Background: YM155, a survivin suppressant, has been shown anticancer efficacy in various cancer cells.
Results: Human gastric cancer cells differentially displayed the sensitivity to YM155 dependent to degradation of cIAP1.
Conclusion: Cellular inhibitor of apoptosis protein 1 (cIAP1) interacts with survivin to control their mutual stability and determine sensitivity to YM155.
Significance: The regulation of cIAP1 enhances the efficacy for YM155 treatment in gastric cancer.
Keywords: Anticancer Drug; Apoptosis; Gastric Cancer; Survivin; Ubiquitylation (Ubiquitination); YM155, cIAP1
Abstract
YM155, which blocks the expression of survivin, a member of the inhibitor of apoptosis (IAP) family, induces cell death in a variety of cancer types, including prostate, bladder, breast, leukemia, and non-small lung cancer. However, the mechanism underlying gastric cancer susceptibility and resistance to YM155 is yet to be specified. Here, we demonstrate that cIAP1 stability dictates resistance to YM155 in human gastric cancer cells. Treatment of human gastric cancer cells with YM155 differentially induced cell death dependent on the stability of cIAP1 as well as survivin. Transfection with cIAP1 expression plasmids decreased cell sensitivity to YM155, whereas knockdown of endogenous cIAP1 using RNA interference enhanced sensitivity to YM155. In addition, double knockdown of survivin and cIAP1 significantly induced cell death in the YM155-resistant cell line, MKN45. We also showed that YM155 induced autoubiquitination and proteasome-dependent degradation of cIAP1. Surprisingly, survivin affected the stability of cIAP1 through binding, contributing to cell sensitivity to YM155. Thus, our findings reveal that YM155 sensitizes human gastric cancer cells to apoptotic cell death by degrading cIAP1, and furthermore, cIAP1 in gastric cancer cells may act as a PD marker for YM155 treatment.
Introduction
Gastric cancer is a major health problem worldwide. In 2011, ∼989,600 new cases (∼7.8% of all cancers) and 738,000 deaths (∼9.7% of all cancers) occurred worldwide (1). Asian and South American countries have higher incidence rates of gastric cancer than Western Europe and the United States. Unfortunately, most gastric cancer patients still present at an advanced stage, and, although there have been advances in diagnostic and treatment strategies, the mortality rates remain poor. Conventional therapies, such as radiotherapy, surgery, and chemotherapy, for the treatment of gastric cancer patients can improve survival. However, numerous gastric cancer patients either do not respond to these therapies or relapse after surgery.
Targeted therapies have been utilized as a treatment strategy for many solid tumors, including colorectal, breast, lung, and renal cancers (2–5). Target therapeutic advances in gastric cancer partially increased ORR and OS rates through the inhibition of cell proliferation and angiogenesis and changes to the tumor microenvironment (6). Recently, targeted therapies for gastric cancer can be divided into two categories (7): those that target the epidermal growth factor receptor family and those that target the vascular endothelial growth factor (VEGF). However, the survival rate of gastric cancer patients receiving these therapeutic strategies remains poor.
YM155, a small imidazolium-based compound, has recently been reported to suppress the expression of survivin through the repression of its promoter activity (8). YM155 may also suppress survivin expression through disrupting the binding of SP1, a zinc finger transcription factor, to the survivin promoter (9). An initial screen was performed to identify YM155 in the human cervical cancer cell line HeLa and in Chinese hamster ovary cells (8). Until now, many reports have shown the efficacy of YM155 alone and in combinational application against cultured cancer cells, including breast, NSCLC, pancreas, prostate, glioblastoma, and head and neck cancers (8, 10–14). However, the mechanism relating to SP1 and YM155 in cancer cells has remained unclear. Moreover, the inhibitory mechanism of YM155 targeting survivin on the growth of various cancer cells is yet unclear.
In this investigation, the inhibitory mechanisms of YM155 treatment against human gastric cancer cells were studied. We first tested whether YM155-induced apoptosis was associated with inhibition of survivin expression. We next focused on the cIAP14 change in anti-apoptotic proteins that related to YM155-induced apoptosis. To our knowledge, this is the first report demonstrating a relationship between survivin and cIAP1 in gastric cancer cells after YM155 treatment.
EXPERIMENTAL PROCEDURES
Cell Cultures
KATOIII, AGS, MKN1, MKN28, MKN45, MKN74, NCI-N87, and 293T cells were purchased from American Type Culture Collection (ATCC, Manassas, VA). All gastric cancer cells or 293T human embryonic kidney cells were maintained with RPMI 1640 medium (GIBCO BRL, Grand Island, NY) or Dulbecco's modified Eagle's medium (DMEM, GIBCO BRL) containing 10% fetal bovine serum (FBS, GIBCO BRL) and 100 μg/ml of penicillin/streptomycin in a 5% CO2 incubator at 37 °C. YM155, a survivin suppressant, and MG132, a proteasome inhibitor, were purchased from Sigma.
Cell Death and Annexin V Staining Assay
Cells were treated with YM155 at the indicated doses for 48 h, and cell death was determined using the trypan blue exclusion method. Cells were seeded in 60-mm dishes at a density of 5 × 105 cells for the annexin V staining assay. Then, cells were treated with YM155 at the indicated doses for 48 h. The cells were washed with PBS, trypsinized, stained with an annexin V staining solution (BD Biosciences, San Jose, CA), and analyzed with a fluorescence-activated cell sorter (BD Biosciences).
Plasmids, siRNAs, and Transfections
Survivin and cIAP1 cDNAs were purchased from Origene (Rockville, MD). Survivin T34A, C84A, T34A/C84A, or cIAP1 H588A mutants were mutated using the polymerase chain reaction (PCR)-based QuikChange Site-directed Mutagenesis Kit (Intron Biotechnology, Seoul, Republic of Korea). The survivin deletion mutant (ΔBIR) was subcloned into a GFP-tagged pCMV plasmid and the cIAP1 RING deletion mutants (ΔRING) or CARD-RING deletion mutants (ΔCARD-RING) were subcloned into a HA-tagged pcDNA3.1 plasmid. The following sequences were used for siRNA transfection: scramble, 5′-GCGCAUUCCAGCUUACGUAUU; survivin, 5′-UGUAGAGAUGCGGUGGUCVUUUU-3′; and cIAP1, 5′-AACATAGTAGCTTGTTCAGTG-3′. All DNA and siRNA transfections were performed using Lipofectamine 2000 (Invitrogen) according to the manufacturer's protocol.
Western Blot Assays
Total cellular proteins (20 μg/well) were resolved on 8–15% SDS-PAGE gels for Western blot analysis and then transferred to Immobilon-P membranes (EMD Millipore, Billerica, MA). The membranes were blocked with 5% nonfat skim milk in Tris-buffered saline (TBS-T; 20 mm Tris-HCl, pH 7.4, 150 mm NaCl, 0.1% Tween 20) and then incubated with the following antibodies: anti-GFP, anti-HA, anti-ubiquitin, anti-GST, anti-His, and anti-β-actin (Santa Cruz Biotechnology, Santa Cruz, CA); anti-survivin, anti-caspase-3, anti-caspase-8, and anti-caspase-9 (Cell Signaling, Beverley, CA); and anti-cIAP1 (Abcam, Cambridge, MA) at 4 °C for 12 h. Primary antibodies were detected using goat anti-mouse or goat anti-rabbit horseradish peroxidase-conjugated secondary antibodies and visualized via chemiluminescence detection (Amersham Biosciences).
