23 gene transcription. Our observations are consistent with the findings that transgenic overexpression of HMW-FGF-2, the ligand for nuclear FGFR1, stimulates FGF-23 expression in bone and that HMW-FGF-2 is increased in bone of adult Hyp mice (26), that the conditional deletion of FGFR1 in osteocytes reduces FGF-23 in the Hyp mouse models of increased FGF-23 (32), and the deletion of HMW-FGF-2 in mice reduces FGF-23 expression in vivo (48).
Second, by showing an additional role of paracrine/autocrine FGFR1 membrane signaling, our data confirm that local matrix-derived factors play an essential role in the regulation of FGF-23 gene transcription. Indeed, local FGFs and FGFR signaling are important in skeletal development and physiology (49–51). These local factors likely coordinate FGF-23 transcription to match bone influx and efflux of phosphorus with the overall mineral metabolism. Thus, membrane and integrative nuclear FGFR signaling may provide a physiological “axis mundi” for integration of multiple factors regulating FGF-23 in bone.
Third, defining cis-elements in the proximal FGF-23 promoter that are necessary for FGF-23 gene transcription provides a molecular framework for understanding how diverse local and systemic pathways act coordinately to regulate FGF-23 promoter activity. For example, calcium has recently been shown to regulate FGF-23 expression in bone (11). Our findings suggest that this may be mediated through calcineurin/NFAT pathways. In addition, circulating FGF-23 concentrations decrease rapidly and disproportionately to reductions in parathyroid hormone levels after successful renal transplantation (52). Involvement of NFAT in FGF-23 gene transcriptions suggests that treatment with calcineurin inhibitors to prevent transplant rejection may have a direct role in suppressing FGF-23 gene transcription, a possibility requiring further study. Our findings may also explain the variable and inconsistent actions of phenylthiohydantoin to stimulate FGF-23 gene transcription (1). Indeed, phenylthiohydantoin, which is coupled to both intracellular calcium and cAMP signaling, induces FGF-23 transcription in UMR106 osteoblast-like cells via activation of nuclear receptor-related protein 1 (Nurr1) (53). Our results showing that forskolin alone is insufficient to activate the proximal FGF-23 promoter and that cAMP and HMW-FGF-2 have synergistic effects, and data showing the cooperation of nuclear FGFR1 and Nurr1 in the activation of neuronal genes, such as tyrosine hydroxylase (54), predict that phenylthiohydantoin actions might be influenced by the activity of FGFR1 in bone.
There are gaps in our knowledge of how LMW-FGF-2 and HMW-FGF-2 are regulated in response to systemic and local factors that impact FGF-23 production and bone remodeling. One possibility is that alteration in the bioactivity of LMW-FGF-2 stored in the extracellular matrix is a mechanism whereby the extracellular protein, dentin matrix protein-1 (DMP1), and the transmembrane enzymes, phosphate regulating gene with homologies to endopeptidases on the X chromosome (PHEX) and ectonucleotide pyrophosphatase/phosphordiesterase 1 (ENPP1), regulate FGF-23 gene transcription (10). Consistent with this possibility, LMW-FGF-2 administration in vivo induces hypophosphatemia and impairs matrix mineralization (55, 56). The Hyp mouse model of X-linked hypophosphatemic rickets (XLH) also has elevated expression of HMW-FGF-2 in osteoblasts, but the mechanisms linking PHEX and HMW-FGF-2 expression remain unknown.
Interestingly, HMW-FGF-2 is regulated by inflammation and oxidative stress, which are additional factors implicated in the regulation of FGF-23 (57). In this regard, HMW-FGF-2 is regulated by H-ras, cytokines, such as interleukin 1β and tumor necrosis factor-α, heat shock, and oxidative stress (33). Activating mutations of HRAS also lead to hypophosphatemia in association with skeletal dysplasia in subjects with epidermal nevus syndrome (58). It would be of interest to know if HMW-FGF-2 is involved in this response. Finally, iron deficiency and inflammation also play a role in regulating FGF-23 expression (57, 59). Oxidative stress may stimulate FGF-23 gene transcription through hypoxia inducible factor-1α, a factor that also regulates HMW-FGF-2 (59). Further studies are needed to determine whether regulation of FGF-23 by inflammation and oxidative stress involves HMW-FGF-2 (33).
