Summary
Epigenetic regulation of key transcriptional programs is a critical mechanism that controls hematopoietic development, and, thus, aberrant expression patterns or mutations in epigenetic regulators occur frequently in hematologic malignancies. We demonstrate that the Polycomb protein L3MBTL1, which is monoallelically deleted in 20q- myeloid malignancies, represses the ability of stem cells to drive hematopoietic-specific transcriptional programs by regulating the expression of SMAD5 and impairing its recruitment to target regulatory regions. Indeed, knockdown of L3MBTL1 promotes the development of hematopoiesis and impairs neural cell fate in human pluripotent stem cells. We also found a role for L3MBTL1 in regulating SMAD5 target gene expression in mature hematopoietic cell populations, thereby affecting erythroid differentiation. Taken together, we have identified epigenetic priming of hematopoietic-specific transcriptional networks, which may assist in the development of therapeutic approaches for patients with anemia.
Highlights
-
•
L3MBTL1 is a chromatin-binding protein that represses SMAD5 expression
-
•
Lack of L3MBTL1 primes the hematopoietic development of pluripotent stem cells
-
•
L3MBTL1 regulates erythroid differentiation
Nimer, Perna, and colleagues report that the Polycomb protein L3MBTL1, deleted in 20q- myeloid malignancies, plays a regulatory role during the differentiation of human iPS cells. Upon lentiviral knockdown of L3MBTL1, iPS cells stay poised for hematopoietic development and upregulate BMP/SMAD target gene expression, leading to impaired neural development.
Introduction
L3MBTL1 is the human homolog of the Drosophila tumor suppressor gene, l(3)mbt (Wismar et al., 1995). The L3MBTL1 gene is located on the long arm of chromosome 20, within the region that is commonly deleted in hematologic malignancies (Bench et al., 2000, 2004). The crystal structure of the three MBT domains in human L3MBTL1 exhibited a triple-bladed propeller-like structure (Wang et al., 2003), and subsequent studies showed that L3MBTL1 binds to mono- and di-methylated lysines on histones H1 (H1K26) and H4 (H4K20) via the second MBT repeat (Kalakonda et al., 2008; Li et al., 2007). Upon recruitment to the chromatin, L3MBTL1 generally functions as a chromatin compactor and transcriptional repressor (Boccuni et al., 2003; Kalakonda et al., 2008; Trojer et al., 2007). Despite its role in affecting chromatin structure, the function of L3MBTL1 in human hematopoiesis had largely remained elusive. Our group and others have demonstrated that knockdown (KD) of L3MBTL1 results in the enhanced erythroid differentiation of human hematopoietic stem/progenitor cells (HSPCs) (Aziz et al., 2013; Perna et al., 2010), which suggests that haploinsufficiency of L3MBTL1 contributes to 20q- myeloproliferative neoplasms.
In the human embryonic stem cells (hESCs), depletion of L3MBTL1 leads to spontaneous trophoblastic differentiation, a phenotype that mirrors BMP4-treated hESCs (Hoya-Arias et al., 2011). BMP4 is a critical signaling molecule (Kawabata et al., 1998), directing the hematopoietic fate from mesoderm during development (Larsson and Karlsson, 2005; Lengerke et al., 2008; Lohmann and Bieker, 2008; McReynolds et al., 2007; Nostro et al., 2008; Pimanda et al., 2007; Zafonte et al., 2007). In particular, exogenous BMP4 can augment the hematopoietic differentiation of hESCs, and substantially increase the proportion of committed hematopoietic cells generated from induced pluripotent stem cells (iPSCs) (Hong et al., 2011). It is now believed that a mixture of trophoblast- and mesoderm-committed cells emerges in response to BMP4 exposure (Bernardo et al., 2011). BMP4 engages the BMP receptor, promoting the interaction between its two subunits (BMPR-IA or IB and II) and triggering the phosphorylation of SMAD1, SMAD5, or SMAD8 prior to their association with SMAD4 and their translocation to the nucleus (Massagué and Chen, 2000).
