Abstract
Many hormonal and environmental signals regulate common cellular and developmental processes in plants. While the molecular pathways that transduce these signals have each been studied in detail, how these pathways are wired into regulatory networks to provide the coordinated responses has remained an outstanding question. Recent studies of the brassinosteroid signaling network have revealed extensive signal integration through direct interactions between components of different signaling pathways. In particular, a circuit of interacting transcription regulators integrates many signaling pathways to enable coordinated and coherent regulation of seedling morphogenesis by hormonal and environmental signals. The recent studies support an emerging theme that complex networks of highly integrated signaling pathways underlie the high levels of developmental plasticity and environmental adaptability of plants.
Introduction
While discovered for its prominent role in promoting cell elongation, brassinosteroid (BR) actually regulates diverse developmental and physiological processes, including seed germination, seedling photomorphogenesis, stomata differentiation, organ boundary formation, flowering, male fertility, and plant responses to biotic and abiotic stresses [1-5]. In maize, BR also plays a role in sex differentiation [6]. Genetic and biochemical studies in Arabidopsis have elucidated in molecular details a complete BR signal transduction cascade from perception by the BRASSINOSTEROID INSENSITIVE1 (BRI1) receptor kinase at the cell surface to transcriptional regulation of thousands of nuclear genes by the BRASSINAZOLE RESISTENT (BZR) family transcription factors [1]. How this BR signaling pathway regulates the diverse developmental and physiological processes has become an outstanding question in the BR field. Likewise, many signaling pathways have been studied in detail, but separately, and how signaling pathways are integrated to ensure coordinated and coherent responses is becoming a prominent question in plant biology. Recent studies have revealed major mechanisms of BR crosstalk with other signaling pathway and the BR signaling network serves as a model for understanding the mechanisms of signal integration in plants.
BR signaling pathway: a brief account
According to the crystal structure reported recently [7,8], BR directly binds to the extracellular domain of BRI1 at a pocket formed by the folding of the island loop onto a region of the leucine-rich repeat (LRR) module, and this creates a surface for dimerization with the co-receptor kinase BRI1-ASSOCIATED RECEPTOR KINASE1 (BAK1) or its homolog SOMATIC EMBRYOGENESIS RECEPTOR KINASE 1 (SERK1). As such BR acts as a “molecular glue” to bring together BRI1 and BAK1. Sequential transphosphorylation between the kinase domains of BRI1 and BAK1 activates the kinases, and BRI1 in turn phosphorylates members of two groups of plasma membrane-anchored cytoplasmic kinases, BRASSINOSTEROID-SIGNALLING KINASE1 (BSK1) and CONSTITUTIVE DIFFERENTIAL GROWTH1 (CDG1). CDG1, and likely also BSK1, phosphorylates the BRI1-SUPPRESSOR1 (BSU1) phosphatase, which then dephosphorylates and inactivates the GSK3-like kinase BRASSINOSTEROID INSENSITIVE2 (BIN2) [9]. When the BR levels are low, BIN2 phosphorylates two homologous transcription factors, BRASSINAZOLE RESISTANT1 (BZR1) and BZR2 (also named BRI1-EMS-SUPPRESSOR1, BES1), to inhibit their nuclear localization and DNA-binding activity. When the BR levels are high, BIN2 is inactivated, and BZR1 and BZR2 are dephosphorylated by PROTEIN PHOSPHATASE 2A (PP2A). Unphosphorylated BZR1 and BZR2 accumulate in the nucleus and bind to the promoters of target genes to confer BR-responsive gene expression [1].
BZR1 and BZR2/BES1 regulate overlapping sets of target genes, which include large numbers of genes with structural and metabolic functions such as cell wall biogenesis, as well as genes with regulatory functions, such as components of signal transduction pathways and key developmental regulators [10,11]. Although earlier studies of different target genes suggested opposite transcriptional activities, the genome-wide analysis showed that BZR1 and BZR2/BES1 regulate shared target genes in similar manner and they both activate and repress roughly equal numbers of target genes [10,11]. The transcriptional activities of BZR1 and BZR2/BES1 are modulated or mediated by interactions with other transcription factors and histone-modifying enzymes, such as the TOPLESS repressor and histone deacetylases [3,12,13]. BR can regulate specific developmental processes through BZR1 target genes, such as the CUP-SHAPED COTYLEDON (CUC) genes involved in organ boundary formation [4]. BR also regulates development through crosstalk mediated by direct interactions between the components of BR pathway and other signaling pathways. Below, we review recent progress in the studies of crosstalks between BR and other signaling pathways, focusing on the molecular mechanisms of direct interactions between components of different signal transduction pathways.
