Abstract
Apico-basal polarity is the defining characteristic of epithelial cells. In Drosophila, apical membrane identity is established and regulated through interactions between the highly conserved Par complex (Bazooka/Par3, atypical protein kinase C and Par6), and the Crumbs complex (Crumbs, Stardust and PATJ). It has been proposed that Bazooka operates at the top of a genetic hierarchy in the establishment and maintenance of apico-basal polarity. However, there is still ambiguity over the correct sequence of events and cross-talk with other pathways during this process. In this study, we reassess this issue by comparing the phenotypes of the commonly used baz4 and baz815-8 alleles with those of the so far uncharacterized bazXR11 and bazEH747 null alleles in different Drosophila epithelia. While all these baz alleles display identical phenotypes during embryonic epithelial development, we observe strong discrepancies in the severity and penetrance of polarity defects in the follicular epithelium: polarity is mostly normal in bazEH747 and bazXR11 while baz4 and baz815-8 show loss of polarity, severe multilayering and loss of epithelial integrity throughout the clones. Further analysis reveals that the chromosomes carrying the baz4 and baz815-8 alleles may contain additional mutations that enhance the true baz loss-of-function phenotype in the follicular epithelium. This study clearly shows that Baz is dispensable for the regulation of polarity in the follicular epithelium, and that the requirement for key regulators of cell polarity is highly dependent on developmental context and cell type.
Keywords: Polarity, Par complex, Epithelia
Introduction
The defining characteristic of epithelial cells in all metazoans is apico-basal polarity. Its establishment and maintenance is essential for multiple cellular processes including cell-cell adhesion, cell-matrix adhesion, protein trafficking, cell shape and the regulation of growth and apoptosis. Given its vital role in cellular homeostasis, it comes as no surprise that defects in cell polarity can cause severe cellular disorders that have been directly linked to numerous diseases ranging from cancer to kidney dysfunction to blindness (Martin-Belmonte and Perez-Moreno, 2012; Tepass, 2012; Rodriguez-Boulan and Macara, 2014).
Apico-basal polarity in epithelial cells manifests upon the assembly of polarized cytoskeletal networks, polarized trafficking of vesicles and cargo and the localization of adhesion complexes to specific cortical locations. This causes division of the epithelial plasma membrane into distinct apical, basal, and lateral domains, typically enriched for specific phospholipid components and distinct, yet highly conserved, protein complexes (Johnson and Wodarz, 2003; McCaffrey and Macara, 2011; Tepass, 2012; Rodriguez-Boulan and Macara, 2014).
In Drosophila epithelial cells, the apical membrane is subdivided into a free apical membrane and a slightly basal subapical region (SAR) (Bilder et al., 2000; Tepass et al., 2001; St Johnston and Ahringer, 2010; Laprise and Tepass, 2011). The SAR is occupied by the Crumbs complex, composed of Crumbs (Crb), Stardust (Sdt), PATJ and Lin7, and the Par complex consisting of atypical protein kinase C (aPKC), Par6 and Bazooka/Par3 (Baz). These protein complexes are crucial for the establishment and maintenance of the apical plasma membrane domain (Bilder et al., 2003; Tanentzapf and Tepass, 2003; Harris and Tepass, 2008; Franz and Riechmann, 2010). The Drosophila basolateral plasma membrane is subdivided into the adherens junctions (AJs) or zonula adherens (ZA), the lateral membrane and the basal membrane. The AJs are key mediators of intercellular adhesion and lie just basal to the SAR. The core of the AJs is formed by the Cadherin-Catenin complex, composed of DE-cadherin (DE-cad), Armadillo/beta-catenin (Arm) and alpha-catenin. In addition, Baz as well as the immunoglobulin like adhesion molecule Echinoid (Ed) and its intracellular actin binding partner Canoe (Cno) localize to the AJs (Müller and Wieschaus, 1996; Wei et al., 2005; Harris and Tepass, 2010; Desai et al., 2013).
A second type of intercellular junction, the septate junction (SJ), also localizes to the lateral membrane. Associated with the SJ are the tumor suppressor proteins Lethal giant larvae (Lgl), Discs large (Dlg), Scribble (Scrib) and Fasciclin III (Bilder et al., 2000; Bilder et al., 2003; Tanentzapf and Tepass, 2003). The basal membrane is characterized by the localization of extracellular matrix receptors, namely integrins and Dystroglycan (Dg) (Tanentzapf et al., 2000; Schneider et al., 2006; Denef et al., 2008). Decades of research have revealed that these different regulatory protein complexes interact in an elaborate, yet highly conserved feedback loop to establish and maintain epithelial polarity (Bilder et al., 2003; St Johnston and Ahringer, 2010; Laprise and Tepass, 2011; Rodriguez-Boulan and Macara, 2014).
Epithelia in Drosophila can be distinguished into primary epithelia which derive from the embryonic blastoderm epithelium, and secondary epithelia which are generated by mesenchymal-epithelial transitions (Tepass et al., 2001). Epithelium formation in the Drosophila embryo occurs through a modified form of cytokinesis termed “cellularization”. Following fertilization, the Drosophila embryo undergoes 13 rounds of nuclear divisions without cytokinesis to form a syncytium comprised of roughly 6000 nuclei. Most of these nuclei align just below the embryonic surface where they become surrounded by plasma membrane invaginations to generate a uniform, highly polarized epithelium (Tepass et al., 2001; Harris, 2012; Choi et al., 2013). Several studies have shown that Baz plays a key role in the establishment and maintenance of apico-basal polarity during cellularization (Bilder et al., 2003; Harris and Peifer, 2004; Harris, 2012; Choi et al., 2013). At the onset of cellularization, localization of Baz to the apical circumference is mediated by Dynein and is mutually dependent on the actin-junctional linker, Cno. During cellularization, the localization and formation of AJs, as well as the apical localization of aPKC, Par6 (Harris and Peifer, 2005; Harris and Peifer, 2007) and Crb (Bilder et al., 2003) require Baz function. Baz is also crucial for zygotic epithelial development, as in its absence neuroectodermal cells lose apico-basal polarity, resulting in the formation of large holes in the ventral epidermis (Harris and Tepass, 2008).
While processes identical to cellularization have not been described in mammals, several similarities exist between the polarization of mammalian cells and secondary epithelia in Drosophila (Goldstein and Macara, 2007; St Johnston and Ahringer, 2010; Tepass, 2012; Rodriguez-Boulan and Macara, 2014). The follicular epithelium (FE) of Drosophila is an excellent example of a secondary epithelium. Somatic stem cells present in the germarium of the ovary divide asymmetrically to generate mesenchymal progenitors which generate FE cells via a mesenchymal-to-epithelial transition (Margolis and Spradling, 1995; Wu et al., 2008). Unlike the cellularizing embryo, which relies exclusively on apical cues for its polarization, the polarization of FE cells depends on apical, lateral and basal cues (Tanentzapf et al., 2000).
In spite of the phenotypic disparities between the developing FE and the cellularizing embryo, in-depth studies reveal stark similarities in the underlying molecular mechanisms and protein interactions regulating the polarization of both tissues (Tanentzapf et al., 2000; Franz and Riechmann, 2010; Morais-de-Sá et al., 2010; Rodriguez-Boulan and Macara, 2014). As in the cellularizing embryo, Baz plays a key role in the polarization of FE cells (Franz and Riechmann, 2010). During the early stages of polarization, Baz localizes to both the AJs and the apical membrane (Morais-de-Sá et al., 2010). At the AJs, it binds to Arm and Ed (Wei et al., 2005) and is required for their correct positioning (Franz and Riechmann, 2010; Morais-de-Sá et al., 2010). In the absence of Arm, Baz continues to localize to the cortex but extends ectopically into the basolateral domain and is distributed in clumps along the apical membrane, as opposed to forming a uniform belt (Franz and Riechmann, 2010). Thus, Baz and AJ components are mutually dependent on each other for positioning to their appropriate membrane domains.
Aside from these interactions, Baz also binds to Sdt, aPKC and Par6 and recruits them together with Crb to the apical domain as the development proceeds (Franz and Riechmann, 2010; Krahn et al., 2010b; Morais-de-Sá et al., 2010). Clonal analysis reveals that in early and late stage baz mutant FE clones, aPKC, Par6, Crb and Sdt fail to localize cortically (Franz and Riechmann, 2010; Morais-de-Sá et al., 2010). On the other hand, while apical localization of Baz is lost in early stage crb, aPKC and par6 mutant clones, the AJ pool remains unaffected (Morais-de-Sá et al., 2010). Thus, according to the current model for establishment of apico-basal polarity in the FE, Baz functions at the top of a genetic hierarchy in the specification of the epithelial apical membrane (Franz and Riechmann, 2010).
As epithelia mature, apical Baz localization is gradually lost with the remaining protein localizing predominantly to the AJs while aPKC, Par6, Sdt and Crb colocalize at the SAR (Morais-de-Sá et al., 2010). Recent studies have shown that exclusion of Baz from the SAR is a key step in the establishment and maintenance of the apical membrane. Binding of Baz to aPKC and Sdt is inhibited by the phosphorylation of Baz by aPKC at Ser-980, while Crb outcompetes Baz for binding to Par6 (Morais-de-Sá et al., 2010; Walther and Pichaud, 2010; Krahn et al., 2010b; Laprise and Tepass, 2011). These dynamic and multifaceted interactions function together to facilitate the displacement of Baz from the SAR and promote its localization to the ZA (Harris and Peifer, 2005; McGill et al., 2009; Krahn et al., 2010b; Morais-de-Sá et al., 2010; Walther and Pichaud, 2010; Laprise and Tepass, 2011). The essential requirement for the exclusion of Baz from the SAR is reflected in the severe polarity defects caused by expression of a version of Baz that cannot be phosphorylated at Ser-980 and thus remains bound to aPKC and Sdt. FE cells expressing such a mutated version of Baz show persistent apical colocalization of Baz with aPKC, Par6, Crb and Sdt as well as relocalization of AJ proteins to the apical membrane (Krahn et al., 2010b; Morais-de-Sá et al., 2010).
Aside from its prominent role in the establishment and maintenance of apico-basal polarity in Drosophila epithelial cells, Baz plays key roles in various other cell types. It is required for the polarization of the developing oocyte and maintenance of oocyte cell fate (Huynh et al., 2001; Pinal et al., 2006; Becalska and Gavis, 2010). In neuroblasts, it functions in a complex with aPKC, Par6 and Inscuteable (Insc) to promote asymmetric cell division through the establishment of cortical polarity and regulation of spindle orientation (Wodarz et al., 1999; Schober et al., 1999; Wodarz et al., 2000; Petronczki and Knoblich, 2001; Rolls et al., 2003; Atwood et al., 2007; Hartenstein and Wodarz, 2013).
Most studies investigating the function of Baz in Drosophila development, including those characterizing its function in cellularization (Müller and Wieschaus, 1996; Bilder et al., 2003; Tanentzapf and Tepass, 2003; Harris and Peifer, 2004), zygotic epithelial development (Müller and Wieschaus, 1996; Bilder et al., 2003; Tanentzapf and Tepass, 2003; Harris and Tepass, 2008), embryonic neuroblasts (Schober et al., 1999; Wodarz et al., 1999; Atwood et al., 2007), oocyte polarity (Cox et al., 2001; Huynh et al., 2001; Doerflinger et al., 2010; Becalska and Gavis, 2010) and FE development (Abdelilah-Seyfried et al., 2003; Franz and Riechmann, 2010; Morais-de-Sá et al., 2010) have been conducted using the baz4 (also known as bazxi106) and the baz815-8 alleles. These alleles were considered null alleles, as they contain stop codons causing deletion of more than two thirds of the Baz protein (Krahn et al., 2010a).