Regardless, LWM- and HWM-FGF-2, which can exhibit different biological functions in bone, both regulate FGF-23 promoter activity. This may allow FGFR1 to control FGF-23 expression under different developmental and physiological conditions. For example, FGF-2 overexpression in transgenic mice causes dysmorphic skeletal syndromes associated with high rates of osteoblast proliferation, suggesting that the membrane FGFR pathway may have greater importance in regulating FGF-23 expression in high bone turnover states (26). On the other hand, HMW-FGF-2 may have a growth arrest-inducing role (60), consistent with the possibility that INFS signaling may play an important role in non-replicating osteocytes.
We have previously reported that osteocyte-specific deletion of FGFR1 results in a 50% reduction in FGF-23 expression in a Hyp mouse model of excess FGF-23 production (32), suggesting that other FGFRs might also be regulating FGF-23 transcription. Existing data suggests that neither FGFR3 nor FGFR4 are involved in up-regulation of FGF-23 in osteoblasts, because these two receptors are expressed in low levels in the osteoblast cell lines that we used to test FGF-23 promoter activity, and more importantly, global ablation of FGFR3 and FGFR4 in Hyp mice results in increased, not decreased, FGF-23 expression (61). Our studies do not preclude, however, a role of membrane FGFR2 signaling in the regulation of FGF-23 gene transcription, because FGFR2 is expressed in osteoblasts and is known to regulate bone formation (62). However, like FGFR3 and FGFR4, the transmembrane domain of FGFR2 lacks the characteristics of the FGFR1 transmembrane domain that facilitates the nuclear translocation of FGFR1 (47). Indeed, comparison of the effects of FGFR1 and FGFR2 on FGF-23 gene transcription showed that FGFR1 had a significantly greater effect, possibly due to its dual role to stimulate both membrane and integrative nuclear signaling. This interpretation is supported by the attenuation of FGFR1 function by substituting the transmembrane domain of FGFR4 in the FGFR1–4 chimeric construct.
Our investigations are potentially limited by the use of only the proximal promoter of FGF-23 to define the molecular pathways linking FGFR1 and LMW- and HMW-FGF-2. This approach would miss additional potential enhancers and transcriptional binding sites located at considerable distances from the transcription start site. Nevertheless, 1,25(OH)2D treatment of URM-106 osteoblasts resulted in enhancement in histone H4 acetylation in the proximal FGF-23 promoter between −57 and 259 bp (44), suggesting that the proximal promoter is important in FGF-23 gene transcription. Chip-seq investigations in human and mouse osteoblasts will be required to completely define the NFAT, ETS-1, CREB, and other potential binding sites that mediate FGFR1 regulation of FGF-23 gene transcription.
In conclusion, FGFR1, which is involved in the regulation of bone development and remodeling (51, 63), and FGF-2 (both LMW and HMW forms), which are produced by the osteoblast lineage and stored in bone matrix, regulate FGF-23 promoter activity in osteoblasts through NFAT and CREB-dependent membrane and INFS FGFR1 signaling. Further studies are needed to define how alterations in bone metabolism and mineralization differentially regulate LMW- and HWM-FGF-2.
Acknowledgments
We acknowledge Dr. Michal Stachowiak, State University of New York, Buffalo, NY, for providing FGFR1, FGFR1/4, FGFR1 (TK−), FGFR1(SP−/NLS), FGFR1 (TK−/SP−/NLS), and HMW-FGF-2 plasmid DNA for this study. Human FGFR1, FGFR2, FGFR3, FGFR4, and vector DNA were provided by the DNASU Plasmid Repository.
This work was supported, in whole or in part, by National Institutes of Health Grant 5R01AR045955 (to L. D. Q.).
- LMW
- low molecular weight
- HMW
- high molecular weight
- FSK
- forskolin Hyp, hypophosphatemic mouse XLH homologue
- NLS
- nuclear localization sequence
- CREB
- cAMP-response element-binding protein
- INFS
- integrative FGFR1 nuclear pathway
- PIC
- protease inhibitor mixture
- BisTris
- 2-[bis(2-hydroxyethyl)amino]-2-(hydroxymethyl)propane-1,3-diol
- PLCγ
- phospholipase Cγ
- SP
- secretory peptide
- CsA
- cyclosporin A
- NFAT
- nuclear factor of activated T cells.