Immunoprecipitation and GST Pulldown Assay
For the in vivo binding assay, cell lysates (0.5 mg) were incubated with anti-survivin (or GFP) or anti-cIAP1 (or HA) antibodies at 4 °C for 12 h. The mixture was added to protein G Plus-Sepharose beads (Santa Cruz Biotechnology) and then incubated for an additional 2 h at 4 °C. The immunoprecipitates were washed with Nonidet P-40 RIPA lysis buffer, boiled in 2× SDS sample buffer, and then analyzed with anti-survivin (or GFP) or anti-cIAP1 (or HA) antibodies. Purified His-survivin, GST-cIAP1, and GST proteins were purchased from Abnova (Taipei, Taiwan) for in vitro GST pulldown assays. Briefly, 500 ng of GST or GST-cIAP1 proteins were incubated with 500 ng of His-survivin protein in reaction buffer (20 mm Tris-HCl, pH 7.5, and 120 mm NaCl) at 30 °C for 1 h, added to GST pulldown buffer (20 mm Tris-HCl, pH 8.0, 500 mm NaCl, 1% Triton X-100, 0.02% bovine serum albumin, and 5 mm 2-mercaptoethanol) to terminate the reaction and then glutathione-Sepharose beads were added (Cell Signaling Technology) for another 1 h at 4 °C. The mixtures were washed five times with GST pulldown buffer and heated with 2× SDS sample buffer. The binding of survivin and cIAP1 was analyzed via Western blotting with anti-His and anti-GST antibodies, respectively.
In Vivo and in Vitro Ubiquitination Assay
For the in vivo ubiquitination assays, cell lysates were precipitated using anti-cIAP1 or anti-HA antibodies at 4 °C for 12 h and then added to protein G-Sepharose beads for another 2 h. The precipitates were prepared for Western blot analysis using anti-ubiquitin antibody. For the in vitro cIAP1 ubiquitination assay, 500 ng of purified GST-cIAP1 proteins were incubated with 8 ng of human E1, 500 ng of human His-UbcH5a, 2 mm Mg-ATP, and 5 μg of ubiquitin (Boston Biochem Inc., Cambridge, MA) in ubiquitination reaction buffer (50 mm Tris-HCl, pH 7.4, 50 mm NaCl) and treated with 10 or 20 nm YM155 for 1 h at 37 °C. The mixtures were pulled down using a GST buffer and then analyzed by Western blot assay using an anti-ubiquitin antibody.
Surface Plasmon Resonance Analysis
ProteonTM XPR36 (Bio-Rad) was used to determine the binding of YM155 to cIAP1. GST and GST-cIAP1 proteins were captured on a Proteon GLH sensor chip (Bio-Rad). GST or GST-cIAP1 proteins were captured to 2600 or 7000 response units after immobilization, respectively. YM155 was injected at various concentrations at a flow rate of 100 μl/min for 60 s and allowed to dissociate for an additional 300 s.
Native PAGE
Native gels were prepared using an 8–15% acrylamide mixture without SDS. Cell lysates (30 μg/well) were loaded onto native gels without heating, and run in Tris glycine electrophoresis buffer (47 mm Tris base, 364 mm glycine) without SDS for 12 h on ice at 30 V.
Non-radioactive Pulse-Chase Assay
Newly synthesized survivin or cIAP1 protein was labeled using the Click-it metabolic labeling reagents (Invitrogen). Briefly, KATOIII cells were transfected with survivin or cIAP1 expressing plasmids for 48 h and then cells were depleted with methionine-free RPMI 1640 medium for 1 h. The cells were incubated with methionine-free RPMI 1640 medium containing 50 μm l-azidohomoalanine, a methionine analog (Invitrogen), for 4 h. The cells were washed with PBS followed by the addition of complete media. The cells were then chased for the indicated times. l-Azidohomoalanine incorporated protein was biotinylated using the Click-it protein reaction buffer kit (Invitrogen). The biotinylated proteins were precipitated using anti-biotin and then analyzed by Western blotting using anti-survivin or anti-cIAP1 antibodies.
Statistical Analyses
All data were statistically analyzed using a two-tailed Student's t test. The significance in the text was verified by p values, and p values <0.05 were deemed significant.
RESULTS
Human Gastric Cancer Cells Display Differential Sensitivity to YM155-induced Apoptotic Cell Death
It has recently been reported that YM155 inhibits cell growth and proliferation in various tumor cells (15). Treatment of p53-mutant or -deficient prostate cancer cells with YM155 decreases survivin expression (16). In mesothelioma cells, YM155 inhibits MCL-1 expression as well as survivin (17). Thus, YM155 has an anti-tumor effect on various cancer cells. However, the mechanism that regulates survivin and sensitivity to YM155 in human gastric cancer cells remain largely unexplored.
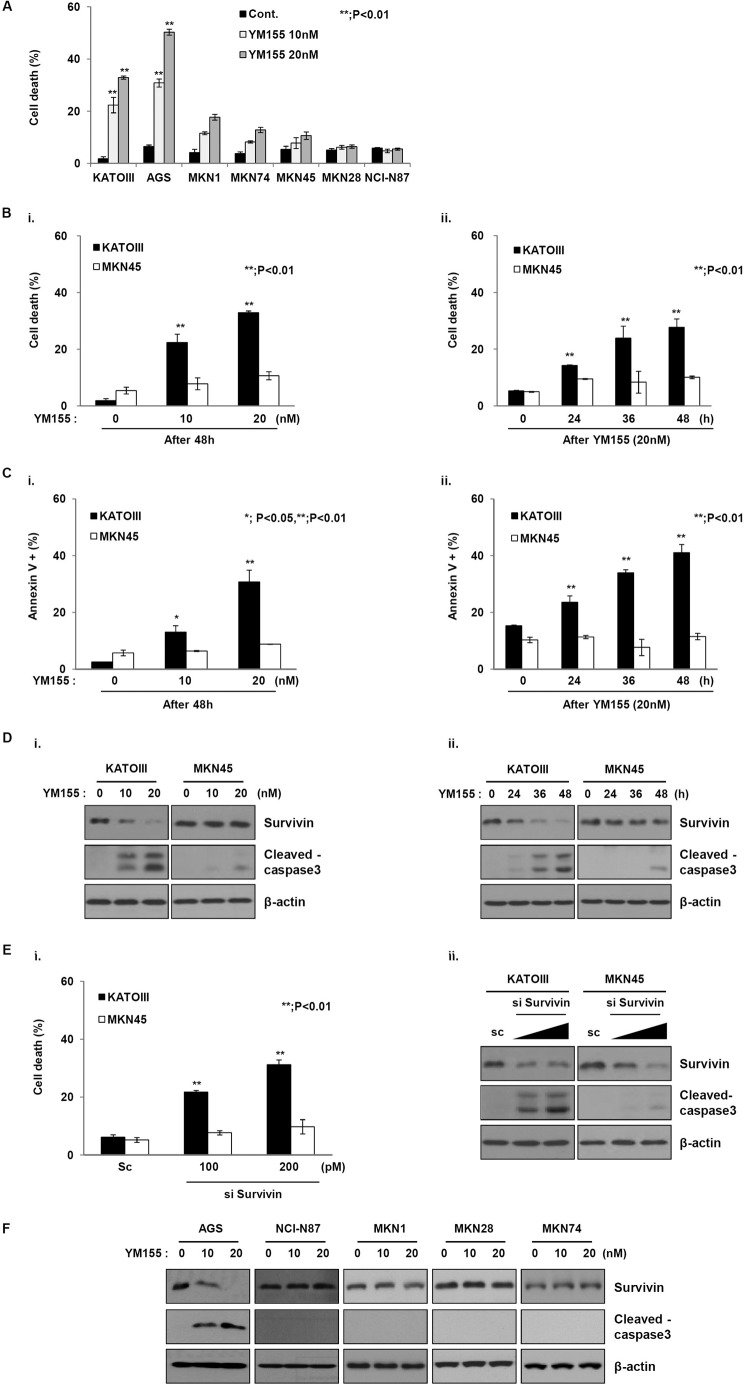
To this aim, we analyzed the inhibitory effect of YM155 on multiple gastric cancer cells (Fig. 1A). The viability of most gastric cancer cells was not affected, with the exception of KATOIII and AGS cells followed by YM155 treatment. To analyze the effect of YM155, two human gastric cancer cell lines with high expression levels of survivin were selected. First, the inhibitory effect of YM155 on these cells was analyzed. Each cell line was treated with different doses of YM155 for different times (Fig. 1). Interestingly, differences in sensitivity to YM155 were observed in the cancer cells (Fig. 1B). Consistently, increased annexin V staining was observed in KATO III cells after exposure to YM155, but not in MKN45 cells (Fig. 1C). To investigate whether the difference in sensitivity to YM155 was a result of different levels of survivin, we analyzed survivin levels after exposure to YM155. The survivin levels in KATO III cells, which were sensitive to YM155, decreased in a dose- and time-dependent manner in response to YM155. However, survivin levels in MKN45 cells, which were resistant to YM155, were not changed (Fig. 1D). In addition, YM155 induced the cleavage of pro-caspase-3 to yield active caspase-3 in KATO III cells, but not in MKN45 cells. These results indicate that human gastric cancer cells display differential sensitivities to YM155.