In this study, we reassess the function of Baz in various developmental contexts using two previously generated yet phenotypically uncharacterized alleles, bazEH747 and bazXR11. These two alleles are bona fide null alleles that do not give rise to any detectable Baz protein. We find that all baz alleles show similar defects in all developmental contexts assessed, except for in the FE. Here, bazEH747 and bazXR11 FE clones display normal apico-basal polarity unlike baz4 and baz815-8 clones, which frequently lose polarity and cause gaps in the epithelium. Our data indicates that the baz4 and baz815-8 chromosomes may carry additional mutations which strongly enhance the baz null phenotype. In contrast to previous reports, we show that Baz is not required for the correct positioning of AJs or the apical localization of aPKC, Par6, Sdt or Crb in the FE. Furthermore, apical plasma membrane domain formation also occurs normally in its absence. A genetic interaction screen reveals that the baz null FE phenotype is strongly enhanced by a reduction in Crb levels.
bazEH747 and bazXR11 FE clones show highly penetrant posterior follicular cell (PFC) multilayering, but the severity of multilayering is much milder than in clones for baz4 and baz815-8. PFC multilayering is commonly observed in mutants affecting Hippo and Notch signaling, pointing to a potential involvement of Baz in these signaling pathways.
Overall, our results infer an auxiliary role for Baz in the polarization of FE cells and challenge the current model, which places it at the top of a genetic hierarchy in this process.
MATERIALS AND METHODS
Fly stocks and genetics
The following mutant alleles were used in this study: baz4 FRT19A (gift from Chris Doe), bazEH747 FRT19A (Eberl and Hilliker, 1988), bazXR11 FRT19A (Ralf Stanewsky, unpublished), baz815–8 FRT19A (McKim et al., 1996), aPKCK06403 FRT42B (Wodarz et al., 2000; Rolls et al., 2003), crb11A22 FRT82B (Tepass and Knust, 1993), lgl4 (Hanratty, 1984), par1W3 FRT42B (Shulman et al., 2000), scrib2 FRT82B (Bilder et al., 2003), PP2AGE16781 (GenExel) (Krahn et al., 2009), PTENdj189 (Gao et al., 2000), shgR69 FRT42B (Godt and Tepass, 1998), FRT19A, Ubi-GFP FRT19A; e22c-gal4 UAS-Flp, e22c-gal4 UAS-Flp; FRT82B Ubi-GFP (gift from Trudi Schupbach), par6Δ226 FRT19A and cdc424 FRT19A (Jones and Metzstein, 2011), ed1x5 FRT40A (Wei et al., 2005), cnoR2 FRT82B (Sawyer et al., 2009), UAS-baz-GFP, UAS-baz-Δ312-1464-GFP, UAS-baz-Δ1-311-GFP (Benton and St Johnston, 2003), FRT19A tubP-Gal80LL1 hsFLP; tubP-gal4 UAS-mCD8-GFP (gift from Heinrich Reichert), yki-RNAi v104523 (Vienna Drosophila Resource Center), baz-GFP-trap CC01941 (Buszczak et al., 2007), tj-GAL4 (Hayashi et al., 2002). The following stocks were obtained from the Bloomington Stock Center: hsFlp122 FRT19A H2AvD-GFP (#32045), FRT19A (#1709), FRT19A ovoD (#23880), ovo-flp (#8705), GFP-RNAi (#41556). The following lines were generated for this study: bazEH747 FRT19A;;UAS-baz-GFP, bazEH747 FRT19A;;UAS-baz-Δ1-311-GFP, bazEH747 FRT19A;;UAS-baz-Δ312-1464-GFP, and baz4 FRT19A;;UAS-baz-GFP.
Generation of antibodies against Baz, Par6 and Lgl
An antibody directed against the three PDZ domains of Baz was generated by immunizing guinea pigs with a GST fusion protein containing amino acids 309–747 of Baz (Eurogentec, Seraing, Belgium). An antibody directed against Par6 was generated by immunizing rats with the peptide CHHQQAASNASTIMASDVKDGVLHL (Eurogentec, Seraing, Belgium). An antibody directed against Lgl was generated by immunizing guinea pigs with the peptides KGQQPSADRHRLQKDC and CNKIGTPKTAPEESQF (Eurogentec, Seraing, Belgium).
Fixation of Drosophila embryos, larval brains and adult ovaries
Embryos were collected overnight at 25°C and dechorionated in bleach for 5 min. Embryos were washed with distilled water, transferred into a 1:1 mixture of heptane and 4% formaldehyde in PBS buffer and were then fixed by vigorous shaking for 30 min. Formaldehyde and PBS were removed and methanol was added to form a 1:2 mixture of heptane and methanol. Embryos were then devitellinized by vigorous shaking for 30 sec. After removing most of the liquid, devitellinized embryos were rinsed twice with methanol, incubated in methanol for 10 min and then rinsed twice with ethanol and stored at −20°C in ethanol for future use. For adult ovaries and larval brains, flies were reared at 25°C, dissected and fixed in Phosphate buffered saline (pH 7.4) (PBS), and fixed in 4% formaldehyde/PBS for 20 min. Fixed samples were washed 3 times in 0.1% Triton X-100/PBS (PBT) for 20 minutes and ovaries were then dissociated by pipetting up and down using a 1 ml pipette tip.
Immunostaining of Drosophila embryos, larval brains and adult ovaries
Fixed ovaries or brains were blocked in 5% Normal Horse Serum (NHS)/1% Triton X-100/PBS for 30 min. Blocked ovaries and brains were rinsed 3 times in PBS to remove excess Triton X-100 and then incubated overnight at 4°C in 5% NHS/PBT with primary antibodies. Embryos were blocked in 5% NHS/PBT for 30 min and then incubated overnight at 4°C in 5% NHS/PBT with primary antibodies. Samples were washed 3 times for 20 min in PBT at room temperature. Samples were then blocked with 5% NHS/PBT for 30 min and incubated with secondary antibodies and DAPI in 5% NHS/PBT at 1:500 for 2 hours at room temperature. Secondary antibody solution was then removed and samples were washed 3 times for 20 min with PBT. Samples stained with fluorescent-conjugated secondary antibodies were mounted in Vectashield (Vector Laboratories, Burlingame, CA, USA) or Mowiol. Confocal microscopy was performed using a Zeiss LSM510 Meta confocal microscope. Images were processed on Photoshop CS3 (Adobe, San Jose, CA, USA) and assembled using Illustrator CS3 (Adobe, San Jose, CA, USA). The primary antibodies used were rabbit anti-Baz N-term (1:1000, Wodarz et al., 1999), guinea pig anti-Baz PDZ (1:1000), rat anti-Par6 (1:500), guinea pig anti-Mira (1:1000, Kim et al., 2009), guinea pig anti-Lgl (1:500), rat anti-Crb (1:500, gift from U. Tepass), mouse anti-Sdt (1:25, gift from E. Knust), rabbit anti-Cno (1:500, gift from M. Pfeifer), rabbit anti-lacZ (1:1000, MP Biosciences), rabbit anti-aPKC (1:500, Santa Cruz Biotechnology), mouse anti-GFP A11120 (1:1000, Molecular Probes), rabbit anti-GFP A11122 (1:1000, Molecular Probes). The following antibodies were obtained from the Developmental Studies Hybridoma Bank: rat anti-DE-cadherin DCAD2 (1:500), mouse anti-Dlg 4F3 (1:25), mouse anti-Orb 4H8 (1:20) and mouse anti-Armadillo N2 7A1 (1:50).
Generation of FE clones
For negatively and positively marked FE clones generated under the control of hsFlp, animals of the appropriate genotype were heat shocked 5 days after egg laying (AEL) for 2 hours at 37°C on two consecutive days. Newly emerged flies were transferred to yeasted vials and allowed to mate for 4–5 days before dissection. To generate targeted mosaics in the FE, UAS-Flp was expressed under the control of e22c-Gal4 which is expressed in the somatic follicular stem cells. Newly emerged flies were transferred to yeasted vials and allowed to mate for 10 days before dissection. To generate maternal/zygotic clones, the dominant female sterile technique was used. When using hsFlp to drive recombination, animals of the appropriate genotype were heat shocked for 2 hours at 37°C 2 days in a row 1–2 days AEL and adult females of the desired genotype were collected. When using Ovo-Flp to drive recombination, flies of the appropriate genotype were crossed and adult females of the correct genotype were selected.
SDS PAGE and western blot
Tissue was homogenized with a fitted pestle in a microcentrifuge tube in Tris-Sodium-Triton buffer (50 mM Tris pH 6.8, 150 mM NaCl, 1% Triton X-100) (TNT) and the concentration was quantified using the BCA Protein Assay Kit (Pierce, Thermo Fisher Scientific, MA, USA) according to the manufacturers instructions. Equal amounts of protein were diluted in 2× Laemmli Buffer (100 mM Tris pH 6.8, 200 mM DTT, 20% glycerol, 0.01% bromophenol blue and 4% SDS), boiled for 5 minutes, and subjected to electrophoresis in an SDS polyacrylamide gel for 60 min at 200 V using a BioRad electrophoresis apparatus. Proteins were transferred onto a nitrocellulose membrane for 60 min at 100 V; membranes were then blocked in 5% skim milk in TBST (Tris-buffered saline, 0.05% Tween-20) for 1 hour at room temperature and incubated with appropriate antibodies at 4°C overnight. Membranes were washed with TBST and incubated with appropriate horseradish peroxidase-labeled secondary antibody (Jackson ImmunoResearch, PA, USA) at a dilution of 1:5000 for 1 hour at room temperature. Membranes were then washed 3 times for 20 min in TBST. For chemiluminescent protein detection, membranes were incubated with BM Chemiluminescent substrate (Roche Diagnostics, Basel, Switzerland), exposed to photographic film and developed using Kodak developer. Blots were stripped by incubating them in stripping buffer (62.5 mM Tris-HCl pH 6.7, 100 mM 2-Mercaptoethanol, 2% SDS) at 60°C for 45 min. Blots were washed thoroughly in ddH2O, blocked in 5% skim milk in TBST for 1 hour and reprobed with appropriate antibodies.
Data analysis
All data are reported as Mean ± S.D. of three independent experiments. Statistical analysis was conducted using R version 3.1.1; significance testing was performed with one-way ANOVA and post-hoc multiple testing was with Tukey's test.
RESULTS
FE phenotypes of baz mutant clones are allele-specific
It was previously reported that large baz mutant clones positioned along the lateral FE lose integrity, which results in the formation of gaps and discontinuities, thus exposing the underlying nurse cells. On the other hand, baz PFC mutant clones were reported to show strong multilayering (Abdelilah-Seyfried et al., 2003). Our results confirmed these findings with regards to baz4 and baz815-8 FE clones which often form large holes in the lateral FE and display strong multilayering of the PFCs (Figs 1 and 2). On the contrary, large holes or discontinuities were rarely observed in bazEH747 and bazXR11 lateral FE clones (Fig. 1), whereas careful examination of PFC clones revealed a mild, yet highly penetrant multilayering phenotype (Fig. 2). In contrast to PFC clones for baz4 and baz815-8, which show severe multilayering throughout the clone, multilayering in bazEH747 and bazXR11 PFC clones is normally restricted to only a few cells (Fig. 2).
