Summary
Flavodoxin replaced stress-sensitive ferredoxin throughout the evolution of phototrophs, then disappeared before plant emergence. Phylogenomic and biochemical data now permit to envisage how this adaptive trait was lost from the plant world.
Key words: Electron transfer, environmental stress, evolution, flavodoxin, ferredoxin, iron limitation.
Abstract
Ferredoxins are electron shuttles harbouring iron–sulfur clusters that connect multiple oxido-reductive pathways in organisms displaying different lifestyles. Some prokaryotes and algae express an isofunctional electron carrier, flavodoxin, which contains flavin mononucleotide as cofactor. Both proteins evolved in the anaerobic environment preceding the appearance of oxygenic photosynthesis. The advent of an oxygen-rich atmosphere proved detrimental to ferredoxin owing to iron limitation and oxidative damage to the iron–sulfur cluster, and many microorganisms induced flavodoxin expression to replace ferredoxin under stress conditions. Paradoxically, ferredoxin was maintained throughout the tree of life, whereas flavodoxin is absent from plants and animals. Of note is that flavodoxin expression in transgenic plants results in increased tolerance to multiple stresses and iron deficit, through mechanisms similar to those operating in microorganisms. Then, the question remains open as to why a trait that still confers plants such obvious adaptive benefits was not retained. We compare herein the properties of ferredoxin and flavodoxin, and their contrasting modes of expression in response to different environmental stimuli. Phylogenetic analyses suggest that the flavodoxin gene was already absent in the algal lineages immediately preceding land plants. Geographical distribution of phototrophs shows a bias against flavodoxin-containing organisms in iron-rich coastal/freshwater habitats. Based on these observations, we propose that plants evolved from freshwater macroalgae that already lacked flavodoxin because they thrived in an iron-rich habitat with no need to back up ferredoxin functions and therefore no selective pressure to keep the flavodoxin gene. Conversely, ferredoxin retention in the plant lineage is probably related to its higher efficiency as an electron carrier, compared with flavodoxin. Several lines of evidence supporting these contentions are presented and discussed.
Functional equivalence between ferredoxins and flavodoxins: how, why, and what for?
Electron shuttling as the currency for energy flow through cellular metabolism
Electron transfer reactions are the primary energy-transduction processes in all living organisms, and life on Earth most certainly began with the evolution of a limited number of metabolic routes coupled to oxido-reductive chemistry (Kim et al., 2013). Reducing equivalents are ferried across networks of redox enzymes to the major metabolic sinks via a small set of diffusible electron carriers. They act as electronic switches between cellular sources of reducing power (i.e. light-driven reactions, pyridine nucleotides, sugars, etc) and electron-consuming routes and processes. The most extensively used of these electron shuttles is ferredoxin (Fd), a small mobile metalloprotein containing a [2Fe–2S] cluster as prosthetic group (Hase et al., 2006). Fds are found throughout the tree of life, with different isoforms located in mitochondria and chloroplasts of eukaryotes. In organisms displaying oxygenic photosynthesis (plants, algae, and cyanobacteria), Fd collects reducing equivalents generated during photosynthetic electron transport, and delivers them to a plethora of metabolic, regulatory, dissipative, and developmental processes. A substantial fraction of photoreduced Fd is employed for NADP+ reduction in an electron-hydride exchange reaction catalysed by the flavoenzyme ferredoxin-NADP+ reductase (FNR) (Carrillo and Ceccarelli, 2003; Ceccarelli et al., 2004). The NADPH thus formed provides the reducing power required for the regenerative step of the Calvin cycle (i.e. conversion of 1,3-bisphosphoglycerate into glyceraldehyde 3-phosphate), and for other biosynthetic, regulatory, and protective pathways. Reduced Fd also delivers electrons for nitrogen and sulfur assimilation; amino acid, fatty acid, and secondary metabolism; antioxidant defence; reductive activation of enzymes, etc. A comprehensive list of Fd-dependent reactions and redox partners in photosynthetic organisms is shown in Table 1.
Table 1.
List of Fd- and Fld-dependent reactions and redox partners in plastids and cyanobacteria
| Protein partner | Function | Metabolic pathway | Organisms | References |
|---|---|---|---|---|
| Ferredoxin | ||||
| Photosystem I (PSI) | Photosynthetic electron transport | Photosynthesis | Cyanobacteria Algae Plants |
Meimberg and Mühlenhoff (1999); Sétif (2001)
Sétif (2001) Sétif (2001) |
| PGR5, FNR, and PSI | Cyclic electron flow | Photosynthesis | Cyanobacteria Algae Plants |
Yeremenko et al. (2005)
Hanke and Mulo (2013) Hanke and Mulo (2013) |
| Ferredoxin–NADP+ reductase (FNR) | NADP+ reduction | Photosynthesis | Cyanobacteria | Hase et al. (2006); Hanke and Mulo (2013) |
| Algae | Peden et al. (2013) | |||
| Plants | Hase et al. (2006); Hanke and Mulo (2013) | |||
| NADPH oxidation | Heterotrophic pathways | Apicomplexa Plants |
Ceccarelli et al. (2004)
Onda et al. (2000) |
|
| Nitrogenase | N2 fixation | Nitrogen assimilation | Cyanobacteria | Masepohl et al. (1997) |
| Nitrate reductase | Reduction of NO3 – to NO2 – | Nitrogen assimilation | Cyanobacteria | Hase et al. (2006); Hanke and Mulo (2013) |
| Nitrite reductase | Reduction of NO2 – to NH4 + | Nitrogen assimilation | Cyanobacteria Algae Plants |
Hase et al. (2006); Hanke and Mulo (2013)
Vigara et al. (1998); Terauchi et al. (2009) Hanke and Mulo (2013) |
| Glutamine synthetase | Combination of NH4 + with glutamate to form glutamine | Nitrogen assimilation | Algae | Vigara et al. (1998) |
| Glutamate-oxoglutarate amino transferase (GOGAT) | Glutamate synthesis | Amino acid synthesis | Cyanobacteria Algae Plants |
Hase et al. (2006)
Hase et al. (2006) Hase et al. (2006) |
| Hydrogenase | H2 formation/H2 oxidation | Hydrogen metabolism | Cyanobacteria Algae |
Gutekunst et al. (2014)
Peden et al. (2013) |
| Sulfite reductase (SiR) | Reduction of SO3 2– to H2S | Sulfur assimilation | Plants | Hase et al. (2006); Hanke and Mulo (2013) |
| Ferredoxin–thioredoxin reductase (FTR) |
Thioredoxin (Trx) reduction | Redox regulation+ | Cyanobacteria Algae Plants |
Hase et al. (2006); Hanke and Mulo (2013)
Hase et al. (2006) Hase et al. (2006); Hanke and Mulo (2013) |
| Fatty acid desaturase (FAD) | Double bond formation in fatty acids | Lipid metabolism | Cyanobacteria Algae Plants |
Hanke and Mulo (2013)
Peden et al. (2013) Hanke and Mulo (2013) |
| Monodehydroascorbate reductase (MDHAR) | Ascorbate regeneration | Antioxidant defence | Plants | Hanke and Mulo (2013) |
| Heme oxygenase and phytochromobilin synthase | Phytochromobilin* synthesis | Development | Plants | Hanke and Mulo (2013) |
| Heme oxygenase and ferredoxin-dependent bilin reductases | Bilin pigment synthesis | Development | Cyanobacteria | Hanke and Mulo (2013) |
| Flavodoxin | ||||
| PSI | Photosynthetic electron transport | Photosynthesis | Cyanobacteria Algae |
Meimberg and Mühlenhoff (1999); Sétif (2001)
Peleato et al. (1994) |
| FNR | NADP+ reduction | Photosynthesis | Cyanobacteria Algae |
Sancho (2006)
Peleato et al. (1994) |
| FNR and PSI | Cyclic electron flow | Photosynthesis | Cyanobacteria | Hagemann et al. (1999) |
| Nitrite reductase | Reduction of NO2 – to NH4 + | Nitrogen assimilation | Algae | Vigara et al. (1998) |
| Glutamine synthetase | Combination of NH4 + with glutamate to form glutamine | Nitrogen assimilation | Algae | Vigara et al. (1998) |
| Hydrogenase | H2 formation/H2 oxidation | Hydrogen metabolism | Cyanobacteria | Gutekunst et al. (2014) |
+Reduced Trx activates a number of chloroplast enzymes of the Calvin cycle, the malate valve, lipid and starch metabolism, translation, and antioxidant defence.
*Plant chromophore of the light sensor phytochrome.
Many prokaryotes (including cyanobacteria) and some algae express an isofunctional electron carrier, flavodoxin (Fld), a soluble flavoprotein that contains flavin mononucleotide (FMN) as prosthetic group instead of an iron–sulfur centre (Sancho, 2006). Fld properties as redox shuttle largely match those of Fd, being able to replace the metalloprotein in many reactions (Table 1). The subcellular location of Fld also corresponds to that of Fd, namely, the bacterial cytosol and algal chloroplasts (La Roche et al., 1996). When both carriers are present in the same genome, the Fld gene is typically induced as an adaptive resource under environmental or nutritional hardships that compromise Fd expression or activity (i.e. iron limitation). Given the key position occupied by Fd in the network of electron distribution, its decline affects vast portions of central metabolism, as well as defensive, regulatory, and developmental processes (reviewed in Zurbriggen et al., 2008). Fld induction mitigates these effects, permitting survival and reproduction under the adverse condition, although the relevance of Fld replacement varies among species. In the cyanobacterium Synechocystis sp. PCC 6803, for instance, disruption of the fld gene has no fatal consequences, even under iron limitation (Kutzki et al., 1998), whereas the gene encoding the main Fd isoform was found to be essential in spite of Fld induction (Poncelet et al., 1998). However, several exceptions to this rule are documented in algae and cyanobacteria (see below), and a few Fld-specific pathways have been described in prokaryotes. Indeed, Fld is an essential gene in Escherichia coli and Helycobacter pylori, whereas Fd is not (Zheng et al., 1999; Freigang et al., 2002; Puan et al., 2005).
Both Fd and Fld are able to mediate NADP+ reduction as substrates of FNR (Nogués et al., 2005). This reaction proceeds backwards, from NADPH to oxidized Fd/Fld, in non-photosynthetic cells and tissues (Onda et al., 2000; Ceccarelli et al., 2004). NADPH is the normal reductant in heterotrophic microorganisms, mitochondria, and non-photosynthetic plastids, but carbohydrates can also be used as electron donors to reduce Fd/Fld by enzymes such as the pyruvate–Fd reductase of E. coli (Blaschkowski et al., 1982). Flds are not found in plants or animals, except as domains in larger enzymes, usually oxidoreductases, where they play specific roles different from electron shuttling (Sancho, 2006).
Comparative properties of ferredoxins and flavodoxins
Many aspects of Fd structure and function have been extensively reviewed by Hase et al. (2006) and by Hanke and Mulo (2013). Therefore, we will provide herein only a brief summary of Fd properties with reference to those of Fld. The reader is referred to the above-mentioned articles for a more comprehensive description of Fd biochemistry. These electron shuttles employ iron–sulfur clusters of different stoichiometry as prosthetic groups, with the photosynthetic Fds harbouring a [2Fe–2S] centre (Hase et al., 2006). The irons are bridged by the two sulfur atoms and bind to the sulfide side-chains of four highly conserved cysteines (Fig. 1A). The FMN group of Fld is non-covalently bound to the apoprotein, with the isoalloxazine moiety sandwiched between the coplanar aromatic side chains of a tyrosine and a tryptophan (Fig. 1A). Fds have a Mw of ~12kDa (Hase et al., 2006), whereas Flds are slightly larger, with Mw=15–22kDa (Sancho, 2006).
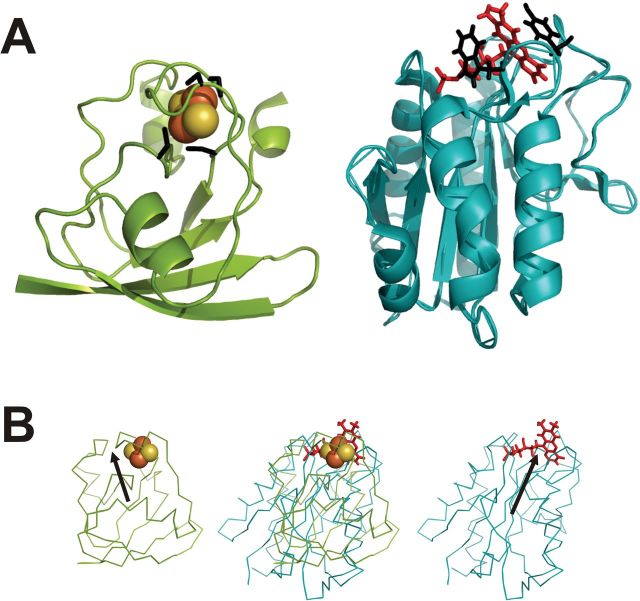
Fig. 1.
Comparative structures of Anabaena ferredoxin and flavodoxin. (A) Ribbon diagrams of Fd (left) and Fld (right) showing interactions of the prosthetic groups at the active sites. The Fe–S cluster of Fd is represented as red-orange balls, and the FMN of Fld as red sticks. The side chains of the binding cysteines of Fd and of the stacking aromatic amino acids of Fld are coloured as black sticks. (B) Fd and Fld can be compared on the basis of their surface electrostatic potentials. The left and right panels show the Anabaena Fd and Fld, respectively, as Cα-traces. Prosthetic groups are highlighted. The two proteins are aligned according to the procedure derived by Ullmann et al. (2000). In the central picture, the structures are superimposed, showing co-localization of the redox centres. For clarity reasons, the vectors corresponding to the dipolar moments (black arrows) are drawn from positive to negative, opposite to conventional representations.