FIGURE 1.
YM155 differentially induces cell death in gastric cancer cells. A, mutiple gastric cancer cells were treated with YM155 for 48 h at the indicated doses. Cell death was evaluated by a trypan blue exclusion assay. B, (i) KATOIII and MKN45 cells were treated with YM155 at the indicated doses for 48 h, or (ii) YM155 (20 nm) at the indicated times. Cell death rate was determined using the trypan blue exclusion method. The error bars represent the mean ± S.D. of three experiments in triplicate. **, p < 0.01. C, (i) KATOIII and MKN45 cells were treated with YM155 at the indicated doses for 48 h, or (ii) YM155 (20 nm) at the indicated times. Cell death was analyzed by annexin V staining using flow cytometry. D, (i) KATOIII and MKN45 cells were treated with YM155 at 10 and 20 nm for 48 h, and the cell lysates were analyzed using Western blotting with anti-survivin and anti-cleaved caspase-3 antibodies; β-actin was used as a loading control. (ii) YM155 (20 nm) cells treated for 48 h were used for Western blotting using anti-survivin, anti-cleaved caspase-3, and the loading control β-actin. E, the cells were transfected with scramble or survivin siRNA for 48 h. (i) Cell death was determined using the trypan blue exclusion method. The graph represents the mean ± S.D. of three separate experiments in triplicate; *, p < 0.01. (ii) The cell lysates were used for Western blot analysis to determine the expressions of survivin, cleaved caspase-3, and β-actin. F, cells were treated with YM155 for 48 h at the indicated doses and then analyzed by Western blot using anti-survivin and anti-cleaved caspase-3. β-Actin was used as a loading control.
Next, we examined the effects of survivin silencing via small interfering RNA (siRNA) on two gastric cancer cell lines, KATO III and MKN45 (Fig. 1E). The knockdown effects of the survivin siRNA on two gastric cancer cells were confirmed. Dead cell populations were dose-dependently increased in KATO III cells after YM155 treatment. However, although the siRNA decreased survivin levels, MKN45 cell death did not increase after exposure to YM155. Consistently, caspase-3 cleavage was significantly induced in KATO III cells, whereas that in MKN45 cells was weakly observed. Moreover, the levels of survivin were clearly decreased in only sensitive AGS cells, whereas survivin levels in the other cells did not decrease (Fig. 1F). These results suggest that YM155 may induce cell death through the regulation of an alternative molecule as well as survivin in human gastric cancer cells.
YM155 Targets cIAP1as Well as Survivin
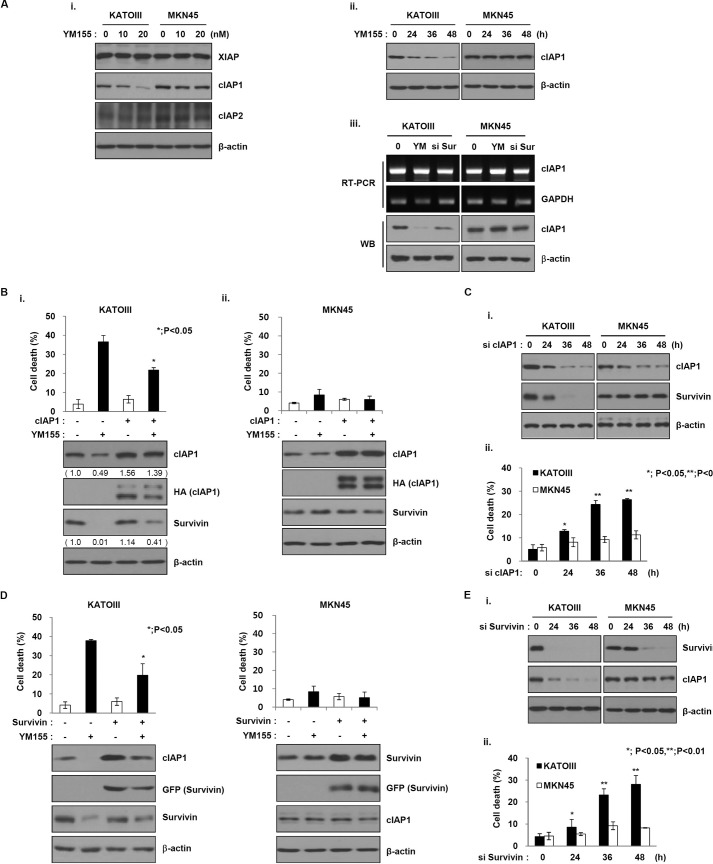
As shown in Fig. 1E, YM155 may target another factor as well as survivin. It was recently reported that YM155 inhibits XIAP in pancreatic cancer cells and cIAP2 in cancer cells (18). To confirm this hypothesis, we initially analyzed the effects of YM155 on XIAP, cIAP1, and 2 other IAP family members. Surprisingly, the levels of cIAP1 protein were significantly decreased in KATO III cells after treatment with YM155, whereas that in MKN45 cells was not decreased (Fig. 2A, i and ii). In addition, XIAP and cIAP2 levels did not change in either cancer cell line, implying that difference in sensitivity to YM155 may relate to cIAP1 as well as survivin. Also, YM155 induced the decrease of cIAP1 expression through protein levels but not mRNA levels (Fig. 2A, iii). Accordingly, we further analyzed the effects of cIAP1 on sensitivity to YM155. Therefore, we first constructed a plasmid expressing HA-tagged cIAP1 cDNA. Transfection with a construct expressing cIAP1 cDNA decreased YM155-induced apoptotic cell death in KATOIII cells but not MKN45 cells (Fig. 2B). Moreover, cIAP1 inhibited the YM155-induced decrease in survivin. In contrast, cIAP1 knockdown via siRNA decreased survivin levels in KATO III cells after exposure to YM155, but not in MKN45 cells (Fig. 2C, i). The dead cell population was also increased in KATO III cells that expressed cIAP1 siRNA (Fig. 2C, ii), indicating that YM155 targets cIAP1 and affects survivin in YM155-sensitive gastric cancer cells.
FIGURE 2.
