Summary
We showed the differential distribution of a subset of specific AGPs along the pollen tube growth pathway until it reached the embryo sac inside the ovule of Arabidopsis thaliana.
Key words: Arabinogalactan proteins, female gametophyte, funiculus, pistil, pollen tube guidance, transmitting tract.
Abstract
Arabinogalactan proteins (AGPs) are heavily glycosylated proteins existing in all members of the plant kingdom and are differentially distributed through distinctive developmental stages. Here, we showed the individual distributions of specific Arabidopsis AGPs: AGP1, AGP9, AGP12, AGP15, and AGP23, throughout reproductive tissues and indicated their possible roles in several reproductive processes. AGP genes specifically expressed in female tissues were identified using available microarray data. This selection was confirmed by promoter analysis using multiple green fluorescent protein fusions to a nuclear localization signal, β-glucuronidase fusions, and in situ hybridization as approaches to confirm the expression patterns of the AGPs. Promoter analysis allowed the detection of a specific and differential presence of these proteins along the pathway followed by the pollen tube during its journey to reach the egg and the central cell inside the embryo sac. AGP1 was expressed in the stigma, style, transmitting tract, and the chalazal and funiculus tissues of the ovules. AGP9 was present along the vasculature of the reproductive tissues and AGP12 was expressed in the stigmatic cells, chalazal and funiculus cells of the ovules, and in the septum. AGP15 was expressed in all pistil tissues, except in the transmitting tract, while AGP23 was specific to the pollen grain and pollen tube. The expression pattern of these AGPs provides new evidence for the detection of a subset of specific AGPs involved in plant reproductive processes, being of significance for this field of study. AGPs are prominent candidates for male–female communication during reproduction.
Introduction
All flowering plants share a common characteristic that distinguishes them from all other organisms that reproduce sexually: double fertilization (Raghavan, 2003). During this process, two male sperm cells are delivered to the female gametophyte—the embryo sac—where one fuses with the egg and the other fuses with the central cell, giving rise to the embryo and the endosperm, respectively (Russell, 1992). In order for the sperm cells to be delivered into the embryo sac, several events need to occur, which implicates tightly regulated interactions between the female sporophytic tissues and the male gametophyte. Once the pollen grain is in contact with the stigmatic cells, it germinates, producing the pollen tube (Kandasamy et al., 1994), which will deliver the two sperm cells to their final destination (Faure et al., 2002; Dresselhaus and Franklin-Tong, 2013). In the majority of seed plants, the pollen tube grows through the stigmatic cells, into the style, and across the extracellular matrix of the transmitting tissue in a very precise way, never losing its focus, to reach the embryo sac. Once at the placenta, it makes a quick turn and grows on the surface of the funiculus until reaching the ovule opening, the micropyle (Hülskamp et al., 1995). After growing through the micropyle, the pollen tube enters the female gametophyte, interacts with one of the two synergid cells and bursts, releasing the two sperm cells that will fuse with the central and the egg cell, ultimately giving rise to the seed and assuring the perpetuation of the next generation (Johnson and Preuss, 2002; Lord and Russell, 2002; Raghavan, 2003; Berger et al., 2008; Sprunck, 2010; Palanivelu and Tsukamoto, 2012).
During the course of all these processes, numerous cell–cell communication events must take place between different cell types. Mainly, recognition signals and attracting signals have to be sent and perceived by the female tissues and the male tissues of the plant, and vice versa, in order for a successful fertilization to occur (Dresselhaus, 2006). To date, despite all the efforts carried out in this field of study, little information is available about which molecules function as signalling or receptor molecules.
Arabinogalactan proteins (AGPs) constitute a large family of hydroxyproline-rich proteins that are highly glycosylated and structurally complex (Showalter, 2001). AGPs are widely distributed in the plant kingdom, being ubiquitously present in land plants and also in the Bryophyta Physcomitrella patens (Lee et al., 2005; Fu et al., 2007) and in all Hepatophyta (Basile et al., 1989), including basal angiosperms (Costa et al., 2013b ) and many algae, indicating an ancient origin for these proteins (Popper et al., 2011).
They are found in distinct developmental stages and cell, tissue, and organ types, being mostly abundant in cell walls, plasma membranes, and extracellular secretions (Majewska-Sawka and Nothnagel, 2000). AGPs are typically divided into four subgroups according to their polypeptide core characteristics: the classical AGPs, which possess an N-terminal signal peptide that is removed in the mature protein, a proline/hydroxyproline-rich domain and a C-terminal signal for the addition of a glycosylphophatidylinositol (GPI) anchor; the arabinogalactan (AG) peptides, structurally similar to the classical AGPs but with a smaller protein backbone, consisting of 10–13 aa; the lysine-rich AGPs, with one or more lysine domains; and the fasciclin-like AGPs (FLAs) with one or more fasciclin-like domains in their polypeptide core (Schultz et al., 2002; Johnson et al., 2003).
AGPs have been implicated in many important processes for plant development and growth, such as cell expansion, proliferation and differentiation, cell–cell recognition, somatic embryogenesis, pollen tube growth, programmed cell death, seed germination, and resistance to infection (Majewska-Sawka and Nothnagel, 2000). Most AGPs are predicted to be anchored to the membrane by a GPI anchor (Borner et al., 2002; Schultz et al., 2004), which provides a way for the AGPs to function as signalling molecules. After comparisons with GPI -anchored proteins from animal cells, two mechanisms were proposed for AGP-mediated signalling: the first consisted of the cleavage of the GPI anchor by specific phospholipases (C and D) that would release the glycoprotein into the extracellular matrix, making it able to act as a signal itself or to be subject to further processing, generating different signals; the other mechanism proposed that AGPs could interact with other proteins and activate downstream signal transduction pathways (Gaspar et al., 2001; Schultz et al., 2004). Besides the hint given by the presence of the GPI anchor, implying a signalling role for these proteins, the prominent carbohydrate content surrounding the core protein also led to some assumptions about their involvement in signalling mechanisms. The importance of sugars as signalling molecules in plants is well known, and, according to some authors, the varied carbohydrate moieties of AGPs might be released via cleavage by specific enzymes (Showalter, 2001). The generated oligosaccharides might function as signalling molecules by binding to specific membrane receptors and activating specific signal transduction systems (Showalter, 2001). The fact that AGPs can act as chitinase substrates, being able to stimulate somatic embryogenesis, reinforces this hypothesis, although it has not yet been demonstrated whether this is an effect of the released oligosaccharides or the modified AGP (Van Hengel et al., 2001).
AGPs have long been suggested to play important roles in sexual plant reproduction. Earlier studies have shown the developmentally regulated enrichment of AGPs in the extracellular matrix of the transmitting tract of several species such as Gladiolus gandavensis, Lilium longiflorum, Nicotiana alata, and Lycopersicon peruvianum (Hoggart and Clarke, 1984; Sedgley et al., 1985; Webb and Williams, 1988; Gane et al., 1995). AGPs have also been implicated in pollen tube growth from the stigma to the ovules in Amaranthus hypochondriacus, Actinidia deliciosa, Catharanthus roseus, and Nicotiana tabacum (Coimbra and Salema, 1997; Cheung et al., 1995; Coimbra and Duarte, 2003). These studies were carried out using the β-glycosyl Yariv reagent that binds specifically to AGPs, precipitating them (Yariv et al., 1967), or using monoclonal antibodies that identify only the glycosidic epitopes of AGPs (Pennell et al., 1989, 1991; Knox et al., 1991). These two approaches have given us information about the distribution and localization of AGPs (Coimbra et al., 2007) and clues about their possible roles (Gao and Showalter, 2002; Sardar et al., 2006), although they allow only the detection of general AGPs and not a specific AGP. The recent discovery that the Yariv reagent binds specifically to the β-1,3-galacto-oligosaccharides of AGPs (Kitazawa et al., 2013) may bring new insights to the possible mode of action of AGP oligosaccharides as signalling molecules. It will be interesting to check whether this particular oligosaccharide is important for many of the physiological processes impaired when Yariv was used in different studies, or if Yariv only hampers the ability of AGPs to function by precipitating them.
Here, we report the use of several constructs to explore the tissue and cell-specific promoter activity of specific AGPs. We have focused on those AGPs that are particularly present along the pollen tube pathway and other female reproductive tissues, according to the available microarray data. With this, we aimed to complement work that has already been done by our group describing AGPs as molecular markers of different stages of Arabidopsis sexual reproductive processes (Coimbra et al., 2007).
Materials and methods
Plant material and growth conditions
Arabidopsis thaliana (L.) Heynh. seeds, ecotype Columbia were obtained from the Nottingham Arabidopsis Stock Centre, UK. Plants were sown on soil, kept for 2 d at 4 °C in the dark to induce stratification, and afterwards they were grown at 22 °C under a short-day photoperiod (9/15h light/dark cycles) for 4 weeks, followed by a long-day photoperiod (16/8h light/dark cycles) to induce flowering, with 60% relative humidity. For phosphinothricin acetyltransferase selection, the seedlings were sprayed with 200mg l–1 of glufosinate ammonium (BASTA®; Bayer Crop Science) supplemented with 0.1% Tween 20 three or four times every 2 d, over a 10-day period.