REFERENCES
- 1. Quarles L. D. (2012) Skeletal secretion of FGF-23 regulates phosphate and vitamin D metabolism. Nat. Rev. Endocrinol. 8, 276–286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Liu S., Zhou J., Tang W., Jiang X., Rowe D. W., Quarles L. D. (2006) Pathogenic role of Fgf23 in Hyp mice. Am. J. Physiol. Endocrinol. Metab. 291, E38-E49 [DOI] [PubMed] [Google Scholar]
- 3. Shimada T., Kakitani M., Yamazaki Y., Hasegawa H., Takeuchi Y., Fujita T., Fukumoto S., Tomizuka K., Yamashita T. (2004) Targeted ablation of Fgf23 demonstrates an essential physiological role of FGF23 in phosphate and vitamin D metabolism. J. Clin. Invest. 113, 561–568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Shimada T., Mizutani S., Muto T., Yoneya T., Hino R., Takeda S., Takeuchi Y., Fujita T., Fukumoto S., Yamashita T. (2001) Cloning and characterization of FGF23 as a causative factor of tumor-induced osteomalacia. Proc. Natl. Acad. Sci. U.S.A. 98, 6500–6505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. White K. E., Carn G., Lorenz-Depiereux B., Benet-Pages A., Strom T. M., Econs M. J. (2001) Autosomal-dominant hypophosphatemic rickets (ADHR) mutations stabilize FGF-23. Kidney Int. 60, 2079–2086 [DOI] [PubMed] [Google Scholar]
- 6. Shimada T., Yamazaki Y., Takahashi M., Hasegawa H., Urakawa I., Oshima T., Ono K., Kakitani M., Tomizuka K., Fujita T., Fukumoto S., Yamashita T. (2005) Vitamin D receptor-independent FGF23 actions in regulating phosphate and vitamin D metabolism. Am. J. Physiol. Renal Physiol. 289, F1088–1095 [DOI] [PubMed] [Google Scholar]
- 7. Tomiyama K., Maeda R., Urakawa I., Yamazaki Y., Tanaka T., Ito S., Nabeshima Y., Tomita T., Odori S., Hosoda K., Nakao K., Imura A. (2010) Relevant use of Klotho in FGF19 subfamily signaling system in vivo. Proc. Natl. Acad. Sci. U.S.A. 107, 1666–1671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Beckman M. J., Tadikonda P., Werner E., Prahl J., Yamada S., DeLuca H. F. (1996) Human 25-hydroxyvitamin D3–24-hydroxylase, a multicatalytic enzyme. Biochemistry 35, 8465–8472 [DOI] [PubMed] [Google Scholar]
- 9. Hoenderop J. G., Chon H., Gkika D., Bluyssen H. A., Holstege F. C., St-Arnaud R., Braam B., Bindels R. J. (2004) Regulation of gene expression by dietary Ca2+ in kidneys of 25-hydroxyvitamin D3–1 α-hydroxylase knockout mice. Kidney Int. 65, 531–539 [DOI] [PubMed] [Google Scholar]
- 10. Martin A., Liu S., David V., Li H., Karydis A., Feng J. Q., Quarles L. D. (2011) Bone proteins PHEX and DMP1 regulate fibroblastic growth factor Fgf23 expression in osteocytes through a common pathway involving FGF receptor (FGFR) signaling. FASEB J. 25, 2551–2562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. David V., Dai B., Martin A., Huang J., Han X., Quarles L. D. (2013) Calcium regulates FGF-23 expression in bone. Endocrinology 154, 4469–4482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Liu S., Tang W., Zhou J., Stubbs J. R., Luo Q., Pi M., Quarles L. D. (2006) Fibroblast growth factor 23 is a counter-regulatory phosphaturic hormone for vitamin D. J. Am. Soc. Nephrol. 17, 1305–1315 [DOI] [PubMed] [Google Scholar]
- 13. Kolek O. I., Hines E. R., Jones M. D., LeSueur L. K., Lipko M. A., Kiela P. R., Collins J. F., Haussler M. R., Ghishan F. K. (2005) 1α,25-Dihydroxyvitamin D3 upregulates FGF23 gene expression in bone: the final link in a renal-gastrointestinal-skeletal axis that controls phosphate transport. Am. J. Physiol. Gastrointest. Liver Physiol. 289, G1036–1042 [DOI] [PubMed] [Google Scholar]