YM155 targets cIAP as well as survivin. A, (i) KATOIII and MKN45 cells were treated with YM155 at the indicated doses for 48 h, and the cell lysates were analyzed by Western blotting using anti-XIAP, cIAP1, and cIAP2 and the loading control β-actin. (ii) The cells were treated with YM155 (20 nm) at the indicated times and analyzed by Western blotting with a cIAP-1 antibody; β-actin was used as a loading control. (iii) KATOIII and MKN45 cells were treated with YM155 (20 nm) or transfected with survivin siRNA for 48 h. Cell lysates were prepared for RT-PCR analysis or Western blot analysis. B and D, KATOIII (i) and MKN45 cells (ii) were transfected with a cIAP-1-expressing plasmid for 48 h (B) or a survivin-expressing plasmid (D) and then treated with YM155 (20 nm) for another 48 h. The cell death rate was measured using the trypan blue exclusion assay. cIAP1 and survivin expression levels were analyzed by Western blotting (WB) using anti-cIAP1 or survivin antibodies. C and E, cells were transfected with cIAP1 siRNA (C) or survivin siRNA (E) at the indicated times. C and E, (i) cell lysates were analyzed for the expressions of cIAP1 and survivin via Western blot analysis. (ii) The percentage of cell death was determined using the trypan blue exclusion method. The data are the mean ± S.D.; *, p < 0.05; **, p < 0.01.
Based on the above data, we analyzed the effects of survivin on sensitivity to YM155 and cIAP1. To this end, we constructed a plasmid expressing GFP-tagged survivin cDNA. Survivin expression inhibited the YM155-induced decrease in cIAP1 protein levels and cell death in KATO III cells but not MKN45 cells (Fig. 2D). Conversely, knockdown of survivin decreased cIAP1 levels, but not in MKN45 cells (Fig. 2E, i). Consistently, cell death was also increased in KATO III cells only (Fig. 2E, ii), suggesting that YM155 induces a decrease of cIAP1 as well as survivin. However, YM155 does not decrease cIAP1 levels in YM155-resistant cells.
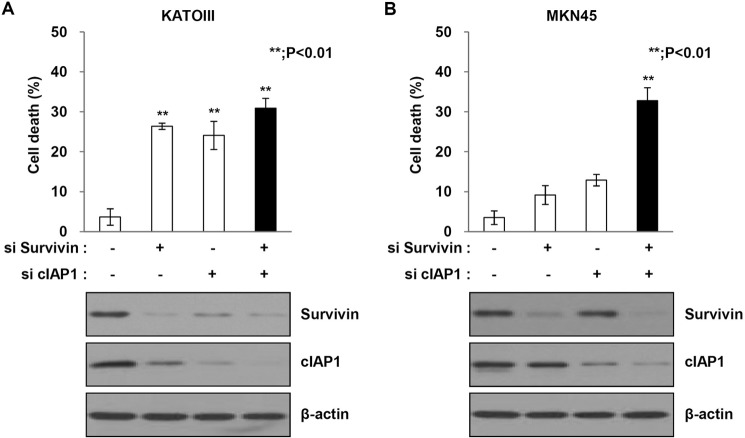
Double Knockdown of Survivin and cIAP1 Induces Cell Death in YM155-resistant Cells
As shown in Figs. 1 and 2, YM155 did not decrease survivin or cIAP1 levels in MKN45 cells, which are resistant to YM155. Furthermore, single knockdown of survivin or cIAP1 did not induce cell death or a decrease in protein levels in MKN45 cells. Accordingly, we analyzed the effects of both survivin and cIAP1 siRNA on the YM155-resistant cell line MKN45. Double knockdown of survivin and cIAP1 significantly induced cell death in MKN45 cells (Fig. 3). Our results strongly suggest that sensitivity to YM155 depends on decreased survivin and cIAP1 protein levels in human gastric cancer cells.
FIGURE 3.
Double knockdown of survivin and cIAP1 induces cell death in YM155-resistant MKN45 cells. A and B, KATOIII (A) and MKN45 (B) cells were co-transfected with survivin-siRNA and cIAP1 siRNA for 48 h, and then cell death was determined using the trypan blue exclusion method. Cell lysates were subjected to Western blot analysis with anti-survivin and anti-CIAP1 and the loading control β-actin. The data are the mean ± S.D.; **, p < 0.01.
Survivin and cIAP1 Affect the Protein Stability of Each Other
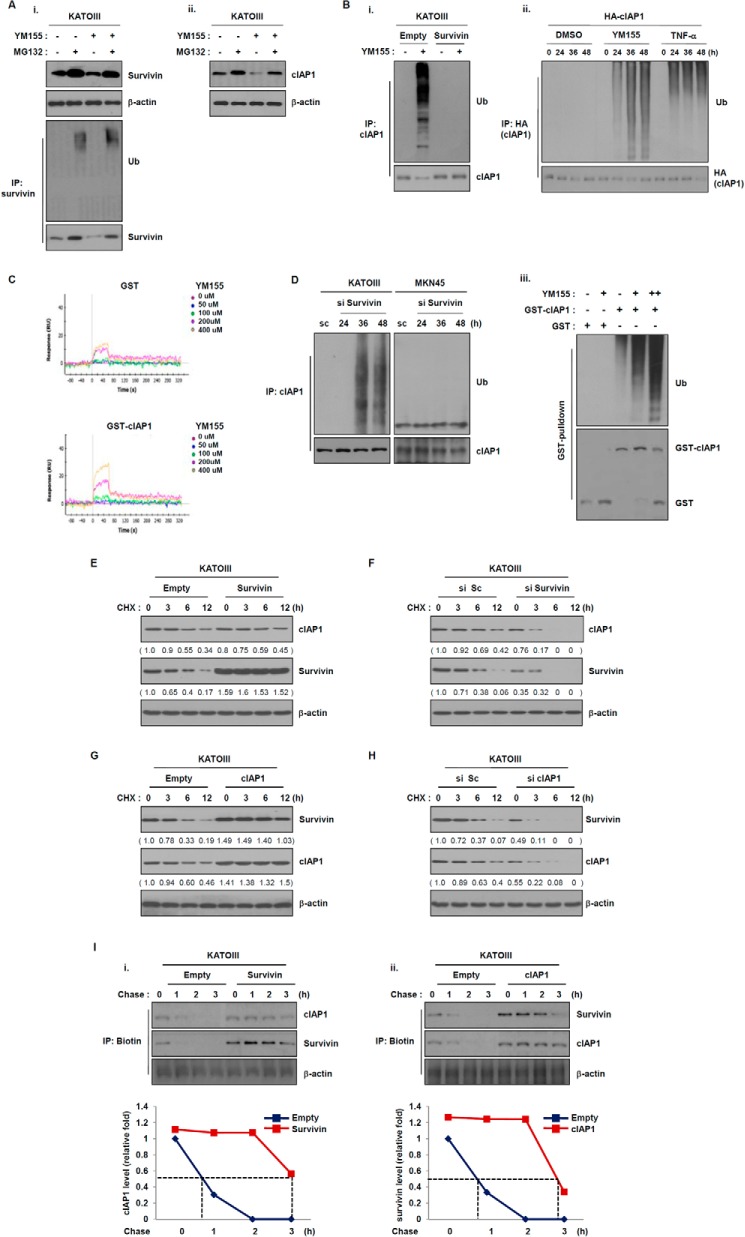
As shown in Fig. 2, survivin affected the expression of cIAP1 protein. Conversely, cIAP1 affected the expression of survivin protein. Thus, the proteins affected the expression of each other. Expression of survivin inhibited the YM155-induced decrease of cIAP1 protein levels. This result implies that survivin may affect cIAP1 protein stability. To confirm this hypothesis, we have performed the in vivo ubiquitination assays using ubiquitin chain-specific antibodies to analyze the effects of YM155 on survivin ubiquitination. Cells were treated with MG132 followed by YM155 treatment. Cell lysates were prepared for Western blot analysis and immunoprecipitated using an anti-survivin antibody. The level of survivin protein clearly increased in response to MG132. Consistently, survivin ubiquitination was also induced (Fig. 4A, i). Also, we analyzed whether the degradation of cIAP1 protein after exposure to YM155 was proteasome dependent using the proteasome inhibitor, MG132. Treatment of the YM155-sensitive cell line, KATO III, with MG132 significantly increased cIAP1 protein levels (Fig. 4A, ii), whereas cIAP1 levels in MKN45 cells were not affected by MG132 treatment followed by YM155 (data not shown). Consistent with these data, endogenous cIAP1 was ubiquitinated in response to YM155 in a time-dependent manner such as TNF-α treatment (Fig. 4B, i and ii). We further analyzed the effect of YM155 on cIAP1 ubiquitination through a in vitro ubiquitination assay. Ubiquitination of purified GST-cIAP1 gradually increased after exposure to YM155 in a dose-dependent manner (Fig. 4B, iii). We analyzed the interaction between YM155 and cIAP1 for the effect of YM155 on cIAP1 ubiquitination. We performed a surface plasmon resonance assay. A peak of YM155-bound cIAP1 protein was observed by surface plasmon resonance analyses of YM155 that was incubated with purified GST-cIAP1 protein (Fig. 4C). These results suggest that YM155 induces the abrogation of cIAP1 autoinhibition through a direct binding interaction. We next analyzed whether cIAP1 ubiquitination was affected by survivin expression; we transfected KATO III cells with survivin-cDNA. Ectopic expression of survivin did not induce cIAP1 ubiquitination (Fig. 4B, i). Conversely, the knockdown of survivin led to cIAP1 ubiquitination (Fig. 4D). These results suggest that survivin inhibits cIAP1 ubiquitination and its subsequent proteasome-mediated degradation.