Integration of BR, auxin, and phytochrome pathways through interaction of BZR, ARF, and PIF factors
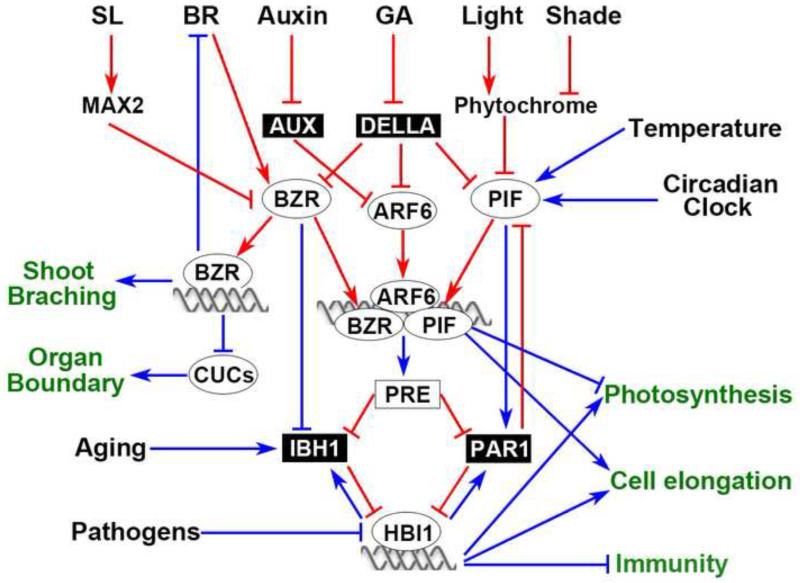
Seedling morphogenesis is controlled by a number of endogenous and environmental signals, including BR, auxin, gibberellin (GA), the circadian clock, light, and temperature. BR, and specifically the activity of BZR factors, is essential for seedling etiolation in the dark. BZR1/2 regulates light responses in at least two ways: it transcriptionally controls the expression levels of many light-signaling components, and it interacts with phytochrome-interacting factors (PIFs) [1]. The BZR1/2-mediated transcriptional repression of light signaling components, including phytochrome and light activated transcription factors GATA2/4, BZR1–1D SUPPRESSOR1 (BZS1), and GOLDEN2-LIKE 2 (GLK2) [10,11,14,15], contribute quantitatively to BR modulation of light sensitivity, but cannot explain the essential role of BR in etiolation in the dark, where photoreceptors and light signaling pathways are inactive. This essential role of BR in etiolation appears to be mediated by BZR1/2 as an essential partner of PIFs, which are basic helix-loop-helix (bHLH) transcription factors that accumulate in the dark or shade but become hyper-phosphorylated and degraded upon interaction with light-activated phytochromes [16]. BZR1 and PIFs are genetically interdependent for promoting cell elongation and etiolation, and they directly interact and co-regulate a large number of shared target genes to promote cell elongation and suppress photomorphogenesis [17]. Such a model of functional interdependence between BZR1/2 and PIFs provides a mechanistic explanation for the antagonistic relationship between BR and light signals in regulating seedling morphogenesis (Figure 1). This model also explains the essential roles of BR in plant responses to additional signals that modulate PIF levels. In addition to light, temperature and the circadian clock also control the levels of PIF proteins transcriptionally [18]. Transcriptional activation of PIF4 expression is essential for the heat-induced hypocotyl elongation, and this function of PIF4 also requires BR and BZR activity [17] (Figure 1).
Figure 1.
A central growth regulation network integrates hormonal and environmental signals in transcriptional regulation of plant growth and physiology. DNA-binding transcription factors are shown in ovals, and non-DNA-binding factors that inhibit DNA binding factors are shown in black boxes. Red lines show posttranslational activation (arrows) or inhibition (bar end), and blue lines show transcriptional activation (arrows) or inhibition (bar end).
Auxin and BR are known to be interdependent and synergistic in promoting Arabidopsis hypocotyl elongation, and they induce highly overlapping transcriptional responses [19,20]. Auxin sensitivity, as well as auxin level, is modulated by light, temperature, and circadian rhythm, through PIFs [21]. While BZR1- and PIF-mediated transcriptional regulation of auxin biosynthetic and signaling genes have been observed [10,22], a recent study revealed more direct roles of PIF factors and BZR1 in auxin response [23]. Identification of genome-wide targets of ARF6 revealed over 50% overlaps between the ARF6 target genes and the targets of BZR1 or PIF4 [23]. BZR1 and PIF4 interact with ARF6 directly and enhance its binding to the shared promoters. Genetic analyses confirmed interdependent relationships between these factors in activating shared target genes and promoting hypocotyl elongation. The study supports a model that the Arabidopsis hypocotyl elongation is regulated through cooperative interactions among BZR1, ARF6, and PIF4 at the promoters of overlapping target genes, many of which encode cell wall proteins involved in cell expansion [23]. Thus, the BZR-ARF-PIF module elegantly explains the co-regulation of shoot cell elongation by BR, auxin, and phytochrome. Furthermore, additional signals regulate plant growth by modulating the activities of these factors.
Integration of gibberellin signal through DELLA repression of BZR, PIF, and ARF factors
GA binds to its receptor GID1 to induce ubiquitination and degradation of the DELLA proteins. Interestingly, DELLA proteins interact with and inhibit DNA binding activities of not only PIFs and BZR1, but also ARF6 [24-27]. Thus, GA promotes cell elongation largely by releasing the DELLA-mediated repression of PIFs, BZR1 and ARF6 [26-28]. GA/DELLA regulation of all three components of the BZR-ARF-PIF module potentially provides balanced and quantitative modulation of the outputs of the BZR-ARF-PIF circuit (Figure 1).
Integration of the strigolactone signaling pathway through MAX2-BZR interaction
Strigolactone (SL) suppresses lateral shoot branch growth and also promotes photomorphogenesis [29,30]. A recent study provided evidence that MORE AXILLARY GROWTH2 (MAX2), an F-box ubiquitin E3 ligase required for SL signaling, mediates ubiquitination and degradation of BZR proteins [31]. Yeast two-hybrid and in vivo assays showed MAX2 interaction with BZR1 and BZR2/BES1. SL treatment caused BZR2/BES1 degradation in a MAX2-dependent manner, and SL inhibition of hypocotyl elongation is abolished in both max2 and the dominant bes1-D mutant. Consistent with PIF-dependent function of BZR in photomorphogenesis, the pif-q mutant is hypersensitive to SL [32]. Interestingly, the bes1-D mutant showed increased branching, and knockdown of BES1 suppressed the more-axillary-growth phenotype of max2. It was proposed that the SL/MAX2-dependent degradation of BZR factors mediates SL regulation of both photomorphogenesis and axillary branch growth [31] (Figure 1).