Construct generation and plant transformation
Genomic regions corresponding to the promoters of five AGPs: AGP1, AGP9, AGP12, AGP15, and AGP23, were amplified using Phusion DNA polymerase (Thermo Scientific), with the primer pairs described in Supplementary Table S1 (at JXB online). The promoter regions were always amplified from the end of the untranslated region of the most proximal gene upstream of the respective AGP until its own start codon. For the genes with promoter regions of more than 3000bp, genomic fragments of about 3000–3300bp positioned upstream of the start codon of the AGP of interest were amplified. The PCR products were cloned into pENTR™/D-TOPO (Invitrogen). The resulting promoter fragments were subsequently transferred into a Gateway-compatible version (Zheng et al., 2011) of the pGreenII-based vector NLS:3GFP:NOSt (Takada and Jürgens, 2007), termed pGII_GW:NLS:3GFP:NOSt. For AGP1, AGP15, and AGP23 β-glucuronidase (GUS) constructs, the respective promoter fragments were cloned into the binary vector pBGWFS7 (Karimi et al., 2002). All constructs were confirmed by DNA sequencing. The pGreenII-based expression vectors were introduced into Agrobacterium tumefaciens GV3101 harbouring the pGreenII helper plasmid pSOUP. All other expression vectors were delivered into Agrobacterium tumefaciens GV3101 (pMP90RK). They were all then used to transform Arabidopsis thaliana (Col-0) by the floral dip method (Clough and Bent, 1998).
Preparation of plant material for microscopy
Pistils kept in 50mM sodium phosphate buffer (pH 7.5) were dissected under a stereomicroscope (model C-DSD230; Nikon) using hypodermic needles (0.4×20mm; Braun). The opened carpels and the ovules that remained attached to the septum were maintained in mounting medium and covered with a cover slip.
Confocal laser-scanning microscopy
A Zeiss Axiovert 200M inverted microscope equipped with a confocal laser-scanning module (LSM 510 META) was used for confocal laser-scanning microscopy. Green fluorescent protein (GFP) was excited by 488nm and detected with a BP 505–550 filter. Optical sections were generally between 0.40 and 0.50 μm each, observed at ×20, ×40 or ×63 magnification. Histology mounting medium Fluoroshield™ with 4′,6-diamidino-2-phenylindole (DAPI; Sigma) was used in order to detect the nuclei in the pollen grains. Images were captured and processed using an AxioCam HRc camera, Zeiss LSM 510 META software and a Zeiss LSM image browser version 3.5.0.359.
Detection of GUS activity
GUS assays were performed on inflorescences as described by Liljegren et al. (2000), overnight. After chemical GUS detection, the samples were incubated in clearing solution [160g of chloral hydrate (Sigma-Aldrich), 100ml of water, and 50ml of glycerol] and incubated at 4 °C overnight. The next day, inflorescences were dissected under a stereomicroscope (model C-DSD230; Nikon) and observed under a microscope. A Zeiss AxioImager AZ microscope equipped with differential interference contrast optics was used. Images were captured with a ZeissAxiocam MRc3 camera using Zen Imaging Software.
Phylogenetic analysis
To generate a phylogenetic tree for the AGP genes, the amino acid sequences of AGP-coding sequences were aligned using Clustal W (Thompson et al., 1994) and manually edited using Jalview to reduce gaps (Clamp et al., 2004). A neighbour-joining (NJ) (Saitou and Nei, 1987) tree was generated using the MEGA4 program (Tamura et al., 2007). The bootstrap values were obtained by 10 000 repetitions. Simultaneously, a maximum parsimony tree was generated using the same AGP amino acid sequences, and a NJ tree was also produced using only the three most conserved blocks of AGP amino acid sequences.
Preparation of plant material for RNA extraction
Arabidopsis pistils from wild-type plants were emasculated 1 d before anthesis and collected 2 d after the emasculation procedure. Pollen from wild-type Arabidopsis recently opened flowers was collected according to Costa et al. (2013a). Arabidopsis seeds were sown in half-strength Murashige and Skoog medium, complemented with 0.7% agar. Agar plates were kept for 2 d at 4 °C in the dark to induce stratification, and subsequently were transferred to a growth chamber at 22 °C under a long-day regime (16h light/8h dark), with irradiance of 130 μmol m–2 s–1 and 60% relative humidity. Seedlings were collected 4–5 d after germination.
RNA extraction, cDNA synthesis and real-time reverse transcription (RT)-PCR
Total RNA from emasculated pistils, pollen, and seedlings was extracted using PureZolTM RNA Isolation Reagent (Bio-Rad) following the manufacturer’s instructions. DNA was removed by a DNase (Thermo Scientific) treatment. The isolated RNA samples were reverse transcribed using a RevertAid First Strand cDNA Synthesis Kit (Thermo Scientific) and oligo(dT)18 primers to initiate the reactions, following the manufacturer’s instructions.
cDNA was amplified using SSoFast™ SYBR® Green Supermix on an iQ5™ Real-Time PCR Detection System (Bio-Rad) using the primers listed in Supplementary Table S2 (at JXB online). Real-time RT-PCRs were run in duplicates. After 3min at 95 °C followed by a 10 s denaturation step at 95 °C, samples were run for 40 cycles of 10 s at 95 °C and 30 s at 60 °C. After each run, a dissociation curve was acquired to check for amplification specificity by heating the samples from 60 to 95 °C. Serial dilutions of pure genomic DNA from Arabidopsis ecotype Columbia were used to set up a calibration curve, which was used to quantify plant DNA in each sample. At the end of the PCR cycles, data were analysed with iQ5 2.0, Standard Edition Optical System Software v2.0.148.060623 (Bio-Rad).
Fluorescence in situ hybridization (FISH)
Genomic DNA was obtained as described by Edwards et al. (1991) and used to amplify the in situ sense and antisense probes for AGP1 and AGP12 using the following primers: AGP1-F 5′-CAAAAACACTCCCAAACCAAA-3′, AGP1-R 5′-CTTCAGT CGGAGAATCGG-3′, AGP12-F 5′-CACAACTCATCATTCGCA CCAAAG-3′ and AGP12-R 5′-GCATCGGAAGTAGGACTT GG-3′. The amplified fragments were cloned in pGEMT-Easy (Promega). Digoxygenin (DIG)-RNA probes were generated by in vitro transcription using a DIG-RNA labelling kit (Roche). The dissected pistils were permeabilized by first dehydrating in a methanol series of increasing concentration and then rehydrating in a methanol series of decreasing concentration. The pistils were then treated with 2% cellulase (Onozuka R-10) for 1h, and then washed and dried. RNA/RNA FISH was performed as described by Testillano and Risueño (2009) using DIG-RNA probes diluted 1:50 in hybridization buffer at 50 °C overnight. Post-hybridization washes were performed in 4× SSC, 2× SSC, and 0.1× SSC. The hybridization signal was detected by incubation with mouse anti-DIG antibodies (diluted 1:5000 in 1% BSA; Sigma) for 90min, followed by incubation with Alexa Fluor 488-conjugated anti-mouse antibody (diluted 1:25 in PBS; Molecular Probes) for 45min. After washing in PBS, sections were counterstained with DAPI, mounted in Mowiol, and observed by confocal microscopy. Controls were performed using the sense probes.
Results
Phylogenetic analysis and AGP distribution across the genome
An alignment of full-length predicted AGP proteins was generated using Clustal W (Thompson et al., 1994) and then manually refined (Fig. 1A). In this study, 13 classical AGPs (AGP1, AGP2, AGP3, AGP4, AGP5, AGP6, AGP7, AGP9, AGP10, AGP11, AGP25, AGP26, and AGP27), 10 AG peptides (AGP12, AGP13, AGP14, AGP15, AGP16, AGP20, AGP21, AGP22, AGP23, and AGP24), and three lysine-rich AGPs (AGP17, AGP18, and AGP19) were considered. For this analysis, only four FLAs were used: FLA18, FLA16, FLA17, and FLA15. These FLAs were chosen randomly and included in the analyses only as outgroup, since they are particularly different from the rest of the family and considered to be chimaeric AGPs (Showalter et al., 2010). The phylogenetic distribution of the selected AGP sequences partially supported the four subgroups of AGPs proposed by previous studies (Schultz et al., 2002; Johnson et al., 2003). The alignments showed a high level of similarity between the predicted amino acid sequences of AGP15, an AG peptide, and AGP1, a classical AGP, as well as the inclusion of the three lysine-rich AGPs in the same branch as the classical AGPs, not supporting the AGP classification currently in use. As expected, the FLAs used in this study aligned together and independently from the other AGPs as a subgroup but were still related to the classical AGP25, AGP26, and AGP27. Comparing the NJ tree generated using the AGPs complete amino acid sequences with the maximum parsimony tree and the NJ tree generated using only the three conserved blocks of AGP amino acid sequences (Supplementary Figs 1 and 2, at JXB online), we could see almost no difference among them. All the main blocks of closely related AGPs remained grouped together in the three different trees, reinforcing the strength of this analysis. These two trees were generated using the three most conserved regions revealed by the AGP multiple alignments (Supplementary Fig. S3, at JXB online). Looking at the AGP gene distribution along the different five Arabidopsis chromosomes (Fig. 1B), there was no evidence of clustering of any specific group of closely related AGP genes or any specific class of AGPs. They seemed to be randomly distributed across the different Arabidopsis chromosomes.