FIGURE 4.
Survivin regulates the stability of cIAP1 in a proteasome-dependent manner. A, KATOIII cells were treated YM155 (20 nm) for 48 h and then incubated with the proteasome inhibitor MG132 (25 μm) for 6 h. (i) Survivin and (ii) cIAP1 expressions were determined by Western blot analysis. B, (i) KATOIII cells were transfected with a GFP-tagged survivin plasmid for 48 h and then treated with YM155 (20 nm) for another 48 h. Cell lysates were immunoprecipitated with anti-cIAP1 and Western blot analysis as performed with an anti-ubiquitin (Ub) antibody. (ii) 293T cells were transfected with HA-tagged cIAP1 plasmid for 24 h and then treated with YM155 (20 nm) or TNF-α (100 ng/ml). The cell lysates were immunoprecipiated with anti-HA antibody. The ubiquitination of cIAP1 by YM155 was determined via Western blot analysis using anti-ubiquitin antibody. (iii) Recombinant GST-cIAP1 proteins were incubated with E1, E2, ATP, YM155 (10 and 20 nm), and then the ubiquitination of cIAP1 by YM155 was determined with GST pulldown analysis. C, direct interaction of YM155 to cIAP1 using surface plasmon resonance. Representative sensograms were obtained from injections of YM155 at the indicated doses. GST serves as a control to GST-cIAP1. D, cells were transfected with survivin siRNA for the indicated times. Cell lysates were prepared for immunoprecipitation (IP), and cIAP1 ubiquitination was detected using an anti-ubiquitin antibody. E and F, KATOIII cells were transfected with a control vector or a survivin expression vector and scramble or survivin siRNA for 48 h and then treated with the protein synthesis inhibitor cycloheximide (CHX) (50 μg/ml) for the indicated times. Survivin and cIAP1 expression levels were determined by Western blot analysis with anti-survivin and anti-cIAP1 antibodies. G and H, KATOIII cells were transfected with a empty or a cIAP1 expressing plasmid and scramble cIAP1 siRNA for 48 h and then treated with cycloheximide for the indicated times. Cell lysates were prepared for Western blot analysis using anti-survivin and anti-cIAP1 antibodies. I, non-radioactive pulse-chase assay of survivin or cIAP1 protein. KATOIII cells were transfected with empty vector and survivin or cIAP1 expressing plasmid for 48 h. The cells were depleted with methionine-free medium for 1 h and then chased for the indicated times. Relative survivin or cIAP1 protein levels were quantified.
On the basis of these results, we further explored the role of survivin in controlling cIAP1 turnover by treating with the protein synthesis inhibitor cycloheximide and measuring changes in exogenously expressed survivin protein levels over time. In a cycloheximide-chase experiment, ectopic expression of GFP-tagged survivin did not induce changes in cIAP1 protein levels in KATO III cells, whereas transfection with an empty vector led to diminished cIAP1 protein levels (Fig. 4E). Conversely, knockdown of survivin significantly decreased cIAP1 protein levels (Fig. 4F), indicating that survivin positively controls the protein stability of cIAP1. In view of these data, we also analyzed the effect of cIAP1 expression on survivin protein stability. Ectopic expression of cIAP1 did not decrease survivin protein levels after treatment with cycloheximide, whereas survivin protein levels were significantly decreased in cells expressing the control vector (Fig. 4G). Conversely, survivin protein levels decreased more than control (Fig. 4H). We additionally performed a non-radioactive pulse-chase assay to re-confirm the effects of cIAP1 or survivin overexpression on protein stability. Cells were transfected with either a survivin or cIAP1 construct or empty vector. After the indicated times, cell lysates were prepared and immunoprecipitated using an anti-biotin antibody. The level of cIAP1 or survivin protein was detected via Western blotting using an anti-cIAP1 or survivin antibody. The overexpression of survivin affected the protein stability of cIAP1, and cIAP1 overexpression also affected survivin protein stability (Fig. 4I). These results indicate that survivin and cIAP1 affect the protein stability of each other.
Survivin Directly Binds to cIAP1
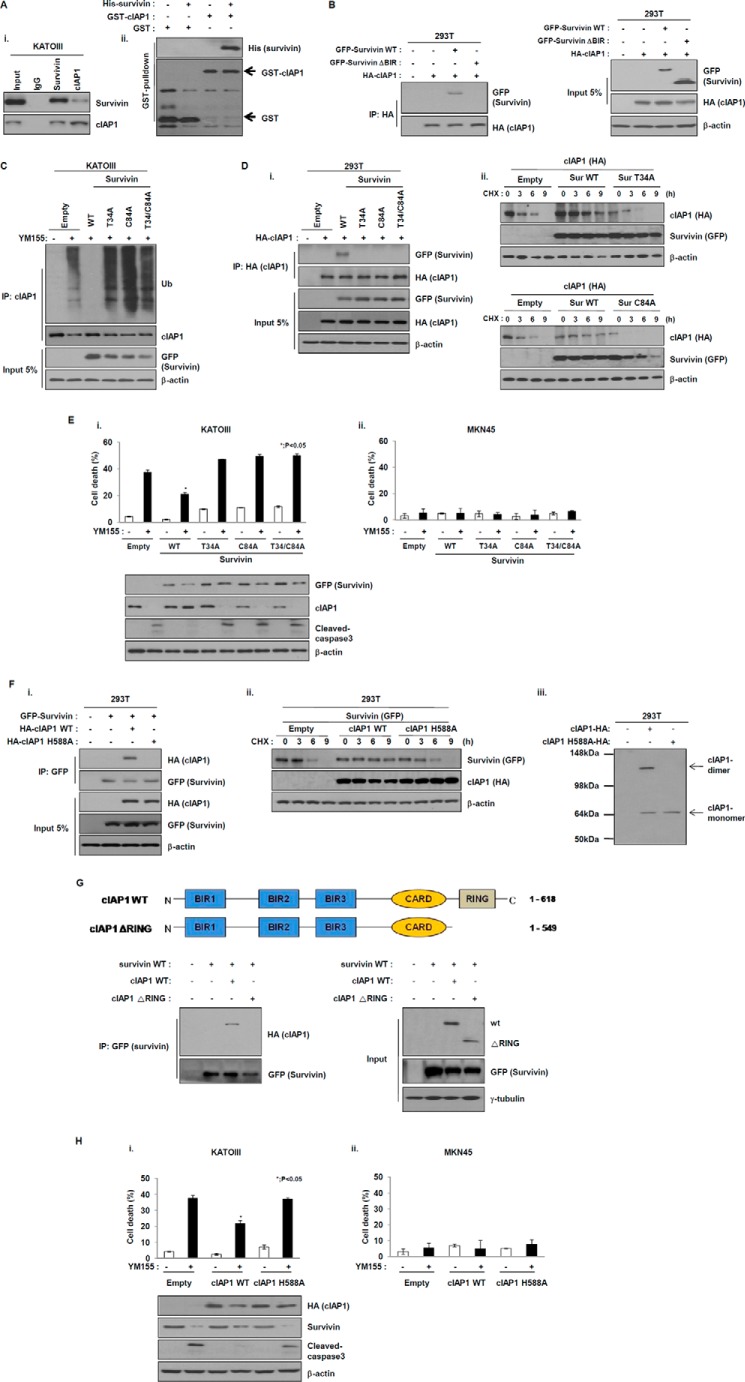
Based on the above data, we examined the direct interaction between endogenous cIAP1 and survivin. Immunoprecipitation and Western blotting analyses revealed both cIAP1-bound survivin and survivin-bound cIAP1 (Fig. 5A, i). To further confirm this, we performed GST pulldown assays. Western blot analyses of purified GST-cIAP1 or GST proteins incubated with purified His-tagged survivin. GST pulldown assays followed by Western blotting detected survivin-bound cIAP1 proteins, but did not detect survivin-bound GST proteins. These results suggest that survivin directly interacts with cIAP1 (Fig. 5A, ii). To further investigate the interaction between cIAP1 and survivin, we constructed HA-tagged cIAP1, GFP-tagged wild-type survivin, and its deleted mutant, δBIR. In 293 cells co-transfected with these plasmids, HA-tagged cIAP1 was detected in GFP-tagged wild-type survivin immunoprecipitates, but not in GFP-tagged BIR-deleted survivin mutant immunoprecipitates (Fig. 5B), indicating that cIAP1 directly interacts