Coupling of the BZR-ARF-PIF module with the tripartite HLH/bHLH module
The promotion of hypocotyl cell elongation by the BZR-ARF-PIF module requires a tripartite helix-loop-helix/basic-helix-loop-helix (HLH/bHLH) module consisting of two classes of non-DNA-binding HLH factors that antagonistically control many DNA-binding bHLH factors [26,33,34]. BZR1, ARF6, and PIF4 directly activate members of the PACLOBUTRAZOLRESISTANCE (PRE) family of HLH factors, which promote plant growth [26]. PREs bind to another group of HLH factors, including ILI1 binding bHLH1 (IBH1), LONG HYPOCOTYL IN FAR-RED (HFR), PHYTOCHROME RAPIDLY REGULATED1/2 (PAR1/2), and ATBS1 INTERACTING FACTOR (AIFs), which inhibit plant growth. Through heterodimerization, HFR and PAR1/2 inhibit DNA binding activities of PIFs [35,36]. As such, activation of PRE expression by the BZR-ARF-PIF module further increases availability of PIFs by sequestrating HFR and PAR1, forming a positive feedback loop. In contrast, activation of PIFs by shade conditions increases transcription of HFR and PAR1 genes, forming a negative feedback loop. Such hormone-dependent positive feedback and hormone-independent negative feedback regulation of PIFs potentially ensures that the responses to shade or darkness are limited by the endogenous signals BR and auxin (Figure 1).
In addition to PIFs, PAR1 also inhibits BR-ENHANCED EXPRESSION 2 (BEE2), which is transcriptionally activated by BR signaling, and BIM1, which interacts with BZR2/BES1. Both BIM1 and BEE2 play a role in promoting hypocotyl elongation [37].
IBH1 inhibits another family of DNA-binding bHLH factors including HOMOLOG OF BEE2 INTERACTING WITH IBH1 (HBI1), BEE2 and three ACTIVATOR FOR CELL ELONGATION factors (ACE1 to ACE3) [33,34]. PRE1 binds to IBH1 to prevent its inhibition of HBI1 and ACEs [33,34]. Genome-wide target gene analyses showed that the HBI1 target genes are mostly (>70%) also direct targets of PIFs, and that HBI1 and PIFs activate many common target genes involved in cell elongation [38]. Like PIF4, HBI1 also interacts with ARF6[23]. But unlike PIFs, HBI1 is not regulated by light, and HBI1 positively regulates many genes encoding chloroplast proteins, suggesting a distinct role in promoting both growth and photosynthesis in light-grown plants [38]. On the other hand, expression levels of HBI1 and BEE family members are repressed by growth-inhibition signals such as pathogen signals and abscisic acid (ABA) [38-40].
Integration with biotic and abiotic stress signals
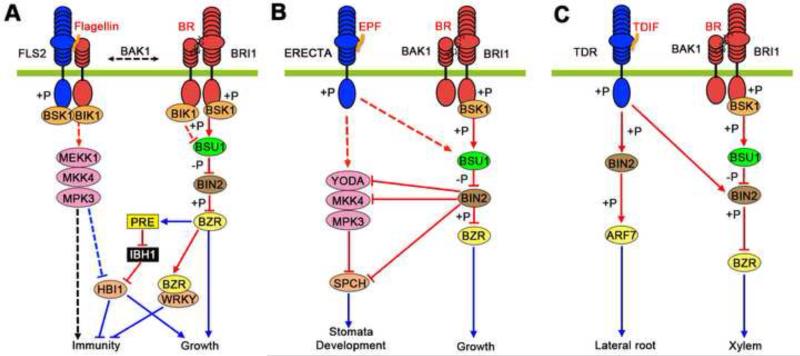
BR and the pathogen associated molecular patterns (PAMPs) antagonistically regulate growth and immunity. Many molecular connections are known between the BR pathway and the pathway activated by the PAMP signal flagellin [41]. First, BRI1 and the flagellin receptor kinase FLAGELLIN-SENSITIVE2 (FLS2) share both co-receptor kinase BAK1 and the substrates BIK1 and BSK1 kinases [42,43] (Figure 2A). The functional significance of these upstream interactions in the BR-PAMP antagonism remains questionable, mostly because flagellin showed no effect on BZR1 phosphorylation and accumulation [44-46]. In contrast, significant crosstalk has been observed at the level of transcriptional regulation [38,39,47].
Figure 2.
Crosstalks between BR/BRI1 and other RLK pathways.
A. Crosstalk between BR/BRI1 and flagellin/FLS2 pathways regulating the tradeoff between growth and immunity.
B. BR/BRI1 crosstalks with the ERECTA-MAPK pathway to regulate stomata development.
C. BR/BRI1 crosstalk with the TDIF/TDR pathway to antagonistically regulate xylem differentiation. TDIF/TDR also regulates BIN2 to promote lateral root development, independent of BR signaling.
Red lines show posttranslational activation (arrows) or inhibition (bar end) by phosphorylation (+p) or dephosphorylation (-p), and blue lines show transcriptional activation (arrows) or inhibition (bar end).