Ectopic BMP signaling activates the primitive erythroid program, while inhibiting the pathway blocks ventral blood island formation in Xenopus (Zhang and Evans, 1996). This suggests that BMP signaling may represent a critical influence on erythroid differentiation, in addition to its role in mesoderm specification (Schmerer and Evans, 2003). In the human adult hematopoietic system, BMP4 mediates regeneration under stress conditions (Lenox et al., 2005; Trompouki et al., 2011) and the differentiation of hematopoietic progenitors into erythroid and myeloid lineages (Detmer and Walker, 2002; Fuchs et al., 2002). BMP signaling also has been implicated in the malignant transformation of HSPCs: the recently discovered, cryptic recurring translocation in pediatric acute megakaryoblastic leukemia, which leads to fusion of the CBFA2T3 and GLIS2 genes, alters the expression of BMP target genes, leading to enhanced self-renewal of HSPCs (Gruber et al., 2012).
Here, we demonstrate that depletion of L3MBTL1 primes human pluripotent stem cells to undergo hematopoietic fate commitment. We observed increased clonogenic hematopoietic potential in the knocked down cells compared to controls and the early emergence of a primitive CD45−CD31+CD34+ cell population thought to be hemogenic precursors. Comprehensive assessment of lineage fates in L3MBTL1-KD pluripotent stem cells showed decreased expression of endodermal- and ectodermal-specific genes. We also found impaired development of neural progenitors by culturing KD embryoid bodies (EBs) with brain-derived neurotrophic factor (BDNF), which was accompanied by increased expression of hematopoietic surface markers, despite the directive cell culture conditions.
The L3MBTL1-KD cells showed an upregulated SMAD5-mediated transcriptional signature, and we found that L3MBTL1 regulates the expression of SMAD5 and impairs its recruitment to target regulatory regions, in both immature and mature hematopoietic cell populations. Via effects on the erythroid-specific transcription factor, EKLF, L3MBTL1, and SMAD5 control the erythroid differentiation of primary cord blood CD34+ cells and hematopoietic cell lines.
Results
KD of L3MBTL1 Primes the Hematopoietic Potential of iPSCs
The generation of iPSC lines has provided opportunities to understand the fundamental processes of human cell fate decisions in the context of tissue regeneration and human disease. We first observed spontaneous downregulation of L3MBTL1 expression in a human iPSC line generated from cord blood cells (iCBCs) (Figures S1A–S1C) upon mesodermal differentiation, suggesting that decreased L3MBTL1 expression is required for mesoderm specification (Figure 1A). To investigate the role of L3MBTL1 on the specification of hematopoiesis during iPSC differentiation, we knocked down L3MBTL1 using H1P-hygro-EGFP+ lentiviral vectors that express small hairpin RNAs (shRNAs) directed against L3MBTL1 (Figure 1B). The subcloned green fluorescent protein (GFP)+ colonies showed a marked decrease in L3MBTL1 expression and retained stem cell morphology and stem-cell-related cell surface markers, including TRA-1-81, KLF4, OCT4, and NANOG expression (Figures S1D–S1G). Expression of mesoderm-specific transcription factors (T, TAL1, LMO2, and RUNX1) was consistently increased in the KD cells, while key markers for endoderm and ectoderm decreased (Figure 1C). We observed downregulation of SOX2 even though the iPSCs maintained self-renewal properties. While consistent with a previous report (Wang et al., 2012), we did find upregulation of SOX3 (data not shown), which also could explain the self-renewal phenotype. Functional assessment of clonogenic hematopoietic progenitors (CFUs) revealed that the lack of L3MBTL1 induced an increase in hematopoietic progenitor cell generation (Figure 1D). The hematopoietic colonies derived from the L3MBTL1-KD cells were also larger in size (data not shown). CD45−CD31+CD34+ cells, which represent a primitive bipotent endothelial-like precursor population that is exclusively responsible for hematopoietic fate (Wang et al., 2004), were also much more abundant within the L3MBTL1-KD cells compared to controls (Figure 1E).