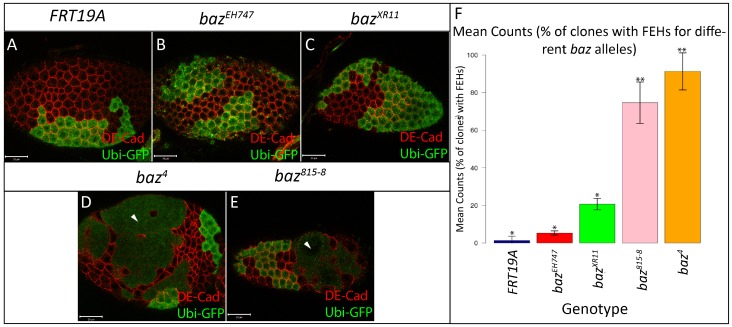
Fig. 1. Different baz alleles display different FE phenotypes.
FE clones marked by absence of GFP were generated using the directed mosaic system by expressing UAS-Flp under the control of e22c-Gal4. FRT19A (A), bazEH747 (B) and bazXR11 (C) mutant FE cells maintain a continuous epithelium while baz4 (D) and baz815-8 (E) mutant cells fail to maintain a continuous epithelium resulting in large holes in the FE, exposing the underlying nurse cells (white arrowheads). (F) Bar graph presenting quantitative analysis of the FEH phenotype. The number of baz4, baz815-8, bazEH747, bazXR11 and FRT19A mosaic follicles with holes in the lateral epithelium were counted and presented as an average percentage from three repetitions of n = 50 (88% in baz4, 75% in baz815-8, 21% in bazXR11, 5% in bazEH747, 1% in FRT19A). At p<0.001, genotypes with same number of stars are not significantly different while those with different number of stars are significantly different. Error bars represent s.d. Scale bars = 20 µm.
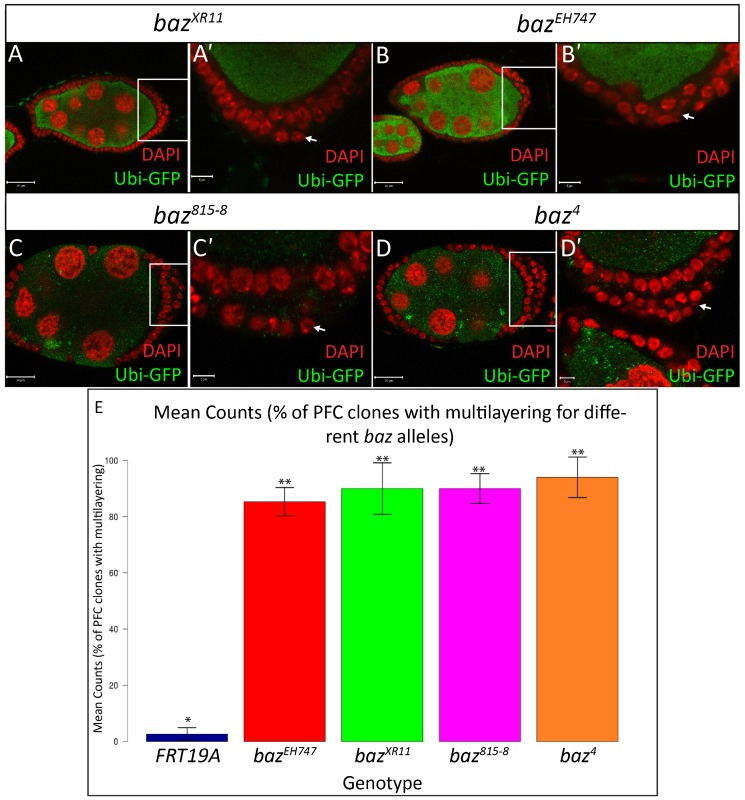
Fig. 2. baz mutant posterior follicle cells show multilayering.
Ovarian follicles in which bazXR11 (A–A′, stage 7), bazEH747 (B–B′, stage 7), baz815-8 (C-C′, stage 8) and baz4 (D–D′, stage 7) mutant cells marked by an absence of Ubi-GFP were generated using the directed mosaic system by expressing UAS-Flp under the control of e22c-gal4. White boxes in A,B,C,D indicate regions shown in A′,B′,C′,D′, respectively. Mosaic follicles were immunostained with GFP and stained with DAPI as indicated. PFCs mutant for bazXR11 (A′), bazEH747 (B′), baz815-8 (C′) and baz4 (D′) show multilayering as visualized by DAPI staining and indicated by arrows. (E) Bar graph presenting quantitative analysis of the PFC multilayering phenotype. The number of baz4, baz815-8, bazEH747, bazXR11, and FRT19A mosaic follicles with PFC multilayering were counted and presented as an average percentage from three repetitions of n = 50 (94% of baz4, 90% of baz815-8, 85% of bazXR11; 90% of bazEH747; 3% of FRT19A). At p<0.001, genotypes with same number of stars are not significantly different while those with different number of stars are significantly different. Error bars represent s.d. Scale bars = 20 µm (A–D); 5 µm (all other scale bars).
We quantitatively assessed the FE hole (FEH) phenotype by counting the number of baz4, baz815-8, bazEH747, bazXR11 and FRT19A control mosaic follicles with lateral epithelial discontinuities. Our analysis revealed a distinct range in the penetrance of the FEH phenotype with baz4 and baz815-8 mosaic follicles showing far higher numbers of FEH compared to bazXR11, bazEH747 and FRT19A control clones (Fig. 1; 88% in baz4, 75% in baz815-8, 21% in bazXR11, 5% in bazEH747, 1% in FRT19A. Percentages are an average from 3 repetitions with n = 50). Post-hoc multiple comparison tests showed that the differences in the penetrance of the FEH phenotype were significant between all genotypes tested albeit at a lower (95%) confidence level for bazEH747 and bazXR11 (Fig. 1F).
Quantitative analysis of the PFC multilayering phenotype revealed a high number of baz4 (94%), baz815-8 (90%), bazXR11 (85%) and bazEH747 (90%) multilayered PFC clones compared to the FRT19A control (3%). Post-hoc multiple comparison tests showed that the differences in the penetrance of the PFC multilayering phenotype in baz4, baz815-8, bazXR11 and bazEH747 clones were significant compared to FRT19A (Fig. 2).
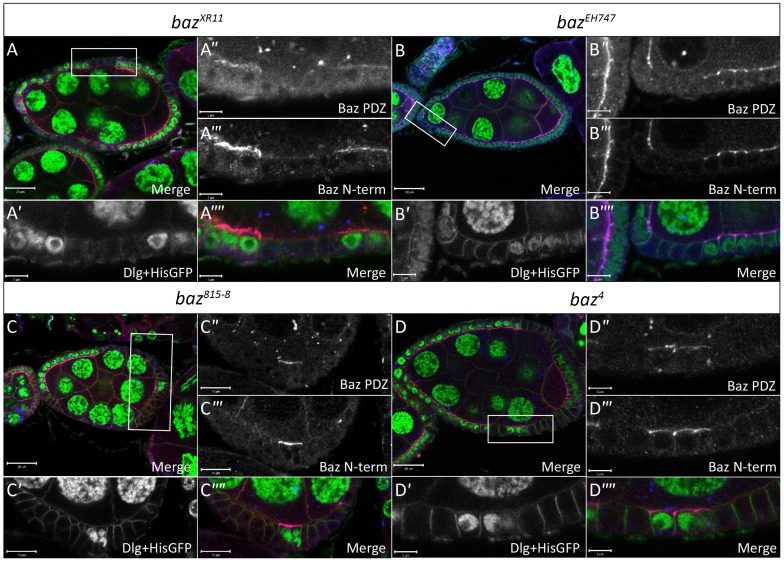
To confirm that the FE defects observed in baz4, baz815-8, bazEH747 and bazXR11 clones were indeed due to an absence of Baz, we conducted immunostaining analyses with an antibody that was raised against the Baz N-terminal domain and another antibody against the Baz Postsynaptic density 95/Dlg/Zonula occludens 1 (PDZ) domains. With both antibodies Baz staining in baz4, baz815-8, bazEH747 and bazXR11 FE clones was below the limit of detection (Fig. 3). Furthermore, all FE defects observed in baz4 (as previously reported by Benton and St Johnston, 2003; Morais-de-Sá et al., 2010) and bazEH747 clones were rescued by overexpression of full length UAS-Baz using the MARCM system (supplementary material Fig. S1).
Fig. 3. baz mutant cells lack Baz immunostaining.
Ovarian follicles in which bazXR11 (A–A″″), bazEH747 (B–B″″), baz815-8 (C–C″″) and baz4 (D–D″″) mutant cells marked by loss of His-GFP were induced by hs-Flp mediated recombination. White boxes in A–D indicate regions shown in A′–A″″, B′–B″″, C′–C″″ and D–D″″, respectively. Mosaic follicles were immunostained with antibodies raised against amino acids 1-297 of the N-terminus of Baz (Baz N-term), amino acids 309-747 of the PDZ domain of Baz (Baz PDZ) and Dlg, which marks the basolateral cell boundaries as indicated. Both the Baz N-term and Baz PDZ antibodies show a complete absence of apical staining in bazXR11 (A″ and A′″), bazEH747 (B″–B′″), baz815-8 (C″–C′″) and baz4 (D″– D′″) mutant FE cells, respectively. Scale bars = 20 µm (A–D); 5 µm (all other scale bars).
In order to obtain additional insight into which of the above mentioned alleles presented a genuine baz loss-of-function FE phenotype, we knocked down baz expression in FE cells by expressing GFP-RNAi in a GFP-baz Trap background under the control of traffic jam-Gal4 (tj-Gal4) (supplementary material Fig. S2). Knockdown of GFP-baz was confirmed by GFP and Baz immunostaining (supplementary material Fig. S2B,B′). These baz deficient FE cells maintained a continuous epithelial sheet and displayed mild multilayering of the PFCs, similar to the phenotypes observed for bazEH747 and bazXR11 FE clones (supplementary material Fig. S2B,C″).
baz4 and baz815-8 mutant animals express truncated Baz protein fragments
Given that we observed a significant difference in the penetrance of the FEH phenotype and severity of the PFC multilayering phenotype between different baz alleles, we sought to elucidate the underlying causes of these discrepancies. Differences in the penetrance of a given phenotype are commonly associated with the “strength” of the alleles in question. “Weak” hypomorphic alleles that retain some function often show relatively milder phenotypes compared to “strong” null alleles or deletion mutants. On the contrary, neomorphic mutations that result in the generation of a gene product with novel biological functions can show a more severe phenotype as compared to a null mutant of the same gene. Therefore, as a first step, we compared the individual characteristics of the aforementioned baz mutations.