Both BZR1 and HBI1 have been shown recently to mediate transcriptional repression of immunity. BZR1 interacts with WRKY40 and activates the expression of several other WRKY transcription factors that inhibit immune responses [47]. However, the lack of a PAMP-induced effect on BZR1 level suggests that PAMP inhibition of BR-induced growth should be mediated by a component parallel or downstream of BZR1 [46]. Indeed transcription of HBI1 is rapidly repressed by PAMP signals such as flagellin and elf18 [38,39]. Overexpression of HBI1 significantly reduces the growth inhibition by flagellin and elf18, indicating that PAMP signaling inhibits growth mainly by repressing HBI1. Surprisingly, the HBI1-overexpression plants also show diminished PAMP-induced defense responses, including reactive oxygen species (ROS) production, defense gene expression, and resistance against pathogen infection [38]. The results demonstrate that HBI1 is activated by growth hormones at the protein level but repressed by PAMP signaling at the RNA level, and HBI1 in turn promotes growth and inhibits immunity. Therefore, HBI1 appears to function as a major node of crosstalk between the hormonal and PAMP signaling pathways [38,39] (Figure 1 and 2A).
Abiotic stresses activate production of ABA, which induces stress responses and inhibits plant growth. BR and ABA antagonize each other in many developmental processes, and various interactions between the two pathways have been observed at molecular levels. First, BR and ABA were shown to induce and repress, respectively, the expression of the BEEs, and genetic analysis supported a role of BEEs in the antagonistic interaction between BR and ABA [40]. Second, ABA was shown to increase phosphorylation of BZR1 [48]. Third, BIN2, the negative regulator of BR signaling, was recently shown to phosphorylate and activate the SnRK2 kinase, a positive regulator of ABA signaling [49]. Fourth, the BR-activated BES1 was shown to transcriptionally repress the ABA-signaling components ABI3 and ABI5 by recruiting the TOPLESS family of repressors [13], which appears to be a general mechanism of BR-induced transcription repression [12]. The relative contributions of these molecular mechanisms to the BR-ABA antagonism require further clarification. Genetic analyses have yielded conflicting results about the ABA-sensitivity phenotypes of the BR-hypersensitive bes1-D mutant [13,49].
Signal crosstalk with other receptor kinase pathways in specific developmental context
In addition to the flagellin/FLS2 pathway, several receptor kinase pathways have recently been shown to interact with the BR/BRI1 pathway, allowing BR regulation of specific differentiation and developmental processes.
The density and distribution of stomata at leaf surface are important for photosynthesis and water use efficiency. Stomata formation is negatively regulated by the ERECTA receptor kinase and a downstream MITOGEN-ACTIVATED PROTEIN (MAP) kinase module that inactivates the bHLH factor SPEECHLESS (SPCH), which promotes stomatal development [50]. BR inhibits stomatal development in leaves through BIN2-mediated phosphorylation and inhibition of the MAP kinase kinase kinase YODA and its substrate MAP kinase kinases (MKK4 and MKK5) [51,52]. BR also positively affects stomatal development in hypocotyl through inhibiting BIN2 phosphorylation and inactivation of SPCH [53]. Thus, BR signaling through BIN2 phosphorylation of several components of the ERECTA-MAPK-SPCH pathway inhibits and promotes stomata differentiation in different organs (Figure 2B). While BR inhibition of stomata formation in leaves is consistent with coordinated inhibition of photomorphogenic development, the biological significance of BR promotion of stomata formation in hypocotyl remains unclear. Whether ERECTA regulates the MAP kinases through BIN2 remains to be elucidated.
Two recent studies demonstrated that direct regulation of BIN2 by a receptor kinase involved in vascular tissue and lateral root development (Figure 2C). The tracheary element differentiation inhibitory factor (TDIF) is a peptide signal that activates the TDIF RECEPTOR (TDR) receptor kinase to inhibit xylem cell differentiation. In contrast, BR promotes xylem differentiation. Kondo et al showed that TDR kinase direct phosphorylates and activates BIN2, leading to phosphorylation and inactivation of BES1 [54]. As such, BR/BRI1 signaling pathway converges with the TDIF/TDR pathway at BIN2 in antagonistic regulation of xylem differentiation [54] (Figure 2C). In contrast, Cho et al, showed that, upon TDR activation, BIN2 phosphorylates the auxin response factor ARF7, to increase ARF7 activity and promote lateral root development. Intriguingly, BR promotes lateral root development independent of BIN2, whereas the TDR-BIN2-ARF7 module promotes lateral root development independent of BR signaling [55] (Figure 2C). How BIN2 performs both BR-dependent and BR-independent function remains unclear.
Conclusions
Integration of signaling pathways is crucial for the robustness of a regulatory system. Recent studies have demonstrated extensive integration of the BR signaling pathway with many other signaling pathways, through molecular interactions at levels of both signal transduction and transcriptional regulation. In particular, the BZR-ARF-PIF module coupled with the bHLH module appears to constitute a central growth regulation (CGR) circuit that integrates major hormonal and environmental signals. The CGR circuit not only reveals a central mechanism of cell elongation regulation but also provides an elegant example of how signaling pathways can be integrated to provide coordinated regulation of a common cellular response. It should be pointed out that the models presented in this review are likely overly simplified, due to space limit of the journal as well as incompleteness of our knowledge. The signal integration and outputs are likely modulated by tissue- and cell type-specific factors. In addition, the cross regulation of hormone levels is an integral part of the regulatory network that is not discussed in this review. A high degree of signal integration and high level of complexity of the growth-regulation networks is expected from the robustness and high plasticity of plant growth and development. A complete understanding of such networks will be important for improving plant productivity.
Highlights.
The BZR-ARF-PIF/DELLA module integrates major hormonal and environmental signals.