with the BIR domain of survivin. Thus, cIAP1 may affect the stability of survivin through interaction with the BIR domain. To confirm this, we first constructed a GFP-tagged survivin T34A mutant (threonine was mutated to alanine in the BIR domain) (19), or C84A mutant (cysteine was mutated to alanine in the BIR domain) (20). cIAP1 ubiquitination was induced independently of the expression of each mutant after exposure to YM155 (Fig. 5C). Consistently, cIAP1 directly interacted with wild-type survivin, but not with other mutants (Fig. 5D, i), implying that cIAP1 may bind to specific amino acid resides that are only present in the BIR domain of survivin. These results suggest that cIAP1 affects the stability of survivin through an interaction with the BIR domain. We additionally analyzed the effect both survivin mutants had on stability of the cIAP1 protein. Each survivin mutant also led to a decrease in ectopic cIAP1 protein levels compared with cells transfected with wild-type survivin after exposure to the protein synthesis inhibitor cycloheximide (Fig. 5D, ii). Based on the above results, whether survivin point mutants specifically affect cell death in KATOIII cells, which are sensitive to YM155, was examined. Cell death was significantly increased in KATOIII cells but not MKN45 cells expressing T34A, C84A, or both, after exposure to YM155, whereas ectopic expression of wild-type survivin partially inhibited YM155-induced cell death (Fig. 5E), indicating that T34A/C84A, which has a point mutation in the BIR domain of survivin, affects YM155-induced cell death. These results suggest that cIAP1 is stabilized through binding to the BIR domain of survivin and thereby can induce resistance to YM155.
FIGURE 5.
Survivin directly interacts with cIAP1. A, (i) KATOIII cell lysates were immunoprecipitated (IP) with anti-survivin, anti-cIAP1, or anti-rabbit IgG, and the precipitates were analyzed for Western blot analysis. (ii) Purified GST or GST-cIAP1 proteins were incubated with recombinant His-survivin proteins and the binding of two proteins was analyzed with GST pulldown analysis. B, 293T cells were co-transfected with survivin wild-type or BIR-deleted survivin and HA-tagged cIAP1 plasmids for 48 h. The cell lysates were prepared for immunoprecipitation. The binding of survivin and cIAP1 was confirmed using anti-HA and anti-GFP antibodies via Western blot analysis. C, KATOIII cells were transfected with survivin wild-type or T34A or C84A or T34A/C84A expressing plasmid for 24 h, treated with YM155 (20 nm) for another 48 h, and then incubated with the proteasome inhibitor MG132 (25 μm) for 6 h. cIAP1 ubiquitination was determined with immunoprecipitation using an anti-cIAP1 antibody by Western blot analysis. D, (i) 293T cells were co-transfected with cIAP1 wild-type and survivin wild-type or T34A or C84A or T34A/C84A vector for 48 h. Cell lysates were prepared for immunoprecipitation, which was performed using an anti-HA antibody and analyzed by Western blot analysis using anti-GFP and anti-HA antibodies. (ii) 293T cells were co-transfected with control or GFP-tagged survivin WT or survivin T34A or survivin C84A and HA-tagged cIAP1 expressing plasmid for 48 h and then treated with the protein synthesis inhibitor cycloheximide (CHX) (50 μg/ml) for the indicated times. The levels of survivin and cIAP1 were evaluated via Western blot analysis using anti-GFP and anti-HA antibodies. E, KATOIII (i) and MKN45 (ii) cells were transfected with GFP-tagged survivin wild-type or T34A or C84A or T34A/C84A expression plasmid for 48 h and then treated with YM155 (20 nm) for another 48 h. Cell death was determined using the trypan blue exclusion method. Expression levels of cIAP1 and cleaved caspase-3 were analyzed by Western blot analysis. The data are the mean ± S.D.; *, p < 0.05. F, (i) 293T cells were co-transfected with GFP-tagged survivin vector and HA-tagged cIAP1 wild-type vector or H588A vector for 48 h, and the cell lysates were immunoprecipitated with an anti-GFP antibody. The precipitates were analyzed by Western blot analysis using anti-HA or anti-GFP antibodies. (ii) 293T cells were transfected with empty vector or GFP-tagged survivin and HA-tagged cIAP1 WT or cIAP1 H588A vector for 48 h and then incubated with cycloheximide (50 μg/ml) at the indicated times. (iii) 293T cells were transfected with empty or HA-tagged cIAP1 WT or cIAP1 H588A plasmid for 48 h and then cell lysates were prepared for determination of cIAP1 monomer or dimer via native gel analysis. G, 293T cells were transfected with GFP-tagged survivin and HA-tagged cIAP1 WT or RING-deletion mutant (ΔRING) for 24 h, cell lysates were immunoprecipitated using anti-GFP antibody and then the immunoprecipitates were analyzed by Western blot using anti-HA or anti-GFP antibodies. H, KATOIII (i) and MKN45 (ii) cells were transfected with GFP-tagged survivin expression plasmid and HA-tagged cIAP1 wild-type or H588A expression plasmid for 24 h and then treated with YM155 (20 nm) for another 48 h. Cell death was determined using the trypan blue exclusion method. Expression levels of cIAP1 and cleaved caspase-3 were analyzed with Western blot analysis. The data are the mean ± S.D.; *, p < 0.05.