Fig. 1.
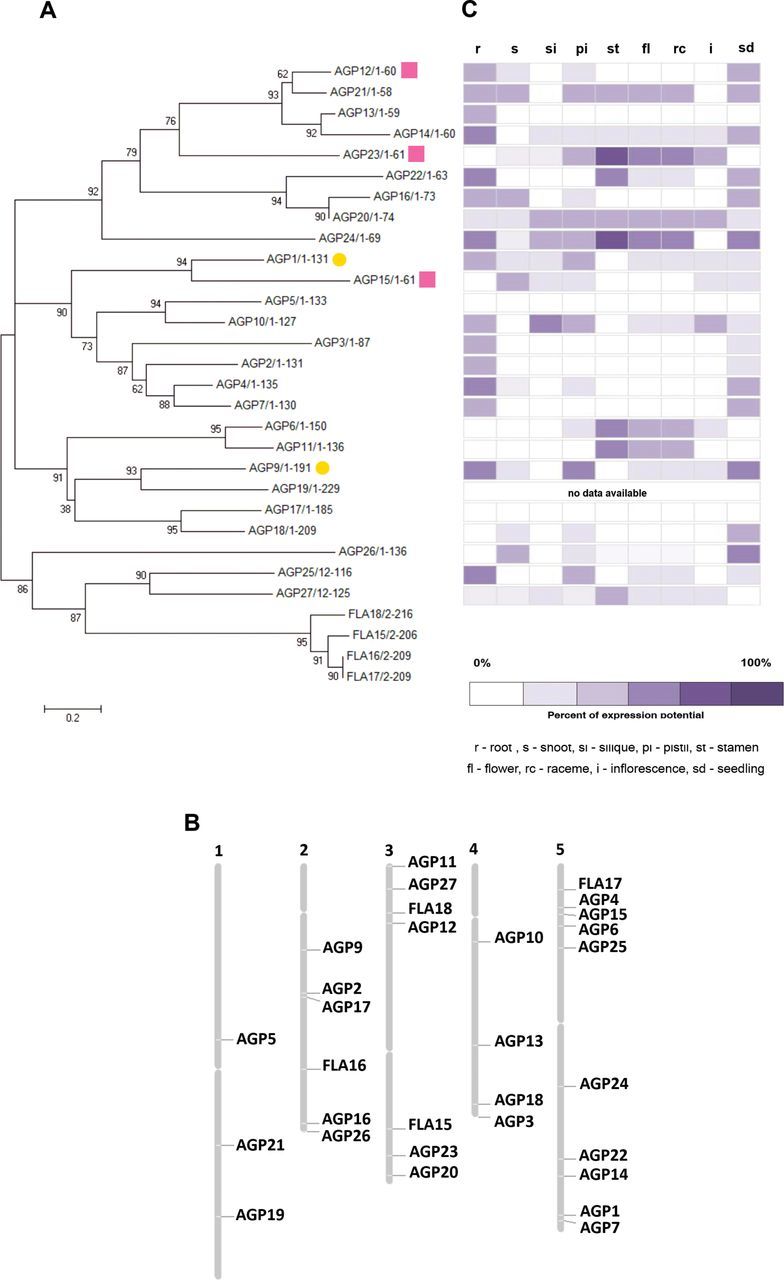
The AGP protein family, gene expression, and AGP gene localization in A. thaliana. (A) Phylogenetic analysis of the AGP family in A. thaliana. To generate the phylogenetic tree for AGPs, all the amino acid sequences of AGP-coding sequences were aligned using Clustal W and manually edited using Jalview to reduce gaps. A NJ tree was generated using the MEGA4 program. The optimal tree with the sum of branch length=14.47033254 is shown. The confidence probability (multiplied by 100) that the interior branch length is greater than 0, as estimated using the bootstrap test (10 000 replicates) is shown next to the branches.The tree is drawn to scale, with branch lengths in the same units as those of the evolutionary distances used to infer the phylogenetic tree. The evolutionary distances were computed using the Poisson correction method and are in the units of the number of amino acid substitutions per site (bar). The analysis involved 30 amino acid sequences. All ambiguous positions were removed for each sequence pair. There were a total of 241 positions in the final dataset. AGPs selected for further analysis are indicated by a yellow circle (classical AGPs) and a pink square (AG peptides). (B) The 26 AGP and four FLA genes were localized on the Arabidospis chromosomes using the Chromosome Map Tool available at The Arabidopsis Information Resource (TAIR: http://www.arabidopsis.org/jsp/ChromosomeMap/tool.jsp). (C) Gene expression patterns for the 26 AGP and four FLA genes obtained using Genevestigator.
AGP gene expression
As a first approach, data from microarray experiments available from online databases such as Genevestigator (https://www.genevestigator.com/gv/; Zimmermann et al., 2004) and the Arabidopsis eFP Browser (http://bar.utoronto.ca/efp/cgi-bin/efpWeb.cgi; Winter et al., 2007) were used to evaluate the distinct levels of AGP gene expression throughout the different plant tissues (Fig. 1C; only Genevestigator data are shown). Eleven AGPs were selected for further analysis: AGP1, AGP4, AGP7, AGP9, AGP10, AGP12, AGP15, AGP16, AGP23, AGP25, and AGP26, most of them based on the presence of their transcripts in pistil tissues and their absence in stamen tissues. In the case of AGP7, although it did not show this pattern of expression, it was selected anyway, based on its predicted high level of amino acid sequence similarity with AGP4. AGP23 was selected as a negative control, since the eFP Browser and the literature data (Costa et al., 2013c ; Nguema-Ona et al., 2013) indicated that it is expressed only in pollen. However, the Genevestigator data also indicated poor expression in female tissues. To check the differences between AGP gene expression levels in these tissues and to validate the microarray-based information, a real-time RT-PCR was performed using emasculated pistils, pollen from flowers at anthesis (stage 13 according to Smyth et al., 1990), and seedling cDNA. The results confirmed the microarray data initially considered (Fig. 2). These analyses confirmed the good quality of the microarray data. In this work, the AGP transcript levels were normalized to ACT8 and RUB1 reference gene levels, and are presented relative to the pollen transcript levels, since the main goal was to determine the AGP genes that are expressed more in the female tissues than in the pollen. AGP7 and AGP23 were downregulated in the pistil tissues when compared with their expression in pollen, while all the other AGPs were upregulated. AGP10, AGP12, and AGP16 were the genes that revealed a higher level of overexpression when compared with their expression in pollen. AGP1, AGP4, AGP15, AGP25, and AGP26 were revealed to be upregulated in the pistils, compared with pollen, but not at such high levels as the genes AGP10, AGP12, and AGP16. From this group of upregulated AGPs, AGP1, AGP9, AGP12, and AGP15 were selected for further analyses.
Fig. 2.
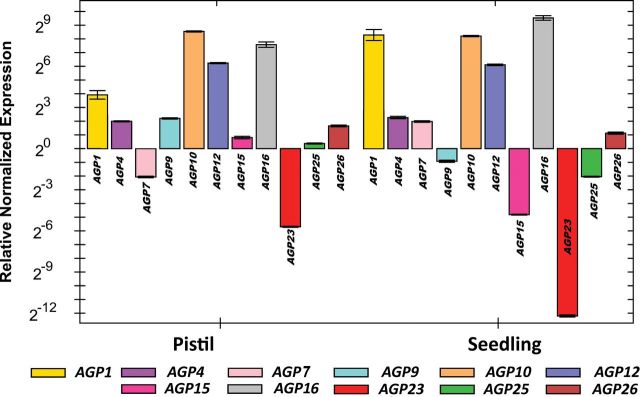
Quantitative PCR relative expression levels of the selected AGP mRNA transcripts in emasculated pistils, pollen, and seedlings of wild-type Arabidopsis plants. The pollen was collected from anthers at stage 12 of flower development according to Smyth et al. (1990). AGP transcript levels were normalized to ACT8 and RUB1 reference gene levels, and are presented relative to the pollen transcript levels. Each bar represents an average of two independent reactions and technical replicates.