From sequence alignments and structural considerations, Flds can be divided into short-chain and long-chain classes, which differ by the presence of a 20-amino acid loop of a so far unknown function (López-Llano et al., 2004). Phylogenetic analyses indicate that the two lineages have diverged once (Sancho, 2006). Only short-chain Flds have been found in Gram-positive bacteria (firmicutes), whereas cyanobacteria and algae harbour members of the long-chain class exclusively (Pérez-Dorado et al., 2013). Other Gram-negative bacteria may contain Flds from both families, occasionally in the same genome. E. coli, for instance, has at least four genes predicted to encode Flds. Two of them (fldA and fldB) belong to the long-chain class, whereas the other two (mioC and yqcA) encode short-chain Flds (Birch et al., 2000). Only members of the long-chain Fld class have been associated with stress protection (Zheng et al., 1999; Sancho, 2006; Moyano et al., 2014).
Both the [2Fe–2S] cluster of Fd and the flavin group of Fld can in principle exchange one or two electrons. However, empirical evidence indicates that they behave as obligatory one-electron carriers, switching between the Fe+3.Fe+3/Fe+3.Fe+2 (Bott, 1999) and the semiquinone/hydroquinone states, respectively (Nogués et al., 2005). These transitions have similar redox potentials (–400 to –430 mV for the photosynthetic isoforms), which allow the two proteins to behave as low potential electron shuttles.
Photosynthetic Fd has been shown to be more efficient than Fld in most reactions assayed in vitro, including NADP+ photoreduction by isolated thylakoids and thioredoxin reduction by Fd-thioredoxin reductase in reconstituted systems (Tognetti et al., 2006), although exceptions to this tendency have been reported. Electron donation to the NiFe-hydrogenase from Synechocystis sp. PCC 6803 proceeds at a higher rate with the endogenous Fld, compared with one of the minor Synechocystis Fd isoforms or the main leaf Fd from spinach (Gutekunst et al., 2014). In most studies, however, Fd proved to be the preferred redox partner. For instance, Fd photoreduction by PSI from Synechococcus sp. PCC 7002 showed a strong preference over Fld (Meimberg and Mühlenhoff, 1999), and the Fd from the green alga Chlorella fusca was more efficient than Fld as electron donor to nitrite reductase and glutamine synthetase, two enzymes involved in nitrogen assimilation (Vigara et al., 1998).
FNR-mediated reactions are the best characterized at the kinetic level, revealing some interesting features. The k cat/K M value of Anabaena Fd was reported to be ~25-fold higher than that of Fld (Medina et al., 1998). The strength of the interaction with Anabaena FNR, as reflected by the Michaelis constants, was similar for both carriers, indicating that the gain in efficiency was at the expense of electron transfer rates (Medina et al., 1998). It is likely that the complex electronic configuration of the flavin is not flexible enough to attain the high electron transfer rates typical of transition metals. Iron ions contain incompletely filled d orbitals which can readily accept electrons from different partners with various geometries, making them particularly versatile in oxido-reductive processes. It is indeed remarkable that some of these electron transfer reactions can be mimicked by the particular arrangement of π-orbitals found in the isoalloxazine ring system.
Functional equivalence without sequence and structural conservation
Fd and Fld do not share any structural similarity, and yet they can engage in essentially the same oxido-reductive reactions. The key to this apparent paradox resides in the very function of these proteins. One of the most desirable features of an electron shuttle is promiscuity, namely, the ability to exchange reducing equivalents with different redox partners. Accordingly, Fd and Fld have been tailored by evolution to be loosely selective in their interactions. Analysis of plant Fd binding sites in various Fd-dependent enzymes revealed no obvious homology (Hase et al., 2006; Hanke and Mulo, 2013). Then, docking of Fd (and Fld) must be determined by general features of the active sites rather than contacts with specific conserved amino acids. The prosthetic groups of both proteins are eccentric and surrounded by patches of negatively charged residues, whereas their enzyme partners harbour a crown of positively charged amino acids around their exposed cofactors (Hase et al., 2006; Hanke and Mulo, 2013). Initial interactions are steered by electrostatic attractions that help to stabilize the binary complexes, and serve to position the corresponding prosthetic groups at the proper distance and geometry to allow direct outer-sphere electron transfer between them (Kurisu et al., 2001). Water exclusion from the hydrophobic area between the two proteins further strengthens binding (Kurisu et al., 2001; Hanke and Mulo, 2013). The charged regions in Fd and Fld are remarkably insensitive to mutations, and different spatial arrangements of the two proteins at the FNR binding site (i.e. rotations) are allowed without losing the ability for electron transfer (Gómez-Moreno et al., 1994; Goñi et al., 2009).
Although Fd and Fld differ in virtually all structural features, they can be aligned on the basis of their Coulomb electrostatic potentials. Applying the Hodgkin index to evaluate their similarity in this sense, Ullmann et al. (2000) obtained a significant overlapping. The cofactors, rather than their centres of mass, coincided in the alignments (Fig. 1B). Both proteins have strong dipolar moments (380–700 Debyes), with the vectors of the negative dipoles pointing toward the active site regions (Fig. 1B). These considerations provide a rationale to understand why Fd and Fld are able to interact with so many different enzymes, and why they can be exchanged as substrates of a given partner without major loss in efficiency.
Ferredoxins and flavodoxins in plants, algae, and cyanobacteria
Ferredoxin accumulation in response to different environmental stimuli
The paralogy of Fd-coding genes varies widely among species (Table 2). Most cyanobacteria contain three or more isoforms, including highly divergent Fds of unknown function such as FdC1 (Poncelet et al., 1998; Voss et al., 2011). Among eukaryotes, glaucophytes and algae from the red-plastid lineage usually have a single Fd, whereas green algae and land plants contain numerous isoforms (Table 2). Six different Fd variants have been described in the unicellular green alga Chlamydomonas reinhardtii, a freshwater chlorophyte (Peden et al., 2013), in the C3 plant Arabidopsis thaliana, with isoforms located in leaves and roots (Hanke et al., 2004; Voss et al., 2011), and in the C4 plant maize, present in root plastids and in mesophyll and bundle sheath chloroplasts (Matsumura et al., 1997; Sakakibara, 2003; Cheng et al., 2008). Additional putative Fds have been retrieved from some sequenced genomes (i.e. maize), but they have not been functionally characterized. In general, the main Fd isoform engaged in photosynthetic electron transport accounts for 80–90% of the total Fd pool in leaves.
Table 2.
Phylogenetic distribution and copy number variation of long-chain Flds and cyanobacterial-/plastidic-type Fds
| Domain of life | Long- chain Fld | Cyanobacterial-/plastidic-like Fd* | |||
|---|---|---|---|---|---|
| Eukarya | Archaeplastida | Glaucophytes | 0 | 1 | |
| Red algae | 0–1 | 1 | |||
| Chlorophytes | 0–2 | 3–6 | |||
| Streptophytes (charophytes + land plants) | 0 | 5–8 | |||
| Protists | Alveolates | Photosynthetic Non-photosynthetic |
1 0 |
1 0–1 |
|
| Rhizaria | Photosynthetic Non-photosynthetic |
1 0 |
1–3 0 |
||
| Stramenopiles | Photosynthetic Non-photosynthetic |
0–2 0 |
1–4 0 |
||
| Haptophytes | 1 | 4 | |||
| Cryptomonads | 0–1 | 1 | |||
| Euglenids | NA | 1 | |||
| Animals | 0 | 0 | |||
| Fungi | 0 | 0 | |||
| Archaea | 0–1# | 0–6 | |||
| Bacteria | Cyanobacteria | 0–5 | 2–8 | ||
| Proteobacteria | 0–2 | 0–2 | |||
| Firmicutes | 0 | 0 | |||
| Actinobacteria | 0 | 0–2 | |||
| Bacteroidetes | 0–4 | 0 | |||
| Chlorobi | 0–2 | 0 | |||
| Chlamydiae | 0 | 0 | |||
| Spirochaetes | 0–1 | 0 | |||
| Tenericutes | 0–1 | 0 | |||
| Fusobacteria | 0–3 | 0 | |||
| Chloroflexi | 0–1 | 0 | |||
| Acidobacteria | 0–1 | 0 | |||
Sources: Integrated Microbial Genomes (IMG) dababase (http://img.jgi.doe.gov; Markowitz et al. 2012) and National Center for Biotechnology Information (NCBI) (http://www.ncbi.nlm.nih.gov).
*The presence of Fd versions with sequence similarity to proteobacterial-/mitochondrial-like Fds is not indicated.
#Long-chain Fld forms found in 3 out of 456 sequenced genomes.
NA, no genomic sequence available
Tissue specificity broadly correlates with labour division and expression patterns. In the case of leaf Fd isoforms, cis-acting elements located upstream of the transcription initiation site provide for moderate light responsiveness (Vorst et al., 1993). However, replacement of these regions by a constitutive promoter does not abolish light induction (Elliott et al., 1989), indicating that Fd expression depends on sequences located in the transcribed portion of the gene and is therefore subject to post-transcriptional regulation. Using chimeric constructs of transcribed and flanking sequences of the Fd gene fused to various reporter genes, Dickey et al. (1992) could identify a region of light responsiveness within the transcription unit itself, extending both up- and downstream of the translation initiation codon. This mode of regulation seems to be an evolutionary novelty, as the vegetative Fd gene of cyanobacteria is light-modulated exclusively through sequences located in the 5′-untranscribed region (Mazouni et al., 2003).
Light also stimulates expression of some Fd isoforms in algae (Lemaire et al., 1999; Whitney et al., 2011). In the diatom Thalassiosira pseudonana, Fd expression is coordinated with a diel cycle, reaching its maximum transcription levels during the light period. However, this oscillating expression pattern is eliminated under continuous light, indicating that it is not under circadian control (Whitney et al., 2011). In contrast, the expression of the photosynthetic Fd in C. reinhartii is regulated by the circadian cycle, as well as by the redox state of the photosynthetic electron transport chain (PETC) (Lemaire et al., 1999).
Non-photosynthetic Fds participate in the assimilation of nitrate as electron donors for nitrite reductase (Terauchi et al., 2009; Peden et al., 2013). Several components of this metabolic pathway are induced in roots when plants are grown on oxidized nitrogen sources (i.e. nitrate), and they are repressed by ammonia (Matsumura et al., 1997; Patterson et al., 2010). Two Fd isoforms are present in maize root plastids; one of them is expressed constitutively (FdIII), whereas the other (FdVI) responds to nitrate (Matsumura et al., 1997). FdVI is also nitrate-induced in mesophyll chloroplasts (Sakakibara, 2003). It is unclear whether this Fd is co-expressed with the photosynthetic isoform in the same cells and chloroplasts, or if it is present in heterotrophic cells of the leaf (Hanke and Mulo, 2013). Some of the non-photosynthetic isoforms have less negative midpoint redox potentials compared with photosynthetic Fds (Hanke et al., 2004; Gou et al., 2006), indicating that they may have a narrower range of suitable electron partners (Peden et al., 2013).
Each of the six Fds of C. reinhardtii harbours a chloroplast-targeting sequence. Expression of the various isoforms responds to different environmental inputs, correlating with their functions. The most abundant among them, the photosynthetic Fd encoded by the petF gene, is induced by light (Terauchi et al., 2009). Its expression is not affected by nitrate, oxidative stress, anoxia, or copper deprivation. One of the minor non-photosynthetic isoforms, Fdx2, is induced by nitrate and H2O2 and is a preferred substrate for nitrite reductase, whereas PetF Fd preferentially interacts with FNR and Fd-thioredoxin reductase (Terauchi et al., 2009; Peden et al., 2013). The results suggest that labour division and differential expression of Fd variants preceded tissue differentiation and the advent of land plants. Interestingly enough, C. reinhardtii lacks Fld (Merchant et al., 2007).
Dinitrogen fixation in microorganisms is an anaerobic process because nitrogenase is very sensitive to oxygen inactivation (Schrautemeier et al., 1995). Nitrogen-fixing cyanobacteria usually express one or more Fd isoforms (generically named FdxH) as dedicated electron donors for the nitrogenase, in addition to the major Fd variants that mediate photosynthesis. Expression of these “diazotrophic” Fds is regulated by different mechanisms depending on the morphological and physiological strategy for dinitrogen fixation adopted by the host. In those cyanobacteria in which heterocysts provide the anaerobic environment, such as Anabaena sp. PCC 7120, an Fd isoform specific for these differentiated cells is expressed under dinitrogen-fixing conditions (Masepohl et al., 1997). In contrast, in the non-heterocystous, filamentous cyanobacterium Plectonema boryanum, which fixes nitrogen only in low oxygen environments, the single FdxH variant accumulates under microaerobic conditions (Schrautemeier et al., 1994). The two types of FdxH isoforms and regulatory mechanisms can be present in the same organism, as it occurs in heterocyst-forming Anabaena variabilis (Schrautemeier et al., 1995). Some non-heterocystous cyanobacteria do not need a microaerobic environment because they temporally separate photosynthesis and dinitrogen fixation, so that the latter process is carried out during the night. Examples of this strategy are Cyanothece sp. ATCC 51142 and Crocosphaera watsonii WH 8501, where an Fd isoform is expressed during the dark period (Stöckel et al., 2008; Shi et al., 2010). All these “diazotrophic” Fds are coded by genes located in nif operons (Schrautemeier et al., 1994, 1995; Welsh et al., 2008; Shi et al., 2010).
Down-regulation of ferredoxin levels by iron deficit and environmental stress
Fd expression is universally repressed under conditions of iron limitation, as revealed by various experimental approaches, including genome-wide microarray analyses in both plants (Thimm et al., 2001) and cyanobacteria (Singh et al., 2003; Thompson et al., 2011). The six Fd isoforms of C. reinhardtii also decline under iron starvation, even though transcription of at least two of the non-photosynthetic isoforms (Fdx3 and Fdx6) was stimulated under these conditions (Terauchi et al., 2009). The transcriptional control of genes related to iron metabolism is carried out by Fur (ferric uptake regulator) in most prokaryotes, but Fd repression in iron-starved cyanobacteria is independent of this mechanism (Ghassemian and Straus, 1996). Instead, it has been reported that iron levels control the stability of Fd mRNA in Synechococcus sp. PCC 7942 and Anabaena sp. PCC 7937 (Bovy et al., 1993a). Cis-acting elements located at the 5′ extreme of the mRNA are involved in this regulation (Bovy et al., 1993b).