BZR-ARF-PIF/DELLA module is coupled with the PRE-IBH1-HBI1 tripartite HLH/bHLH module.
Biotic and abiotic stresses antagonize growth-promoting pathways at multiple levels.
BIN2/GSK3 mediates crosstalks between BR and other receptor kinase pathways.
The TDR receptor kinase phosphorylates BIN2 to regulate xylem and lateral root development.
Acknowledgement
This study was supported by grants from the National Institutes of Health (R01 GM066258) and the Division of Chemical Sciences, Geosciences, and Biosciences, Office of Basic Energy Sciences of the US Department of Energy (DE-FG02-08ER15973) to Z-Y.W., and a FAFU-BFBC postdoctoral fellowship from Fujian Agriculture and Forestry University to W.W.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
• of special interest
•• of outstanding interest
- 1.Wang ZY, Bai MY, Oh E, Zhu JY. Brassinosteroid signaling network and regulation of photomorphogenesis. Annu Rev Genet. 2012;46:701–724. doi: 10.1146/annurev-genet-102209-163450. [DOI] [PubMed] [Google Scholar]
- 2.Zhu JY, Sae-Seaw J, Wang ZY. Brassinosteroid signalling. Development. 2013;140:1615–1620. doi: 10.1242/dev.060590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Guo H, Li L, Aluru M, Aluru S, Yin Y. Mechanisms and networks for brassinosteroid regulated gene expression. Curr Opin Plant Biol. 2013;16:545–553. doi: 10.1016/j.pbi.2013.08.002. [DOI] [PubMed] [Google Scholar]
- 4•.Gendron JM, Liu JS, Fan M, Bai MY, Wenkel S, Springer PS, Barton MK, Wang ZY. Brassinosteroids regulate organ boundary formation in the shoot apical meristem of Arabidopsis. Proc Natl Acad Sci U S A. 2012;109:21152–21157. doi: 10.1073/pnas.1210799110. [The study, together with Ref [5•], uncovered a spatiotemporal function of BR in regulating shoot organ boundary development and shoot architecture.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bell EM, Lin WC, Husbands AY, Yu L, Jaganatha V, Jablonska B, Mangeon A, Neff MM, Girke T, Springer PS. Arabidopsis lateral organ boundaries negatively regulates brassinosteroid accumulation to limit growth in organ boundaries. Proc Natl Acad Sci U S A. 2012;109:21146–21151. doi: 10.1073/pnas.1210789109. [See annotation to Ref. [4•].] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hartwig T, Chuck GS, Fujioka S, Klempien A, Weizbauer R, Potluri DP, Choe S, Johal GS, Schulz B. Brassinosteroid control of sex determination in maize. Proc Natl Acad Sci U S A. 2011;108:19814–19819. doi: 10.1073/pnas.1108359108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Santiago J, Henzler C, Hothorn M. Molecular mechanism for plant steroid receptor activation by somatic embryogenesis co-receptor kinases. Science. 2013;341:889–892. doi: 10.1126/science.1242468. [This study, and Ref. [8], solved the crystal structure of the BR bound extracellular domains of the BRI1-BAK1 receptor complex, demonstrating ligand-induced receptor dimerization.] [DOI] [PubMed] [Google Scholar]
- 8•.Sun Y, Han Z, Tang J, Hu Z, Chai C, Zhou B, Chai J. Structure reveals that BAK1 as a co- receptor recognizes the BRI1-bound brassinolide. Cell Res. 2013;23:1326–1329. doi: 10.1038/cr.2013.131. [See annotation to Ref. [7•].] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kim TW, Guan S, Burlingame AL, Wang ZY. The CDG1 kinase mediates brassinosteroid signal transduction from BRI1 receptor kinase to BSU1 phosphatase and GSK3-like kinase BIN2. Mol Cell. 2011;43:561–571. doi: 10.1016/j.molcel.2011.05.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10•.Sun Y, Fan XY, Cao DM, Tang W, He K, Zhu JY, He JX, Bai MY, Zhu S, Oh E, et al. Integration of brassinosteroid signal transduction with the transcription network for plant growth regulation in Arabidopsis. Dev Cell. 2010;19:765–777. doi: 10.1016/j.devcel.2010.10.010. [This study identified several thousands high-confidence target genes of BZR1 using ChIP-chip.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yu X, Li L, Zola J, Aluru M, Ye H, Foudree A, Guo H, Anderson S, Aluru S, Liu P, et al. A brassinosteroid transcriptional network revealed by genome-wide identification of BESI target genes in Arabidopsis thaliana. Plant J. 2011;65:634–646. doi: 10.1111/j.1365-313X.2010.04449.x. [DOI] [PubMed] [Google Scholar]
- 12••.Oh E, Zhu JY, Ryu H, Hwang I, Wang ZY. TOPLESS mediates brassinosteroid-induced transcriptional repression through interaction with BZR1. Nat Commun. 2014;5:4140. doi: 10.1038/ncomms5140. [This study showed that the BZR1’s EAR motif recruits transcription repressor TOPLESSS and histone deacetylase to the promoters of BR-repressed genes.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13••.Ryu H, Cho H, Bae W, Hwang I. Control of early seedling development by BES1/TPL/HDA19-mediated epigenetic regulation of ABI3. Nat Commun. 2014;5:4138. doi: 10.1038/ncomms5138. [This study showed that the BES1 recruits transcription repressor TOPLESS and histone de acetylase to the promoter of ABI3, to repress ABI3 expression and ABA responses.] [DOI] [PubMed] [Google Scholar]