- 14. Brown W. W., Jüppner H., Langman C. B., Price H., Farrow E. G., White K. E., McCormick K. L. (2009) Hypophosphatemia with elevations in serum fibroblast growth factor 23 in a child with Jansen's metaphyseal chondrodysplasia. J. Clin. Endocrinol. Metab. 94, 17–20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Kawata T., Imanishi Y., Kobayashi K., Miki T., Arnold A., Inaba M., Nishizawa Y. (2007) Parathyroid hormone regulates fibroblast growth factor-23 in a mouse model of primary hyperparathyroidism. J. Am. Soc. Nephrol. 18, 2683–2688 [DOI] [PubMed] [Google Scholar]
- 16. Lavi-Moshayoff V., Wasserman G., Meir T., Silver J., Naveh-Many T. (2010) PTH increases FGF23 gene expression and mediates the high-FGF23 levels of experimental kidney failure: a bone parathyroid feedback loop. Am. J. Physiol. Renal Physiol. 299, F882–889 [DOI] [PubMed] [Google Scholar]
- 17. Sato T., Tominaga Y., Ueki T., Goto N., Matsuoka S., Katayama A., Haba T., Uchida K., Nakanishi S., Kazama J. J., Gejyo F., Yamashita T., Fukagawa M. (2004) Total parathyroidectomy reduces elevated circulating fibroblast growth factor 23 in advanced secondary hyperparathyroidism. Am. J. Kidney Dis. 44, 481–487 [PubMed] [Google Scholar]
- 18. Urakawa I., Yamazaki Y., Shimada T., Iijima K., Hasegawa H., Okawa K., Fujita T., Fukumoto S., Yamashita T. (2006) Klotho converts canonical FGF receptor into a specific receptor for FGF23. Nature 444, 770–77417086194 [Google Scholar]
- 19. Kuro-o M., Matsumura Y., Aizawa H., Kawaguchi H., Suga T., Utsugi T., Ohyama Y., Kurabayashi M., Kaname T., Kume E., Iwasaki H., Iida A., Shiraki-Iida T., Nishikawa S., Nagai R., Nabeshima Y. I. (1997) Mutation of the mouse klotho gene leads to a syndrome resembling ageing. Nature 390, 45–51 [DOI] [PubMed] [Google Scholar]
- 20. Li S. A., Watanabe M., Yamada H., Nagai A., Kinuta M., Takei K. (2004) Immunohistochemical localization of Klotho protein in brain, kidney, and reproductive organs of mice. Cell Struct. Funct. 29, 91–99 [DOI] [PubMed] [Google Scholar]
- 21. Samadfam R., Richard C., Nguyen-Yamamoto L., Bolivar I., Goltzman D. (2009) Bone formation regulates circulating concentrations of fibroblast growth factor 23. Endocrinology 150, 4835–4845 [DOI] [PubMed] [Google Scholar]
- 22. Ben-Dov I. Z., Galitzer H., Lavi-Moshayoff V., Goetz R., Kuro-o M., Mohammadi M., Sirkis R., Naveh-Many T., Silver J. (2007) The parathyroid is a target organ for FGF23 in rats. J. Clin. Invest. 117, 4003–4008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Martin A., David V., Quarles L. D. (2012) Regulation and Function of the FGF23/Klotho Endocrine Pathways. Physiol. Rev. 92, 131–155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. White K. E., Cabral J. M., Davis S. I., Fishburn T., Evans W. E., Ichikawa S., Fields J., Yu X., Shaw N. J., McLellan N. J., McKeown C., Fitzpatrick D., Yu K., Ornitz D. M., Econs M. J. (2005) Mutations that cause osteoglophonic dysplasia define novel roles for FGFR1 in bone elongation. Am. J. Hum. Genet. 76, 361–367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Stevens D. A., Harvey C. B., Scott A. J., O'Shea P. J., Barnard J. C., Williams A. J., Brady G., Samarut J., Chassande O., Williams G. R. (2003) Thyroid hormone activates fibroblast growth factor receptor-1 in bone. Mol. Endocrinol. 17, 1751–1766 [DOI] [PubMed] [Google Scholar]