Plasmid construction and expression in A. thaliana
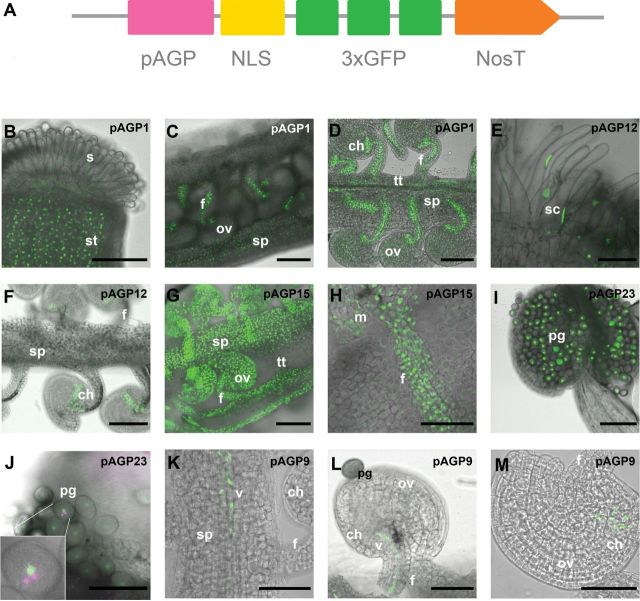
To improve the visualization and to avoid diffuse fluorescent signals in the detection of the promoter activities, the reporter gene NLS:3GFP was used (Takada and Jürgens, 2007). This consists of the simian virus 40 nuclear localization signal (NLS) and three tandem enhanced GFP (3×EGFP) sequences (Fig. 3A). The fluorescent signal should be then targeted to the nuclei, thereby enhancing the sensitivity of the GFP signal. In all the transgenic A. thaliana plants bearing the different pAGP:NLS:3GFP constructs, the GFP reporter expression was limited to the nuclei as expected, as shown in Fig. 3B–M.
Fig. 3.
Schematic representation of the expression cassette used in this study, and the resulting GFP signal shown in Arabidopsis reproductive tissues. (A) Expression cassette showing the relative position of the promoter sequence (pAGP), NLS, a fusion of three GFPs (3×GFP), and the terminator Nos (NosT). (B–D) NLS:3GFP expression driven by the AGP1 promoter in the style tissues (B), the opened pistil, and the funiculus and septum tissues (C), and seen in more detail in the transmitting tissue, funiculus, and the chalazal pole of the ovule (D). (E–F) NLS:3GFP expression under the control of the AGP12 promoter was observed in the stigmatic cells (E) and in the chalazal pole of the ovule (F). (G–H) NLS:3GFP expression driven by the AGP15 promoter was detected in the ovule integuments, funiculus, and septum, but absent from the transmitting tissue (G). In (H) the GFP signal is seen in more detail in the nuclei of the funiculus. (I, J) NLS:3GFP under the control of the AGP23 promoter is absent in all the sporophytic tissues (I), with its expression restricted to the pollen grain, and, as can be seen in the insert in (J), DAPI staining (here in magenta) revealed this expression to be limited to the vegetative cell of the pollen grain; DAPI-stained germinative nuclei are visible (white arrowheads). (K–M) NLS:3GFP signals expressed by the AGP9 promoter. Signals were observed in the vascular bundle of the transmitting tract (K) and funiculus (L) as well as in the chalazal pole of the ovule (M). All the flowers used in these observations were at stages 12 and 13 according to Smyth et al. (1990). ch, Chalaza; f, funiculus; m, micropyle region of the ovule; ov, ovule; pg, pollen grain; s, stigma; sc, stigmatic cell; sp, septum; st, style; tt, transmitting tract; v, vascultature. Bars, 100 μm (B–G, I); 50 μm (H, K–M); 20 μm (J).
AGP differential expression patterns in A. thaliana reproductive tissues
The AGP promoters selected for this study allowed us to detect the different patterns of expression of these proteins in the female reproductive tissues. All the flowers analysed were between stages 12 and 13 according to Smyth et al. (1990). GFP expression driven by the AGP1 promoter was strong in the style tissues (Fig. 3B), septum (Fig. 3C), transmitting tract (Fig. 3D), funiculus that attaches the ovules to the placenta (Fig. 3C, D), and chalazal region of the ovules (Fig. 3D). Weaker GFP expression was detected in the stigmatic cells (Fig. 3B) and the integuments of the ovule (Fig. 3D). The AGP12 promoter guided the expression of GFP strongly to the stigmatic cells (Fig. 3E) and the chalazal pole of the ovules (Fig. 3F). Very weak GFP expression was observed along the internal tissues of the funiculus and septum (Fig. 3F). Plants transgenic for the pAGP15:NLS:3GFP expression cassette exhibited GFP expression in all the female reproductive tissues, except in the transmitting tract cells (Fig. 3G, H). The AGP23 promoter drove GFP expression specifically into the vegetative cell of the pollen grains (Fig. 3I, J). This was clarified by the DAPI staining of the pollen grains, showing that the GFP signal was present only in the nucleus of the vegetative cell and not in the generative cell nuclei, where there was only DAPI staining without any green signal. The AGP9 promoter led to the expression of GFP in the vascular tissues of the pistil transmitting tract, the septum (Fig. 3K), and the funiculus (Fig. 3L), exhibiting very weak expression in the chalazal pole of the ovules (Fig. 3M).
At the same time, pAGP:GUS constructs were also analysed for three AGPs: AGP1, AGP15, and AGP23. For the pAGP1:GUS fusion-expressing plants, a low GUS activity was observed in the stigmatic cells, while a higher GUS activity was detected in the septum, transmitting tract, funiculus, chalaza, and ovule integument cells (Figs. 4A, B). Regarding the plants expressing GUS under the control of the AGP15 promoter, a high GUS activity was detected in almost all the tissues of the pistil, except in the transmitting tract (Fig. 4C, D). As well as the plants expressing the three GFP molecules under the control of the AGP23 promoter, the Arabidopsis plants bearing GUS under the control of this same promoter showed a very specific and high GUS activity in the pollen (Fig. 4E, F). This activity was also observed in the pollen tubes (Fig. 4G), and it was especially high when the pollen tube burst occurred inside the embryo sac (Fig. 4H), staining almost all the embryo sac with a weaker GUS signal. This GUS expression in the embryo sac was never observed when pAGP23:GUS pistils were pollinated with wild-type pollen, only in embryo sacs fertilized with pAGP23:GUS pollen. This indicated that the GUS product present in the maternal embryo sac after fertilization was released by the burst of the pollen tube.
Fig. 4.
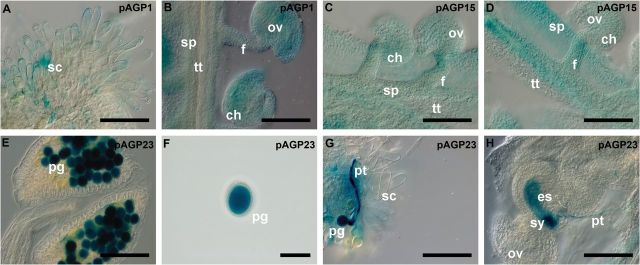
Histochemical localization of GUS activity in transgenic Arabidopsis reproductive tissues expressing the pAGP:GUS fusion genes. (A, B) GUS activity driven by the AGP1 promoter is detected in the stigmatic cells (A) and the transmitting tract, funiculus, and integument cells (B). (C, D) GUS activity driven by the AGP15 promoter observed in the ovule integuments, funiculus, and septum cells. (E–H) Strong GUS activity driven by the AGP23 promoter was identified inside pollen grains (E) and (F) and the growing pollen tube (G). Upon fertilization, inside the embryo sac, a strong staining is observed at the local where the pollen tube bursts (H), followed by a weak staining that spread inside the whole embryo sac (H). Flowers of stages 12 and 13 (Smyth et al., 1990) were used in this study. ch, Chalaza; es, embryo sac; f, funiculus; ov, ovule; pg, pollen grain; pt, pollen tube; sc, stigmatic cell; sp, septum; sy, synergid; tt, transmitting tract. Bars, 100 μm (A–E, G, H); 50 μm (F).
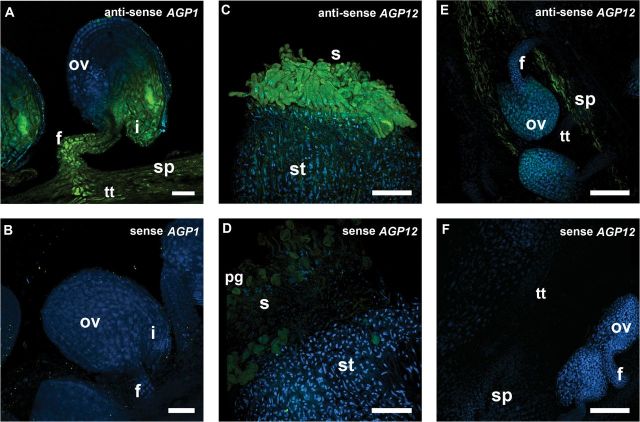
FISH confirms the GFP reporter line patterns of expression
FISH was used to verify whether the GFP signals and GUS activity obtained with the pAGP:3GFP and pAGP:GUS fusions in fact reflected the real AGP gene expression. For this study, FISH was used to analyse two AGP genes: AGP1 and AGP12. Hybridization signals for the AGP1 antisense probe were detected throughout the septum, transmitting tract, and funiculus cells, as well as in the integuments surrounding the micropylar region of the embryo sac (Fig. 5A). The same experiment using the AGP1 sense probe revealed the absence of hybridization signal along all the reproductive tissues (Fig. 5B). With the AGP12 antisense probe, strong hybridization signals were detected in the stigmatic cells (Fig. 5C) and a weaker signal was observed across the style and the septum (Fig. 5E). The corresponding AGP12 sense probe did not show any hybridization signals along the reproductive tissues (Fig. 5D, F).
Fig. 5.