Although the main value of Fld under iron limitation is certainly the taking over of Fd functions as the major electron-distributing hub in chloroplasts and cyanobacteria (Table 1), it is conceivable that this substitution might also contribute to plant welfare by permitting reallocation of the scarce available iron to other metal-dependent routes. The question is then how substantial this contribution could be, and the response differs depending on the organism and/or tissue. In Fe-replete Thalassiosira weissflogii, 30–40% of the cellular iron was found to be associated with Fd (Erdner and Anderson, 1999). As a complete PETC has 22 Fe atoms, this would represent a stoichiometry of 4–5 Fd molecules per chain unit, in fairly good agreement with experimental determinations (Böhme, 1978). The iron share of Fd seems to be lower but still significant in plants (Terry and Abadía, 1986; Shikanai et al., 2013). Stromal iron represents 20–30% of total leaf Fe, with Fd and ferritin as the most abundant metalloproteins (Terry and Abadía, 1986). Interestingly, when iron is withheld from plants, the stromal fraction declines much faster than the thylakoid or extrachloroplastic fractions (Terry and Low, 1982), confirming the high sensitivity of Fd to iron limitation. On the other hand, heterotrophic Fd isoforms account for less than 10% of their leaf counterparts (Voss et al., 2008), indicating that they play a marginal role in cell iron homeostasis.
Oxidative stress and adverse environmental situations (salinity, extreme temperatures, water deficit) lead to down-regulation of Fd levels in both plants and cyanobacteria (Mazouni et al., 2003; Singh et al., 2003; Zimmermann et al., 2004; Tognetti et al., 2006; Terauchi et al., 2009; Ceccoli et al., 2011). Algal Fd isoforms also declined when C. reinhardtii cells were exposed to H2O2 (Terauchi et al., 2009). Transcription of the gene encoding non-photosynthetic Fdx2 increased under oxidative conditions, but this induction was not reflected at the protein level. Moreover, when C. reinhardtii was grown in the presence of nitrate to obtain maximal Fdx2 accumulation, the contents of this isoform still declined after H2O2 treatment (Terauchi et al., 2009).
Expression of Anabaena Fd in tobacco plants under control of a constitutive promoter resulted in the loss of light responsiveness. In contrast, when these transgenic plants were exposed to various stress conditions, the cyanobacterial Fd was down-regulated even faster than the endogenous counterparts (Ceccoli et al., 2012). The results suggest that: (i) repression of the Fd gene by adverse environmental situations also has post-transcriptional components, and (ii) light- and stress-dependent regulation of Fd expression involve different responsive elements (Ceccoli et al., 2012).
Then, data collected from a number of systems indicate that when plants are exposed to situations in which the Fd holoprotein cannot assemble (i.e. iron starvation) or is likely to be destroyed (i.e. oxidative stress), they respond by down-regulating accumulation of the corresponding transcripts. Such a response might seem odd, considering that Fd activity helps to relieve the electron pressure on the PETC and contributes to the antioxidant defence by providing reducing power for dissipative and scavenging processes. However, adoption of this pre-emptive strategy might respond to different imperatives; for instance, to save the energetic cost of synthesizing an apoprotein that cannot be used anyway. An additional advantage in the case of oxidative conditions is that formation of the Fe–S cluster would be prevented by lack of the Fd apoprotein, therefore eliminating a potential target for oxidants that upon destruction could lead to release of free iron and propagation of deadly hydroxyl radicals via Fenton-type reactions (see below). Besides these somehow obvious benefits, recent observations suggest that decline of Fd levels might play yet unknown roles in tolerance to certain environmental onslaughts. Mutant Arabidopsis lines deficient in the major photosynthetic Fd were, as expected, hypersensitive to a short-term treatment with high light (Liu et al., 2013). However, these plants exhibited unpredicted tolerance to prolonged high light exposure compared with WT siblings, apparently profiting from a differential increase in PGR5-dependent cyclic electron flow (Liu et al., 2013). Although it remains to be determined whether these effects could be extended to other stress situations, the results suggest that down-regulation of at least some Fd isoforms might contribute to long-term plant acclimation to adverse environments.
Flavodoxin and ferredoxin expressions display contrasting responses to environmental inputs
In most algal and cyanobacterial species, Fld is not expressed under iron-replete conditions. This restriction has limited studies on the light responsiveness of this protein, which is clearly involved in photosynthesis when present. The only exception is the fld gene from the cyanobacterium Prochlorococcus marinus MED4, which has been shown to be up-regulated under daylight as a typical photosynthetic gene (Zinser et al., 2009). The mechanism of this diel mode of expression is unknown.
In general, the response of Fld expression to environmental stimuli follows a pattern opposite to that exhibited by Fd. In E. coli and other enterobacteria, the fldA and fldB genes are members of the soxRS regulon (Gaudu and Weiss, 2000), which orchestrates the defence of the bacterial cell against oxidants and redox-cyclic compounds (Giró et al., 2006). The global trancriptional regulator Fur also belongs to this regulatory system (Zheng et al., 1999), and its gene is located immediately downstream of that encoding FldA. The SoxS transcription factor binds to the promoter of the fldA gene, leading to the expression of a bicistronic mRNA that encompasses both fldA and fur (Zheng et al., 1999). No SoxR/SoxS homologues have been identified in cyanobacteria and the regulator sustaining the superoxide response is still unclear in these phototrophs (Latifi et al., 2009). On the other hand, the fld gene is part of the Fur regulon in cyanobacteria (González et al., 2014), and FurA is also induced by oxidative stress (López-Gomollón et al., 2009). In fact, Fld induction has been described in cyanobacteria under osmotic (Hagemann et al., 1999; Rai et al., 2014), heat (Kojima et al., 2006), high light (Havaux et al., 2005), and oxidants (Singh et al., 2004), all conditions that lead to Fd down-regulation.
Iron deficit seems to be the most critical imperative that determines Fld adaptive value (Erdner et al., 1999). However, iron responsiveness is not a universal feature of Fld genes. Although most cyanobacteria do respond to iron deficiency by inducing Fld (Singh et al., 2003; Chappell and Webb, 2010), expression of certain Fld isoforms proved insensitive to this nutritional stress. Expression patterns correlate with phylogenetic positions, permitting allocation of cyanobacterial Flds to two different groups. One of them consists of those Flds whose expression is enhanced by iron deficiency (Singh et al., 2003; Chappell and Webb, 2010). They form a highly supported clade in Fld phylogenetic trees (Lin et al., 2009). The other group of Flds do not respond to iron deficiency and are only present in some diazotrophic cyanobacteria. They cluster with the Flds of non-photosynthetic diazotrophic proteobacteria (Lin et al., 2009), suggesting that they were transferred from proteobacteria to cyanobacteria by horizontal gene transfer (HGT).
As indicated previously, several diazotrophic cyanobacteria fix nitrogen in the dark, temporally separating photosynthesis and nitrogen fixation (Compaoré and Stal, 2010). In these organisms, the iron-unresponsive Flds have been observed to increase their abundance during the dark period, similar to the nitrogenase subunits and the “diazotrophic” Fd isoforms (Stöckel et al., 2008; Shi et al., 2010; Saito et al., 2011), suggesting that these Flds might act as electron donors to nitrogenase. The use of Fld in nitrogen fixation has been proposed as part of a general metabolic strategy to spare iron for the iron-rich nitrogenase complex (Saito et al., 2011).
Lack of iron responsiveness has also been observed in eukaryotes. In most algal species containing Fld, the gene is present as a single iron-responsive copy (Li et al., 2004). Exceptions are some diatoms with two homologous genes, only one of them regulated by iron availability (Whitney et al., 2011). A palindromic motif has been identified in the promoter region of low-iron regulated genes, including those coding for the iron-responsive Fld isoforms (Lommer et al., 2012). The role of these cis-acting elements in iron-dependent gene regulation was confirmed by the promoter truncation technique in Phaeodactylum tricornutum (Yoshinaga et al., 2014). On the other hand, iron-insensitive Fld isoforms display diel periodicities, reaching the highest transcript abundances during the dark period (Whitney et al., 2011). The functions and redox partners of these “nocturnal” Flds are unknown.
The role of iron limitation in the spread and disappearance of Fld from the Viridiplantae kingdom will be discussed in a broader context in a forthcoming section.
The limits of flavodoxin distribution
Fds are found in a wide range of organisms pervading all kingdoms. They include cyanobacteria and α-proteobacteria, the types of organisms that gave origin to modern day chloroplasts and mitochondria (Table 2).
In the case of Flds, as already indicated, they are absent from plants and animals, except for the “enslaved” Fld-like domains of complex enzymes (Sancho, 2006). Despite Fld presence in α-proteobacteria, they are not found in mitochondria, suggesting that the original endosymbiont already lacked this gene, or that it was lost very early after integration. The situation in chloroplasts is different. Fld is present in plastids from most major algal taxa (Table 2), including red and green algae, as well as those groups resulting from secondary endosymbiotic events such as dinoflagellates, haptophytes, diatoms, cryptophytes, and chlorarachniophytes (Fukuyama et al., 1990; Erdner et al., 1999; Inda and Peleato, 2002; Li et al., 2004; Jaeckisch et al., 2011; Whitney et al., 2011; Curtis et al., 2012; Read et al., 2013). In contrast, it has not been found in streptophytes, which consist of land plants (embryophytes) and charophytes, a subgroup of freshwater green algae that represent the sister lineage of land plants (www.ncbi.nlm.nih.gov/genomes/PLANTS/PlantList.html; Timme et al., 2012). This distribution suggests that the gene was lost somewhere in the transition between green algae and terrestrial plants.
Transgenic expression of flavodoxin in plants: and yet it works
In vitro experiments showed that cyanobacterial Flds were able to act as substrates for the plant descendants of many Fld-dependent prokaryotic enzymes (Scheller, 1996; Nogués et al., 2004; Tognetti et al., 2006). Anabaena Fld can even exchange electrons with the FNR from mammalian mitochondria (Zöllner et al., 2004), although this reductase is structurally unrelated to plant or cyanobacterial FNRs and has a different evolutionary origin. Taking into account the loose requirements for productive binding of Fld and Fd to diverse redox partners, these results were not entirely unexpected. Then, both biochemical predictions and experimental observations suggested that Fld could function in planta. Confirmation of this hypothesis was obtained by introducing a cyanobacterial Fld gene into model and crop species (Tognetti et al., 2006; Zurbriggen et al., 2010). The transgenic product was targeted to chloroplasts and assembled there with FMN to yield a functional Fld holoprotein (Tognetti et al., 2006).
Still, the possible effects of Fld presence in plant chloroplasts were not obvious. Plants have largely lost the substitutive strategies found in prokaryotes and algae, and respond to adverse environments by deploying complex defence systems involving many genes whose products combat the stress situation at various levels, such as increased repair and scavenging activities, metabolic reprogramming, and optimization of iron uptake (Zurbriggen et al., 2008, 2010; Foyer and Shigeoka, 2011; Kobayashi and Nishizawa, 2012; Munné-Bosch et al., 2013). The existence of these alternative strategies might simply indicate that replacement of Fd in stressed or iron-starved plants has no further adaptive value because other stress-sensitive proteins more critical to plant survival could have appeared along the evolutionary pathway that led to terrestrial plants.
Experimental evaluation of these possibilities revealed, however, that transgenic plants expressing a plastid-targeted Fld displayed increased tolerance, relative to their wild-type (WT) siblings, to multiple environmental adversities such as drought, high light intensities, heat, chilling, ultraviolet radiation, and poisoning with the contact herbicide paraquat (Tognetti et al., 2006; Coba de la Peña et al., 2010). Accumulation of reactive oxygen species (ROS) such as hydrogen or organic peroxides and the superoxide radical, which was prominent in stressed WT plants, was significantly mitigated in the Fld transformants (Tognetti et al., 2006). Complementation of Fd functions by Fld was demonstrated by introducing a plastid-directed Fld into tobacco plants in which Fd expression had been knocked down using RNA antisense or RNA silencing (Blanco et al., 2011). Fd deficiency caused growth arrest, leaf chlorosis, and photosynthetic impairment (Holtgrefe et al., 2003; Hanke and Hase, 2008; Voss et al., 2008; Blanco et al., 2011). Expression of Fld resulted in partial recovery of all these parameters, with nearly WT phenotypes obtained in lines accumulating less than 15% of normal Fd levels (Blanco et al., 2011). Photosynthesis and life were largely based on Fld in these plants.
Fld expression also prevented ROS-triggered localized cell death following inoculation of a non-host pathogen (Zurbriggen et al., 2009), and allowed growth and reproduction in iron-limited soils and media (Tognetti et al., 2007). Interestingly, iron-starved Fld-expressing lines accumulated WT (low) iron levels, and displayed a normal response to Fe deficit, indicating that the presence of Fld did not interfere with processes involved in iron status sensing, uptake, or mobilization (Tognetti et al., 2007). These plants simply lived and reproduced on lower iron quotas. Finally, transformation of either plant or rizhobia with a cyanobacterial Fld gene delayed legume nodule senescence (Redondo et al., 2009; Coba de la Peña et al., 2010), and protected nitrogen fixation activity of nodules exposed to salt or heavy metal toxicity (Coba de la Peña et al., 2010; Shvaleva et al., 2010).