- 26. Xiao L., Naganawa T., Lorenzo J., Carpenter T. O., Coffin J. D., Hurley M. M. (2010) Nuclear isoforms of fibroblast growth factor 2 are novel inducers of hypophosphatemia via modulation of FGF23 and KLOTHO. J. Biol. Chem. 285, 2834–2846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Xiao L., Esliger A., Hurley M. M. (2013) Nuclear fibroblast growth factor 2 (FGF2) isoforms inhibit bone marrow stromal cell mineralization through FGF23/FGFR/MAPK in vitro. J. Bone Miner. Res. 28, 35–45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Liu S., Tang W., Fang J., Ren J., Li H., Xiao Z., Quarles L. D. (2009) Novel regulators of Fgf23 expression and mineralization in Hyp bone. Mol. Endocrinol. 23, 1505–1518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Wöhrle S., Henninger C., Bonny O., Thuery A., Beluch N., Hynes N. E., Guagnano V., Sellers W. R., Hofmann F., Kneissel M., Graus Porta D. (2013) Pharmacological inhibition of fibroblast growth factor (FGF) receptor signaling ameliorates FGF23-mediated hypophosphatemic rickets. J. Bone Miner. Res. 28, 899–911 [DOI] [PubMed] [Google Scholar]
- 30. Wöhrle S., Bonny O., Beluch N., Gaulis S., Stamm C., Scheibler M., Müller M., Kinzel B., Thuery A., Brueggen J., Hynes N. E., Sellers W. R., Hofmann F., Graus-Porta D. (2011) FGF receptors control vitamin D and phosphate homeostasis by mediating renal FGF-23 signaling and regulating FGF-23 expression in bone. J. Bone Miner. Res. 26, 2486–2497 [DOI] [PubMed] [Google Scholar]
- 31. Wu A. L., Feng B., Chen M. Z., Kolumam G., Zavala-Solorio J., Wyatt S. K., Gandham V. D., Carano R. A., Sonoda J. (2013) Antibody-mediated activation of FGFR1 induces FGF23 production and hypophosphatemia. PloS One 8, e57322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Xiao Z., Huang J., Cao L., Liang Y., Han X., Quarles L. D. (2014) Osteocyte-specific deletion of Fgfr1 suppresses FGF23. PloS One 9, e104154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Delrieu I. (2000) The high molecular weight isoforms of basic fibroblast growth factor (FGF-2): an insight into an intracrine mechanism. FEBS Lett. 468, 6–10 [DOI] [PubMed] [Google Scholar]
- 34. Degnin C. R., Laederich M. B., Horton W. A. (2010) FGFs in endochondral skeletal development. J. Cell. Biochem. 110, 1046–1057 [DOI] [PubMed] [Google Scholar]
- 35. Eswarakumar V. P., Lax I., Schlessinger J. (2005) Cellular signaling by fibroblast growth factor receptors. Cytokine Growth Factor Rev. 16, 139–149 [DOI] [PubMed] [Google Scholar]
- 36. Stachowiak M. K., Fang X., Myers J. M., Dunham S. M., Berezney R., Maher P. A., Stachowiak E. K. (2003) Integrative nuclear FGFR1 signaling (INFS) as a part of a universal “feed-forward-and-gate” signaling module that controls cell growth and differentiation. J. Cell. Biochem. 90, 662–691 [DOI] [PubMed] [Google Scholar]
- 37. Itoh N., Ornitz D. M. (2004) Evolution of the Fgf and Fgfr gene families. Trends Genet. 20, 563–569 [DOI] [PubMed] [Google Scholar]
- 38. Quarles L. D., Yohay D. A., Lever L. W., Caton R., Wenstrup R. J. (1992) Distinct proliferative and differentiated stages of murine MC3T3-E1 cells in culture: an in vitro model of osteoblast development. J. Bone Miner. Res. 7, 683–692 [DOI] [PubMed] [Google Scholar]