FISH localization of AGP1 and AGP12 transcripts in Arabidopsis pistil tissues. Merged images of FISH signals (green) and DAPI staining of nuclei (blue) are shown. (A) AGP1 transcripts were detected in the funiculus, transmitting tissue, and integuments. (C, E) AGP12 transcripts were localized in the stigmatic cells and along the septum tissues. (B, D, F) FISH controls with the sense probe for AGP1 in ovules (B) and for AGP12 in stigma (D) and ovules (F). All the flowers used in these observations were at stages 12 and 13 according to Smyth et al. (1990). f, Funiculus; i, integuments; pg, pollen grain; ov, ovule; s, stigma; sp, septum; st, style. Bars, 25 μm (A, B); 75 μm (C–F).
Discussion
AGPs selection
Bioinformatics analyses recently allowed the identification of 64 potential AGPs in Arabidopsis (Showalter et al., 2010). The present work started by analysing 26 of these: those with more information available. However, it is important to keep in mind that for all individual AGPs almost no information is available at the structural level. Sequence comparisons revealed a high level of similarity between amino acid sequences of AGP15, an AG peptide, and AGP1, a classical AGP, as well as the inclusion of the three lysine-rich AGPs in the same branch as the classical AGPs. These results pinpointed the artificial nature of the classification currently in use to organize this family of proteins. The availability of more data regarding AGP expression patterns in different plant species and more information regarding their functions may allow the classification of these proteins based on their functions and localization, rather than on their amino acid sequences similarities. However, there are still some pairs of AGPs that share a high degree of similarity between their amino acid sequences, and, simultaneously, display a similar expression pattern in the reproductive tissues, suggesting that they might act redundantly, such as the AGP16/AGP20, AGP1/AGP15, AGP5/AGP10, and AGP6/AGP11 pairs. AGP6/AGP11 is a pair of redundant AGPs involved in Arabidopsis pollen grain and pollen tube growth and development (Coimbra et al., 2009). A total of 11 AGPs were picked for further analysis: AGP1, AGP4, AGP7, AGP9, AGP10, AGP12, AGP15, AGP16, AGP23, AGP25, and AGP26. This group was selected by an in silico search of AGP genes that could be transcribed preferentially in pistils rather than in the stamens or seedlings. This selection was based on analyses of microarray data available for pistil and stamen tissues obtained from Genevestigator, using the Anatomy tool provided by this service (Zimmermann et al., 2004) and the eFP Browser (Winter et al., 2007). Although AGP18 fits perfectly into this category, it was not selected as it is already well described (Acosta-García and Vielle-Calzada, 2004; Demesa-Arévalo and Vielle-Calzada, 2013). AGP23 was chosen as a control, since it is only transcribed during pollen development (Costa et al., 2013c ; Nguema-Ona et al., 2013). Although microarray data from Genevestigator also predicted its expression in whole flowers and pistils, our real-time RT-PCR data confirmed that AGP23 was detected only in pollen, being highly downregulated in pistils and seedlings.
The validation of this selection through real-time RT-PCR allowed us to limit the number of AGPs selected for further analysis to four: AGP1, AGP9, AGP12, and AGP15. AGP9 and AGP15 are upregulated in the pistil and downregulated in the seedlings, and were selected for this reason. AGP1 was also selected, although its transcripts show a higher upregulation in seedlings than in emasculated pistils, because it is phylogenetically close to AGP15. AGP12 was chosen as one of the most upregulated AGPs in the pistil.
Regarding AGP gene localization in the Arabidopsis chromosomes, it was clear that the AGPs were randomly distributed over the Arabidopsis genome. This was the case for AGP16 and AGP20, located, respectively, on chromosomes 2 and 3, and also for AGP6, located on chromosome 5, and AGP11, on chromosome 3, two AGPs that have already been shown to act redundantly (Coimbra et al., 2009). This is probably due to duplications in the genome, since most of these genes are included in segments of the respective chromosomes that have been subject to large duplications events (Blanc et al., 2000). This is consistent with the prediction that genetic redundancy may occur as a consequence of gene duplication (Kafri et al., 2009). Only the pairs of the most similar AGPs, AGP4/AGP7 and AGP1/AGP15, have their genes positioned in the same chromosome but in opposed regions. It is plausible that some of the AGP genes acquired a certain degree of specialization, and are now expressed in different tissues under different conditions.
AGP expression in the reproductive tissues
The results obtained in this work confirmed the specific and differential pattern of expression of AGPs predicted previously by immunolocalization studies, where several monoclonal antibodies, recognizing distinctive AGP glycosidic epitopes revealed the presence of these proteins throughout diverse tissues in different developmental stages in Arabidopsis (Coimbra et al., 2007). These results not only confirmed and complemented this previous study but also improved the information already available about AGP distribution throughout the reproductive tissues by identifying specific AGPs present in these tissues. In the study by Coimbra et al. (2007), no antibody labelling was detected in the stigmatic cells, which, as was shown here, are rich at least in AGP1 and AGP12. Also, in the same study, no antibody labelling was detected in the funiculus of the ovules, whereas, in this study, the presence of several AGPs, such as AGP1, AGP12, and AGP15, was revealed in this tissue. This work illustrates the usefulness of these techniques in contrast to the use of monoclonal antibodies to detect AGPs. As expected from quantitative PCR data, AGP23 was expressed only in pollen grains and pollen tubes. Although the microarray data available from Genevestigator indicated that AGP23 should be present in pistils, this was not observed here. The analysis of transgenic Arabidopsis plants carrying the pAG23:GUS and the pAGP23:NLS:3GFP constructs revealed that both reporters were detected in pollen, proving that AGP23 is specific to the pollen vegetative cell. The prediction of potential of expression of AGP23 in flowers and pistils was most probably due to the high levels of AGP23 expression in pollen grains contained in the samples used for those studies. Concerning the pistil, the manipulation of these tissues is complicated if the flowers are not in the correct stage of development, as it is easy to get pollen contamination in the stigma, misleading to some false-positive expression. A summary of the different approaches used to localize these AGPs and their differential patterns of expression in the reproductive tissues is shown in Fig. 6.
Fig. 6.
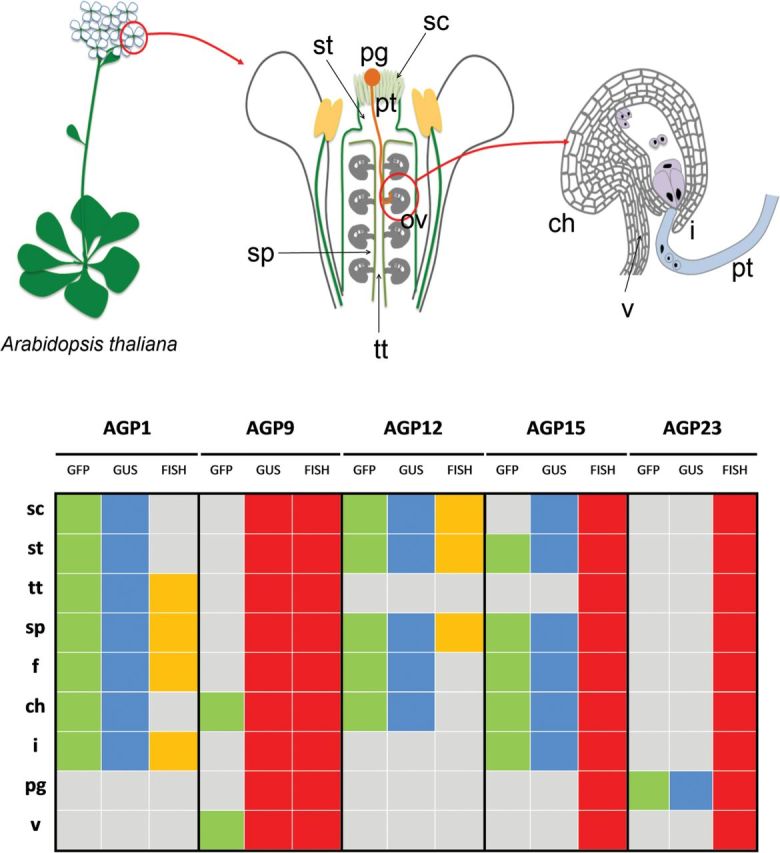
A schematic representation of the reproductive structures and tissues of A. thaliana and the distribution of the five AGPs analysed in this study throughout the different tissues, regarding the different techniques used. GFP presence, green; GUS presence, blue; FISH positive, yellow; experiment not performed, red; absence of signal, grey. ch, Chalaza; f, funiculus; i, integuments; pg, pollen grain; sc, stigmatic cell; sp, septum; st, style; tt, transmitting tract; v, vasculature.