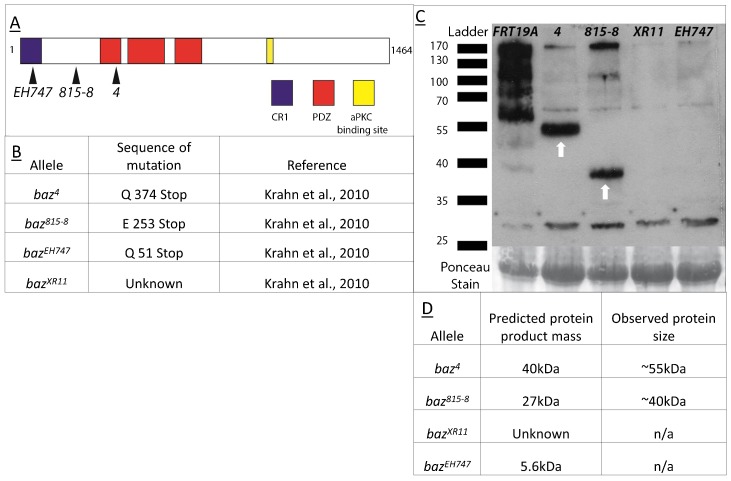
The baz4 (Wieschaus et al., 1984) and bazEH747 (Eberl and Hilliker, 1988) alleles were generated by Ethyl Methanesulfonate (EMS) mutagenesis, while the baz815-8 (McKim et al., 1996) and bazXR11 (Kuchinke et al., 1998; Ralf Stanewsky, unpublished) alleles were induced by X-ray mutagenesis (Fig. 4A,B). Based on sequencing data for the different baz alleles (Krahn et al., 2010a), we found that the position of the mutagenesis-induced stop codons in the baz4, baz815-8 and bazEH747 alleles predicts that each of these alleles should produce truncated Baz proteins of different lengths (Fig. 4B,D). The bazXR11 allele does not contain any mutation in the coding region, pointing to a mutation in a regulatory element that prevents transcription or translation of the baz locus (Krahn et al., 2010a). Hypothetically, the baz4 allele should generate a 40 kDa protein consisting of the first 374 amino acids of Baz, spanning the Conserved Region 1 (CR1) domain and a large portion of PDZ domain 1, while the baz815-8 allele should produce a 27 kDa truncated protein consisting of the first 253 amino acids, also spanning the CR1 domain (Fig. 4A,B,D). The bazEH747 allele should encode a 5.6 kDa, 51 amino acid fragment of the N-terminal region of Baz and should span only a portion of the CR1 domain (Fig. 4A,B,D).
Fig. 4. Schematic representation and sequence characteristics of baz mutant alleles.
(A) Schematic representation of Baz protein with the CR1 domain shown in blue, PDZ domains in red and the aPKC binding site in yellow with black arrows indicating the approximate location of the stop codons present in the respective mutant alleles. (B) Table containing sequence details for the different baz mutant alleles. (C) Western Blot analysis of protein extracts from maternal zygotic baz4, baz815-8, bazXR11, bazEH747, and FRT19A mutant embryos. Bands at 55 and 40 kDa (white arrows) are observed in baz4 and baz815-8. Faint bands are observed in all baz allele lanes at 170 kDa. (D) Summary of results of Western blot analysis of protein extracts from maternal zygotic baz4, baz815-8, bazXR11, bazEH747 with a comparison to predicted protein product mass for each allele.
To determine whether these truncated proteins affect the penetrance of the baz mutant FEH phenotype as well as the severity of PFC multilayering, we sought to establish whether they were indeed being stably expressed, or being post-translationally degraded. Western blot analysis of protein extracts from maternal zygotic (m/z) mutant embryos for the baz4, baz815-8, bazEH747, bazXR11 and FRT19A as control revealed that truncated proteins were being stably expressed in the case of the baz4 and baz815-8 alleles, but not in case of the bazEH747 and bazXR11 alleles. Interestingly, these truncated proteins showed an upward shift in weight of approximately 15 kDa compared to the predicted sizes, which may be caused by some form of post-translational modification (Fig. 4C). Despite the detection of the truncated Baz proteins in Western blots of baz4 and baz815-8 embryos derived from germ line clones, we could not detect these Baz fragments by confocal microscopy, most likely because of their diffuse cytoplasmic distribution. This interpretation is supported by the diffuse cytoplasmic distribution of an overexpressed N-terminal fragment of Baz consisting of amino acids 1-311 (supplementary material Fig. S3; Benton and St Johnston, 2003).
Expression of Baz N-terminal fragments does not interfere with FE integrity
The above data indicate a correlation between the presence of Baz N-terminal fragments with a minimum length of 253 amino acids and strong penetrance of the FEH phenotype as well as severe PFC multilayering. This could in turn lead one to speculate that baz4 and baz815-8 are neomorphic alleles and that the above mentioned Baz N-terminal fragments may be responsible for the high penetrance of the FEH phenotype and strong PFC multilayering, potentially through destabilization of epithelial polarity (Mizuno et al., 2003).
To assess these possibilities, we used the MARCM system (Lee and Luo, 2001) to express a UAS-bazΔ312-1464-GFP construct in bazEH747 FE clones. As shown in supplementary material Fig. S3, there were no adverse effects on the integrity of the FE due to expression of a 311 amino acid N-terminal fragment of Baz. There was no statistical difference between the number of FEHs in bazEH747, bazEH747;UAS-bazΔ312-1464-GFP, and FRT19A mosaic follicles (supplementary material Fig. S3B). Furthermore, the severity of PFC multilayering was also not increased. These results indicate that the strong penetrance of the FEH phenotype in baz4 and baz815-8 mosaic follicles is unlikely to be due to the presence of Baz N-terminal fragments. This interpretation is supported by the fact that overexpression of UAS-bazΔ312-1464-GFP in a wild type background did not affect cell polarity or viability (data not shown; Benton and St Johnston, 2003).
The bazEH747 FE phenotype is enhanced by mutations for known polarity regulators
The above evidence indicated that baz4 and baz815-8 are not neomorphic alleles. An alternative explanation for the high penetrance of the FEH phenotype and strong PFC multilayering could be that second site mutations that enhance the baz mutant phenotype are present on the baz4 and baz815-8 chromosomes.
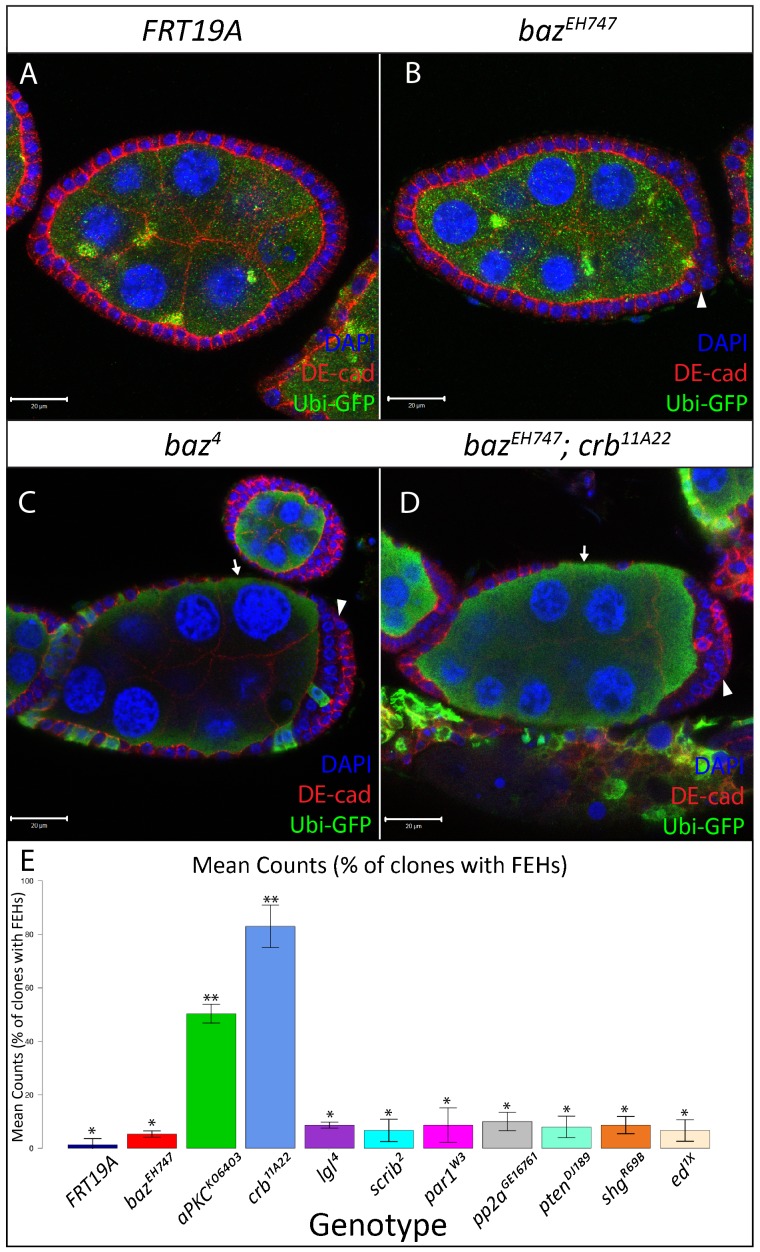
We thus conducted a small-scale genetic interaction screen to test whether removing one copy of genes known to function in cell polarity enhances the bazEH747 FEH and PFC multilayering phenotypes. Quantitative analysis of the results of this screen revealed a stark increase in the penetrance of the FEH phenotype and severity of PFC multilayering in bazEH747 clones that are simultaneously heterozygous for crb11A22 or aPKCk06403 (Fig. 5). Post-hoc multiple comparison tests showed that differences in the penetrance of the FEH phenotype were significant between bazEH747; crb11A22 and all other genotypes, and bazEH747; aPKCk06403 and all other genotypes (Fig. 5). Intriguingly, bazEH747 mosaic follicles heterozygous for crb11A22 or aPKCk06403 (data not shown) were phenotypically indistinguishable from baz4 mosaic follicles (Fig. 5). These results raise the possibility that the chromosomes carrying the baz4 and baz815-8 alleles may carry second site mutations that synergistically enhance the baz loss-of-function phenotype.
Fig. 5. FE defects in bazEH747 mosaic ovaries are enhanced by loss of one copy of crb or aPKC.
FE clones for FRT19A (A), bazEH747 (B), baz4 (C), bazEH747; crb11A22/+ (D), marked by absence of Ubi-GFP, were generated using the directed mosaic system by expressing UAS-Flp under the control of e22c-gal4. Lateral FE cells mutant for bazEH747 (B) look identical to FRT19A (A) control epithelia and form a continuous epithelium over the underlying nurse cells. However, mild multilayering of the FE is visible at the posterior pole of bazEH747 mosaic follicles (B, white arrowhead) as opposed to FRT19A control mosaic follicles (A). The bazEH747 mutant phenotype is strongly enhanced in a crb11A22 heterozygous mutant background (D) and appears identical to the baz4 (C) FE mutant phenotype with discontinuities in the lateral FE (white arrows) and strong multilayering at the posterior poles (white arrowheads). (E) Bar graph presenting quantitative analysis of the FEH phenotype from the baz interaction screen. All clones analyzed are hemizygous mutant for bazEH747 and heterozygous for the respective second mutation. The number of mosaic follicles with FEHs were counted and presented as an average percentage from 3 repetitions of n = 50 for each genotype listed (1% of FRT19A, 5% of bazEH747, 50% of bazEH747;aPKCK06403/+, 83% of bazEH747;crb11A22/+, 9% of bazEH747;lgl4/+, 7% of bazEH747;scrib2/+, 9% of bazEH747;par1W3/+, 10% of bazEH747;pp2aGE16781/+, 8% of bazEH747;ptenDJ189/+, 9% of bazEH747; shgR69B/+, 7% of bazEH747;ed1x5/+). At p<0.001, genotypes with same number of stars are not significantly different while those with different number of stars are significantly different. Error bars represent s.d. Scale bars = 20 µm.
Comparison of the bazEH747 and baz4 phenotypes in other developmental contexts
Given that both bazEH747 and baz4 show distinct phenotypes in the FE, we next looked into whether these alleles show different phenotypes in other tissue specific contexts. We compared the cellularization phenotypes displayed by baz4 and bazEH747 maternal zygotic embryos. Previous studies using the baz4 allele proposed that baz is epistatic to other members of the Par complex and the Crb complex during cellularization (Bilder et al., 2003; Harris and Peifer, 2004). Our analysis revealed that both baz4 and bazEH747 maternal zygotic mutants show similar phenotypes during cellularization, with aPKC and DE-cad failing to localize to their normal positions (supplementary material Fig. S4).