For all types of stresses assayed, the effect of Fld was dose-dependent, saturated at a certain concentration threshold, and then became detrimental to plant fitness (Ceccoli et al., 2012). Also, chloroplast location of the expressed product was mandatory. Transgenic plants that accumulated high Fld amounts in the cytosol displayed WT levels of stress tolerance (Tognetti et al., 2006, 2007; Redondo et al., 2009). The main conclusion drawn from these studies was that Fld contributed to the welfare of stressed plants by restoring chloroplast redox homeostasis compromised by stress-dependent Fd decline. The presence of the flavoprotein prevented electron misrouting and ROS formation, and favoured delivery of reducing equivalents to productive metabolic, regulatory and dissipative pathways (Zurbriggen et al., 2008). A model summarizing these findings is depicted in Fig. 2. The importance of Fld activity to bypass acceptor side limitation at the reducing end of PSI (namely, Fd and NADP+ shortage) was confirmed by introducing a second Fld reduction system. As NADPH build-up at the expense of NADP+ is another unwanted consequence of stress episodes, overexpression of a soluble FNR seemed to be the logical choice to use the excess of NADPH as electron source for Fld reduction. Double-transgenic lines expressing cyanobacterial Fld and FNR had a higher NADP+/NADPH ratio, and exhibited increased tolerance to paraquat-mediated oxidative stress relative to Fld-only siblings (Fig. 2) (Giró et al., 2011).
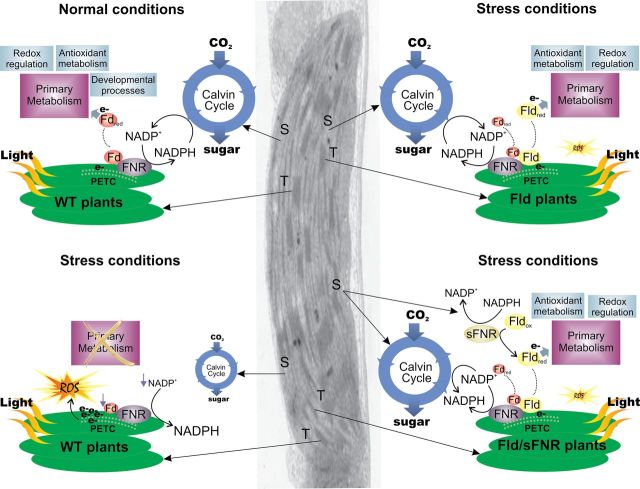
Fig. 2.
Proposed model for the protective mechanism of Fld in plastids of transgenic plants. A transmission electron micrograph of a chloroplast is used to illustrate the places (thylakoids or stroma) where the various reactions are expected to occur. The situation in WT plants under normal growth conditions is shown in the top left panel, with Fd acting as the regular shuttle between the PETC and electron-accepting processes of the chloroplast. Stress conditions in WT lines (bottom left) lead to Fd down-regulation, NADP+ exhaustion, and ROS build-up. Most central metabolic routes, including the Calvin cycle, are inhibited, whereas a few, such as respiration, are unaffected or even induced (not shown). Fld expression in stressed plants (top right) bypass Fd limitation, reactivating electron delivery to metabolic sinks. Simultaneous expression of Fld plus a soluble FNR (sFNR) in stressed plants (bottom right) further improves NADP(H) turnover and distribution of reducing equivalents to productive routes, resulting in increased tolerance to environmental hardships. T, thylakoids; S, stroma; ox, oxidized; red, reduced. Other abbreviations are given in the text.
The rise and fall of flavodoxin in photosynthetic organisms
Before flavodoxins, ferredoxins
The origin of life on Earth is placed at about 3,500 Mya in an anaerobic environment, and essential enzymatic mechanisms and biochemical pathways evolved in the absence of any selective pressure to avoid reactivity with oxygen or oxygen derivatives (Lane et al., 2013). Whole genome analyses of phylogenetically diverse microorganisms suggest that the earliest proteins incorporated metals, and that metal usage evolved over time in accordance with its availability, which was in turn dependent on the redox state of the environment (Dupont et al., 2006; Lane et al., 2013). Iron is, by far, the most abundant transition metal on Earth (Taylor, 1964), and its bioavailability in the primitive Archaean and early Proterozoic oceans is reflected by its wide use as a catalyst for oxido-reductive proteins and enzymes. Indeed, iron is identified as a cofactor in two thirds of the oxidoreductases present in the PDB and Swiss-Prot datasets (Lane et al., 2013). Fe-containing proteins can use the metal in a mineral form (as in superoxide dismutases), coordinated to imidazole nitrogens in porphyrins (haems), or complexed to sulfides in iron–sulfur clusters.
Several lines of evidence support the notion that Fds are among the oldest iron–sulfur proteins on Earth. First, Fe+2 and sulfide were thought to be plentiful in the anaerobic Archaean ocean, and unlike organic prosthetic groups, they can spontaneously form iron–sulfur centres and assemble into extant polypeptide structures. Indeed, analogous complexes can be created in vitro by incubating ferrous and sulfide salts with organic thiolates (Imlay, 2006). It has even been speculated that the earliest biologically relevant redox reactions occurred on the surface of iron–sulfur mineral deposits associated with hydrothermal vents (Wächtershäuser, 2007). Second, Fe–S groups exhibit great chemical versatility: they can accept and donate electrons in a range of oxido-reductive processes, act as Lewis acids during dehydration of carbonyl compounds in hydro-lyases, and mediate derivatization of aliphatic metabolites by radical-based mechanisms (Imlay, 2006). Third, phylogenomic analysis of protein architecture indicates that the Fd fold is very ancient. All Fds have a simple, conserved fold made up of ~60 amino acids that binds the Fe–S cluster and presumably evolved from an early gene duplication event of a 30-residue sequence, a “protoferredoxin” composed of a primeval subset of amino acids (Beinert, 1990; Darimont and Sterner, 1994; Eck and Dayhoff, 1966; Trifonov, 2000). Reconstruction of a phylogenomic tree of protein fold architecture using data from a domain census in almost 200 sequenced genomes of the three superkingdoms placed Fd as the fifth oldest domain on Earth (starting with the P-loop-containing NTP hydrolases), and second among redox-related domains, after the NAD(P)+-binding Rossmann fold (Caetano-Anollés et al., 2007).
These remarkable traits favoured dispersion of organisms containing iron–sulfur proteins throughout the anaerobic world, and placed these cofactors among the earliest catalysts. Other proteins containing identical [2Fe–2S] clusters, such as the Rieske proteins with their higher midpoint potentials, most probably evolved after Fd (Kim et al., 2013).
If Fe–S clusters are indeed evolutionary relics of earlier biochemistries and prebiotic chemistries, it could be that the most ancient among extant organisms contained more Fe–S proteins. This contention has been put to test by Major et al. (2004) and Sousa et al. (2013), using 120 and 1606 bacterial genomes, respectively. They observed that methanogens, acetogens (clostridia), and sulphate reducers, which are considered the life forms that more closely resemble ancient organisms, contained the highest numbers of Fe–S proteins. These organisms thrive in the deep biosphere, where, as on the early Earth, there are limited ways to make a living.
The preceding discussion was intended to explain why iron–sulfur proteins were widespread when, approximately 2,700 Mya, cyanobacteria evolved oxygenic photosynthesis, relieving these prokaryotes from the need of external electron donors. Oxygen concentrations remained low over the following two billion years or so, limited by both the scarcity of oceanic phosphorous to support ATP synthesis, and by oxygen removal through reaction with dissolved ferrous and sulfide ions (Bjerrum and Canfield, 2002). The sharp rise of the oxygen levels at the brink of the Precambrian (~800 Mya) led to one of the most catastrophic evolutionary stresses of biotic history.
As aerobes kept many of the catabolic and biosynthetic pathways present in their anaerobic ancestors, maintenance of most iron–sulfur protein families was ensured. At the same time, oxygen build-up negatively affected the function of these metalloproteins in a number of ways. First, spin-pairing rules dictate that molecular oxygen accepts electrons one at a time rather than in pairs, discouraging reaction with most organic biomolecules but facilitating oxidation of transition metals, which are good univalent electron donors. As a consequence, oxygen oxidized ferrous iron in the environment to its ferric form, which rapidly precipitated as ferric polyhydroxides or formed insoluble complexes with anionic salts. The upshot was that as oxygen accumulated, iron decreased its bioavailability and became a limiting nutrient in most aerobic habitats (Imlay, 2006).
Second, partial reduction of oxygen generates superoxide, H2O2, and other ROS, oxidants, which display still higher reactivity than oxygen. Even under optimal growth conditions, a fraction of the electrons moving through the photosynthetic or respiratory chains is adventitiously delivered to oxygen with concomitant ROS generation. This fraction increases significantly under adverse environmental conditions (Mittler et al., 2004).
Iron–sulfur centres are vulnerable to ROS attack to various extents, depending on solvent exposure and the polypeptide environment surrounding the cluster. Oxidation yields unstable intermediates that quickly decompose, resulting in protein inactivation and iron release. Elevated concentrations of free iron can wreak cellular havoc and lead to oxidative damage by engaging in Fenton-type reactions with hydrogen peroxide to generate the extremely toxic hydroxyl radical (Imlay, 2006). This process is generally self-propagating. In photosynthetic organisms, for instance, stress-dependent Fd down-regulation leads to over-reduction of the PETC owing to shortage of electron acceptors, and under such circumstances the electron surplus can be passed straight to oxygen resulting in runaway ROS generation (Holtgrefe et al., 2003; Nogués et al., 2004; Voss et al., 2008; Blanco et al., 2011).
Then, the type of iron–sulfur chemistry that modern aerobes inherited from their anaerobic ancestors does not suit well an oxygen-rich world. Air-thriving organisms tackled these problems at various levels by developing more sophisticated antioxidant and repair systems, and by reprogramming vast portions of metabolism from growth to defence (Foyer and Shigeoka, 2011; Munné-Bosch et al., 2013). As the oceans became progressively iron-deficient, several mechanisms were also developed by marine microorganisms to overcome this limitation, such as high surface-to-volume ratio to aid nutrient uptake (Chisholm, 1992), a more extensive machinery for metal capture and storage (Toulza et al., 2012), and a decrease of iron-rich PSI (12 iron atoms per complex) in favour of PSII (2–3 iron atoms per complex) (Strzepek and Harrison, 2004). The many adjustments made were expensive and bestowed only a limited capacity to tolerate this threat. It is doubtful that iron–sulfur clusters could have emerged as central catalysts had life originally evolved in an aerobic environment. Largely because of their reliance on these cofactors, aerobes remain vulnerable to iron restriction and oxidative stress.
As a consequence of these combined challenges, there was also intense selective pressure to replace oxidant-sensitive, iron-dependent proteins by oxidant-resistant, iron-free isofunctional counterparts (Palenik et al., 2006; Allen et al., 2008; Toulza et al., 2012). Known examples are the use of the copper protein plastocyanin instead of cytochrome c 6 (a hemoprotein), of cobalt-containing ribonucleotide reductase in place of the Fe-dependent isoenzyme, and of Cu/Zn-, Mn- and Ni-containing superoxide dismutases (De la Rosa et al., 2006; Palenik et al., 2007). Within this context, substitution of labile Fd acquired paramount importance.
Presumably a first step toward this direction was the development of more oxygen-tolerant Fd versions by improving protection of their Fe–S centres from oxidants. Some of the changes undergone by these proteins can be recognized by comparing extant aerobic and anaerobic isoforms. In typical anaerobic Fds, the bridging µ–sulfido atoms of the cluster are partially solvent-exposed. In contrast, the iron and sulfur atoms of the [2Fe–2S] centres of aerobic Fds are better shielded by the protein (Jagannathan and Golbeck, 2009). In spite of these improvements, oxygen tolerance was not complete and the question of iron shortage remained unsolved. Fld was the ultimate solution, as it is oxygen-insensitive and does not require iron.
The origin of flavodoxins
Expression of Fld to replace Fd is generally regarded as one of the most crucial factors determining the colonization of iron-poor waters by phytoplankton (Rocap et al., 2003), and evaluation of metagenomic data confirmed that algae and cyanobacteria lacking this flavoprotein are usually confined to iron-rich coastal/freshwater habitats (Toulza et al., 2012, see below). The importance of Fld in the dynamics of sea ecology is reflected by its use as a proxy for iron stress in the oceans (Erdner et al., 1999). As the role of Fld as a backup of Fd seems to be a response to aerobiosis, it is natural to assume that this flavoprotein evolved well after Fd. Surprisingly, this does not seem to be the case.
Phylogenomic evaluation of protein-fold architecture actually placed the Fld structure among the nine most ancestral and widely shared folds, appearing immediately after Fd (Caetano-Anollés et al., 2007). According to these data, the ancient folds represent architectures of fundamental importance encoded in a genetic core that can be tracked back to the universal ancestor of the three superkingdoms of life (Harris et al., 2003). They appeared in a relatively short time-frame, long before diversification of prokaryotes (Caetano-Anollés et al., 2007). Then, early Flds either played roles independent from Fd substitution or responded to environmental cues not related to aerobiosis. Most likely, they were recruited to replace iron- and oxygen-sensitive Fd at a later stage, after oxygen build-up. The widespread presence of Fld in anaerobes and the existence of Fld-specific metabolic routes (Freigang et al., 2002; Puan et al., 2005) provide circumstantial evidence to this tenet.