Figure 1.
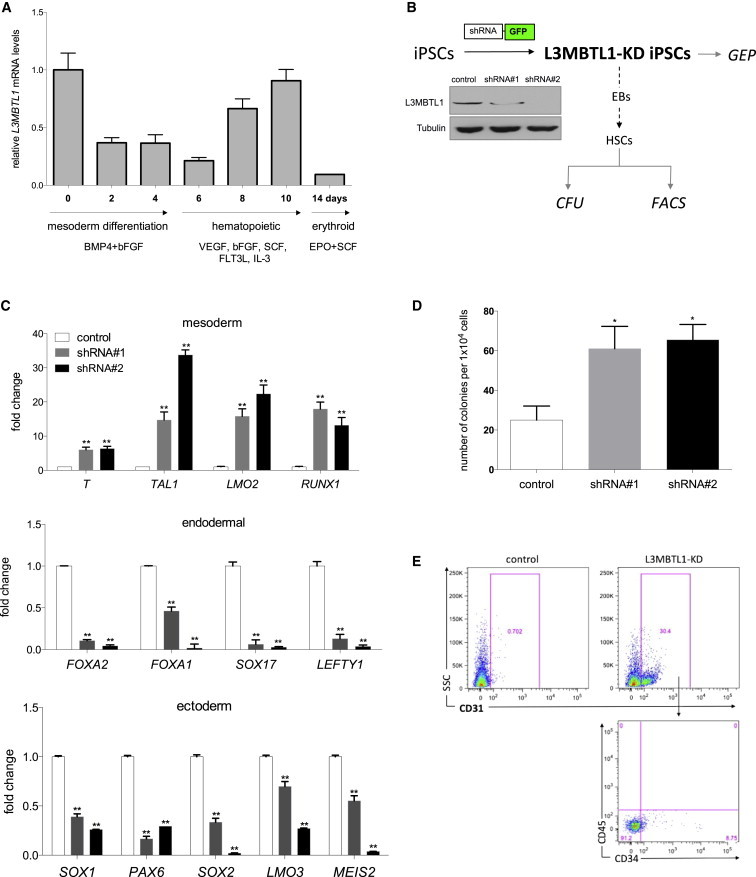
KD of L3MBTL1 Primes the Hematopoietic Potential of iPSCs
(A) Endogenous L3MBTL1 expression was assessed in iPSCs at different time points during the mesodermal, hematopoietic, and erythroid differentiation using quantitative real-time PCR. The data represent the mean ± SD of the three independent experiments.
(B) Strategy diagram. L3MBTL1 expression is efficiently knocked down as assessed by western blot assay in undifferentiated iPSCs. Tubulin served as the loading control.
(C) KD of L3MBTL1 increases the expression of mesodermal-specific transcription factors while it decreases expression of key markers of endodermal and ectodermal lineages, as shown in L3MBTL1-KD undifferentiated iPSCs by qPCR compared to controls. Data indicate the relative expression level of the gene of interest, normalized by GAPDH. The reported fold changes have been calculated by ΔΔCt analysis versus control cells. The data represent the mean ± SD of the three independent experiments. ∗∗p < 0.01 by Student’s t test.
(D) KD of L3MBTL1 increases CFU capacity of iPSC-derived HSCs. Cells from day 10 hEBs were plated in methylcellulose and colonies were scored after 15 days. The data represent the mean ± SD of the three independent experiments. ∗p < 0.05 by Student’s t test.
(E) KD of L3MBTL1 promotes the emergence of early hemogenic precursor cells. EBs were harvested at day 10, prior to CD45 emergence, and analyzed by flow cytometry for CD31 and CD34 expression.
Refer to Figure S1 for the characterization of the iPSC line generated from cord blood CD34+ cells.
Taken together, these findings indicate that lowering L3MBTL1 expression primes hematopoietic development in iPSCs, inducing changes in gene expression that regulate lineage fate divisions driven by specific differentiation-promoting conditions.