The FISH data obtained for AGP1 and AGP12 were partially consistent with the promoter analysis results shown for these two AGPs. The GFP expression driven by the AGP1 and AGP12 promoters revealed the presence of GFP signal in the chalazal tissues of the ovule, and, surprisingly, this was not observed in FISH results. This technique implies the analysis of whole-ovule amounts, making the tissue permeabilization more difficult in order for the probe to reach the most internal cell layers of the ovules, as is the case of the chalazal region (García-Aguilar et al., 2005; Hejátko et al., 2006). Still, we are aware that some regulatory elements of these two promoters might be missing, thus leading to the AGP misexpression in the chalazal tissues. Besides having regulatory sequences within the promoter itself, in eukaryotes there may be regulatory elements located tens of thousands of base pairs away from the start site, in introns, or even downstream from the coding sequence of the gene (Korkuć et al., 2014). Also, AGP1 transcripts were not detected in the stigmatic cells or in the style by FISH analysis. It is important to underline the fact that the microarray data used and the FISH technique were performed with whole organs, while the promoter analysis referred to a spatial–temporal analysis, which was much more detailed. The older immunolocalization studies (Junqueira, 2007) did not detect the glycosidic AGP epitopes in the chalazal tissues. Although we are aware that the antibodies used identify only sugar epitopes from all AGPs, we may conclude, with some caution, that the accordance between the immunolocalization data and FISH results fortifies confidence in the use of antibodies to determine AGP localization.
AGP1 and AGP12 expression in the stigmatic cells suggested the possible involvement of this protein in pollen–stigmatic cells interactions, and acquisition of pollen grain competence to initiate pollen tube growth. Losada and Herrero (2012) indicated a role for AGPs in supporting pollen tube germination, suggesting that the secretion of AGPs can be associated with the acquisition of stigma receptivity in apple flower. The same mechanism may occur with AGP12 and AGP1 in Arabidopsis. Also, in the early divergent angiosperm Trithuria, immunocytochemistry results suggest AGPs to be involved in attracting the pollen tubes through the stigmatic cuticle, as in most evolved angiosperms (Prychid et al., 2011), reinforcing our hypothesis.
The presence of AGP1 and AGP15 in the main female reproductive tissues through which the pollen tube grows until it reaches the embryo sac – the stigma, style, transmitting tract, septum, and funiculus – strengthens the putative role of AGPs in pollen tube growth and fertilization. Many early studies implied AGPs from the female tissues as playing major roles in reproductive processes (Du et al., 1994; Cheung et al., 1995; Cheung and Wu, 1999; Wu et al., 2000; Coimbra et al., 2007). For example, TTS proteins (AGPs from Nicotiana tabacum) were shown to attract and promote pollen tube growth either in vivo or in vitro, nutritionally supporting its growth and providing it with guidance cues (Cheung et al., 1995; Wu et al., 2000). Wu et al. (1995) also revealed that the carbohydrate part of these TTS proteins forms an increasing gradient from the top to the bottom of the Nicotiana style, by the action of specific pollen tube hydrolases, which may have a chemotropic effect on growing pollen tubes. In Arabidopsis, the transmitting tract begins at the style between the stigma–style boundaries, extending to the base of the ovary (Crawford and Yanofsky, 2008). AGP1 is mainly present along this transmitting tract, while AGP15 is mostly present at the septum surrounding the transmitting tract. Since these two proteins are closely related to each other, this fortifies their possible redundant function in these tissues. agp1 null mutants were analysed (data not shown) but revealed no visible phenotype. Most probably a double agp1 agp15 mutant is needed to access their precise function. These AGPs might act in these tissues in a similar manner to the TTS proteins in Nicotiana. A study of the NTT gene in Arabidopsis has indirectly implied the involvement of AGPs in pollen tube guidance through the transmitting tract (Crawford et al., 2007). The ntt mutants lacked a functional transmitting tract and exhibited a reduced staining for acidic polysaccharides. Crawford et al. (2007) speculated that AGPs, acidic glycoproteins that are a main component of the transmitting tract, might be reduced in these mutants. It will be extremely interesting to check whether there is a control of AGP expression by this NTT zinc-finger transcription factor.
AGP1, AGP9, and more strongly AGP12, showed expression at the chalazal tissues of Arabidopsis ovules and at the cells located on the top of the vascular supply coming from the funiculus, as well as along this tissue. It is known that the main nutrient uptake into the endosperm occurs via the chalazal pole, with this being important for nutrient transfer from the maternal parent to the developing embryo (Debeaujon et al., 2003; Ingram, 2010). This may indicate the possible participation of these glycoproteins in nutrition or signalling between the vasculature and the embryo sac, endosperm, or embryo, being quickly mobilized. The incomplete correlation between GFP and GUS activity driven by the AGP12 and AGP1 promoters in this region and their transcript expression revealed the importance of analysing these AGPs at the protein level in future studies.
For double fertilization to take place, the pollen tube must travel a long and challenging pathway in order to reach its final destination: the micropylar entrance to the embryo sac, where it will discharge, through one of the two synergids, two immotile sperm cells to fertilize the egg cell and the central cell, giving rise to the embryo and the endosperm, respectively, initiating a new generation (Márton and Dresselhaus, 2010). Along this narrow road, the pollen tube lengthens through a mucilage-rich extracellular matrix from the stigmatic cells, along the specialized transmitting tract cells, funiculus, and ovary integuments (Webb and Williams, 1988; Lennon et al., 1998). Although most of these studies showed that this extracellular matrix tract, through which the pollen tube travels, is rich in AGPs and pectins, to date only some specific molecules have been shown to function as pollen tube growth enhancers such as GABA in Arabidopsis (Palanivelu et al., 2003) and chemocyanin in Lilium longiflorum (Kim et al., 2003).
The results shown in this study support previous work where AGPs were proposed to be part of this pathway and to sustain pollen tube growth (Clarke et al., 1979; Herrero and Dickinson, 1979; Gell et al., 1986; Cheung et al., 1995). AGP1, AGP12, and AGP15 (Fig. 6) are located along all these tissues and might well contribute to pollen tube growth from the top of the stigma to the base of the pistil, into the ovules, either by nutritionally supporting their growth, facilitating their movement, guiding them to their targets, or even by making them competent for pollen tube reception by the embryo sac. These hypotheses need further studies to fully assign AGP functions in these tissues, most probably involving studies with double or triple null mutants. It is interesting to note that we identified AGPs along the entire pollen tube pathway (stigma, style, and transmitting tract), showing that AGPs are most probably essential for all the different steps of pollen tube growth through the pistil. The molecular mechanism of action of AGPs and how they interact with other cell-wall and cell components is still elusive, although some enlightenment has recently been given to this matter (Costa et al., 2013a ). One possibility may be related to the most recent finding that AGPs can act as calcium reservoirs, making calcium available temporarily in a developmental way (Lamport and Várnai, 2012). The importance of calcium in sexual plant reproduction is well known (Ge et al., 2007). One of the key characteristics of growing pollen tubes is a tip-focused calcium gradient maintained by the influx of extracellular calcium through calcium channels active at the extreme end of the growing tip (Feijó et al., 1995). AGPs may be regulating in some way the release of calcium along the pollen tube pathway, making calcium available for the pollen tubes to grow. Most likely, different AGPs play several different roles during different steps of the reproductive process, according to their localization and timing of expression (Fig. 6). Our results support and improve the study of these enigmatic and inscrutable glycoproteins in the sexual plant reproductive process, opening doors for new pathways for the study of specific AGPs. Also, this type of analysis overcomes the main difficulty regarding the older immunolocalization AGP studies made by the use of monoclonal antibodies that detected only the glycosidic epitope of the AGPs, instead allowing the identification of a specific AGP in plant tissues.
Supplementary Material
Acknowledgements
This work was financed by FEDER through the COMPETE programme, and by Portuguese National funds through FCT – Fundação para a Ciência e Tecnologia (Project PTDC/AGR-GPL/115358/2009) and from an FCT PhD grant SFRH/BD/60995/2009 awarded to AMP. This project also benefited from financial support from the COST Action FA0903: ‘Harnessing Plant Reproduction for Crop Improvement’. We would like to thank Mily Ron from UC-Berkeley, Plant Gene Expression Center, for kindly sharing with us the pGII_GW:NLS:3GFP:NOSt destination vector.
Glossary
Abbreviations:
- AG
arabinogalactan
- AGP
arabinogalactan protein
- DAPI
4′,6-diamidino-2-phenylindole
- DIG
digoxygenin
- FISH
fluorescence in situ hybridization
- FLA
fasciclin-like AGPs
- GPI
glycosylphophatidylinositol
- GUS
β-glucuronidase
- NJ
neighbour joining
- NLS
nuclear localization signal
- RT-PCR
reverse transcription-PCR.