Flavodoxin entered the algal world and never came out; the phylogenetic patchwork
A simplified phylogenetic tree of photosynthetic eukaryotes is shown in Fig. 3. The initial endosymbiotic event that gave origin to all algal lineages is estimated to have occurred ~1,600 Mya (Hedges et al., 2004; Yoon et al., 2004). Glaucophytes were probably the first to diverge from the common ancestor, followed by the split between red and green algae, a few hundred million years after the primary endosymbiosis (Hedges et al., 2004; Yoon et al., 2004). Members of these latter lineages subsequently became the subjects of secondary and tertiary endosymbiosis (Fig. 3), spreading the algal heritage throughout all kinds of habitats. Viridiplantae separated ~1,000 Mya in two lineages: Chlorophyta and Streptophyta, which followed radically different evolutionary trajectories (Hedges et al., 2004). Chlorophytes, which comprise the majority of green algal species, radiated in marine and coastal environments, whereas streptophytes, which include land plants (Embryophyta) and a paraphyletic assembly of green algae (charophytes), evolved largely in freshwater. Ancestral charophytes colonized the dry land ~450 Mya, giving rise to land plants (Fig. 3), which have dominated the terrestrial environments since then, and some have even become secondarily adapted to freshwater or marine habitats (Waycott et al., 2002; Chambers et al., 2008). Monophyly of Embryophyta is supported by both morphological and molecular comparisons (Kenrick and Crane, 1997). The two most complex groups of the Charophyta, the coleochaetales and the charales (stoneworts), both of which are characterized by true multicellular organization and oogamous sexual reproduction, have been variously invoked as sisters to land plants (Bowman, 2013), as well as the structurally simpler Zygnematophyceae (Ruhfel et al., 2014). Within the embryophytes, liverworts are the most basal group, followed by mosses, and then hornworts, and vascular plants sharing a sister-group relationship (Wellman, 2014). Vascular plants appeared ~410 Mya and then diverged into several lineages, only two of which survive: the euphyllophytes (ferns and seed plants) and the lycophytes (Kenrick and Crane, 1997).
Fig. 3.
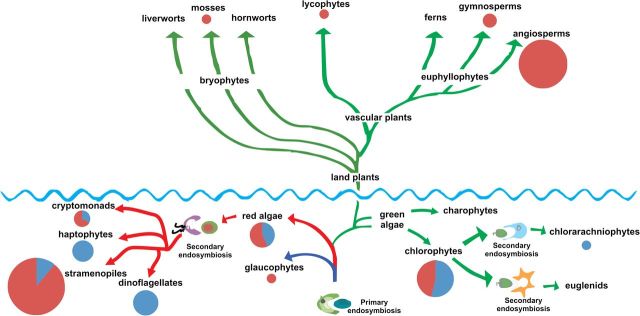
An evolutionary framework of Fld distribution among photosynthetic eukaryotes. A simplified phylogenetic tree was constructed using data from Bowman (2013) and Keeling (2013). Circle sizes represent the absolute number of species analysed in each branch, whereas colours indicate the fraction of species containing (blue) or not (purple) the fld gene.
Prevalence of the fld gene in each taxon of the photosynthetic eukaryotes is indicated in Fig. 3 by size and colour codes. The gene is found in all major algal taxa including green and red algae, as well as those groups derived from them through secondary and tertiary endosymbiosis. Notorious exceptions are glaucophytes and euglenids, although only one sequenced genome is available from the former group (Cyanophora paradoxa) and none from the latter. On the other hand, analysis of genome databases confirmed the absence of this flavoprotein in algal species from most taxa. Within Viridiplantae, all the organisms in which the presence of Fld was confirmed belong to the chlorophytes. No charophyte genome is yet available, and no Fld sequence could be retrieved from the expressed sequence tag collections of this group of algae (Timme et al., 2012). The absence of the fld gene in the genomes of land plants is confirmed in the more than 30 fully sequenced genomes (www.ncbi.nlm.nih.gov/genomes/PLANTS/PlantList.html). They include the lycophyte Selaginella moellendorffii, a “primitive” vascular plant, as well as the moss Physcomitrella patens, a bryophyte, phylogenetically closer to the first green algal lineage that successfully colonized terrestrial habitats.
Although the “Middle Age” of plant evolution (from charales and choleochaetales to gymnosperms) is largely empty of hard data, the absence of Fld sequences in that vast range of species suggest that the gene was lost earlier, most likely when streptophytes split from the chlorophytes (Fig. 3). More extensive genetic and genomic analyses of the “missing links” (charophytes, liverworts, hornworts, ferns) will be needed to elucidate this question.
Geographical distribution: environmental constraints
Although each environment has complex chemistry and physics, it is accepted that iron levels tend to be high in freshwater and coastal habitats, owing to suspended sediments and aerial inputs from land (Jickells et al., 2005). In contrast, iron deficiency is predicted in as much as 40% of the open ocean, notably in the Southern Ocean as well as in the equatorial and north Pacific (Moore et al., 2001). Iron utilization also posed a serious challenge to plants after land colonization, but of a different nature. Iron is the fifth most abundant element, and the problem of iron acquisition in soil is not of paucity but of availability. The main forms of iron in soils are ferric oxides, which are sparingly soluble at neutral pH and even less in alkaline media (Guerinot and Yi, 1994; Kobayashi and Nishizawa, 2012). It is worth noting that alkaline calcareous soils represent about one-third of the planet’s cultivable land (Guerinot and Yi, 1994).
The phylogenetic distribution described in the previous section indicates that retention of the fld gene along the path of evolution has been disparate, even among species of the same genera, such as in the chlorophytes Bathycoccus (Monier et al., 2012; Moreau et al., 2012; Vaulot et al., 2012) and Ostreococcus (Derelle et al., 2006; Palenik et al., 2007; http://genome.jgi-psf.org/OstRCC809_2).
As already indicated, Fld presence shows a clear bias towards iron-deficient habitats (Erdner et al., 1999). Conversely, Fld absence is a common feature in species isolated from iron-rich environments. In fact, all freshwater algal species with completely sequenced genomes lack the gene (Merchant et al., 2007; Blanc et al., 2010; Prochnik et al., 2010; Curtis et al., 2012; Price et al., 2012; Bogen et al., 2013). The same pattern holds true for cyanobacteria (Toulza et al., 2012).
The enigma of flavodoxin disappearance from the plant genome: a hypothesis
The preceding discussion suggests that the major adaptive advantage of Fld in marine habitats seems to be increased tolerance to iron starvation, a frequent stress in open oceans. Indeed, iron deficiency is regarded as a main selective pressure which determined the fate of many genes in phototrophs (Rocap et al., 2003; Palenik et al., 2006). It is therefore surprising that Fd was retained in all photosynthetic eukaryotes in spite of its lability to iron starvation and oxidants, whereas Fld disappeared from the plant genome. The causes underlying this outcome are unclear, and the life history of other Fe-containing proteins (and their replacements) has been entirely different. For instance, cytochrome c 6 was in the end displaced by plastocyanin (De la Rosa et al., 2006), and Cu/Zn-containing superoxide dismutases eventually became the dominant isoforms of this type of enzymes in eukaryotes (Miller, 2012).
On the one side, the reasons for Fd conservation along the Viridiplantae lineage are probably related to its catalytic efficiency. Although Fld might be the preferred or specific electron donor in a few heterotrophic pathways (Freigang et al., 2002; Puan et al., 2005; Gutekunst et al., 2014), Fd consistently displayed higher rates of electron transfer in photosynthesis-related reactions (Vigara et al., 1998; Medina et al., 1998; Meimberg and Mühlenhoff, 1999; Tognetti et al., 2006). Possible reasons for this kinetic advantage have been discussed previously. Photosynthetic electron transport is usually one order of magnitude faster than most heterotrophic oxido-reductive processes, and increases in catalytic efficiency have also been observed for FNR after recruitment into the PETC (Carrillo and Ceccarelli, 2003). Then, if Fd did confer the highest rates of electron distribution to photosynthetic acceptors under iron-replete conditions, it could still hold enough selective value to warrant retention in the genomes of phototrophs, including those which spend most of their lifetime in iron-deficient habitats. Indeed, available evidence indicates that photosynthetic organisms become non viable below a certain threshold of Fd content (Mazouni et al., 2003; Holtgrefe et al., 2003), even in the presence of Fld (Poncelet et al., 1998; Blanco et al., 2011). The results suggest that Fld migh help to alleviate the symptoms of iron limitation and other environmental hardships, but that some residual Fd activity (or isoform) is required to ensure survival and reproduction. In other words, Fld does not provide full complementation of Fd in photosynthetic organisms. Although no definite proof for this contention is yet available, the evidence collected from mutant microorganisms and knocked-downed plants strongly support the essentiality of Fd for phototrophs.
The reasons behind Fld loss from the Viridiplantae are more obscure. Functional arguments cannot be invoked as the experimental evidence indicates that Fld expression in plants still confers selective advantages (Tognetti et al., 2006). Disappearance of this valuable asset from the plant genome might be related to ecological adaptations to iron bioavailability and the successive stages of land colonization (Fig. 4). It is accepted that terrestrial plants evolved from coastal/freshwater macroalgae (charales, choleochaetales). We propose that these ancestors, thriving in an environment in which iron was both abundant and readily accessible, had no need to induce Fld expression to replace Fd (Fig. 4). Under such conditions, selection pressures for Fld retention as an adaptive trait might have been relaxed. Indeed, the geographical distribution of marine phototrophs shows that iron-rich coastal and freshwater regions are preferentially populated by Fld-lacking algae and cyanobacteria (Fig. 3), increasing the probability that plants evolved from algal precursors that had already lost the Fld gene. According to this hypothesis, the absence of Fld form the Embryophyta would be the result of a founder effect.
Fig. 4.
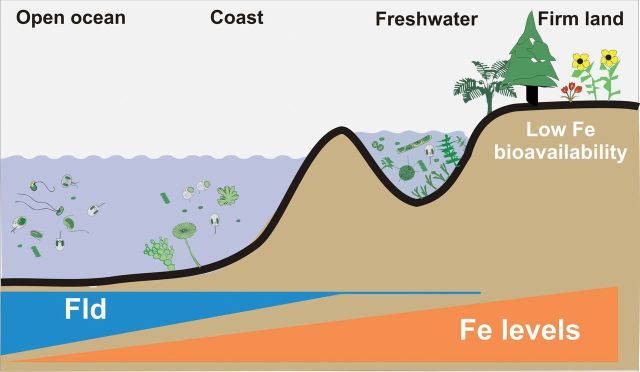
The flavodoxin gene was lost along the evolution of Viridiplantae during the colonization of terrestrial habitats by streptophyte algae. The hypothesis assumes that plants originated from freshwater streptophytes which already lacked Fld. Cyanobacteria and algae thriving in iron-deficient oceanic environments usually contained the fld gene, whereas coastal and freshwater counterparts evolved in a habitat in which iron was plentiful and bioavailable, and the role of Fld as a backup of Fd was presumably not required. Typical cyanobacteria, algae, and plants are depicted schematically. The negative correlation between iron levels and Fld presence is shown in the bottom. They are intended to represent tendencies and do not depict actual iron/Fld contents.
After colonization of the firm land, plants faced a novel type of challenge: how to get iron from an environment in which the metal was plentiful but not readily available. They adapted to the new situation by deploying a multigenic response in which many genes were recruited to operate at various levels, including rhizosphere acidification, reduction of Fe+3 to Fe+2, metal chelation, and metal transport, all of them contributing to optimization of iron uptake (Kobayashi and Nishizawa, 2012). This strategy was successful enough to allow spreading of plant lineages throughout most of the planet. It is somehow surprising, however, that they were not able to recover the fld gene by, for instance, HGT from contemporary algae or bacteria. It should be borne in mind, however, that the evolution of land plants has been largely associated with expansion and diversification of existing gene families as the result of large-scale gene or whole-genome duplication events (Richardt et al., 2007; Rensing et al., 2008), rather than incorporation of novel genes and functions (Bock, 2010). In fact, examples of HGT to multicellular eukaryotes are rare (Rumpho et al., 2008; Bock, 2010), and this limitation might have prevented recovery of an adaptive trait that provides ample stress tolerance when present in plants (Tognetti et al., 2006, 2007). Although this conjecture is consistent with current observations, experimental validation is lacking and further research will be required to properly address this issue.
Note added in proof
In support of our hypothesis of an early disappearance of the flavodoxin gene in the Streptophyta, the first sequenced genome of a charophyte alga has been just published (Hori et al., 2014), and it does not contain flavodoxin.
Acknowledgements
AFL and NC are staff members from the National Research Council (CONICET, Argentina), and JJPK is a doctoral fellow of the same Institution. Authors are Faculty members of the Molecular Biology (JJPK, NC) and Biophysics (AFL) Units, Biochemistry School, UNR (Facultad de Ciencias Bioquímicas y Farmacéuticas, Universidad Nacional de Rosario, Argentina). Part of the research reported in this review was supported by grant PICT-2011–2621 from ANPCYT, Argentina.
Glossary
Abbreviations:
- Fd
ferredoxin
- Fld
flavodoxin
- FMN
flavin mononucleotide
- FNR
ferredoxin–NADP+ reductase
- HGT
horizontal gene transfer
- PETC
photosynthetic electron transport chain
- ROS
reactive oxygen species
- WT
wild-type.