Impaired Development of Neural Progenitors in L3MBTL1-KD Pluripotent Stem Cells
To assess the neural differentiation potential of the L3MBTL1-KD cells, we generated EBs and cultured them in BDNF-containing conditions that support the development of neural progenitor cells (Figure 2A, top). While control EBs developed into Nestin+ neural precursors and Tuj1+ differentiated neurons, L3MBTL1-KD cells showed impaired neural lineage differentiation (Figure 2A, bottom and right). The impaired generation of neural precursors is not dependent on decreased cell proliferation or increased apoptosis, as we found no significant change in the expression of Ki-67 and Annexin V (Figures S2A and S2B).
Figure 2.
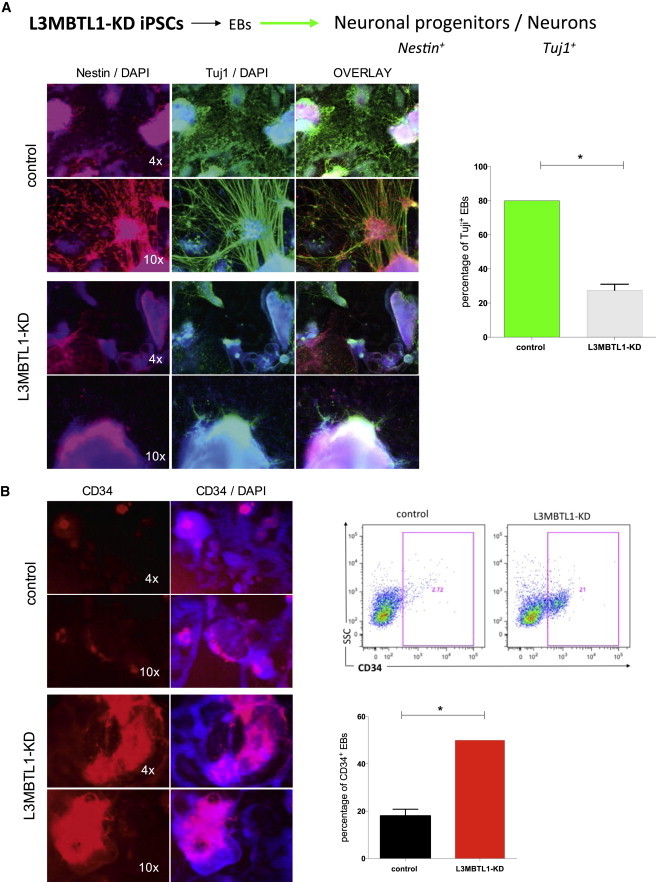
Impaired Development of Neural Progenitors in L3MBTL1-KD Pluripotent Stem Cells
(A) KD of L3MBTL1 impairs the ability to form neuronal progenitor cells and neurons. Nestin and Tuj1 expression were evaluated in EBs by IF. DAPI served as control. (Top) The first line shows multiple EBs at lesser magnification (4×); the second line shows a single EB at 10× magnification. (Bottom) The impairment in neural progenitor formation of the L3MBTL1-KD EBs. The graph on the right shows the percentage of EBs containing Tuj1+ cells. The data represent the mean ± SD of the three independent experiments. ∗p < 0.05 by Student’s t test.
(B) KD of L3MBTL1 enhances CD34 expression. (Top left) The first line shows few positive CD34+ EBs at low magnification (4×) in the controls; the second line shows a single EB at higher magnification (10×). (Bottom left) Increased CD34 staining in the L3MBTL1-KD EBs at 4× and 10× magnification is apparent. (Top right) Increased expression of CD34 marker (21%) in the L3MBTL1-KD cells compared to controls (3%) by FACS is shown. (Bottom right) The graph gives the percentage of EBs containing CD34+ cells. The data represent the mean ± SD of the three independent experiments. ∗p < 0.05 by Student’s t test. See also Figure S2.