- 14.Fan XY, Sun Y, Cao DM, Bai MY, Luo XM, Yang HJ, Wei CQ, Zhu SW, Chong K, Wang ZY. BZS1, a B-box protein, promotes photomorphogenesis downstream of both brassinosteroid and light signaling pathways. Mol Plant. 2012;5:591–600. doi: 10.1093/mp/sss041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Luo XM, Lin WH, Zhu S, Zhu JY, Sun Y, Fan XY, Cheng M, Hao Y, Oh E, Tian M, et al. Integration of light-and brassinosteroid-signaling pathways by a GATA transcription factor in Arabidopsis. Dev Cell. 2010;19:872–883. doi: 10.1016/j.devcel.2010.10.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ni W, Xu SL, Chalkley RJ, Pham TN, Guan S, Maltby DA, Burlingame AL, Wang ZY, Quail PH. Multisite light-induced phosphorylation of the transcription factor PIF3 is necessary for both its rapid degradation and concomitant negative feedback modulation of photoreceptor phyB levels in Arabidopsis. Plant Cell. 2013;25:2679–2698. doi: 10.1105/tpc.113.112342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17••.Oh E, Zhu JY, Wang ZY. Interaction between BZR1 and PIF4 integrates brassinosteroid and environmental responses. Nat Cell Biol. 2012;14:802–809. doi: 10.1038/ncb2545. [The study discovered BZR1-PIF4 interaction as a key mechanism of integrating BR signaling with light and temperature responses. The study identified a large gene set co-regulated BZR1 and PIF4.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Leivar P, Quail PH. PIFs: pivotal components in a cellular signaling hub. Trends Plant Sci. 2011;16:19–28. doi: 10.1016/j.tplants.2010.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Goda H, Shimada Y, Fujioka S, Yoshida S. Classification of brassinosteroid-regulated genes based on expression profiles in bri1 and in response to a protein kinase inhibitor, staurosporin. Biosci Biotechnol Biochem. 2004;68:1605–1607. doi: 10.1271/bbb.68.1605. [DOI] [PubMed] [Google Scholar]
- 20.Nemhauser JL, Mockler TC, Chory J. Interdependency of brassinosteroid and auxin signaling in Arabidopsis. PLoS Biol. 2004;2:E258. doi: 10.1371/journal.pbio.0020258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.de Lucas M, Prat S. PIFs get BRright: PHYTOCHROME INTERACTING FACTORs as integrators of light and hormonal signals. New Phytol. 2014;202:1126–1141. doi: 10.1111/nph.12725. [DOI] [PubMed] [Google Scholar]
- 22.Hornitschek P, Kohnen MV, Lorrain S, Rougemont J, Ljung K, Lopez-Vidriero I, Franco-Zorrilla JM, Solano R, Trevisan M, Pradervand S, et al. Phytochrome interacting factors 4 and 5 control seedling growth in changing light conditions by directly controlling auxin signaling. Plant J. 2012;71:699–711. doi: 10.1111/j.1365-313X.2012.05033.x. [DOI] [PubMed] [Google Scholar]
- 23••.Oh E, Zhu JY, Bai MY, Arenhart RA, Sun Y, Wang ZY. Cell elongation is regulated through a central circuit of interacting transcription factors in the Arabidopsis hypocotyl. Elife. 2014;e03031 doi: 10.7554/eLife.03031. [This study identified large number of genomic targets of ARF6, and showed co-regulation of gene expression and cell elongation through direct interaction of ARF6 with BZR1 and PIF4, and DELLA, components of the BR, phytochrome, and GA pathways. The study reveals a central mechanism of cell elongation regulation in plants.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24•.de Lucas M, Daviere JM, Rodriguez-Falcon M, Pontin M, Iglesias-Pedraz JM, Lorrain S, Fankhauser C, Blazquez MA, Titarenko E, Prat S. A molecular framework for light and gibberellin control of cell elongation. Nature. 2008;451:480–484. doi: 10.1038/nature06520. [This study and Ref [24] showed DELLA interaction with PIFs.] [DOI] [PubMed] [Google Scholar]
- 25•.Feng S, Martinez C, Gusmaroli G, Wang Y, Zhou J, Wang F, Chen L, Yu L, Iglesias-Pedraz JM, Kircher S, et al. Coordinated regulation of Arabidopsis thaliana development by light and gibberellins. Nature. 2008;451:475–479. doi: 10.1038/nature06448. [See annotation to Ref. [23•].] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26••.Bai MY, Shang JX, Oh E, Fan M, Bai Y, Zentella R, Sun TP, Wang ZY. Brassinosteroid, gibberellin and phytochrome impinge on a common transcription module in Arabidopsis. Nat Cell Biol. 2012;14:810–817. doi: 10.1038/ncb2546. [Together with Ref. [26•• and 27••] this study showed DELLA-BZR-PIF module as a key mechanism for integrating BR, GA and phytochrome pathways.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- ••27.Li QF, Wang C, Jiang L, Li S, Sun SS, He JX. An interaction between BZR1 and DELLAs mediates direct signaling crosstalk between brassinosteroids and gibberellins in Arabidopsis. Sci Signal. 2012;5:ra72. doi: 10.1126/scisignal.2002908. [See annotation to Ref. [25••].] [DOI] [PubMed] [Google Scholar]