- 39. Shalhoub V., Ward S. C., Sun B., Stevens J., Renshaw L., Hawkins N., Richards W. G. (2011) Fibroblast growth factor 23 (FGF23) and α-klotho stimulate osteoblastic MC3T3.E1 cell proliferation and inhibit mineralization. Calcif. Tissue Int. 89, 140–150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Gee K., Angel J. B., Mishra S., Blahoianu M. A., Kumar A. (2007) IL-10 regulation by HIV-Tat in primary human monocytic cells: involvement of calmodulin/calmodulin-dependent protein kinase-activated p38 MAPK and Sp-1 and CREB-1 transcription factors. J. Immunol. 178, 798–807 [DOI] [PubMed] [Google Scholar]
- 41. Miraoui H., Marie P. J. (2010) Fibroblast growth factor receptor signaling crosstalk in skeletogenesis. Sci. Signal. 3, re9. [DOI] [PubMed] [Google Scholar]
- 42. Crabtree G. R., Olson E. N. (2002) NFAT signaling: choreographing the social lives of cells. Cell 109, S67-S79 [DOI] [PubMed] [Google Scholar]
- 43. Wasylyk B., Hagman J., Gutierrez-Hartmann A. (1998) Ets transcription factors: nuclear effectors of the Ras-MAP-kinase signaling pathway. Trends Biochem. Sci. 23, 213–216 [DOI] [PubMed] [Google Scholar]
- 44. Saini R. K., Kaneko I., Jurutka P. W., Forster R., Hsieh A., Hsieh J. C., Haussler M. R., Whitfield G. K. (2013) 1,25-Dihydroxyvitamin D3 regulation of fibroblast growth factor-23 expression in bone cells: evidence for primary and secondary mechanisms modulated by leptin and interleukin-6. Calcif. Tissue Int. 92, 339–353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Peng H., Myers J., Fang X., Stachowiak E. K., Maher P. A., Martins G. G., Popescu G., Berezney R., Stachowiak M. K. (2002) Integrative nuclear FGFR1 signaling (INFS) pathway mediates activation of the tyrosine hydroxylase gene by angiotensin II, depolarization and protein kinase C. J. Neurochem. 81, 506–524 [DOI] [PubMed] [Google Scholar]
- 46. Dunham-Ems S. M., Lee Y. W., Stachowiak E. K., Pudavar H., Claus P., Prasad P. N., Stachowiak M. K. (2009) Fibroblast growth factor receptor-1 (FGFR1) nuclear dynamics reveal a novel mechanism in transcription control. Mol. Biol. Cell 20, 2401–2412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Stachowiak M. K., Birkaya B., Aletta J. M., Narla S. T., Benson C. A., Decker B., Stachowiak E. K. (2015) “Nuclear FGF receptor-1 and CREB binding protein: an integrative signaling module.” J. Cell. Physiol. 230, 989–1002 [DOI] [PubMed] [Google Scholar]
- 48. Homer-Bouthiette C., Doetschman T., Xiao L., Hurley M. M. (2014) Knockout of nuclear high molecular weight FGF2 isoforms in mice modulates bone and phosphate homeostasis. J. Biol. Chem. 289, 36303–36314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Marie P. J. (2012) Fibroblast growth factor signaling controlling bone formation: an update. Gene 498, 1–4 [DOI] [PubMed] [Google Scholar]
- 50. Du X., Xie Y., Xian C. J., Chen L. (2012) Role of FGFs/FGFRs in skeletal development and bone regeneration. J. Cell. Physiol. 227, 3731–3743 [DOI] [PubMed] [Google Scholar]
- 51. Ornitz D. M., Marie P. J. (2002) FGF signaling pathways in endochondral and intramembranous bone development and human genetic disease. Genes Dev. 16, 1446–1465 [DOI] [PubMed] [Google Scholar]