As shown in Fig. 5A, cIAP1 also directly interacted with survivin. In view of these data, the interaction of survivin with cIAP1 was further investigated. To this end, we first constructed a plasmid for HA-tagged cIAP1 cDNA and its point mutant, H588A, which is an alanine substitution mutation that is solely present in the RING domain of cIAP1 (21). In 293 cells co-transfected with these plasmids, HA-tagged cIAP1 wild-type was detected in GFP-tagged survivin immunoprecipitates, but not in HA-tagged cIAP1 H588A immunoprecipitates (Fig. 5F, i), indicating that survivin interacts with the RING domain of cIAP1. We further analyzed the effect of the cIAP1 mutant, H588A, on the stability of survivin protein. Consistent with the two survivin mutants, ectopic expression of the cIAP1 mutant, H588A, resulted in decreased levels of ectopic survivin protein compared with cells treated with wild-type cIAP1. Furthermore, dimerization of the cIAP1 mutant, H588A, was not observed (Fig. 5F, ii and iii). These results suggest that survivin may interact with the RING domain of cIAP1 and subsequently increase its stability. In addition, we analyzed the RING domain of cIAP1 responsible for the interaction with survivin. To this end, we constructed an HA-tagged mutant construct (delta RING cIAP1) lacking the RING domain of cIAP1. Cells were co-transfected with wild-type survivin and HA-tagged wild-type cIAP1 or δRING cIAP1. Wild-type survivin was readily detected via Western blotting following incubation with extracts from cells expressing wild-type cIAP1, but not δRING cIAP1 (Fig. 5G). Thus, the RING domain of cIAP1 is crucial for the interaction with survivin as well as dimerization. Transfection of HA-tagged cIAP1 H588A in KATOIII cells but not MKN45 cells led to cell death after exposure to YM155, unlike in cells expressing wild-type cIAP1 (Fig. 5H), indicating that the stabilization of cIAP1 by survivin affects YM155-induced cell death. Taken together, these results provide evidence that cIAP1 interacts with the BIR domain of survivin, and survivin binds to the RING domain of cIAP1, and these interactions affect gastric cancer cell sensitivity to YM155.
DISCUSSION
Several studies have elucidated the mechanisms by which YM155, a novel small-molecule survivin suppressant, exerts anticancer effects in a variety of human solid tumors (15). However, the mechanism underlying human cancer susceptibility to YM155 remains to be fully elucidated. Recently, it was reported that YM155 exerts an antiproliferative effect, inducing cell death through suppression of X-linked inhibitor of apoptosis (XIAP) in human pancreatic cancer cells (18). Thus, YM155 may be an effective therapeutic agent, showing that its action on other IAP superfamily members as well as survivin. Here, we present evidence that differences in sensitivity to YM155 depend on the protein stability of cIAP1 as well as survivin in human gastric cancer cells. We further showed that their protein stability is interactively controlled through a direct interaction of survivin with cIAP1.
Over the past decade, targeted therapy for cancer treatment has progressed. Unfortunately, despite a wide variety of approaches, human gastric cancer remains a major challenge with fewer effective targeted therapies than other cancers. Recently, it was reported that YM155 promotes cisplatin-induced apoptosis in gastric cancer (22). Our research focused on the inhibitory effect of YM155 alone on gastric cancer. We investigated whether YM155 could induce apoptotic cell death through the down-regulation of cIAP1 protein as well as survivin in human gastric cancer cells, but not mRNA levels (Fig. 2). In addition, decreased levels of XIAP and cIAP2 were not observed (Figs. 1 and 2). On the basis of these results, we focused on a decrease of cIAP1 protein after exposure to YM155. For this, we first analyzed the stability of cIAP1 and survivin with regard to their expression. As shown in Fig. 4, the decrease in the levels of cIAP1 or survivin by YM155 was blocked by the ectopic expression of each protein. Conversely, survivin knockdown led to cIAP1 ubiquitination, suggesting that survivin and cIAP1 each affect the protein stability of the other and subsequently determine cell sensitivity to YM155.
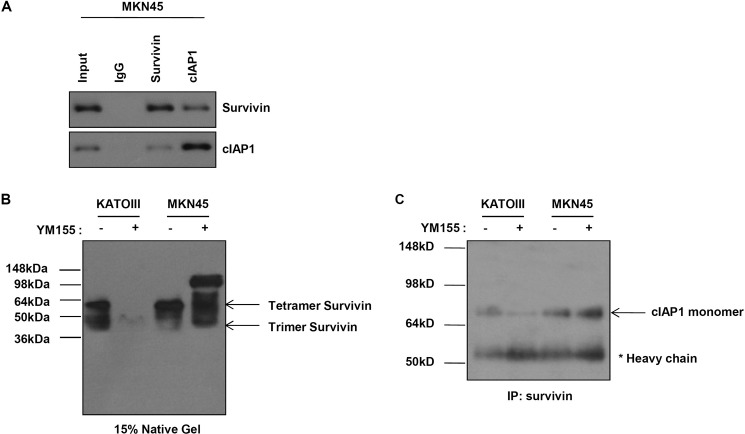
We further confirmed the mechanism underlying the degradation of cIAP1 and survivin by YM155. As shown in Fig. 5, cIAP1 directly interacted with the BIR domain of survivin, and survivin bound to the RING domain of cIAP1. We also analyzed the RING or combined RING-CARD on survivin-bound cIAP1. To this end, we first constructed HA-tagged constructs of the RING domain and the RING-CARD domain. We assessed the levels of survivin-bound cIAP1 after co-transfecting 293 cells with GFP-tagged wild-type survivin and HA-tagged wild-type cIAP1, HA-tagged RING, or RING-CARD. The levels of wild-type survivin-bound cIAP1 clearly decreased in the cells transfected with HA-tagged RING or RING-CARD (data not shown). These results suggest that the RING or combined RING-CARD acts as an antagonist to the survivin-cIAP1 interaction. Interactive control of cIAP1 and survivin affects the sensitivity to YM155. However, these interactions were also observed in MKN45 cells, which are resistant to YM155 (Fig. 6A). A survivin heteromer disappeared in KATOIII cells, which are sensitive to YM155, after exposure to YM155, but an increase of the heteromer was observed in MKN45 cells (Fig. 6B). Also, we analyzed the interaction between survivin and the cIAP1 dimer and monomer. We prepared lysates from YM155-treated cells and immunoprecipitated them using an anti-survivin antibody. Immunoprecipitation and native gel analyses revealed that survivin interacts with the monomer of cIAP1. In addition, treatment of YM155 led to a decrease in the cIAP1 monomer in KATOIII cells, but not in MKN45 cells (Fig. 6C). These results suggest that the survivin-cIAP1 complex is regulated by other factors in MKN45. That is, sensitivity to YM155 determine to the presence of other factor regulators for the survivin-cIAP1 complex. Therefore, knockdown of survivin or cIAP1 induces cell death through each others regulation in KATOIII, which is not regulated by other factors for the survivin-cIAP1 complex, but only double knockdown of survivin and cIAP1 induces cell death in MKN45, which is regulated by other factors for the survivin-cIAP1 complex (Fig. 3). The results coincided with data shown in Fig. 2. Next, we analyzed whether cIAP1 or survivin point mutants specifically affect cell death in MKN45 cells. Cell death was not observed in MKN45 cells expressing survivin T34A, C84A, both, or cIAP1 H588A after exposure to YM155, unlike KATOIII cells (Figs. 4 and 5), implying that there may be another factor that interacts with cIAP1 or/and survivin. Future research will be focused on the detailed mechanisms or unknown molecules that can directly regulate the stability of cIAP1 or/and survivin in human gastric cancer cells that are resistant to YM155.
FIGURE 6.
The survivin heteromer decreases after YM155 in YM155-sensitive KATOIII cells but not MKN45 cells. A, MKN45 cells were immunoprecipitated with anti-survivin or anti-cIAP1 or anti-rabbit IgG antibodies. Interaction of survivin and cIAP1 was determined with Western blot analysis using anti-survivin or anti-cIAP1 antibodies. B, KATOIII and MKN45 cells were treated with YM155 (20 nm) for 48 h. Cell lysates were run on a native gel. Various heteromers of survivin were determined with Western blot analysis using anti-survivin antibody. C, KATOIII and MKN45 cells were treated with YM155 (20 nm) for 48 h and then cell lysates were immunoprecipitated (IP) using an anti-survivin antibody. Determination of the binding of survivin to the monomer or dimer of cIAP1 was determined via native PAGE using anti-cIAP1.