- 28••.Gallego-Bartolome J, Minguet EG, Grau-Enguix F, Abbas M, Locascio A, Thomas SG, Alabadi D, Blazquez MA. Molecular mechanism for the interaction between gibberellin and brassinosteroid signaling pathways in Arabidopsis. Proc Natl Acad Sci U S A. 2012;109:13446–13451. doi: 10.1073/pnas.1119992109. [See annotation to Ref. [25••].] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Stirnberg P, Furner IJ, Ottoline Leyser HM. MAX2 participates in an SCF complex which acts locally at the node to suppress shoot branching. Plant J. 2007;50:80–94. doi: 10.1111/j.1365-313X.2007.03032.x. [DOI] [PubMed] [Google Scholar]
- 30.Shen H, Zhu L, Bu QY, Huq E. MAX2 affects multiple hormones to promote photomorphogenesis. Mol Plant. 2012;5:750–762. doi: 10.1093/mp/sss029. [DOI] [PubMed] [Google Scholar]
- 31•.Wang Y, Sun S, Zhu W, Jia K, Yang H, Wang X. Strigolactone/MAX2-induced degradation of brassinosteroid transcriptional effector BES1 regulates shoot branching. Dev Cell. 2013;27:681–688. doi: 10.1016/j.devcel.2013.11.010. [This study showed strigolactone regulation of the ubiquitination and degradation of BES1 protein through MAX2-BES1 interaction, and a function of BES1 in shoot branching.] [DOI] [PubMed] [Google Scholar]
- 32.Jia KP, Luo Q, He SB, Lu XD, Yang HQ. Strigolactone-regulated hypocotyl elongation is dependent on cryptochrome and phytochrome signaling pathways in Arabidopsis. Mol Plant. 2014;7:528–540. doi: 10.1093/mp/sst093. [DOI] [PubMed] [Google Scholar]
- 33•.Bai MY, Fan M, Oh E, Wang ZY. A triple helix-loop-helix/basic helix-loop-helix cascade controls cell elongation downstream of multiple hormonal and environmental signaling pathways in Arabidopsis. Plant Cell. 2012;24:4917–4929. doi: 10.1105/tpc.112.105163. [This study, together with Ref. [33], showed IBH1’s function as a non-DNA-binding factor that inhibits DNA-binding of the HBI1/ACE family of bHLH factors, demonstrating the PRE IBH1-HBI1/ACE tripartite cascade and its essential function downstream of the BZR PIF/DELLA module.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34•.Ikeda M, Fujiwara S, Mitsuda N, Ohme-Takagi M. A triantagonistic basic helix-loop-helix system regulates cell elongation in Arabidopsis. Plant Cell. 2012;24:4483–4497. doi: 10.1105/tpc.112.105023. [See annotation to Ref. [32••].] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hornitschek P, Lorrain S, Zoete V, Michielin O, Fankhauser C. Inhibition of the shade avoidance response by formation of non-DNA binding bHLH heterodimers. EMBO J. 2009;28:3893–3902. doi: 10.1038/emboj.2009.306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36•.Hao Y, Oh E, Choi G, Liang Z, Wang ZY. Interactions between HLH and bHLH factors modulate light-regulated plant development. Mol Plant. 2012;5:688–697. doi: 10.1093/mp/sss011. [This study demonstrated that direct interaction between PAR1 and PIF4 inhibit PIF4 DNA binding.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cifuentes-Esquivel N, Bou-Torrent J, Galstyan A, Gallemi M, Sessa G, Salla Martret M, Roig-Villanova I, Ruberti I, Martinez-Garcia JF. The bHLH proteins BEE and BIM positively modulate the shade avoidance syndrome in Arabidopsis seedlings. Plant J. 2013;75:989–1002. doi: 10.1111/tpj.12264. [DOI] [PubMed] [Google Scholar]
- 38••.Fan M, Bai MY, Kim JG, Wang T, Oh E, Chen L, Park CH, Son SH, Kim SK, Mudgett MB, et al. The bHLH Transcription Factor HBI1 Mediates the Trade-Off between Growth and Pathogen-Associated Molecular Pattern-Triggered Immunity in Arabidopsis. Plant Cell. 2014;26:828–841. doi: 10.1105/tpc.113.121111. [This study, together with Ref [38], showed the function of HBI1 in promoting growth and inhibiting immunity, and thus the tradeoff between BR-promoted growth and PAMP-induced immunity. This study also showed that HBI1 has overlapping yet distinct functions compared to PIFs.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39••.Malinovsky FG, Batoux M, Schwessinger B, Youn JH, Stransfeld L, Win J, Kim SK, Zipfel C. Antagonistic Regulation of Growth and Immunity by the Arabidopsis Basic Helix- Loop-Helix Transcription Factor HOMOLOG OF BRASSINOSTEROID ENHANCED EXPRESSION2 INTERACTING WITH INCREASED LEAF INCLINATION1 BINDING bHLH1. Plant Physiol. 2014;164:1443–1455. doi: 10.1104/pp.113.234625. [See annotation to Ref. [37••].] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Friedrichsen DM, Nemhauser J, Muramitsu T, Maloof JN, Alonso J, Ecker JR, Furuya M, Chory J. Three redundant brassinosteroid early response genes encode putative bHLH transcription factors required for normal growth. Genetics. 2002;162:1445–1456. doi: 10.1093/genetics/162.3.1445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wang W, Wang ZY. At the intersection of plant growth and immunity. Cell Host Microbe. 2014;15:400–402. doi: 10.1016/j.chom.2014.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42•.Lin W, Lu D, Gao X, Jiang S, Ma X, Wang Z, Mengiste T, He P, Shan L. Inverse modulation of plant immune and brassinosteroid signaling pathways by the receptor- like cytoplasmic kinase BIK1. Proc Natl Acad Sci U S A. 2013;110:12114–12119. doi: 10.1073/pnas.1302154110. [This study showed that BIK1 is a substrate of BRI1 but plays an inhibitory role in BR responses.