References
- Acosta-García G, Vielle-Calzada J-P. 2004. A classical arabinogalactan protein is essential for the initiation of female gametogenesis in Arabidopsis . Plant Cell 16, 2614–2628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basile DV, Kushner BK, Basile MR. 1989. A new method for separating and comparing arabinogalactan proteins for the chemosystematics of the Hepaticae. The Bryologist 92, 164–169. [Google Scholar]
- Berger F, Hamamura Y, Ingouff M, Higashiyama T. 2008. Double fertilization: caught in the act. Trends in Plant Science 13, 437–443. [DOI] [PubMed] [Google Scholar]
- Blanc G, Barakat A, Guyot R, Cooke R, Delseny M. 2000. Extensive duplication and reshuffling in the Arabidopsis genome. Plant Cell 12, 1093–1101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borner GHH, Sherrier DJ, Stevens TJ, Arkin IT, Dupree P. 2002. Prediction of glycosylphosphatidylinositol-anchored proteins in Arabidopsis. A genomic analysis. Plant Physiology 129, 486–499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung AY, Wang H, Wu HM. 1995. A floral transmitting tissue-specific glycoprotein attracts pollen tubes and stimulates their growth. Cell 82, 383–393. [DOI] [PubMed] [Google Scholar]
- Cheung AY, Wu HM. 1999. Arabinogalactan proteins in plant sexual reproduction. Protoplasma 208, 87–98. [Google Scholar]
- Clamp M, Cuff J, Searle SM, Barton GJ. 2004. The Jalview Java alignment editor. Bioinformatics 20, 426–427. [DOI] [PubMed] [Google Scholar]
- Clarke A, Gleeson P, Harrison S, Knox RB. 1979. Pollen–stigma interactions: identification and characterization of surface components with recognition potential. Proceedings of the National Academy of Sciences, USA 76, 3358–3362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough SJ, Bent AF. 1998. Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana . The Plant Journal 16, 735–743. [DOI] [PubMed] [Google Scholar]
- Coimbra S, Almeida J, Junqueira V, Costa ML, Pereira LG. 2007. Arabinogalactan proteins as molecular markers in Arabidopsis thaliana sexual reproduction. Journal of Experimental Botany 58, 4027–4035. [DOI] [PubMed] [Google Scholar]
- Coimbra S, Costa M, Jones B, Mendes MA, Pereira LG. 2009. Pollen grain development is compromised in Arabidopsis agp6 agp11 null mutants. Journal of Experimental Botany 60, 3133–3142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coimbra S, Duarte C. 2003. Arabinogalactan proteins may facilitate the movement of pollen tubes from the stigma to the ovules in Actinidia deliciosa and Amaranthus hypocondriacus . Euphytica 133, 171–178. [Google Scholar]
- Coimbra S, Salema R. 1997. Immunolocalization of arabinogalactan proteins in Amaranthus hypocondriacus L. ovules. Protoplasma 199, 75–82. [Google Scholar]
- Costa M, Nobre MS, Becker JD, Masiero S, Amorim MI, Pereira LG, Coimbra S. 2013. a Expression-based and co-localization detection of Arabinogalactan protein 6 and Arabinogalactan protein 11 interactors in Arabidopsis pollen and pollen tubes. BMC Plant Biology 13, 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa M, Pereira AM, Rudall PJ, Coimbra S. 2013. b Immunolocalization of arabinogalactan proteins (AGPs) in reproductive structures of an early-divergent angiosperm, Trithuria (Hydatellaceae). Annals of Botany 111, 183–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa M, Pereira LG, Coimbra S. 2013. c Growth media induces variation in cell wall associated gene expression in Arabidopsis thaliana pollen tube. Plants 2, 429–440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crawford B, Ditta G, Yanofsky M. 2007. The NTT gene is required for transmitting-tract development in carpels of Arabidopsis thaliana . Current Biology 17, 1101–1108. [DOI] [PubMed] [Google Scholar]
- Crawford BC, Yanofsky MF. 2008. The formation and function of the female reproductive tract in flowering plants. Current Biology 18, R972–R978. [DOI] [PubMed] [Google Scholar]
- Debeaujon I, Nesi N, Perez P, Devic M, Grandjean O, Caboche M, Lepiniec L. 2003. Proanthocyanidin-accumulating cells in Arabidopsis testa: regulation of differentiation and role in seed development. Plant Cell 15, 2514–2531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demesa-Arévalo E, Vielle-Calzada J-P. 2013. The classical arabinogalactan protein AGP18 mediates megaspore selection in Arabidopsis . Plant Cell 25, 1274–1287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dresselhaus T. 2006. Cell–cell communication during double fertilization. Current Opinion in Plant Biology 9, 41–47. [DOI] [PubMed] [Google Scholar]
- Dresselhaus T, Franklin-Tong N. 2013. Male–female crosstalk during pollen germination, tube growth and guidance, and double fertilization. Molecular Plant 6, 1018–1036. [DOI] [PubMed] [Google Scholar]
- Du H, Simpson RJ, Moritz RL, Clarke AE, Bacic A. 1994. Isolation of the protein backbone of an arabinogalactan-protein from the styles of Nicotiana alata and characterization of a corresponding cDNA. Plant Cell 6, 1643–1653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards K, Johnstone C, Thompson C. 1991. A simple and rapid method for the preparation of plant genomic DNA for PCR analysis. Nucleic Acids Research 19, 1349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faure JE, Rotman N, Fortune P, Dumas C. 2002. Fertilization in Arabidopsis thaliana wild type: developmental stages and time course. The Plant Journal 30, 481–488. [DOI] [PubMed] [Google Scholar]
- Feijó JA, Malhó R, Obermeyer G. 1995. Ion dynamics and its possible role during in vitro pollen germination and tube growth. Protoplasma 187, 155–167. [Google Scholar]
- Fu H, Yadav MP, Nothnagel EA. 2007. Physcomitrella patens arabinogalactan proteins contain abundant terminal 3-O-methyl- l -rhamnosyl residues not found in angiosperms. Planta 226, 1511–1524. [DOI] [PubMed] [Google Scholar]
- Gane AM, Clarke AE, Bacic A. 1995. Localisation and expression of arabinogalactan-proteins in the ovaries of Nicotiana alata Link and Otto. Sexual Plant Reproduction 8, 278–282. [Google Scholar]
- Gao M, Showalter AM. 2002. Yariv reagent treatment induces programmed cell death in Arabidopsis cell cultures and implicates arabinogalactan protein involvement. The Plant Journal 19, 321–331. [DOI] [PubMed] [Google Scholar]
- García-Aguilar M, Dorantes-Acosta A, Pérez-España V, Vielle-Calzada J-P. 2005. Whole-mount in situ mRNA localization in developing ovules and seeds of Arabidopsis . Plant Molecular Biology Reporter 23, 279–289. [Google Scholar]
- Gaspar Y, Johnson KL, McKenna JA, Bacic A, Schultz CJ. 2001. The complex structures of arabinogalactan-proteins and the journey towards understanding function. Plant Molecular Biology 47, 161–176. [PubMed] [Google Scholar]
- Ge LL, Tian HQ, Russell SD. 2007. Calcium function and distribution during fertilization in Angiosperms. American Journal of Botany 94, 1046–1060. [DOI] [PubMed] [Google Scholar]
- Gell AC, Bacic A, Clarke AE. 1986. Arabinogalactan-proteins of the female sexual tissue of Nicotiana alata: I. Changes during flower development and pollination. Plant Physiology 82, 885–889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hejátko J, Blilou I, Brewer PB, Friml J, Scheres B, Benková E. 2006. In situ hybridization technique for mRNA detection in whole mount Arabidopsis samples. Nature Protocols 1, 1939–1946. [DOI] [PubMed] [Google Scholar]
- Herrero M, Dickinson HG. 1979. Pollen–pistil incompatibility in Petunia hybrida: changes in the pistil following compatible and incompatible intraspecific crosses. Journal of Cell Science 36, 1–18. [DOI] [PubMed] [Google Scholar]
- Hoggart RM, Clarke AE. 1984. Arabinogalactans are common components of Angiosperm styles. Phytochemistry 23, 1571–1573. [Google Scholar]
- Hülskamp M, Schneitz K, Pruitt RE. 1995. Genetic evidence for a long-range activity that directs pollen tube guidance in Arabidopsis . Plant Cell 7, 57–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingram GC. 2010. Family life at close quarters: communication and constraint in angiosperm seed development. Protoplasma 247, 195–214. [DOI] [PubMed] [Google Scholar]
- Johnson KL, Jones BJ, Bacic A, Schultz CJ. 2003. The fasciclin-like arabinogalactan proteins of Arabidopsis. A multigene family of putative cell adhesion molecules. Plant Physiology 133, 1911–1925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson MA, Preuss D. 2002. Plotting a course: multiple signals guide pollen tubes to their targets. Developmental Cell 2, 273–281. [DOI] [PubMed] [Google Scholar]
- Junqueira V. 2007. Imunolocalização de proteínas arabinogalactânicas no gineceu de Arabidopsis thaliana Wt e nos mutantes mur1, mur4 e reb1–1. Masters thesis, Faculty of Sciences, University of Porto, Portugal. [Google Scholar]
- Kafri R, Springer M, Pilpel Y. 2009. Genetic redundancy: new tricks for old genes. Cell 136, 389–392. [DOI] [PubMed] [Google Scholar]
- Kandasamy MK, Nasrallah J B, Nasrallah ME. 1994. Pollen–pistil interactions and developmental regulation of pollen tube growth in Arabidopsis . Development 120, 3405–3418. [Google Scholar]
- Karimi M, Inzé D, Depicker A. 2002. GATEWAY vectors for Agrobacterium-mediated plant transformation. Trends in Plant Science 7, 193–195. [DOI] [PubMed] [Google Scholar]