To determine whether L3MBTL1-KD EBs still express hematopoietic markers under culture conditions that specifically support neural development, we first assessed CD34 expression using immunofluorescence (IF) and flow cytometry. We found increased CD34 expression in the KD EBs under conditions that generally promote neural fate (Figure 2B). Given the general impairment of neural differentiation, the expression of CD31 and TAL1, which are expressed in hemato-endothelial progenitors (Real et al., 2012) but also increased in the KD EBs (Figure S2C), suggests the presence of hemogenic precursors. It is possible that the TAL1+ cells in the L3MBTL1-KD are undifferentiated ESCs within a mixture of cell populations that could not undergo neural development under BDNF-mediated culture conditions.
Overall, these data indicate that KD of L3MBTL1 promotes hematopoietic development while impairing neural development.
L3MBTL1 Transcriptionally Represses SMAD5
To investigate the mechanisms underlying the observed effects of L3MBTL1 on stem cell biology, we utilized an unbiased, genome-wide approach to analyze the gene expression profile (GEP) of two independent clones of undifferentiated L3MBTL1-KD iPSCs. We found increased expression of several BMP/SMAD target genes, as well as downregulation of negative regulators of BMP signaling (Figures 3A and 3B). ID2 and ID3 were among the upregulated genes, which are direct targets of BMP4 and regulate HSPC fate decisions (Hong et al., 2011; van Galen et al., 2014). HHEX also was upregulated; it is known to regulate globin gene expression during ontogeny and its promoter contains a 71 nucleotide BMP-responsive element (BRE) (Zhang et al., 2002). We also found increased expression of ZFP36L2, an RNA-binding protein required for BFU-E self-renewal (Zhang et al., 2013) (data not shown). Expression of SMAD7, which inhibits TGFβ-related signaling (Nakao et al., 1997), was downregulated, as were LEFTY1 and LEFTY2, which function as extracellular antagonists of Nodal signaling (Yeo and Whitman, 2001).
Figure 3.
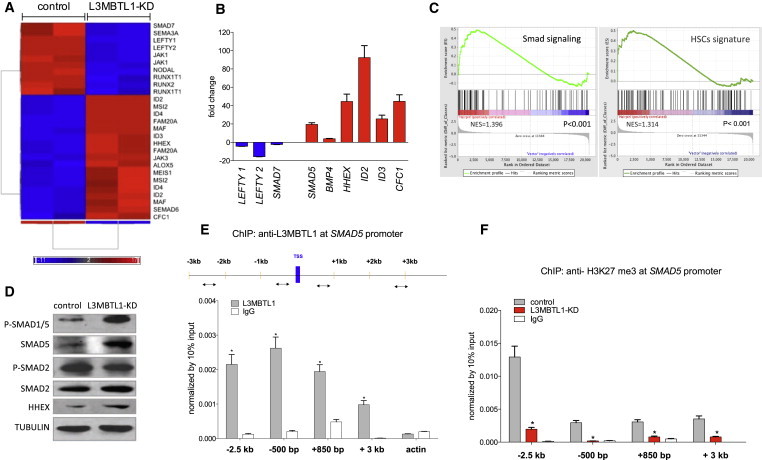
L3MBTL1 Transcriptionally Represses SMAD5
(A) GEP of undifferentiated L3MBTL1-KD iPSCs, compared to controls, based on microarray analysis. We utilized independent clones of iPSCs (generated from cord blood CD34+ cells), which we independently infected with lentiviral vectors expressing shRNAs against L3MBTL1.
(B) mRNA expression levels of several upregulated and downregulated genes, identified by microarray analysis, was confirmed by qPCR. Data were normalized by GAPDH expression and shown as shRNA versus control. The data represent the mean ± SD of the three independent experiments.
(C) GSEA for the combined set of SMAD and hematopoietic stem cells genes compared with the differentially expressed genes in L3MBTL1-KD iPSCs.
(D) Expression levels of phospho SMAD1/5, total SMAD5, phospho SMAD2, total SMAD2, and HHEX were evaluated in L3MBTL1-KD and control iPSCs by western blot. Tubulin served as the loading control.