- 52. Wesseling-Perry K., Pereira R. C., Tsai E., Ettenger R., Jüppner H., Salusky I. B. (2013) FGF23 and mineral metabolism in the early post-renal transplantation period. Pediat. Nephrol. 28, 2207–2215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Meir T., Durlacher K., Pan Z., Amir G., Richards W. G., Silver J., Naveh-Many T. (2014) Parathyroid hormone activates the orphan nuclear receptor Nurr1 to induce FGF23 transcription. Kidney Int. 86, 1106–1115 [DOI] [PubMed] [Google Scholar]
- 54. Baron O., Förthmann B., Lee Y. W., Terranova C., Ratzka A., Stachowiak E. K., Grothe C., Claus P., Stachowiak M. K. (2012) Cooperation of nuclear fibroblast growth factor receptor 1 and Nurr1 offers new interactive mechanism in postmitotic development of mesencephalic dopaminergic neurons. J. Biol. Chem. 287, 19827–19840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Nauman E. A., Sakata T., Keaveny T. M., Halloran B. P., Bikle D. D. (2003) bFGF administration lowers the phosphate threshold for mineralization in bone marrow stromal cells. Calcif. Tissue Int. 73, 147–152 [DOI] [PubMed] [Google Scholar]
- 56. Liang H., Pun S., Wronski T. J. (1999) Bone anabolic effects of basic fibroblast growth factor in ovariectomized rats. Endocrinology 140, 5780–5788 [DOI] [PubMed] [Google Scholar]
- 57. Munoz Mendoza J., Isakova T., Ricardo A. C., Xie H., Navaneethan S. D., Anderson A. H., Bazzano L. A., Xie D., Kretzler M., Nessel L., Hamm L. L., Negrea L., Leonard M. B., Raj D., Wolf M., and Chronic Renal Insufficiency, C. (2012) Fibroblast growth factor 23 and inflammation in CKD. Clin. J. Am. Soc. Nephrol. 7, 1155–1162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Avitan-Hersh E., Tatur S., Indelman M., Gepstein V., Shreter R., Hershkovitz D., Brick R., Bergman R., Tiosano D. (2014) Postzygotic HRAS mutation causing both keratinocytic epidermal nevus and thymoma and associated with bone dysplasia and hypophosphatemia due to elevated FGF23. J. Clin. Endocrinol. Metab. 99, E132-E136 [DOI] [PubMed] [Google Scholar]
- 59. Farrow E. G., Yu X., Summers L. J., Davis S. I., Fleet J. C., Allen M. R., Robling A. G., Stayrook K. R., Jideonwo V., Magers M. J., Garringer H. J., Vidal R., Chan R. J., Goodwin C. B., Hui S. L., Peacock M., White K. E. (2011) Iron deficiency drives an autosomal dominant hypophosphatemic rickets (ADHR) phenotype in fibroblast growth factor-23 (Fgf23) knock-in mice. Proc. Natl. Acad. Sci. U.S.A. 108, E1146-E1155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Valtink M., Knels L., Stanke N., Engelmann K., Funk R. H., Lindemann D. (2012) Overexpression of human HMW FGF-2 but not LMW FGF-2 reduces the cytotoxic effect of lentiviral gene transfer in human corneal endothelial cells. Invest. Ophthalmol. Vis. Sci. 53, 3207–3214 [DOI] [PubMed] [Google Scholar]
- 61. Liu S., Vierthaler L., Tang W., Zhou J., Quarles L. D. (2008) FGFR3 and FGFR4 do not mediate renal effects of FGF23. J. Am. Soc. Nephrol. 19, 2342–2350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Yu K., Xu J., Liu Z., Sosic D., Shao J., Olson E. N., Towler D. A., Ornitz D. M. (2003) Conditional inactivation of FGF receptor 2 reveals an essential role for FGF signaling in the regulation of osteoblast function and bone growth. Development 130, 3063–3074 [DOI] [PubMed] [Google Scholar]
- 63. Jacob A. L., Smith C., Partanen J., Ornitz D. M. (2006) Fibroblast growth factor receptor 1 signaling in the osteo-chondrogenic cell lineage regulates sequential steps of osteoblast maturation. Dev. Biol. 296, 315–328 [DOI] [PMC free article] [PubMed] [Google Scholar]