References
- Allen AE, LaRoche J, Maheswari U, Lommer M, Schauer N, López PJ, Finazzi G, Fernie AR, Bowler C. 2008. Whole-cell response of the pennate diatom Phaeodactylum tricornutum to iron starvation. Proceedings of the National Academy of Sciences, USA 105, 10438–10443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beinert H. 1990. Recent developments in the field of iron–sulfur proteins. The FASEB Journal 4, 2483–2491. [DOI] [PubMed] [Google Scholar]
- Birch OM, Hewitson KS, Fuhrmann M, Burgdorf K, Baldwin JE, Roach PL, Shaw NM. 2000. MioC is an FMN-binding protein that is essential for Escherichia coli biotin synthase activity in vitro . Journal of Biological Chemistry 275, 32277–32280. [DOI] [PubMed] [Google Scholar]
- Bjerrum CJ, Canfield DE. 2002. Ocean productivity before about 1.9 Gyr ago limited by phosphorus adsorption onto iron oxides. Nature 417, 159–162. [DOI] [PubMed] [Google Scholar]
- Blanc G, Duncan G, Agarkova I, Borodovsky M, Gurnon J, Kuo A, Lindquist E, Lucas S, Pangilinan J, Polle J. 2010. The Chlorella variabilis NC64A genome reveals adaptation to photosymbiosis, coevolution with viruses, and cryptic sex. The Plant Cell 22, 2943–2955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanco NE, Ceccoli RD, Segretin ME, Poli HO, Voss I, Melzer M, Bravo-Almonacid FF, Scheibe R, Hajirezaei M-R, Carrillo N. 2011. Cyanobacterial flavodoxin complements ferredoxin deficiency in knocked‐down transgenic tobacco plants. The Plant Journal 65, 922–935. [DOI] [PubMed] [Google Scholar]
- Blaschkowski HP, Knappe J, Ludwig-Festl M, Neuer G. 1982. Routes of flavodoxin and ferredoxin reduction in Escherichia coli . European Journal of Biochemistry 123, 563–569. [PubMed] [Google Scholar]
- Bock R. 2010. The give-and-take of DNA: horizontal gene transfer in plants. Trends in Plant Science 15, 11–22. [DOI] [PubMed] [Google Scholar]
- Bogen C, Al-Dilaimi A, Albersmeier A, Wichmann J, Grundmann M, Rupp O, Lauersen KJ, Blifernez-Klassen O, Kalinowski J, Goesmann A. 2013. Reconstruction of the lipid metabolism for the microalga Monoraphidium neglectum from its genome sequence reveals characteristics suitable for biofuel production. BMC Genomics 14, 926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Böhme H. 1978. Quantitative determination of ferredoxin, ferredoxin-NADP+ reductase and plastocyanin in spinach chloroplasts. European Journal of Biochemistry 83, 137–141. [DOI] [PubMed] [Google Scholar]
- Bott AW. 1999. Redox properties of electron transfer metalloproteins. Current Separations 18, 47–54. [Google Scholar]
- Bovy A, de Vrieze G, Lugones L, van Horssen P, van den Berg C, Borrias M, Weisbeek P. 1993a. Iron-dependent stability of the ferredoxin I transcripts from the cyanobacterial strains Synechococcus species PCC 7942 and Anabaena species PCC 7937. Molecular Microbiology 7, 429–439. [DOI] [PubMed] [Google Scholar]
- Bovy A, de Kruif J, de Vrieze G, Borrias M, Weisbeek P. 1993b. Iron-dependent protection of the Synechococcus ferredoxin I transcript against nucleolytic degradation requires cis-regulatory sequences in the 5’ part of the messenger RNA. Plant Molecular Biology 22, 1047–1065. [DOI] [PubMed] [Google Scholar]
- Bowman JL. 2013. Walkabout on the long branches of plant evolution. Current Opinion in Plant Biology 16, 70–77. [DOI] [PubMed] [Google Scholar]
- Caetano-Anollés G, Kim HS, Mittenthal JE. 2007. The origin of modern metabolic networks inferred from phylogenomic analysis of protein architecture. Proceedings of the National Academy of Sciences, USA 104, 9358–9363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrillo N, Ceccarelli EA. 2003. Open questions in ferredoxin-NADP+ reductase catalytic mechanism. European Journal of Biochemistry 270, 1900–1915. [DOI] [PubMed] [Google Scholar]
- Ceccarelli EA, Arakaki AK, Cortez N, Carrillo N. 2004. Functional plasticity and catalytic efficiency in plant and bacterial ferredoxin-NADP(H) reductases. Biochimica et Biophysica Acta 1698, 155–165. [DOI] [PubMed] [Google Scholar]
- Ceccoli RD, Blanco NE, Medina M, Carrillo N. 2011. Stress response of transgenic tobacco plants expressing a cyanobacterial ferredoxin in chloroplasts. Plant Molecular Biology 76, 535–544. [DOI] [PubMed] [Google Scholar]
- Ceccoli RD, Blanco NE, Segretin ME, Melzer M, Hanke GT, Scheibe R, Hajirezaei M-R, Bravo-Almonacid FF, Carrillo N. 2012. Flavodoxin displays dose-dependent effects on photosynthesis and stress tolerance when expressed in transgenic tobacco plants. Planta 236, 1447–1458. [DOI] [PubMed] [Google Scholar]
- Coba de la Peña T, Redondo FJ, Manrique E, Lucas MM, Pueyo JJ. 2010. Nitrogen fixation persists under conditions of salt stress in transgenic Medicago truncatula plants expressing a cyanobacterial flavodoxin. Plant Biotechnology Journal 8, 954–965. [DOI] [PubMed] [Google Scholar]
- Compaoré J, Stal LJ. 2010. Oxygen and the light–dark cycle of nitrogenase activity in two unicellular cyanobacteria. Environmental Microbiology 12, 54–62. [DOI] [PubMed] [Google Scholar]
- Curtis BA, Tanifuji G, Burki F, Gruber A, Irimia M, Maruyama S, Arias MC, Ball SG, Gile GH, Hirakawa Y. 2012. Algal genomes reveal evolutionary mosaicism and the fate of nucleomorphs. Nature 492, 59–65. [DOI] [PubMed] [Google Scholar]
- Chambers P, Lacoul P, Murphy K, Thomaz S. 2008. Global diversity of aquatic macrophytes in freshwater. Hydrobiologia 595, 9–26. [Google Scholar]
- Chappell PD, Webb EA. 2010. A molecular assessment of the iron stress response in the two phylogenetic clades of Trichodesmium . Environmental Microbiology 12, 13–27. [DOI] [PubMed] [Google Scholar]
- Cheng Y-Q, Liu Z-M, Xu J, Zhou T, Wang M, Chen Y-T, Li H-F, Fan Z-F. 2008. HC-Pro protein of sugar cane mosaic virus interacts specifically with maize ferredoxin-5 in vitro and in planta . Journal of General Virology 89, 2046–2054. [DOI] [PubMed] [Google Scholar]
- Chisholm SW. 1992. Phytoplankton size. In: Falkowski PG, Woodhead AD, Vivirito K, eds. Primary Productivity and Biogeochemical Cycles in the Sea. New York: Springer, 213–237. [Google Scholar]
- Darimont B, Sterner R. 1994. Sequence, assembly and evolution of a primordial ferredoxin from Thermotoga maritima . EMBO Journal 13, 1772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De la Rosa MA, Molina-Heredia FP, Hervás M, Navarro JA. 2006. Convergent evolution of cytochrome c 6 and plastocyanin. In: Golbeck JH, ed. Photosystem I: The Light-Driven Plastocyanin:Ferredoxin Oxidoreductase. The Netherlands: Springer, 683–696. [Google Scholar]
- Derelle E, Ferraz C, Rombauts S, Rouzé P, Worden AZ, Robbens S, Partensky F, Degroeve S, Echeynié S, Cooke R. 2006. Genome analysis of the smallest free-living eukaryote Ostreococcus tauri unveils many unique features. Proceedings of the National Academy of Sciences, USA 103, 11647–11652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickey LF, Gallo-Meagher M, Thompson W. 1992. Light regulatory sequences are located within the 5’ portion of the Fed-1 message sequence. EMBO Journal 11, 2311–2317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dupont CL, Yang S, Palenik B, Bourne PE. 2006. Modern proteomes contain putative imprints of ancient shifts in trace metal geochemistry. Proceedings of the National Academy of Sciences, USA 103, 17822–17827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eck RV, Dayhoff MO. 1966. Evolution of the structure of ferredoxin based on living relics of primitive amino acid sequences. Science 152, 363–366. [DOI] [PubMed] [Google Scholar]
- Elliott RC, Dickey LF, White MJ, Thompson WF. 1989. Cis-acting elements for light regulation of pea ferredoxin I gene expression are located within transcribed sequences. The Plant Cell 1, 691–698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erdner DL, Anderson DM. 1999. Ferredoxin and flavodoxin as biochemical indicators of iron limitation during open-ocean iron enrichment. Limnology and oceanography 44, 1609–1615. [Google Scholar]
- Erdner DL, Price NM, Doucette GJ, Peleato ML, Anderson DM. 1999. Characterization of ferredoxin and flavodoxin as markers of iron limitation in marine phytoplankton. Marine Ecology. Progress series 184, 43–53. [Google Scholar]
- Foyer CH, Shigeoka S. 2011. Understanding oxidative stress and antioxidant functions to enhance photosynthesis. Plant Physiology 155, 93–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freigang J, Diederichs K, Schafer KP, Welte W, Paul R. 2002. Crystal structure of oxidized flavodoxin, an essential protein in Helicobacter pylori . Protein Science 11, 253–261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukuyama K, Wakabayashi S, Matsubara H, Rogers LJ. 1990. Tertiary structure of oxidized flavodoxin from an eukaryotic red alga Chondrus crispus at 2.35-Å resolution. Localization of charged residues and implication for interaction with electron transfer partners. Journal of Biological Chemistry 265, 15804–15812. [PubMed] [Google Scholar]
- Gaudu P, Weiss B. 2000. Flavodoxin mutants of Escherichia coli K-12. Journal of Bacteriology 182, 1788–1793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghassemian M, Straus NA. 1996. Fur regulates the expression of iron-stress genes in the cyanobacterium Synechococcus sp. strain PCC 7942. Microbiology 142, 1469–1476. [DOI] [PubMed] [Google Scholar]
- Giró M, Carrillo N, Krapp AR. 2006. Glucose 6-phosphate dehydrogenase and ferredoxin-NADP(H) reductase contribute to damage repair during the soxRS response of Escherichia coli . Microbiology 152, 1119–1128. [DOI] [PubMed] [Google Scholar]
- Giró M, Ceccoli RD, Poli HO, Carrillo N, Lodeyro AF. 2011. An in vivo system involving co-expression of cyanobacterial flavodoxin and ferredoxin–NADP+ reductase confers increased tolerance to oxidative stress in plants. FEBS Open Bio 1, 7–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gómez-Moreno C, Medina M, Hurley J, Cusanovich M, Markley J, Cheng H, Xia B, Chae Y, Tollin G. 1994. Protein engineering for the elucidation of the mechanism of electron transfer in redox proteins. Biochemical Society Transactions 22, 796–800. [DOI] [PubMed] [Google Scholar]
- González A, Angarica VE, Sancho J, Fillat MF. 2014. The FurA regulon in Anabaena sp. PCC 7120: in silico prediction and experimental validation of novel target genes. Nucleic Acids Research 42, 4833–4846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goñi G, Herguedas B, Hervás M, Peregrina JR, De la Rosa MA, Gómez-Moreno C, Navarro JA, Hermoso JA, Martínez-Júlvez M, Medina M. 2009. Flavodoxin: a compromise between efficiency and versatility in the electron transfer from photosystem I to ferredoxin-NADP+ reductase. Biochimica et Biophysica Acta 1787, 144–154. [DOI] [PubMed] [Google Scholar]
- Gou P, Hanke GT, Kimata-Ariga Y, Standley DM, Kubo A, Taniguchi I, Nakamura H, Hase T. 2006. Higher order structure contributes to specific differences in redox potential and electron transfer efficiency of root and leaf ferredoxins. Biochemistry 45, 14389–14396. [DOI] [PubMed] [Google Scholar]
- Guerinot ML, Yi Y. 1994. Iron: nutritious, noxious, and not readily available. Plant Physiology 104, 815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutekunst K, Chen X, Schreiber K, Kaspar U, Makam S, Appel J. 2014. The bidirectional NiFe-hydrogenase in Synechocystis sp. PCC 6803 is reduced by flavodoxin and ferredoxin and is essential under mixotrophic, nitrate-limiting conditions. Journal of Biological Chemistry 289, 1930–1937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagemann M, Jeanjean R, Fulda S, Havaux M, Joset F, Erdmann N. 1999. Flavodoxin accumulation contributes to enhanced cyclic electron flow around photosystem I in salt‐stressed cells of Synechocystis sp. strain PCC 6803. Physiologia Plantarum 105, 670–678. [Google Scholar]
- Hanke G, Mulo P. 2013. Plant type ferredoxins and ferredoxin-dependent metabolism. Plant, Cell and Environment 36, 1071–1084. [DOI] [PubMed] [Google Scholar]
- Hanke GT, Hase T. 2008. Variable photosynthetic roles of two leaf‐type ferredoxins in Arabidopsis, as revealed by RNA interference. Photochemistry and Photobiology 84, 1302–1309. [DOI] [PubMed] [Google Scholar]
- Hanke GT, Kimata-Ariga Y, Taniguchi I, Hase T. 2004. A post genomic characterization of Arabidopsis ferredoxins. Plant Physiology 134, 255–264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris JK, Kelley ST, Spiegelman GB, Pace NR. 2003. The genetic core of the universal ancestor. Genome Research 13, 407–412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hase T, Schürmann P, Knaff DB. 2006. The interaction of ferredoxin with ferredoxin-dependent enzymes. In: Golbeck JH, ed. Photosystem I: The Light-Driven Plastocyanin:Ferredoxin Oxidoreductase. The Netherlands: Springer, 477–498. [Google Scholar]
- Havaux M, Guedeney G, Hagemann M, Yeremenko N, Matthijs HC, Jeanjean R. 2005. The chlorophyll-binding protein IsiA is inducible by high light and protects the cyanobacterium Synechocystis PCC6803 from photooxidative stress. FEBS Letters 579, 2289–2293. [DOI] [PubMed] [Google Scholar]