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43•.Shi H, Shen Q, Qi Y, Yan H, Nie H, Chen Y, Zhao T, Katagiri F, Tang D. BR-SIGNALING KINASE1 physically associates with FLAGELLIN SENSING2 and regulates plant innate immunity in Arabidopsis. Plant Cell. 2013;25:1143–1157. doi: 10.1105/tpc.112.107904. [This study showed a role of the BR-signaling kinase in flagellin signaling.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chinchilla D, Shan L, He P, de Vries S, Kemmerling B. One for all: the receptor- associated kinase BAK1. Trends Plant Sci. 2009;14:535–541. doi: 10.1016/j.tplants.2009.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Belkhadir Y, Jaillais Y, Epple P, Balsemao-Pires E, Dangl JL, Chory J. Brassinosteroids modulate the efficiency of plant immune responses to microbe-associated molecular patterns. Proc Natl Acad Sci U S A. 2012;109:297–302. doi: 10.1073/pnas.1112840108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Albrecht C, Boutrot F, Segonzac C, Schwessinger B, Gimenez-Ibanez S, Chinchilla D, Rathjen JP, de Vries SC, Zipfel C. Brassinosteroids inhibit pathogen-associated molecular pattern-triggered immune signaling independent of the receptor kinase BAK1. Proc Natl Acad Sci U S A. 2012;109:303–308. doi: 10.1073/pnas.1109921108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47••.Lozano-Duran R, Macho AP, Boutrot F, Segonzac C, Somssich IE, Zipfel C. The transcriptional regulator BZR1 mediates trade-off between plant innate immunity and growth. Elife. 2013;2:e00983. doi: 10.7554/eLife.00983. [This study reported direct BZR1 regulation of genes involved in immunity.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhang S, Cai Z, Wang X. The primary signaling outputs of brassinosteroids are regulated by abscisic acid signaling. Proc Natl Acad Sci U S A. 2009;106:4543–4548. doi: 10.1073/pnas.0900349106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49••.Cai Z, Liu J, Wang H, Yang C, Chen Y, Li Y, Pan S, Dong R, Tang G, Barajas-Lopez Jde D, et al. GSK3-like kinases positively modulate abscisic acid signaling through phosphorylating subgroup III SnRK2s in Arabidopsis. Proc Natl Acad Sci U S A. 2014;111:9651–9656. doi: 10.1073/pnas.1316717111. [This study showed that the BIN2/GSK3 kinases phosphorylate the SnRK2s to activate ABA responses.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Dong J, Bergmann DC. Stomatal patterning and development. Curr Top Dev Biol. 2010;91:267–297. doi: 10.1016/S0070-2153(10)91009-0. [DOI] [PubMed] [Google Scholar]
- 51••.Kim TW, Michniewicz M, Bergmann DC, Wang ZY. Brassinosteroid regulates stomatal development by GSK3-mediated inhibition of a MAPK pathway. Nature. 2012;482:419–422. doi: 10.1038/nature10794. [This study showed that the BIN2/GSK3 kinase phosphorylates the MAP kinase kinase kinase YODA to inhibit stomata development.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52•.Khan M, Rozhon W, Bigeard J, Pflieger D, Husar S, Pitzschke A, Teige M, Jonak C, Hirt H, Poppenberger B. Brassinosteroid-regulated GSK3/Shaggy-like kinases phosphorylate mitogen-activated protein (MAP) kinase kinases, which control stomata development in Arabidopsis thaliana. J Biol Chem. 2013;288:7519–7527. doi: 10.1074/jbc.M112.384453. [Consistent with Ref [51••], this study showed that BIN2/GSK3 kinase phosphorylates the MAP kinase kinase MKK4 and MKK5 to inhibit stomata development.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53•.Gudesblat GE, Schneider-Pizon J, Betti C, Mayerhofer J, Vanhoutte I, van Dongen W, Boeren S, Zhiponova M, de Vries S, Jonak C, et al. SPEECHLESS integrates brassinosteroid and stomata signalling pathways. Nat Cell Biol. 2012;14:548–554. doi: 10.1038/ncb2471. [This study showed BR promotion of stomata development in hypocotyls through BIN2 phosphorylation of SPCH.] [DOI] [PubMed] [Google Scholar]
- 54••.Kondo Y, Ito T, Nakagami H, Hirakawa Y, Saito M, Tamaki T, Shirasu K, Fukuda H. Plant GSK3 proteins regulate xylem cell differentiation downstream of TDIF-TDR signalling. Nat Commun. 2014;5:3504. doi: 10.1038/ncomms4504. [This study reported direct regulation of BIN2 by the TDIF-TDR receptor kinase, leading to inactivation of BES1 and inhibition of xylem formation. The BR and TDR pathways converge though antagonistic regulation of BIN2, to co-regulate xylem differentiation.] [DOI] [PubMed] [Google Scholar]
- 55•.Cho H, Ryu H, Rho S, Hill K, Smith S, Audenaert D, Park J, Han S, Beeckman T, Bennett MJ, et al. A secreted peptide acts on BIN2-mediated phosphorylation of ARFs to potentiate auxin response during lateral root development. Nat Cell Biol. 2014;16:66–76. doi: 10.1038/ncb2893. [This study showed that BIN2 phosphorylation and activation of ARF7 promotes lateral root growth, downstream of TDIF-TDR but independent of BR signaling.] [DOI] [PubMed] [Google Scholar]