The baz gene was initially identified by its strong embryonic phenotype characterized by the formation of large holes in the ventral epidermis (Wieschaus et al., 1984; Müller and Wieschaus, 1996; Bilder et al., 2003; Harris and Tepass, 2008). Our comparison of zygotic epithelial development in baz4 and bazEH747 zygotic mutant embryos by immunostaining analysis revealed indistinguishable phenotypes characterized by large holes in the ventral epidermis in stage 13 embryos of both genotypes (supplementary material Fig. S5).
Baz has also been shown to function in oocyte development, where it is required for the oocyte to maintain its DNA in a haploid karyosome, in comparison to the 15 polyploid nurse cells with which it shares the egg chamber (Cox et al., 2001; Huynh et al., 2001). Furthermore, the oocyte fails to enrich Orb protein past stage 3 in the absence of Baz. Our comparison of the baz4 and bazEH747 alleles in this developmental context revealed identical defects during oocyte development with both baz4 and bazEH747 mutant oocyte nuclei becoming polyploid and failing to maintain Orb enrichment past stage 3 (supplementary material Fig. S6).
Lastly, we sought to compare the phenotypes of the baz4 and bazEH747 alleles in mitotic neuroblasts. We attempted to generate and compare baz4 and bazEH747 mutant neuroblasts in late stage 3rd instar larvae brains using the MARCM technique but were unable to identify baz4 or bazEH747 GFP marked larval mutant neuroblasts as compared to the FRT19A control (supplementary material Table S1). However, GFP marked neural progenitors were regularly identified in both baz4 and bazEH747 mosaic brains, raising the possibility that both baz4 and bazEH747 mutant neuroblasts are unable to maintain stem cell fate or undergo apoptosis.
Overall, the results of these experiments indicate that although the phenotypes for the baz4 and bazEH747 alleles differ dramatically in the FE, there are no discrepancies in cellularizing embryos, embryonic epithelia, oocytes or neuroblasts.
Re-assessment of the function of Baz in FE cell polarity
As mentioned before, results from previous studies have led to the conclusion that Baz functions as a key regulator of cell polarity in Drosophila FE cells (Abdelilah-Seyfried et al., 2003; Franz and Riechmann, 2010; Morais-de-Sá et al., 2010). However, as the majority of these studies were conducted using the baz4 and baz815-8 alleles, whose strong FE phenotype differ from the true baz loss of function FE phenotype, the possibility arises that the true nature of the relationship between Baz and other polarity regulators may have been obscured. We thus reassessed these relationships by comparing the localization of components of the different polarity complexes in bazEH747, bazXR11, baz4 and baz815-8 lateral FE clones by immunostaining analysis.
We started by comparing the effects of the different alleles on the localization of aPKC and DE-cad in the lateral FE. Neither bazXR11 (Fig. 6A–A″″) nor bazEH747 (Fig. 6B–B″″) clones showed any difference in the localization of aPKC or DE-cad compared to neighboring wild type cells. Furthermore, baz GFP RNAi knockdown FE cells showed normal apical localization of aPKC and basolateral localization of Lgl (supplementary material Fig. S2C–C′″).
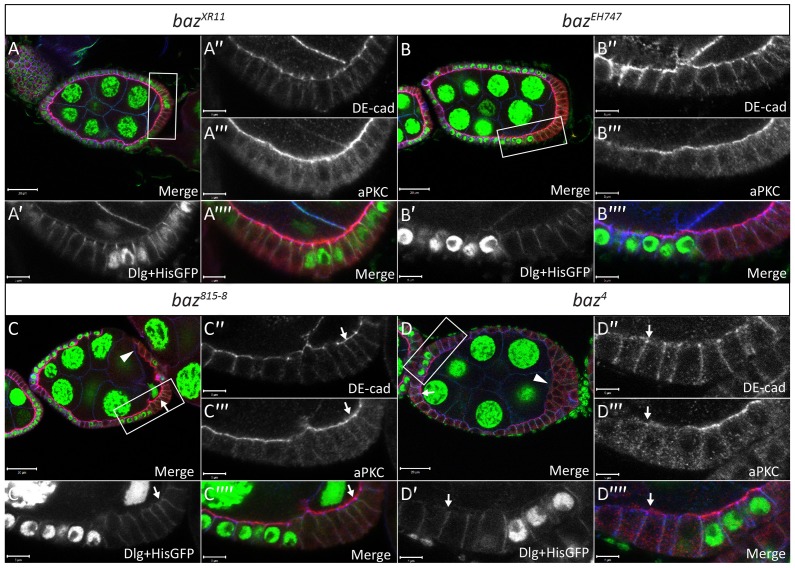
Fig. 6. Localization of DE-cad and aPKC are unaffected in bazXR11 and bazEH747 FE clones.
Ovarian follicles in which bazXR11 (A–A″″), bazEH747 (B–B″″), baz815-8 (C–C″″) and baz4 (D-D″″) mutant cells were induced by hs-Flp mediated recombination and marked by loss of His-GFP. White boxes in A,B,C,D indicate regions shown in A′–A″″, B′–B″″, C′–C″″ and D–D″″, respectively. Follicles were stained with DE-cad, aPKC and Dlg as indicated. Neither DE-cad nor aPKC apical localization is affected in bazXR11 (A″ and A′″) or bazEH747 (B″ and B′″) mutant cells. Many baz815-8 mutant cells show wild type localization of aPKC and DE-cad as indicated by arrows (C–C″″). Some baz815-8 mutant cells in which cell shape has been compromised appear to lose apical aPKC and junctional DE-cad staining as indicated by an arrowhead (C). Many baz4 mutant FE cells retain junctional DE-cad localization even in the absence of apical aPKC localization as indicated by arrows (D–D″″). Others show a loss of both apical aPKC and junctional DE-cad as indicated by an arrowhead (D). Scale bars = 20 µm (A–D); 5 µm (all other scale bars).
On the other hand, both baz815-8 (Fig. 6C–C″″) and baz4 FE (Fig. 6D–D″″) clones displayed a range of defects in the localization of aPKC and DE-cad as well as cell shape. In line with previous studies, baz4 and baz815-8 mutant lateral FE cells often displayed a flat cell shape with a stretched appearance (Fig. 6C, arrowhead) and showed a loss of apical aPKC and junctional DE-cad (Fig. 6C,D arrowheads) (Abdelilah-Seyfried et al., 2003; Morais de Sá et al., 2010). Additionally, we often observed intermediate phenotypes in baz815-8 (data not shown) and baz4 clones where mutant cells retained a relatively normal shape and displayed junctional DE-cad localization (Fig. 6D″, arrows) but strongly diminished apical aPKC localization (Fig. 6D′″, arrows). Furthermore, several baz815-8 (Fig. 6C″,C′″) and baz4 (data not shown) clones were present that displayed wild type cell shape with normal DE-cad and aPKC localization. Also, there were no baz815-8 or baz4 clones in which aPKC localized to the apical membrane but DE-cad failed to localize to the apical junctions.
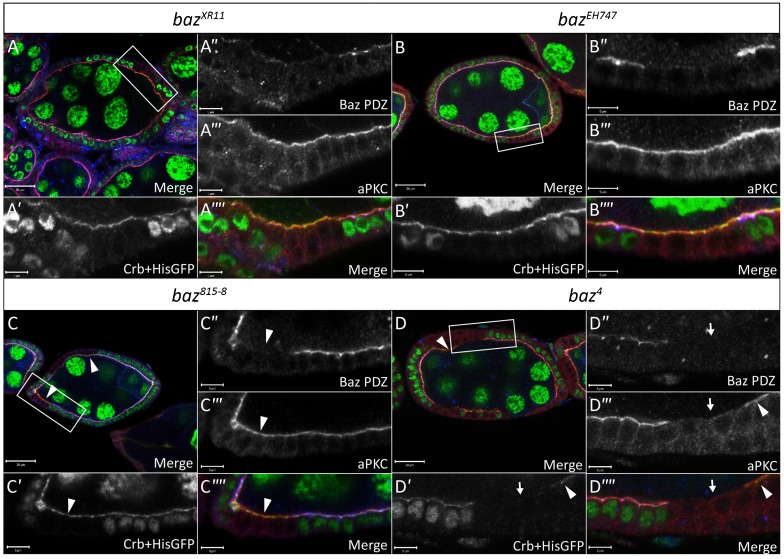
We next analyzed the effects of the different baz alleles on the localization of Crb. Our analysis of bazXR11 and bazEH747 FE clones revealed that loss of Baz had no effect on the localization of Crb (Fig. 7A,B″″). On the other hand, analysis of Crb localization in baz4 and baz815-8 FE clones yielded results similar to those for aPKC localization. In accordance with previous reports, Crb apical staining was often absent in baz4 (Fig. 7D–D″″, arrows) and baz815-8 (data not shown) mutant FE clones. However, we also observed many baz815-8 and baz4 mutant cells where Crb immunostaining appeared normal (Fig. 7C–C″″, arrowheads). Furthermore, unlike aPKC and DE-cad, aPKC and Crb localization appeared to be linked as all mutant cells that maintained apical Crb also maintained apical aPKC, while all those that showed a loss of apical Crb also showed a concomitant loss of aPKC.
Fig. 7. Localization of Crb is unaffected in bazXR11 and bazEH747 FE clones.
Ovarian follicles in which bazXR11 (A–A″″), bazEH747 (B–B″″), baz815-8 (C–C″″) and baz4 (D–D″″) mutant cells were induced by hs-Flp mediated recombination and marked by loss of His-GFP and Baz immunostaining. White boxes in A,B,C,D indicate regions shown in A′–A″″, B′–B″″, C′–C″″ and D–D″″, respectively. Follicles were stained with anti-Baz-PDZ, aPKC and Crb antibodies as indicated. Baz is clearly absent from bazXR11 (A″), bazEH747 (B″), baz815-8 (C″) and baz4 (D″) mutant cells, respectively. No effect is seen on aPKC and Crb localization at the apical membrane in bazXR11 (A′ and A′″) and bazEH747 (B′ and B′″) mutant cells. Furthermore, no bazXR11 (A) or bazEH747 (B) mutant cells lose epithelial morphology. baz4 (D′ and D′″) mutant cells show coincident loss of aPKC and Crb from the apical membrane as indicated by white arrows. However, apical aPKC and apical Crb staining is still visible in many baz815-8 (C–C″″) and baz4 (D–D″″) mutant cells as indicated by white arrowheads. Scale bars = 20 µm (in A–D); 5 µm (all other scale bars).
We also analyzed the distribution of Par6 and Sdt, members of the Par complex and Crumbs complex, respectively, in bazEH747 and baz815-8 FE clones (Fig. 8A,B). Again, we observed no effect on the localization of these proteins in bazEH747 FE clones. However, immunostaining for Par6 and Sdt in baz815-8 FE clones (Fig. 8A,B) and baz4 FE clones (data not shown) showed results similar to those obtained for aPKC and Crumbs.