- Kim S, Mollet J-C, Dong J, Zhang K, Park S-Y, Lord EM. 2003. Chemocyanin, a small basic protein from the lily stigma, induces pollen tube chemotropism. Proceedings of the National Academy of Sciences, USA 100, 16125–16130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitazawa K, Tryfona T, Yoshimi Y, et al. 2013. β-Galactosyl Yariv reagent binds to the β-1,3-galactan of arabinogalactan proteins. Plant Physiology 161, 1117–1126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knox JP, Linstead PJ, Peart J, Cooper C, Roberts K. 1991. Developmentally regulated epitopes of cell surface arabinogalactan proteins and their relation to root tissue pattern formation. The Plant Journal 1, 317–326. [DOI] [PubMed] [Google Scholar]
- Korkuć P, Schippers JHM, Walther D. 2014. Characterization and identification of cis-regulatory elements in Arabidopsis based on single-nucleotide polymorphism information. Plant Physiology 164, 181–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamport DT, Várnai P. 2012. Periplasmic arabinogalactan glycoproteins act as a calcium capacitor that regulates plant growth and development. New Phytologist 197, 58–64. [DOI] [PubMed] [Google Scholar]
- Lee KJD, Sakata Y, Mau S-L, Pettolino F, Bacic A, Quatrano RS, Knight CD, Knox JP. 2005. Arabinogalactan proteins are required for apical cell extension in the moss Physcomitrella patens . Plant Cell 17, 3051–3065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lennon KA, Roy S, Hepler PK, Lord EM. 1998. The structure of the transmitting tissue of Arabidopsis thaliana (L.) and the path of pollen tube growth. Sexual Plant Reproduction 11, 49–59. [Google Scholar]
- Liljegren SJ, Ditta GS, Eshed Y, Savidge B, Bowman JL, Yanofsky MF. 2000. SHATTERPROOF MADS-box genes control seed dispersal in Arabidopsis. Nature 404, 766–770. [DOI] [PubMed] [Google Scholar]
- Lord EM, Russell SD. 2002. The mechanisms of pollination and fertilization in plants. Annual Reviews of Cell and Developmental Biology 18, 81–105. [DOI] [PubMed] [Google Scholar]
- Losada JM, Herrero M. 2012. Arabinogalactan-protein secretion is associated with the acquisition of stigmatic receptivity in the apple flower. Annals of Botany 110, 573–584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majewska-Sawka A, Nothnagel EA. 2000. The multiple roles of arabinogalactan proteins in plant development. Plant Physiology 122, 3–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Márton ML, Dresselhaus T. 2010. Female gametophyte-controlled pollen tube guidance. Biochemical Society Transactions 38, 627–30. [DOI] [PubMed] [Google Scholar]
- Nguema-Ona E, Vicré-Gibouin M, Cannesan M-A, Driouich A. 2013. Arabinogalactan proteins in root–microbe interactions. Trends in Plant Science 18, 440–449. [DOI] [PubMed] [Google Scholar]
- Palanivelu R, Brass L, Edlund AF, Preuss D. 2003. Pollen tube growth and guidance is regulated by POP2, an Arabidopsis gene that controls GABA levels. Cell 114, 47–59. [DOI] [PubMed] [Google Scholar]
- Palanivelu R, Tsukamoto T. 2012. Pathfinding in angiosperm reproduction: pollen tube guidance by pistils ensures successful double fertilization. WIREs Developmental Biology 1, 96–113. [DOI] [PubMed] [Google Scholar]
- Pennell RI, Janniche L, Kjellbom P, Scofield GN, Peart JM, Roberts K. 1991. Developmental regulation of a plasma membrane arabinogalactan protein epitope in oilseed rape flowers. Plant Cell 3, 1317–1326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pennell RI, Knox JP, Scofield GN, Selvendran RR, Roberts K. 1989. A family of abundant plasma membrane-associated glycoproteins related to the arabinogalactan proteins is unique to flowering plants. Journal of Cell Biology 108, 1967–1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popper ZA, Michel G, Hervé C, Domozych DS, Willats WGT, Tuohy MG, Kloareg B, Stengel DB. 2011. Evolution and diversity of plant cell walls: from algae to flowering plants. Annual Review of Plant Biology 62, 567–590. [DOI] [PubMed] [Google Scholar]
- Prychid CJ, Sokoloff DD, Remizowa MV, Tuckett RE, Yadav SR, Rudall PJ. 2011. Unique stigmatic hairs and pollen-tube growth within the stigmatic cell wall in the early-divergent angiosperm family Hydatellaceae. Annals of Botany 108, 599–608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raghavan V. 2003. Some reflections on double fertilization, from its discovery to the present. New Phytologist 159, 565–583. [DOI] [PubMed] [Google Scholar]
- Russell SD. 1992. Double fertilization. International Review of Cytology 140, 357–388. [Google Scholar]
- Saitou N, Nei M. 1987. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Molecular Biology and Evolution 4, 406–425. [DOI] [PubMed] [Google Scholar]
- Sardar HS, Yang J, Showalter AM. 2006. Molecular interactions of arabinogalactan proteins with cortical microtubules and F-actin in Bright Yellow-2 tobacco cultured cells. Plant Physiology 142, 1469–1479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultz CJ, Ferguson KL, Lahnstein J, Bacic A. 2004. Post-translational modifications of arabinogalactan-peptides of Arabidopsis thaliana . Journal of Biological Chemistry 279, 45503–45511. [DOI] [PubMed] [Google Scholar]
- Schultz CJ, Rumsewicz MP, Johnson KL, Jones BJ, Gaspar YM, Bacic A. 2002. Using genomic resources to guide research directions: the arabinogalactan protein gene family as a test case. Plant Physiology 129, 1448–1463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sedgley M, Blesing MA, Bonig I, Anderson MA, Clarke AE. 1985. Arabinogalactan-proteins are localized extracellularly in the transmitting tissue of Nicotiana alata link and otto, an ornamental tobacco. Micron and Microscopica Acta 16, 247–254. [Google Scholar]
- Showalter AM. 2001. Arabinogalactan-proteins: structure, expression and function. Cellular and Molecular Life Sciences 58, 1399–1417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Showalter AM, Keppler B, Lichtenberg J, Gu D, Welch LR. 2010. A bioinformatics approach to the identification, classification, and analysis of hydroxyproline-rich glycoproteins. Plant Physiology 153, 485–513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smyth DR, Bowman JL, Meyerowitz EM. 1990. Early flower development in Arabidopsis. Plant Cell 2, 755–767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sprunck S. 2010. Let′s get physical: gamete interaction in flowering plants. Biochemical Society Transactions 38, 635–640. [DOI] [PubMed] [Google Scholar]
- Takada S, Jürgens G. 2007. Transcriptional regulation of epidermal cell fate in the Arabidopsis embryo. Development 134, 1141–1150. [DOI] [PubMed] [Google Scholar]
- Tamura K, Dudley J, Nei M, Kumar S. 2007. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Molecular Biology and Evolution 24, 1596–1599. [DOI] [PubMed] [Google Scholar]
- Testillano PS, Risueño MC. 2009. Tracking gene and protein expression during microspore embryogenesis by Confocal Laser Scanning Microscopy. In: Touraev A, Forster BP, Jain SM, eds. Advances in haploid production in higher plants. The Netherlands: Springer, 339–347. [Google Scholar]
- Thompson JD, Higgins DG, Gibson TJ. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Research 22, 4673–4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Hengel AJ, Tadesse Z, Immerzeel P, Schols H, Van Kammen A, de Vries SC. 2001. N-Acetylglucosamine and glucosamine-containing arabinogalactan proteins control somatic embryogenesis. Plant Physiology 125, 1880–1890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webb MC, Williams EG. 1988. The pollen tube pathway in the pistil of Lycopersicon peruvianum . Annals of Botany 61, 415–423. [Google Scholar]
- Winter D, Vinegar B, Nahal N, Ammar R, Wilson V, Provart N. 2007. An ‘Electronic Fluorescent Pictograph’ Browser for exploring and analyzing large-scale biological data sets. PLoS ONE 2, e718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H, Wong E, Ogdahl J, Cheung AY. 2000. A pollen tube growth-promoting arabinogalactan protein from Nicotiana alata is similar to the tobacco TTS protein. The Plant Journal 22, 165–176. [DOI] [PubMed] [Google Scholar]
- Wu HM, Wang H, Cheung AY. 1995. A pollen tube growth stimulatory glycoprotein is deglycosylated by pollen tubes and displays a glycosylation gradient in the flower. Cell 82, 395–403. [DOI] [PubMed] [Google Scholar]
- Yariv J, Lis H, Katchalski E. 1967. Precipitation of arabic acid and some seed polysaccharides by glycosylphenylazo dyes. Biochemical Journal 105, 1C–2C. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng B, Chen X, McCormick S. 2011. The anaphase-promoting complex is a dual integrator that regulates both microRNA-mediated transcriptional regulation of cyclin B1 and degradation of cyclin B1 during Arabidopsis male gametophyte development. Plant Cell 23, 1033–1046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmermann P, Hirsch-Hoffmann M, Henning L, Gruissem W. 2004. GENEVESTIGATOR. Arabidopsis microarray database and analysis toolbox. Bioinformatics 136, 2621–2632. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.