- Hedges SB, Blair JE, Venturi ML, Shoe JL. 2004. A molecular timescale of eukaryote evolution and the rise of complex multicellular life. BMC Evolutionary Biology 4, 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holtgrefe S, Bader KP, Horton P, Scheibe R, von Schaewen A, Backhausen JE. 2003. Decreased content of leaf ferredoxin changes electron distribution and limits photosynthesis in transgenic potato plants. Plant Physiology 133, 1768–1778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hori K, Maruyama F, Fujisawa T, et al. 2014. Klebsormidium flaccidum genome reveals primary factors for plant terrestrial adaptation. Nature Communications, 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imlay JA. 2006. Iron–sulphur clusters and the problem with oxygen. Molecular Microbiology 59, 1073–1082. [DOI] [PubMed] [Google Scholar]
- Inda L, Peleato ML. 2002. Immunoquantification of flavodoxin and ferredoxin from Scenedesmus vacuolatus (Chlorophyta) as iron-stress molecular markers. European Journal of Phycology 37, 579–586. [Google Scholar]
- Jaeckisch N, Yang I, Wohlrab S, Glöckner G, Kroymann J, Vogel H, Cembella A, John U. 2011. Comparative genomic and transcriptomic characterization of the toxigenic marine dinoflagellate Alexandrium ostenfeldii . PLoS One 6, e28012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jagannathan B, Golbeck JH. 2009. Understanding of the binding interface between PsaC and the PsaA/PsaB heterodimer in photosystem I. Biochemistry 48, 5405–5416. [DOI] [PubMed] [Google Scholar]
- Jickells T, An Z, Andersen KK, Baker A, Bergametti G, Brooks N, Cao J, Boyd P, Duce R, Hunter K. 2005. Global iron connections between desert dust, ocean biogeochemistry, and climate. Science 308, 67–71. [DOI] [PubMed] [Google Scholar]
- Keeling PJ. 2013. The number, speed, and impact of plastid endosymbioses in eukaryotic evolution. Annual Review of Plant Biology 64, 583–607. [DOI] [PubMed] [Google Scholar]
- Kenrick P, Crane PR. 1997. Origin and Early Diversification of Land Plants : Smithsonian Institution Press. [Google Scholar]
- Kim JD, Senn S, Harel A, Jelen BI, Falkowski PG. 2013. Discovering the electronic circuit diagram of life: structural relationships among transition metal binding sites in oxidoreductases. Philosophical Transactions of the Royal Society B: Biological Sciences 368, 20120257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi T, Nishizawa NK. 2012. Iron uptake, translocation, and regulation in higher plants. Annual Review of Plant Biology 63, 131–152. [DOI] [PubMed] [Google Scholar]
- Kojima K, Suzuki-Maenaka T, Kikuchi T, Nakamoto H. 2006. Roles of the cyanobacterial isiABC operon in protection from oxidative and heat stresses. Physiologia Plantarum 128, 507–519. [Google Scholar]
- Kurisu G, Kusunoki M, Katoh E, Yamazaki T, Teshima K, Onda Y, Kimata-Ariga Y, Hase T. 2001. Structure of the electron transfer complex between ferredoxin and ferredoxin–NADP+ reductase. Nature Structural and Molecular Biology 8, 117–121. [DOI] [PubMed] [Google Scholar]
- Kutzki C, Masepohl B, Böhme H. 1998. The isiB gene encoding flavodoxin is not essential for photoautotrophic iron limited growth of the cyanobacterium Synechocystis sp. strain PCC 6803. FEMS Microbiology Letters 160, 231–235. [DOI] [PubMed] [Google Scholar]
- La Roche J, Boyd PW, McKay RML, Geider RJ. 1996. Flavodoxin as an in situ marker for iron stress in phytoplankton. Nature 382, 802–805. [Google Scholar]
- Lane N, Martin WF, Raven JA, Allen JF. 2013. Energy, genes and evolution: introduction to an evolutionary synthesis. Philosophical Transactions of the Royal Society B: Biological Sciences 368, 20120253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Latifi A, Ruiz M, Zhang CC. 2009. Oxidative stress in cyanobacteria. FEMS Microbiology Reviews 33, 258–278. [DOI] [PubMed] [Google Scholar]
- Lemaire SD, Stein M, Issakidis-Bourguet E, Keryer E, Benoit V, Pineau B, Gérard-Hirne C, Miginiac-Maslow M, Jacquot J-P. 1999. The complex regulation of ferredoxin/thioredoxin-related genes by light and the circadian clock. Planta 209, 221–229. [DOI] [PubMed] [Google Scholar]
- Li X, Yakunin AF, McKay RML. 2004. Fe-responsive accumulation of redox proteins ferredoxin and flavodoxin in a marine cryptomonad. European Journal of Phycology 39, 73–82. [Google Scholar]
- Lin S, Sandh G, Zhang H, Cheng J, Perkins K, Carpenter EJ, Bergman B. 2009. Two flavodoxin genes in Trichodesmium (Oscillatoriales, Cyanophyceae): Remarkable sequence divergence and possible functional diversification. Journal of Experimental Marine Biology and Ecology 371, 93–101. [Google Scholar]
- Liu J, Wang P, Liu B, Feng D, Zhang J, Su J, Zhang Y, Wang JF, Wang HB. 2013. A deficiency in chloroplastic ferredoxin 2 facilitates effective photosynthetic capacity during long-term high light acclimation in Arabidopsis thaliana . The Plant Journal 76, 861–874. [DOI] [PubMed] [Google Scholar]
- Lommer M, Specht M, Roy A-S, Kraemer L, Andreson R, Gutowska MA, Wolf J, Bergner SV, Schilhabel MB, Klostermeier UC. 2012. Genome and low-iron response of an oceanic diatom adapted to chronic iron limitation. Genome Biology 13, R66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- López-Gomollón S, Sevilla E, Bes MT, Peleato ML, Fillat MF. 2009. New insights into the role of Fur proteins: FurB (All2473) from Anabaena protects DNA and increases cell survival under oxidative stress. Biochemical Journal 418, 201–207. [DOI] [PubMed] [Google Scholar]
- López-Llano J, Maldonado S, Jain S, Lostao A, Godoy-Ruiz R, Sánchez-Ruiz JM, Cortijo M, Fernández-Recio J, Sancho J. 2004. The long and short flavodoxins: II. The role of the differentiating loop in apoflavodoxin stability and folding mechanism. Journal of Biological Chemistry 279, 47184–47191. [DOI] [PubMed] [Google Scholar]
- Major TA, Burd H, Whitman WB. 2004. Abundance of 4Fe–4S motifs in the genomes of methanogens and other prokaryotes. FEMS Microbiology Letters 239, 117–123. [DOI] [PubMed] [Google Scholar]
- Markowitz VM, Chen IMA, Palaniappan K, et al. 2012. IMG: the integrated microbial genomes database and comparative analysis system. Nucleic acids research 40, 115–122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masepohl B, Scholisch K, Gorlitz K, Kutzki C, Böhme H. 1997. The heterocyst-specific fdxH gene product of the cyanobacterium Anabaena sp. PCC 7120 is important but not essential for nitrogen fixation. Molecular and General Genetics 253, 770–776. [DOI] [PubMed] [Google Scholar]
- Matsumura T, Sakakibara H, Nakano R, Kimata Y, Sugiyama T, Hase T. 1997. A nitrate-inducible ferredoxin in maize roots (genomic organization and differential expression of two nonphotosynthetic ferredoxin isoproteins). Plant Physiology 114, 653–660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazouni K, Domain F, Chauvat F, Cassier-Chauvat C. 2003. Expression and regulation of the crucial plant‐like ferredoxin of cyanobacteria. Molecular Microbiology 49, 1019–1029. [DOI] [PubMed] [Google Scholar]
- Medina M, Martínez-Júlvez M, Hurley JK, Tollin G, Gómez-Moreno C. 1998. Involvement of glutamic acid 301 in the catalytic mechanism of ferredoxin-NADP+ reductase from Anabaena PCC 7119. Biochemistry 37, 2715–2728. [DOI] [PubMed] [Google Scholar]
- Meimberg K, Mühlenhoff U. 1999. Laser-flash absorption spectroscopy study of the competition between ferredoxin and flavodoxin photoreduction by Photosystem I in Synechococcus sp. PCC 7002: Evidence for a strong preference for ferredoxin. Photosynthesis Research 61, 253–267. [Google Scholar]
- Merchant SS, Prochnik SE, Vallon O, Harris EH, Karpowicz SJ, Witman GB, Terry A, Salamov A, Fritz-Laylin LK, Maréchal-Drouard L. 2007. The Chlamydomonas genome reveals the evolution of key animal and plant functions. Science 318, 245–250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller AF. 2012. Superoxide dismutases: ancient enzymes and new insights. FEBS Letters 586, 585–595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mittler R, Vanderauwera S, Gollery M, van Breusegem F. 2004. Reactive oxygen gene network of plants. Trends in Plant Science 9, 490–498. [DOI] [PubMed] [Google Scholar]
- Monier A, Welsh RM, Gentemann C, Weinstock G, Sodergren E, Armbrust E, Eisen JA, Worden AZ. 2012. Phosphate transporters in marine phytoplankton and their viruses: cross‐domain commonalities in viral‐host gene exchanges. Environmental Microbiology 14, 162–176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore JK, Doney SC, Glover DM, Fung IY. 2001. Iron cycling and nutrient-limitation patterns in surface waters of the World Ocean. Deep Sea Research Part II: Topical Studies in Oceanography 49, 463–507. [Google Scholar]
- Moreau H, Verhelst B, Couloux A, Derelle E, Rombauts S, Grimsley N, van Bel M, Poulain J, Katinka M, Hohmann-Marriott MF. 2012. Gene functionalities and genome structure in Bathycoccus prasinos reflect cellular specializations at the base of the green lineage. Genome Biology 13, R74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moyano AJ, Tobares RA, Rizzi YS, Krapp AR, Mondotte JA, Bocco JL, Saleh M-C, Carrillo N, Smania AM. 2014. A long-chain flavodoxin protects Pseudomonas aeruginosa from oxidative stress and host bacterial clearance. PLoS Genetics 10, e1004163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munné-Bosch S, Queval G, Foyer CH. 2013. The impact of global change factors on redox signaling underpinning stress tolerance. Plant Physiology 161, 5–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nogués I, Tejero J, Hurley JK, et al. 2004. Role of the C-terminal tyrosine of ferredoxin-nicotinamide adenine dinucleotide phosphate reductase in the electron transfer processes with its protein partners ferredoxin and flavodoxin. Biochemistry 43, 6127–6137. [DOI] [PubMed] [Google Scholar]
- Nogués I, Pérez-Dorado I, Frago S, Bittel C, Mayhew SG, Gómez-Moreno C, Hermoso JA, Medina M, Cortez N, Carrillo N. 2005. The ferredoxin–NADP(H) reductase from Rhodobacter capsulatus: molecular structure and catalytic mechanism. Biochemistry 44, 11730–11740. [DOI] [PubMed] [Google Scholar]
- Onda Y, Matsumura T, Kimata-Ariga Y, Sakakibara H, Sugiyama T, Hase T. 2000. Differential interaction of maize root ferredoxin:NADP+ oxidoreductase with photosynthetic and non-photosynthetic ferredoxin isoproteins. Plant Physiology 123, 1037–1046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palenik B, Ren Q, Dupont CL, Myers GS, Heidelberg JF, Badger JH, Madupu R, Nelson WC, Brinkac LM, Dodson RJ. 2006. Genome sequence of Synechococcus CC9311: insights into adaptation to a coastal environment. Proceedings of the National Academy of Sciences, USA 103, 13555–13559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palenik B, Grimwood J, Aerts A, Rouzé P, Salamov A, Putnam N, Dupont C, Jorgensen R, Derelle E, Rombauts S. 2007. The tiny eukaryote Ostreococcus provides genomic insights into the paradox of plankton speciation. Proceedings of the National Academy of Sciences, USA 104, 7705–7710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patterson K, Cakmak T, Cooper A, Lager I, Rasmusson AG, Escobar MA. 2010. Distinct signalling pathways and transcriptome response signatures differentiate ammonium‐and nitrate‐supplied plants. Plant, Cell and Environment 33, 1486–1501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peden EA, Boehm M, Mulder DW, Davis R, Old WM, King PW, Ghirardi ML, Dubini A. 2013. Identification of global ferredoxin interaction networks in Chlamydomonas reinhardtii . Journal of Biological Chemistry 288, 35192–35209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peleato ML, Ayora S, Inda L, Gómez-Moreno C. 1994. Isolation and characterization of two different flavodoxins from the eukaryote Chlorella fusca . Biochemical Journal 302, 807–811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pérez-Dorado I, Bortolotti A, Cortez N, Hermoso JA. 2013. Structural and phylogenetic analysis of Rhodobacter capsulatus NifF: uncovering general features of nitrogen-fixation (nif)-flavodoxins. International Journal of Molecular Sciences 14, 1152–1163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poncelet M, Cassier-Chauvat C, Leschelle X, Bottin H, Chauvat F. 1998. Targeted deletion and mutational analysis of the essential (2Fe–2S) plant‐like ferredoxin in Synechocystis PCC6803 by plasmid shuffling. Molecular Microbiology 28, 813–821. [DOI] [PubMed] [Google Scholar]
- Price DC, Chan CX, Yoon HS, Yang EC, Qiu H, Weber AP, Schwacke R, Gross J, Blouin NA, Lane C. 2012. Cyanophora paradoxa genome elucidates origin of photosynthesis in algae and plants. Science 335, 843–847. [DOI] [PubMed] [Google Scholar]
- Prochnik SE, Umen J, Nedelcu AM, Hallmann A, Miller SM, Nishii I, Ferris P, Kuo A, Mitros T, Fritz-Laylin LK. 2010. Genomic analysis of organismal complexity in the multicellular green alga Volvox carteri . Science 329, 223–226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puan KJ, Wang H, Dairi T, Kuzuyama T, Morita CT. 2005. fldA is an essential gene required in the 2-C-methyl-D-erythritol 4-phosphate pathway for isoprenoid biosynthesis. FEBS Letters 579, 3802–3806. [DOI] [PubMed] [Google Scholar]
- Rai S, Agrawal C, Shrivastava AK, Singh PK, Rai L. 2014. Comparative proteomics unveils cross species variations in Anabaena under salt stress. Journal of Proteomics 98, 254–270. [DOI] [PubMed] [Google Scholar]