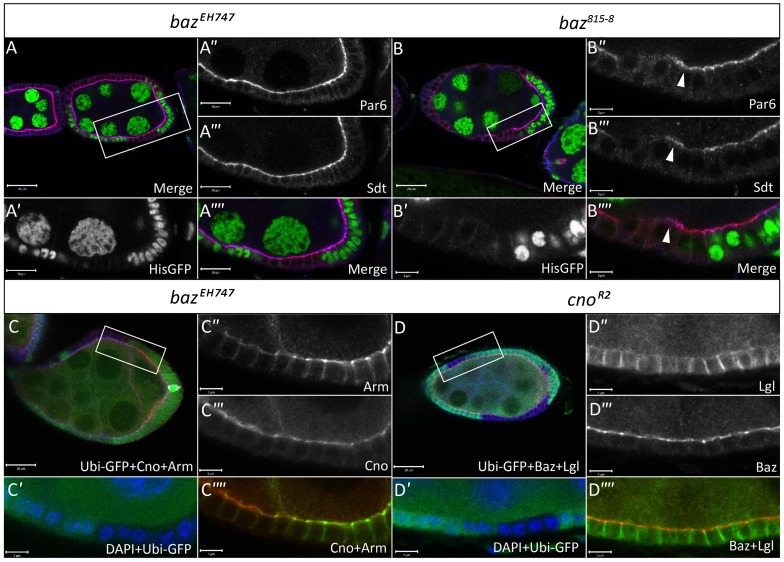
Fig. 8. Baz and Cno do not regulate the localization of one another in the FE.
Ovarian follicles in which, bazEH747 (A–A″″, Stage 6) and baz815-8 (B–B″″, Stage 8) clones induced by hs-Flp mediated recombination are marked by loss of His-GFP. White boxes in A,B indicate regions shown in A′–A″″ and B′–B″″, respectively. Follicles were stained with antibodies against Par6 and Sdt as indicated. No effect is seen on Par6 and Sdt localization at the apical membrane in bazEH747 (A″,A′″) mutant cells. On the other hand, baz815-8 (B″,B′″) mutant cells show coincident loss of Par6 and Sdt from the apical membrane. However, apical Par6 and Sdt staining is still visible in many baz815-8 mutant cells (B″–B″″) as indicated by white arrowheads. (C–C″″) FE clones for bazEH747 (stage 8) marked by absence of GFP and generated using the directed mosaic system by expressing UAS-Flp under the control of e22c-Gal4, immunostained with Cno, Arm and GFP antibodies as indicated. (D–D″″) cnoR2 clones (stage 7) immunostained with Baz, Lgl and GFP antibodies as indicated, marked by absence of GFP, were generated using the directed mosaic system by expressing UAS-Flp under the control of e22c-Gal4. White boxes in C and D indicate regions shown in C′–C″″ and D′–D″″, respectively. No effect is seen on Cno or Arm apical localization in bazEH747 mutant FE clones (C–C″″) or Baz localization in cnoR2 mutant FE clones (D–D″″). Scale bars = 20 µm (A–D); 5 µm (all other scale bars).
A recent study has shown that during cellularization, Cno functions to localize Baz to the apical membrane and vice versa (Choi et al., 2013). However, up till now, no studies examining the relationship between Baz and the Ed/Cno complex in the FE have been reported. We thus examined Cno localization in bazEH747 FE clones (Fig. 8C) and Baz localization in cnoR2 FE clones (Fig. 8D). No effect was seen on the apical localization of Cno in the absence of baz (Fig. 8C). FE clones for cno occasionally showed mild defects in FE development similar to the bazEH747 phenotype (data not shown). However, the vast majority of cno clones showed no polarity defects and displayed normal Baz localization (Fig. 8D).
DISCUSSION
Discrepancies between baz alleles and elucidation of the true baz loss-of-function FE phenotype
Previous studies using the baz4 and baz815-8 alleles reported that baz is essential for polarization of the FE. baz4 and baz815-8 clones show large holes in the FE and strong multilayering of the PFCs. It was believed that baz functions at the top of a genetic hierarchy to specify the apical membrane as neither Par6, aPKC, Crb nor Sdt show apical localization in baz4 and baz815-8 mutant clones, whereas Baz cortical localization persists in par6, aPKC, crb and sdt mutant clones (Franz and Riechmann, 2010; Morais-de-Sá et al., 2010). On the contrary, our results from analyzing the bazEH747 and bazXR11 alleles indicate that baz does not play an essential role in polarization of the FE, as defects observed in bazEH747 and bazXR11 FE clones were statistically insignificant compared to FRT19A control clones. Furthermore, the localization of aPKC, Crb, Par6 and Sdt was unaffected in bazEH747 FE clones. Following the observation of this discrepancy in phenotypes, we attempted to answer two important and interrelated questions. Firstly, which of these alleles represents the true baz loss-of-function phenotype and secondly, what is the underlying cause of the differences observed between these alleles.
Our immunostaining analysis revealed that Baz protein was absent from FE clones for all baz alleles examined. This immunostaining data was further corroborated by our western blot analysis, which revealed the absence of Baz protein in bazEH747 and bazXR11 and the presence of truncated Baz proteins in baz4 and baz815-8 protein extracts. Both the mild multilayering phenotype of the bazEH747 and bazXR11 alleles as well as the strong multilayering phenotype and FEHs of the baz4 and baz815-8 alleles can be rescued by overexpression of UAS-Baz-GFP, indicating that a loss of full-length Baz underlies both the strong and mild FE phenotypes. Based on these results, it can be postulated that the baz4 and baz815-8 lines carry enhancer mutations or that they function as neomorphic alleles due to the expression of truncated Baz protein fragments. Alternatively, the bazXR11 and bazEH747 lines may carry additional suppressor mutations. However, the phenotype generated by GFP-RNAi directed against baz-GFP Trap, which occurs in the absence of truncated Baz protein fragments or any potential enhancer or suppressor mutations present in the aforementioned lines, strongly resembles the bazEH747 and bazXR11 phenotype. Thus, these data argue in favor of bazXR11 and bazEH747 representing the true baz loss-of-function phenotype in the FE.
We next sought to determine whether the underlying cause of the severe phenotype displayed by the baz4 and baz815-8 alleles is due to the production of truncated Baz protein fragments that interfere with polarity or the presence of additional enhancer mutations in these lines. Expression of a Baz N-terminal fragment in a bazEH747 mutant background failed to enhance the mild baz loss-of-function phenotype, indicating that baz4 and baz815-8 are unlikely to be neomorphic alleles. On the other hand, removal of one copy of crb or aPKC from bazEH747 FE clones resulted in severe defects that phenocopy baz4 and baz815-8 FE clones. These data provide strong evidence that the strains carrying the baz4 and baz815-8 alleles contain additional mutations that enhance the true baz loss-of-function phenotype. As the baz4 and baz815-8 alleles were generated by EMS and X-ray mutagenesis respectively, it appears likely that the respective chromosomes accumulated additional mutations. However, given that the FE defects observed in baz4 clones can be fully rescued by expression of full length Baz (supplementary material Fig. S1; Benton and St Johnston, 2003; Morais-de-Sá et al., 2010), the high penetrance of the FEH phenotype and severe PFC multilayering is likely to be the result of a synergistic enhancement of the baz mutant phenotype by additional mutations which do not result in FE defects on their own. Given that sdt and par6, two known baz interactors are also located on the first chromosome, one could speculate that hypomorphic mutations in either of these genes could result in the strong penetrance of FE phenotypes observed in baz4 and baz815-8 clones.
Baz is required for establishment of polarity during cellularization, but is dispensable for polarity in the FE
The vast majority of bazEH747 and bazXR11 FE clones showed normal polarization of the FE and maintained aPKC, Par6, Crb and Sdt localization. On the other hand, bazEH747 m/z mutant embryos were unable to establish apico-basal polarity during cellularization, reflected in their failure to localize DE-cad and aPKC to their appropriate locations. From these results, it can be concluded that baz plays an essential role in the establishment and maintenance of polarity during cellularization, but not in the FE. This raises the important question as to why the requirement for baz function differs in these two developmental contexts.
One explanation for this discrepancy could be that Baz functions redundantly with another protein in the establishment and maintenance of apico-basal polarity in the FE as compared to the embryo. Several lines of evidence indicate that crb, which also displays severe defects in the apico-basal polarity in the embryo, but mild defects in the FE, may function redundantly with baz in certain developmental contexts (Morais-de-Sá et al., 2010; Walther and Pichaud, 2010; Fletcher et al., 2012; Thompson et al., 2013). For example, during FE development, Baz initially colocalizes apically with aPKC and Par6 but is gradually replaced by Crb and restricted basally to the AJs (Franz and Riechmann, 2010), indicating that Crb can function redundantly with Baz in the maintenance of polarity. Our experiments revealed that the bazEH747 FE phenotype is strongly enhanced by the loss of one copy of crb while a previous report from Morais-de-Sá et al., 2010. (Morais-de-Sá et al., 2010) showed that overexpression of Baz can rescue crb mutant FE defects. These data taken together could lead one to speculate that Crb and Baz can substitute for each other in the establishment and maintenance of apico-basal polarity in the FE. However, Crb expression and localization to the apical membrane of FE cells occurs at stage 1, sometime after Baz, aPKC and Par6 are detectable in region 2B of the germarium (Franz and Riechmann, 2010). Thus, Crb is unlikely to substitute for Baz in establishing polarity in the FE.
Another potential explanation for the divergent requirements for baz in the establishment of epithelial polarity during cellularization vs FE development is the difference in the underlying polarity cues operating in these tissues. As mentioned earlier, FE cells are derived from somatic stem cells in the ovarian niche and make use of basal, lateral and apical cues to establish polarity as they undergo a mesenchymal-epithelial transition. For example, contact between polarizing FE precursors and the underlying basal lamina is sufficient for the specification of the basal membrane, while the germ cells act as a signal for polarization and specification of the apical and lateral membranes (Tanentzapf et al., 2000). On the other hand, the only polarization signal that has been identified during cellularization is the embryonic membrane, which goes on to form the apical membrane once cellularization is complete (Mavrakis et al., 2009; Choi et al., 2013). Therefore, one could speculate that the presence of multiple polarity cues in the FE is sufficient to compensate for an absence of baz during the establishment of polarity. This explanation is further supported by our finding that Cno, which is essential for establishing apico-basal polarity in the cellularizing embryo, displayed only mild polarity defects in the FE, similar to baz. Thus, it can be hypothesized that Baz function in the establishment of FE polarity may be redundant with other basal, lateral and apical polarity cues, while its function in the maintenance of apico-basal polarity could be redundant with Crb.
A potential function for baz in Hippo and Notch signaling
PFC multilayering is a trademark phenotype of several genes involved in Hippo signaling and results from unrestricted cell division (Meignin et al., 2007; Polesello and Tapon, 2007; Genevet et al., 2010; Yue et al., 2012). The high penetrance of this multilayering phenotype in baz PFC clones raises the possibility that Baz too may function in Hippo signaling. Several studies have provided strong evidence showing that known interactors of Baz, namely Crb and aPKC, interact with members of the Hippo signaling pathway to regulate cell division and tissue growth (Chen et al., 2010; Ling et al., 2010; Parsons et al., 2010; Robinson et al., 2010). Thus, it comes as no surprise that the activities of Baz may also impinge on this pathway, either through direct interaction with core Hippo pathway members or indirectly through the regulation of aPKC and Crb in their function as apico-basal polarity regulators.
Regulation of PFC division by Hippo signaling occurs through the Notch pathway (Maitra et al., 2006; Polesello and Tapon, 2007; Yu et al., 2008). Notch is required for the mitotic to endocycle switch at stage 6 by upregulating the zinc finger transcription factor hindsight, which functions to downregulate the homeodomain gene cut (Sun and Deng, 2005; Sun and Deng, 2007). Mislocalization of Notch through disruption of apical basal polarity or defective vesicle sorting has been shown to result in excess PFC proliferation and multilayering (Deng et al., 2001; Yan et al., 2009). Thus, one could speculate that the efficacy of Notch localization and signaling may be attenuated in a baz mutant background, where the systems regulating polarity are highly sensitized, therefore resulting in mild PFC multilayering. Further studies are required to determine how exactly baz functions to restrict PFC proliferation and whether it mediates its effects through Hippo, Notch or other pathways.