Although we clearly established the important role of cIAP1 as well as survivin for the anti-tumor effects of YM155 in human gastric cancer cells, the regulatory mechanisms of cIAP1 and survivin in YM155-resistant cells were not addressed. Our findings highlight the therapeutic potential of YM155 for gastric cancer treatment through the degradation of cIAP1 as well as survivin.
This work was supported by Korea Health 21 R&D Project, Ministry of Health and Welfare and Family Affairs Grant HI06C0868, Republic of Korea, the National Research Foundation of Korea (NRF) funded by Korea government (MEST) Grant 2013R1A2A2A01067394, and National R&D Program for Cancer Control, Ministry of Health & Welfare, Republic of Korea Grant 1420030.
- cIAP1
- cellular inhibitor of apoptosis protein 1
- IAP
- inhibitor of apoptosis protein
- BIR
- baculovirus IAP repeat
- XIAP
- X-linked inhibitor of apoptosis.
REFERENCES
- 1. Jemal A., Bray F., Center M. M., Ferlay J., Ward E., Forman D. (2011) Global cancer statistics. CA Cancer J. Clin. 61, 69–90 [DOI] [PubMed] [Google Scholar]
- 2. Custodio A., Méndez M., Provencio M. (2012) Targeted therapies for advanced non-small-cell lung cancer: current status and future implications. Cancer Treat. Rev. 38, 36–53 [DOI] [PubMed] [Google Scholar]
- 3. Mohamed A., Krajewski K., Cakar B., Ma C. X. (2013) Targeted therapy for breast cancer. Am. J. Pathol. 183, 1096–1112 [DOI] [PubMed] [Google Scholar]
- 4. Singer E. A., Gupta G. N., Srinivasan R. (2012) Targeted therapeutic strategies for the management of renal cell carcinoma. Curr. Opin. Oncol. 24, 284–290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Tabernero J., Salazar R., Casado E., Martinelli E., Gómez P., Baselga J. (2004) Targeted therapy in advanced colon cancer: the role of new therapies. Ann. Oncol. 15, iv55–62 [DOI] [PubMed] [Google Scholar]
- 6. Kasper S., Schuler M. (2014) Targeted therapies in gastroesophageal cancer. Eur. J. Cancer 50, 1247–1258 [DOI] [PubMed] [Google Scholar]
- 7. Yoong J., Michael M., Leong T. (2011) Targeted therapies for gastric cancer: current status. Drugs 71, 1367–1384 [DOI] [PubMed] [Google Scholar]
- 8. Cho Y. Y., Yao K., Kim H. G., Kang B. S., Zheng D., Bode A. M., Dong Z. (2007) Ribosomal S6 kinase 2 is a key regulator in tumor promoter induced cell transformation. Cancer Res. 67, 8104–8112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Cheng Q., Ling X., Haller A., Nakahara T., Yamanaka K., Kita A., Koutoku H., Takeuchi M., Brattain M. G., Li F. (2012) Suppression of survivin promoter activity by YM155 involves disruption of Sp1-DNA interaction in the survivin core promoter. Int. J. Biochem. Mol. Biol. 3, 179–197 [PMC free article] [PubMed] [Google Scholar]
- 10. Nakahara T., Yamanaka K., Hatakeyama S., Kita A., Takeuchi M., Kinoyama I., Matsuhisa A., Nakano K., Shishido T., Koutoku H., Sasamata M. (2011) YM155, a novel survivin suppressant, enhances taxane-induced apoptosis and tumor regression in a human Calu 6 lung cancer xenograft model. Anticancer Drugs 22, 454–462 [DOI] [PubMed] [Google Scholar]
- 11. Yoon D. H., Shin J. S., Jin D. H., Hong S. W., Jung K. A., Kim S. M., Hong Y. S., Kim K. P., Lee J. L., Suh C., Lee J. S., Kim T. W. (2012) The survivin suppressant YM155 potentiates chemosensitivity to gemcitabine in the human pancreatic cancer cell line MiaPaCa-2. Anticancer Res. 32, 1681–1688 [PubMed] [Google Scholar]
- 12. Kaneko N., Yamanaka K., Kita A., Tabata K., Akabane T., Mori M. (2013) Synergistic antitumor activities of sepantronium bromide (YM155), a survivin suppressant, in combination with microtubule-targeting agents in triple-negative breast cancer cells. Biol. Pharm. Bull. 36, 1921–1927 [DOI] [PubMed] [Google Scholar]
- 13. Premkumar D. R., Jane E. P., Foster K. A., Pollack I. F. (2013) Survivin inhibitor YM-155 sensitizes tumor necrosis factor-related apoptosis-inducing ligand-resistant glioma cells to apoptosis through Mcl-1 downregulation and by engaging the mitochondrial death pathway. J. Pharmacol. Exp. Ther. 346, 201–210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Kumar B., Yadav A., Lang J. C., Cipolla M. J., Schmitt A. C., Arradaza N., Teknos T. N., Kumar P. (2012) YM155 reverses cisplatin resistance in head and neck cancer by decreasing cytoplasmic survivin levels. Mol. Cancer Ther. 11, 1988–1998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Rauch A., Hennig D., Schäfer C., Wirth M., Marx C., Heinzel T., Schneider G., Krämer O. H. (2014) Survivin and YM155: how faithful is the liaison? Biochim. Biophys. Acta 1845, 202–220 [DOI] [PubMed] [Google Scholar]
- 16. Yamauchi T., Nakamura N., Hiramoto M., Yuri M., Yokota H., Naitou M., Takeuchi M., Yamanaka K., Kita A., Nakahara T., Kinoyama I., Matsuhisa A., Kaneko N., Koutoku H., Sasamata M., Kobori M., Katou M., Tawara S., Kawabata S., Furuichi K. (2012) Sepantronium bromide (YM155) induces disruption of the ILF3/p54(nrb) complex, which is required for survivin expression. Biochem. Biophys. Res. Commun. 425, 711–716 [DOI] [PubMed] [Google Scholar]
- 17. Tang H., Shao H., Yu C., Hou J. (2011) Mcl-1 downregulation by YM155 contributes to its synergistic anti-tumor activities with ABT-263. Biochem. Pharmacol. 82, 1066–1072 [DOI] [PubMed] [Google Scholar]
- 18. Na Y. S., Yang S. J., Kim S. M., Jung K. A., Moon J. H., Shin J. S., Yoon D. H., Hong Y. S., Ryu M. H., Lee J. L., Lee J. S., Kim T. W. (2012) YM155 induces EGFR suppression in pancreatic cancer cells. PLoS One 7, e38625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Aspe J. R., Wall N. R. (2010) Survivin-T34A: molecular mechanism and therapeutic potential. Onco. Targets Ther. 3, 247–254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Li F., Ambrosini G., Chu E. Y., Plescia J., Tognin S., Marchisio P. C., Altieri D. C. (1998) Control of apoptosis and mitotic spindle checkpoint by survivin. Nature 396, 580–584 [DOI] [PubMed] [Google Scholar]
- 21. Yang Y., Fang S., Jensen J. P., Weissman A. M., Ashwell J. D. (2000) Ubiquitin protein ligase activity of IAPs and their degradation in proteasomes in response to apoptotic stimuli. Science 288, 874–877 [DOI] [PubMed] [Google Scholar]
- 22. Sun X. P., Dong X., Lin L., Jiang X., Wei Z., Zhai B., Sun B., Zhang Q., Wang X., Jiang H., Krissansen G. W., Qiao H., Sun X. (2014) Up-regulation of survivin by AKT and hypoxia-inducible factor 1α contributes to cisplatin resistance in gastric cancer. FEBS J. 281, 115–128 [DOI] [PubMed] [Google Scholar]