- Read BA, Kegel J, Klute MJ, Kuo A, Lefebvre SC, Maumus F, Mayer C, Miller J, Monier A, Salamov A. 2013. Pan genome of the phytoplankton Emiliania underpins its global distribution. Nature 499, 209–213. [DOI] [PubMed] [Google Scholar]
- Redondo FJ, de la Peña TC, Morcillo CN, Lucas MM, Pueyo JJ. 2009. Overexpression of flavodoxin in bacteroids induces changes in antioxidant metabolism leading to delayed senescence and starch accumulation in alfalfa root nodules. Plant Physiology 149, 1166–1178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rensing SA, Lang D, Zimmer AD, Terry A, Salamov A, Shapiro H, Nishiyama T, Perroud P-F, Lindquist EA, Kamisugi Y. 2008. The Physcomitrella genome reveals evolutionary insights into the conquest of land by plants. Science 319, 64–69. [DOI] [PubMed] [Google Scholar]
- Richardt S, Lang D, Reski R, Frank W, Rensing SA. 2007. PlanTAPDB, a phylogeny-based resource of plant transcription-associated proteins. Plant Physiology 143, 1452–1466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rocap G, Larimer FW, Lamerdin J, Malfatti S, Chain P, Ahlgren NA, Arellano A, Coleman M, Hauser L, Hess WR. 2003. Genome divergence in two Prochlorococcus ecotypes reflects oceanic niche differentiation. Nature 424, 1042–1047. [DOI] [PubMed] [Google Scholar]
- Ruhfel BR, Gitzendanner MA, Soltis PS, Soltis DE, Burleigh JG. 2014. From algae to angiosperms: inferring the phylogeny of green plants (Viridiplantae) from 360 plastid genomes. BMC Evolutionary Biology 14, 23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rumpho ME, Worful JM, Lee J, Kannan K, Tyler MS, Bhattacharya D, Moustafa A, Manhart JR. 2008. Horizontal gene transfer of the algal nuclear gene psbO to the photosynthetic sea slug Elysia chlorotica . Proceedings of the National Academy of Sciences, USA 105, 17867–17871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito MA, Bertrand EM, Dutkiewicz S, Bulygin VV, Moran DM, Monteiro FM, Follows MJ, Valois FW, Waterbury JB. 2011. Iron conservation by reduction of metalloenzyme inventories in the marine diazotroph Crocosphaera watsonii. Proceedings of the National Academy of Sciences, USA 108, 2184–2189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakakibara H. 2003. Differential response of genes for ferredoxin and ferredoxin:NADP+ oxidoreductase to nitrate and light in maize leaves. Journal of Plant Physiology 160, 65–70. [DOI] [PubMed] [Google Scholar]
- Sancho J. 2006. Flavodoxins: sequence, folding, binding, function and beyond. Cellular and Molecular Life Sciences 63, 855–864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheller HV. 1996. In vitro cyclic electron transport in barley thylakoids follows two independent pathways. Plant Physiology 110, 187–194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schrautemeier B, Cassing A, Böhme H. 1994. Characterization of the genome region encoding an fdxH-type ferredoxin and a new 2[4Fe–4S] ferredoxin from the nonheterocystous, nitrogen-fixing cyanobacterium Plectonema boryanum PCC 73110. Journal of Bacteriology 176, 1037–1046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schrautemeier B, Neveling U, Schmitz S. 1995. Distinct and differently regulated Mo-dependent nitrogen-fixing systems evolved for heterocysts and vegetative cells of Anabaena variabilis ATCC 29413: characterization of the fdxH1/2 gene regions as part of the nif1/2 gene clusters. Molecular Microbiology 18, 357–369. [DOI] [PubMed] [Google Scholar]
- Sétif P. 2001. Ferredoxin and flavodoxin reduction by photosystem I. Biochimica et Biophysica Acta 1507, 161–179. [DOI] [PubMed] [Google Scholar]
- Shi T, Ilikchyan I, Rabouille S, Zehr JP. 2010. Genome-wide analysis of diel gene expression in the unicellular N2-fixing cyanobacterium Crocosphaera watsonii WH 8501. The ISME Journal 4, 621–632. [DOI] [PubMed] [Google Scholar]
- Shikanai T, Müller-Moulé P, Munekaga Y, Niyogi KK, Pilon M. 2003. PAA1, a P-type ATPase of Arabidopsis, functions in copper transport in chloroplasts. The Plant Cell 15, 1333–1346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shvaleva A, de la Peña TC, Rincón A, Morcillo CN, de la Torre VSG, Lucas MM, Pueyo JJ. 2010. Flavodoxin overexpression reduces cadmium-induced damage in alfalfa root nodules. Plant and Soil 326, 109–121. [Google Scholar]
- Singh AK, McIntyre LM, Sherman LA. 2003. Microarray analysis of the genome-wide response to iron deficiency and iron reconstitution in the cyanobacterium Synechocystis sp. PCC 6803. Plant Physiology 132, 1825–1839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh AK, Li H, Sherman LA. 2004. Microarray analysis and redox control of gene expression in the cyanobacterium Synechocystis sp. PCC 6803. Physiologia Plantarum 120, 27–35. [DOI] [PubMed] [Google Scholar]
- Sousa FL, Thiergart T, Landan G, Nelson-Sathi S, Pereira IA, Allen JF, Lane N, Martin WF. 2013. Early bioenergetic evolution. Philosophical Transactions of the Royal Society B: Biological Sciences 368, 20130088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stöckel J, Welsh EA, Liberton M, Kunnvakkam R, Aurora R, Pakrasi HB. 2008. Global transcriptomic analysis of Cyanothece 51142 reveals robust diurnal oscillation of central metabolic processes. Proceedings of the National Academy of Sciences, USA 105, 6156–6161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strzepek RF, Harrison PJ. 2004. Photosynthetic architecture differs in coastal and oceanic diatoms. Nature 431, 689–692. [DOI] [PubMed] [Google Scholar]
- Taylor S. 1964. Abundance of chemical elements in the continental crust: a new table. Geochimica et Cosmochimica Acta 28, 1273–1285. [Google Scholar]
- Terauchi AM, Lu S-F, Zaffagnini M, Tappa S, Hirasawa M, Tripathy JN, Knaff DB, Farmer PJ, Lemaire SD, Hase T. 2009. Pattern of expression and substrate specificity of chloroplast ferredoxins from Chlamydomonas reinhardtii . Journal of Biological Chemistry 284, 25867–25878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terry N, Abadia J. 1986. Function of iron in chloroplasts. Journal of Plant Nutrition 9, 609–646. [Google Scholar]
- Terry N, Low G. 1982. Leaf chlorophyll content and its relation to the intracellular localization of iron. Journal of Plant Nutrition 5, 301–310 [Google Scholar]
- Thimm O, Essigmann B, Kloska S, Altmann T, Buckhout TJ. 2001. Response of Arabidopsis to iron deficiency stress as revealed by microarray analysis. Plant Physiology 127, 1030–1043. [PMC free article] [PubMed] [Google Scholar]
- Thompson AW, Huang K, Saito MA, Chisholm SW. 2011. Transcriptome response of high- and low-light-adapted Prochlorococcus strains to changing iron availability. The ISME Journal 5, 1580–1594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timme RE, Bachvaroff TR, Delwiche CF. 2012. Broad phylogenomic sampling and the sister lineage of land plants. PLoS One 7, e29696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tognetti VB, Palatnik JF, Fillat MF, Melzer M, Hajirezaei M-R, Valle EM, Carrillo N. 2006. Functional replacement of ferredoxin by a cyanobacterial flavodoxin in tobacco confers broad-range stress tolerance. The Plant Cell 18, 2035–2050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tognetti VB, Zurbriggen MD, Morandi EN, Fillat MF, Valle EM, Hajirezaei M-R, Carrillo N. 2007. Enhanced plant tolerance to iron starvation by functional substitution of chloroplast ferredoxin with a bacterial flavodoxin. Proceedings of the National Academy of Sciences, USA 104, 11495–11500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toulza E, Tagliabue A, Blain S, Piganeau G. 2012. Analysis of the global ocean sampling (GOS) project for trends in iron uptake by surface ocean microbes. PLoS One 7, e30931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trifonov E. 2000. Consensus temporal order of amino acids and evolution of the triplet code. Gene 261, 139–151. [DOI] [PubMed] [Google Scholar]
- Ullmann GM, Hauswald M, Jensen A, Knapp EW. 2000. Structural alignment of ferredoxin and flavodoxin based on electrostatic potentials: Implications for their interactions with photosystem I and ferredoxin‐NADP reductase. Proteins: Structure, Function, and Bioinformatics 38, 301–309. [PubMed] [Google Scholar]
- Vaulot D, Lepere C, Toulza E, De la Iglesia R, Poulain J, Gaboyer F, Moreau H, Vandepoele K, Ulloa O, Gavory F. 2012. Metagenomes of the picoalga Bathycoccus from the Chile coastal upwelling. PLoS One 7, e39648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vigara AJ, Inda LA, Vega JM, Gómez-Moreno C, Peleato ML. 1998. Flavodoxin as an electronic donor in photosynthetic inorganic nitrogen assimilation by iron‐deficient Chlorella fusca cells. Photochemistry and Photobiology 67, 446–449. [Google Scholar]
- Vorst O, Dam F, Weisbeek P, Smeekens S. 1993. Light‐regulated expression of the Arabidopsis thaliana ferredoxin A gene involves both transcriptional and post‐transcriptional processes. The Plant Journal 3, 793–803. [DOI] [PubMed] [Google Scholar]
- Voss I, Goss T, Murozuka E, Altmann B, McLean KJ, Rigby SE, Munro AW, Scheibe R, Hase T, Hanke GT. 2011. FdC1, a novel ferredoxin protein capable of alternative electron partitioning, increases in conditions of acceptor limitation at photosystem I. Journal of Biological Chemistry 286, 50–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voss I, Koelmann M, Wojtera J, Holtgrefe S, Kitzmann C, Backhausen JE, Scheibe R. 2008. Knockout of major leaf ferredoxin reveals new redox‐regulatory adaptations in Arabidopsis thaliana . Physiologia Plantarum 133, 584–598. [DOI] [PubMed] [Google Scholar]
- Wächtershäuser G. 2007. On the chemistry and evolution of the pioneer organism. Chemistry and Biodiversity 4, 584–602. [DOI] [PubMed] [Google Scholar]
- Waycott M, Freshwater DW, York RA, Calladine A, Kenworthy WJ. 2002. Evolutionary trends in the seagrass genus Halophila (Thouars): insights from molecular phylogeny. Bulletin of Marine Science 71, 1299–1308. [Google Scholar]
- Welsh EA, Liberton M, Stöckel J, Loh T, Elvitigala T, Wang C, Wollam A, Fulton RS, Clifton SW, Jacobs JM. 2008. The genome of Cyanothece 51142, a unicellular diazotrophic cyanobacterium important in the marine nitrogen cycle. Proceedings of the National Academy of Sciences, USA 105, 15094–15099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wellman CH. 2014. The nature and evolutionary relationships of the earliest land plants. New Phytologist 202, 1–3. [DOI] [PubMed] [Google Scholar]
- Whitney LP, Lins JJ, Hughes MP, Wells ML, Chappell PD, Jenkins BD. 2011. Characterization of putative iron responsive genes as species-specific indicators of iron stress in thalassiosiroid diatoms. Frontiers in Microbiology 2, 234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeremenko N, Jeanjean R, Prommeenate P, Krasikov V, Nixon PJ, Vermaas WF, Havaux M, Matthijs HC. 2005. Open reading frame ssr2016 is required for antimycin A-sensitive photosystem I-driven cyclic electron flow in the cyanobacterium Synechocystis sp. PCC 6803. Plant and Cell Physiology 46, 1433–1436. [DOI] [PubMed] [Google Scholar]
- Yoon HS, Hackett JD, Ciniglia C, Pinto G, Bhattacharya D. 2004. A molecular timeline for the origin of photosynthetic eukaryotes. Molecular Biology and Evolution 21, 809–818. [DOI] [PubMed] [Google Scholar]
- Yoshinaga R, Niwa-Kubota M, Matsui H, Matsuda Y. 2014. Characterization of iron-responsive promoters in the marine diatom Phaeodactylum tricornutum . Marine Genomics pii: S1874-7787(14)00006-3. [DOI] [PubMed] [Google Scholar]
- Zheng M, Doan B, Schneider TD, Storz G. 1999. OxyR and SoxRS regulation of fur . Journal of Bacteriology 181, 4639–4643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmermann P, Hirsch-Hoffmann M, Hennig L, Gruissem W. 2004. GENEVESTIGATOR. Arabidopsis microarray database and analysis toolbox. Plant Physiology 136, 2621–2632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zinser ER, Lindell D, Johnson ZI, Futschik ME, Steglich C, Coleman ML, Wright MA, Rector T, Steen R, McNulty N. 2009. Choreography of the transcriptome, photophysiology, and cell cycle of a minimal photoautotroph, Prochlorococcus . PLoS One 4, e5135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zöllner A, Nogués I, Heinz A, Medina M, Gómez-Moreno C, Bernhardt R. 2004. Analysis of the interaction of a hybrid system consisting of bovine adrenodoxin reductase and flavodoxin from the cyanobacterium Anabaena PCC 7119. Bioelectrochemistry 63, 61–65. [DOI] [PubMed] [Google Scholar]
- Zurbriggen MD, Carrillo N, Tognetti VB, Melzer M, Peisker M, Hause B, Hajirezaei M-R. 2009. Chloroplast‐generated reactive oxygen species play a major role in localized cell death during the non‐host interaction between tobacco and Xanthomonas campestris pv. vesicatoria . The Plant Journal 60, 962–973. [DOI] [PubMed] [Google Scholar]
- Zurbriggen MD, Hajirezaei M-R, Carrillo N. 2010. Engineering the future. Development of transgenic plants with enhanced tolerance to adverse environments. Biotechnology and Genetic Engineering Reviews 27, 33–56. [DOI] [PubMed] [Google Scholar]
- Zurbriggen MD, Tognetti VB, Fillat MF, Hajirezaei M-R, Valle EM, Carrillo N. 2008. Combating stress with flavodoxin: a promising route for crop improvement. Trends in Biotechnology 26, 531–537. [DOI] [PubMed] [Google Scholar]