CONCLUSIONS
In conclusion, the results of this study provide evidence that chromosomes carrying the commonly used baz4 and baz815-8 alleles may carry additional mutations that enhance the true baz loss-of-function phenotype in the FE. Our results show that contrary to previous studies using the aforementioned alleles, baz is not essential for the establishment and maintenance of FE cell polarity. Our analyses of the true baz loss-of-function phenotype in the FE indicate that baz does not function at the top of a genetic hierarchy in the localization of apical membrane determinants such as Crb and aPKC or AJ components such as DE-cad or β-catenin. This study provides an intriguing example of how the function and requirement for regulatory proteins and their corresponding signaling pathways can vary drastically depending on the developmental context and cell type.
Supplementary Material
Acknowledgments
We thank Ulrich Tepass, Dorothea Godt, Tony Harris, Trudi Schupbach, Mark Peiffer and Eli Knust for suggestions and reagents. We thank the Vienna Drosophila Resource Center and the Bloomington Drosophila Stock Center at the University of Indiana for sending fly stocks and the Developmental Studies Hybridoma Bank at the University of Iowa for sending antibodies. We thank the TRiP at Harvard Medical School (NIH/NIGMS R01-GM084947) for providing transgenic RNAi fly stocks used in this study.
Footnotes
Competing interests: The authors have no competing or financial interests to declare.
Author Contributions: J.S. and A.W. conceived and designed the experiments, J.S., M.H.-C. and M.P.K. performed the experiments, J.S., M.D.T. and A.W. analyzed the data, J.S. and A.W. wrote the paper.
Funding
This work was supported by the Cluster of Excellence and DFG Research Center Nanoscale Microscopy and Molecular Physiology of the Brain.
References
- Abdelilah-Seyfried S., Cox D. N., Jan Y. N. (2003). Bazooka is a permissive factor for the invasive behavior of discs large tumor cells in Drosophila ovarian follicular epithelia. Development 130, 1927–1935 10.1242/dev.00420 [DOI] [PubMed] [Google Scholar]
- Atwood S. X., Chabu C., Penkert R. R., Doe C. Q., Prehoda K. E. (2007). Cdc42 acts downstream of Bazooka to regulate neuroblast polarity through Par-6 aPKC. J. Cell Sci. 120, 3200–3206 10.1242/jcs.014902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becalska A. N., Gavis E. R. (2010). Bazooka regulates microtubule organization and spatial restriction of germ plasm assembly in the Drosophila oocyte. Dev. Biol. 340, 528–538 10.1016/j.ydbio.2010.02.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benton R., St Johnston D. (2003). A conserved oligomerization domain in drosophila Bazooka/PAR-3 is important for apical localization and epithelial polarity. Curr. Biol. 13, 1330–1334 10.1016/S0960-9822(03)00508-6 [DOI] [PubMed] [Google Scholar]
- Bilder D., Li M., Perrimon N. (2000). Cooperative regulation of cell polarity and growth by Drosophila tumor suppressors. Science 289, 113–116 10.1126/science.289.5476.113 [DOI] [PubMed] [Google Scholar]
- Bilder D., Schober M., Perrimon N. (2003). Integrated activity of PDZ protein complexes regulates epithelial polarity. Nat. Cell Biol. 5, 53–58 10.1038/ncb897 [DOI] [PubMed] [Google Scholar]
- Buszczak M., Paterno S., Lighthouse D., Bachman J., Planck J., Owen S., Skora A. D., Nystul T. G., Ohlstein B., Allen A. et al. (2007). The carnegie protein trap library: a versatile tool for Drosophila developmental studies. Genetics 175, 1505–1531 10.1534/genetics.106.065961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C.-L., Gajewski K. M., Hamaratoglu F., Bossuyt W., Sansores-Garcia L., Tao C., Halder G. (2010). The apical-basal cell polarity determinant Crumbs regulates Hippo signaling in Drosophila. Proc. Natl. Acad. Sci. USA 107, 15810–15815 10.1073/pnas.1004060107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi W., Harris N. J., Sumigray K. D., Peifer M. (2013). Rap1 and Canoe/afadin are essential for establishment of apical-basal polarity in the Drosophila embryo. Mol. Biol. Cell 24, 945–963 10.1091/mbc.E12-10-0736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox D. N., Seyfried S. A., Jan L. Y., Jan Y. N. (2001). Bazooka and atypical protein kinase C are required to regulate oocyte differentiation in the Drosophila ovary. Proc. Natl. Acad. Sci. USA 98, 14475–14480 10.1073/pnas.261565198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denef N., Chen Y., Weeks S. D., Barcelo G., Schüpbach T. (2008). Crag regulates epithelial architecture and polarized deposition of basement membrane proteins in Drosophila. Dev. Cell 14, 354–364 10.1016/j.devcel.2007.12.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng W., Althauser C., Ruohola-baker H. (2001). Notch-Delta signaling induces a transition from mitotic cell cycle to endocycle in Drosophila follicle cells. Development 128, 4737–4746. [DOI] [PubMed] [Google Scholar]
- Desai R., Sarpal R., Ishiyama N., Pellikka M., Ikura M., Tepass U. (2013). Monomeric α-catenin links cadherin to the actin cytoskeleton. Nat. Cell Biol. 15, 261–273 10.1038/ncb2685 [DOI] [PubMed] [Google Scholar]
- Doerflinger H., Vogt N., Torres I. L., Mirouse V., Koch I., Nüsslein-Volhard C., St Johnston D. (2010). Bazooka is required for polarisation of the Drosophila anterior-posterior axis. Development 137, 1765–1773 10.1242/dev.045807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eberl D., Hilliker A. (1988). Characterization of X-linked recessive lethal mutations affecting embryonic morphogenesis in Drosophila melanogaster. Genetics 118, 109–120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fletcher G. C., Lucas E. P., Brain R., Tournier A., Thompson B. J. (2012). Positive feedback and mutual antagonism combine to polarize Crumbs in the Drosophila follicle cell epithelium. Curr. Biol. 22, 1116–1122 10.1016/j.cub.2012.04.020 [DOI] [PubMed] [Google Scholar]
- Franz A., Riechmann V. (2010). Stepwise polarisation of the Drosophila follicular epithelium. Dev. Biol. 338, 136–147 10.1016/j.ydbio.2009.11.027 [DOI] [PubMed] [Google Scholar]
- Gao X., Neufeld T. P., Pan D. (2000). Drosophila PTEN regulates cell growth and proliferation through PI3K-dependent and -independent pathways. Dev. Biol. 221, 404–418 10.1006/dbio.2000.9680 [DOI] [PubMed] [Google Scholar]
- Genevet A., Wehr M. C., Brain R., Thompson B. J., Tapon N. (2010). Kibra is a regulator of the Salvador/Warts/Hippo signaling network. Dev. Cell 18, 300–308 10.1016/j.devcel.2009.12.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Godt D., Tepass U. (1998). Drosophila oocyte localization is mediated by differential cadherin-based adhesion. Nature 395, 387–391 10.1038/26493 [DOI] [PubMed] [Google Scholar]
- Goldstein B., Macara I. G. (2007). The PAR proteins: fundamental players in animal cell polarization. Dev. Cell 13, 609–622 10.1016/j.devcel.2007.10.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanratty W. (1984). Isolation and characterization of temperature-sensitive lethal (2) giant larva alleles. I. Mutagenesis and larval pathology. Rouxs Arch. Dev. Biol. 193, 90–97 10.1007/BF00848636 [DOI] [PubMed] [Google Scholar]
- Harris T. J. C. (2012). Adherens junction assembly and function in the drosophila embryo. Int. Rev. Cell Mol. Biol. 293, 45–83 10.1016/B978-0-12-394304-0.00007-5 [DOI] [PubMed] [Google Scholar]
- Harris T. J. C., Peifer M. (2004). Adherens junction-dependent and -independent steps in the establishment of epithelial cell polarity in Drosophila. J. Cell Biol. 167, 135–147 10.1083/jcb.200406024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris T. J. C., Peifer M. (2005). The positioning and segregation of apical cues during epithelial polarity establishment in Drosophila. J. Cell Biol. 170, 813–823 10.1083/jcb.200505127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris T. J. C., Peifer M. (2007). aPKC controls microtubule organization to balance adherens junction symmetry and planar polarity during development. Dev. Cell 12, 727–738 10.1016/j.devcel.2007.02.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris K. P., Tepass U. (2008). Cdc42 and Par proteins stabilize dynamic adherens junctions in the Drosophila neuroectoderm through regulation of apical endocytosis. J. Cell Biol. 183, 1129–1143 10.1083/jcb.200807020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris T. J. C., Tepass U. (2010). Adherens junctions: from molecules to morphogenesis. Nat. Rev. Mol. Cell Biol. 11, 502–514 10.1038/nrm2927 [DOI] [PubMed] [Google Scholar]
- Hartenstein V., Wodarz A. (2013). Initial neurogenesis in Drosophila. Wiley Interdiscip. Rev. Dev. Biol. 2, 701–721 10.1002/wdev.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi S., Ito K., Sado Y., Taniguchi M., Akimoto A., Takeuchi H., Aigaki T., Matsuzaki F., Nakagoshi H., Tanimura T. et al. (2002). GETDB, a database compiling expression patterns and molecular locations of a collection of Gal4 enhancer traps. Genesis 34, 58–61 10.1002/gene.10137 [DOI] [PubMed] [Google Scholar]
- Huynh J. R., Petronczki M., Knoblich J. A., St Johnston D. (2001). Bazooka and PAR-6 are required with PAR-1 for the maintenance of oocyte fate in Drosophila. Curr. Biol. 11, 901–906 10.1016/S0960-9822(01)00244-5 [DOI] [PubMed] [Google Scholar]
- Johnson K., Wodarz A. (2003). A genetic hierarchy controlling cell polarity. Nat. Cell Biol. 5, 12–14 10.1038/ncb0103-12 [DOI] [PubMed] [Google Scholar]
- Jones T. A., Metzstein M. M. (2011). A novel function for the PAR complex in subcellular morphogenesis of tracheal terminal cells in Drosophila melanogaster. Genetics 189, 153–164 10.1534/genetics.111.130351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S., Gailite I., Moussian B., Luschnig S., Goette M., Fricke K., Honemann-Capito M., Grubmüller H., Wodarz A. (2009). Kinase-activity-independent functions of atypical protein kinase C in Drosophila. J. Cell Sci. 122, 3759–3771 10.1242/jcs.052514 [DOI] [PubMed] [Google Scholar]
- Krahn M. P., Klopfenstein D. R., Fischer N., Wodarz A. (2010a). Membrane targeting of Bazooka/PAR-3 is mediated by direct binding to phosphoinositide lipids. Curr. Biol. 20, 636–642 10.1016/j.cub.2010.01.065 [DOI] [PubMed] [Google Scholar]
- Krahn M. P., Bückers J., Kastrup L., Wodarz A. (2010b). Formation of a Bazooka-Stardust complex is essential for plasma membrane polarity in epithelia. J. Cell Biol. 190, 751–760 10.1083/jcb.201006029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krahn M. P., Egger-Adam D., Wodarz A. (2009). PP2A antagonizes phosphorylation of Bazooka by PAR-1 to control apical-basal polarity in dividing embryonic neuroblasts. Dev. Cell 16, 901–908 10.1016/j.devcel.2009.04.011 [DOI] [PubMed] [Google Scholar]
- Kuchinke U., Grawe F., Knust E. (1998). Control of spindle orientation in Drosophila by the Par-3-related PDZ-domain protein Bazooka. Curr. Biol. 8, 1357–1365 10.1016/S0960-9822(98)00016-5 [DOI] [PubMed] [Google Scholar]
- Laprise P., Tepass U. (2011). Novel insights into epithelial polarity proteins in Drosophila. Trends Cell Biol. 21, 401–408 10.1016/j.tcb.2011.03.005 [DOI] [PubMed] [Google Scholar]
- Lee T., Luo L. (2001). Mosaic analysis with a repressible cell marker (MARCM) for Drosophila neural development. Trends Neurosci. 24, 251–254 10.1016/S0166-2236(00)01791-4 [DOI] [PubMed] [Google Scholar]
- Ling C., Zheng Y., Yin F., Yu J., Huang J., Hong Y., Wu S., Pan D. (2010). The apical transmembrane protein Crumbs functions as a tumor suppressor that regulates Hippo signaling by binding to Expanded. Proc. Natl. Acad. Sci. USA 107, 10532–10537 10.1073/pnas.1004279107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maitra S., Kulikauskas R. M., Gavilan H., Fehon R. G. (2006). The tumor suppressors Merlin and Expanded function cooperatively to modulate receptor endocytosis and signaling. Curr. Biol. 16, 702–709 10.1016/j.cub.2006.02.063 [DOI] [PubMed] [Google Scholar]
- Margolis J., Spradling A. (1995). Identification and behavior of epithelial stem cells in the Drosophila ovary. Development 121, 3797–3807. [DOI] [PubMed] [Google Scholar]
- Martin-Belmonte F., Perez-Moreno M. (2012). Epithelial cell polarity, stem cells and cancer. Nat. Rev. Cancer 12, 23–38 10.1038/nrc3169 [DOI] [PubMed] [Google Scholar]
- Mavrakis M., Rikhy R., Lippincott-Schwartz J. (2009). Plasma membrane polarity and compartmentalization are established before cellularization in the fly embryo. Dev. Cell 16, 93–104 10.1016/j.devcel.2008.11.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCaffrey L. M., Macara I. G. (2011). Epithelial organization, cell polarity and tumorigenesis. Trends Cell Biol. 21, 727–735 10.1016/j.tcb.2011.06.005 [DOI] [PubMed] [Google Scholar]
- McGill M. A., McKinley R. F., Harris T. J. (2009). Independent cadherin-catenin and Bazooka clusters interact to assemble adherens junctions. J. Cell Biol. 185, 787–796 10.1083/jcb.200812146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKim K., Dahmus J., Hawley R. (1996). Cloning of the Drosophila melanogaster meiotic recombination gene mei-218: a genetic and molecular analysis of interval 15E. Genetics 144, 215–228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meignin C., Alvarez-Garcia I., Davis I., Palacios I. M. (2007). The salvador-warts-hippo pathway is required for epithelial proliferation and axis specification in Drosophila. Curr. Biol. 17, 1871–1878 10.1016/j.cub.2007.09.062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizuno K., Suzuki A., Hirose T., Kitamura K., Kutsuzawa K., Futaki M., Amano Y., Ohno S. (2003). Self-association of PAR-3-mediated by the conserved N-terminal domain contributes to the development of epithelial tight junctions. J. Biol. Chem. 278, 31240–31250 10.1074/jbc.M303593200 [DOI] [PubMed] [Google Scholar]
- Morais-de-Sá E., Mirouse V., St Johnston D. (2010). aPKC phosphorylation of Bazooka defines the apical/lateral border in Drosophila epithelial cells. Cell 141, 509–523 10.1016/j.cell.2010.02.040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller H. A., Wieschaus E. (1996). armadillo, bazooka, and stardust are critical for early stages in formation of the zonula adherens and maintenance of the polarized blastoderm epithelium in Drosophila. J. Cell Biol. 134, 149–163 10.1083/jcb.134.1.149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parsons L. M., Grzeschik N. A., Allott M. L., Richardson H. E. (2010). Lgl/aPKC and Crb regulate the Salvador/Warts/Hippo pathway. Fly (Austin) 4, 288–293 10.4161/fly.4.4.13116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petronczki M., Knoblich J. A. (2001). DmPAR-6 directs epithelial polarity and asymmetric cell division of neuroblasts in Drosophila. Nat. Cell Biol. 3, 43–49 10.1038/35050550 [DOI] [PubMed] [Google Scholar]
- Pinal N., Goberdhan D. C. I., Collinson L., Fujita Y., Cox I. M., Wilson C., Pichaud F. (2006). Regulated and polarized PtdIns(3,4,5)P3 accumulation is essential for apical membrane morphogenesis in photoreceptor epithelial cells. Curr. Biol. 16, 140–149 10.1016/j.cub.2005.11.068 [DOI] [PubMed] [Google Scholar]
- Polesello C., Tapon N. (2007). Salvador-warts-hippo signaling promotes Drosophila posterior follicle cell maturation downstream of notch. Curr. Biol. 17, 1864–1870 10.1016/j.cub.2007.09.049 [DOI] [PubMed] [Google Scholar]
- Robinson B. S., Huang J., Hong Y., Moberg K. H. (2010). Crumbs regulates Salvador/Warts/Hippo signaling in Drosophila via the FERM-domain protein Expanded. Curr. Biol. 20, 582–590 10.1016/j.cub.2010.03.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez-Boulan E., Macara I. G. (2014). Organization and execution of the epithelial polarity programme. Nat. Rev. Mol. Cell Biol. 15, 225–242 10.1038/nrm3775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rolls M. M., Albertson R., Shih H.-P., Lee C.-Y., Doe C. Q. (2003). Drosophila aPKC regulates cell polarity and cell proliferation in neuroblasts and epithelia. J. Cell Biol. 163, 1089–1098 10.1083/jcb.200306079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawyer J. K., Harris N. J., Slep K. C., Gaul U., Peifer M. (2009). The Drosophila afadin homologue Canoe regulates linkage of the actin cytoskeleton to adherens junctions during apical constriction. J. Cell Biol. 186, 57–73 10.1083/jcb.200904001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider M., Khalil A. A., Poulton J., Castillejo-Lopez C., Egger-Adam D., Wodarz A., Deng W.-M., Baumgartner S. (2006). Perlecan and Dystroglycan act at the basal side of the Drosophila follicular epithelium to maintain epithelial organization. Development 133, 3805–3815 10.1242/dev.02549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schober M., Schaefer M., Knoblich J. A. (1999). Bazooka recruits Inscuteable to orient asymmetric cell divisions in Drosophila neuroblasts. Nature 402, 548–551 10.1038/990135 [DOI] [PubMed] [Google Scholar]
- Shulman J. M., Benton R., St Johnston D. (2000). The Drosophila homolog of C. elegans PAR-1 organizes the oocyte cytoskeleton and directs oskar mRNA localization to the posterior pole. Cell 101, 377–388 10.1016/S0092-8674(00)80848-X [DOI] [PubMed] [Google Scholar]
- St Johnston D., Ahringer J. (2010). Cell polarity in eggs and epithelia: parallels and diversity. Cell 141, 757–774 10.1016/j.cell.2010.05.011 [DOI] [PubMed] [Google Scholar]
- Sun J., Deng W.-M. (2005). Notch-dependent downregulation of the homeodomain gene cut is required for the mitotic cycle/endocycle switch and cell differentiation in Drosophila follicle cells. Development 132, 4299–4308 10.1242/dev.02015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun J., Deng W.-M. (2007). Hindsight mediates the role of notch in suppressing hedgehog signaling and cell proliferation. Dev. Cell 12, 431–442 10.1016/j.devcel.2007.02.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanentzapf G., Tepass U. (2003). Interactions between the crumbs, lethal giant larvae and bazooka pathways in epithelial polarization. Nat. Cell Biol. 5, 46–52 10.1038/ncb896 [DOI] [PubMed] [Google Scholar]
- Tanentzapf G., Smith C., McGlade J., Tepass U. (2000). Apical, lateral, and basal polarization cues contribute to the development of the follicular epithelium during Drosophila oogenesis. J. Cell Biol. 151, 891–904 10.1083/jcb.151.4.891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tepass U. (2012). The apical polarity protein network in Drosophila epithelial cells: regulation of polarity, junctions, morphogenesis, cell growth, and survival. Annu. Rev. Cell Dev. Biol. 28, 655–685 10.1146/annurev-cellbio-092910-154033 [DOI] [PubMed] [Google Scholar]
- Tepass U., Knust E. (1993). Crumbs and stardust act in a genetic pathway that controls the organization of epithelia in Drosophila melanogaster. Dev. Biol. 159, 311–326 10.1006/dbio.1993.1243 [DOI] [PubMed] [Google Scholar]
- Tepass U., Tanentzapf G., Ward R., Fehon R. (2001). Epithelial cell polarity and cell junctions in Drosophila. Annu. Rev. Genet. 35, 747–784 10.1146/annurev.genet.35.102401.091415 [DOI] [PubMed] [Google Scholar]
- Thompson B. J., Pichaud F., Röper K. (2013). Sticking together the Crumbs – an unexpected function for an old friend. Nat. Rev. Mol. Cell Biol. 14, 307–314 10.1038/nrm3568 [DOI] [PubMed] [Google Scholar]
- Walther R. F., Pichaud F. (2010). Crumbs/DaPKC-dependent apical exclusion of Bazooka promotes photoreceptor polarity remodeling. Curr. Biol. 20, 1065–1074 10.1016/j.cub.2010.04.049 [DOI] [PubMed] [Google Scholar]
- Wei S.-Y., Escudero L. M., Yu F., Chang L.-H., Chen L.-Y., Ho Y.-H., Lin C.-M., Chou C.-S., Chia W., Modolell J. et al. (2005). Echinoid is a component of adherens junctions that cooperates with DE-Cadherin to mediate cell adhesion. Dev. Cell 8, 493–504 10.1016/j.devcel.2005.03.015 [DOI] [PubMed] [Google Scholar]
- Wieschaus E., Nusslein-Volhard C., Jurgens G. (1984). Mutations affecting the pattern of the larval cuticle in Drosophila melanogaster. Rouxs Arch. Dev. Biol. 193, 296–307 10.1007/BF00848158 [DOI] [PubMed] [Google Scholar]
- Wodarz A., Ramrath A., Kuchinke U., Knust E. (1999). Bazooka provides an apical cue for Inscuteable localization in Drosophila neuroblasts. Nature 402, 544–547 10.1038/990128 [DOI] [PubMed] [Google Scholar]
- Wodarz A., Ramrath A., Grimm A., Knust E. (2000). Drosophila Atypical Protein Kinase C Associates with Bazooka and Controls Polarity of Epithelia and Neuroblasts. J. Cell Biol. 150, 1361–1374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X., Tanwar P. S., Raftery L. A. (2008). Drosophila follicle cells: morphogenesis in an eggshell. Semin. Cell Dev. Biol. 19, 271–282 10.1016/j.semcdb.2008.01.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan Y., Denef N., Schüpbach T. (2009). The vacuolar proton pump, V-ATPase, is required for notch signaling and endosomal trafficking in Drosophila. Dev. Cell 17, 387–402 10.1016/j.devcel.2009.07.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu J., Poulton J., Huang Y.-C., Deng W.-M. (2008). The hippo pathway promotes Notch signaling in regulation of cell differentiation, proliferation, and oocyte polarity. PLoS One 3, e1761 10.1371/journal.pone.0001761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yue T., Tian A., Jiang J. (2012). The cell adhesion molecule echinoid functions as a tumor suppressor and upstream regulator of the Hippo signaling pathway. Dev. Cell 22, 255–267 10.1016/j.devcel.2011.12.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.