(E) iPSCs were crosslinked with 1% formaldehyde and immunoprecipitated with anti-L3MBTL1 or IgG antibody (as a nonspecific control). Plotted values are relative enrichments (y axis) to 10% input and measured for sites in the SMAD5 promoter and actin (x axis). The data represent the mean ± SD of the three independent experiments. ∗p < 0.05 by Student’s t test.
(F) L3MBTL1-KD and control iPSCs were crosslinked with 1% formaldehyde and immunoprecipitated with an anti-H3K27 tri-methyl antibody or an IgG antibody (as a nonspecific control). Plotted values are relative enrichments (y axis) to 10% input and measured for sites in the SMAD5 promoter (x axis). The data represent the mean ± SD of the three independent experiments. ∗p < 0.05 by Student’s t test. See also Figure S3.
Analysis of curated pathway gene sets by gene set enrichment analysis (GSEA) showed statistically significant (p < 0.0002; FDR < 0.001) enrichment of SMAD targets as well as enrichment of three different sets of hematopoietic stem cell genes (Figures 3C and S3A). We also found enrichment of hematopoietic (non-stem cell) gene sets and enrichment of hematopoietic transcription factor target gene sets (Figures S3B and S3C). These data, which were obtained from the undifferentiated stage of L3MBTL1-KD iPSCs, show activation of a hematopoietic cell transcriptional program, fully compatible with the phenotype we induced by culturing these cells under hematopoietic-specifying culture conditions. Gene ontology analysis also supported these findings (Table S1).
To identify which SMAD-regulated signaling pathways are triggered by L3MBTL1 depletion, we evaluated the phosphorylation of SMAD1/5 and SMAD2/3, which primarily function downstream of the BMP receptors and the TGFβ, Activin, and Nodal receptors, respectively. We found enhanced SMAD1/5 phosphorylation (Figure 3D), indicating that L3MBTL1 titrates BMP signaling.
We also found increased SMAD5 (Figure 3D) and SMAD1 (Figure S3D) expression in the L3MBTL1-KD cells compared to controls. Given the transcriptional repressor functions of L3MBTL1 (Boccuni et al., 2003), we investigated whether L3MBTL1 directly regulates SMAD5 and SMAD1 expression by performing chromatin immunoprecipitation (ChIP) assays. We found recruitment of L3MBTL1 to regulatory regions of the SMAD5 and SMAD1 promoters (Figures 3E and S3E), yet the increase in expression was much higher for SMAD5 than for SMAD1 and correlated with stronger recruitment of L3MBTL1 to the SMAD5 than to the SMAD1 target promoter. Consistent with the increased SMAD5 and SMAD1 expression, ChIP experiments in the L3MBTL1-KD cells showed fewer repressive histone marks (i.e., H3K27me3) at the SMAD5 than at the SMAD1 promoter (Figures 3F and S3F).
Regulation of SMAD5 by L3MBTL1 Occurs in Erythroid Cells
We, and others, have suggested that L3MBTL1 plays an important role in erythroid differentiation of human CD34+ HSPCs (Aziz et al., 2013; Perna et al., 2010). We found that downregulation of L3MBTL1 occurs during normal erythroid differentiation, in a manner similar to that seen during mesodermal differentiation (Figure 1A). We first assessed the effect of L3MBTL1-KD in iPSC-derived CD34+ cells undergoing erythroid differentiation, and observed a marked acceleration in the expression of the erythroid-specific marker GlyA (after 2 days in EPO-induced culture) by flow cytometry in the L3MBTL1-KD cells (Figure 4A). This was followed by a significant increase in the expression of several erythroid-specific transcription factors, especially EKLF (Figure 4B). We also confirmed this phenotype in β-thalassemic iPSCs (Papapetrou et al., 2011), which we lentivirally infected to express shRNAs targeting L3MBTL1 (Figure S4A). Similar to the CB-iPSC line, knocking down L3MBTL1 in thalassemic erythroid progeny also increased EKLF expression (Figure S4B) and increased fetal globin gene expression compared to controls (Figure S4C).
Figure 4.
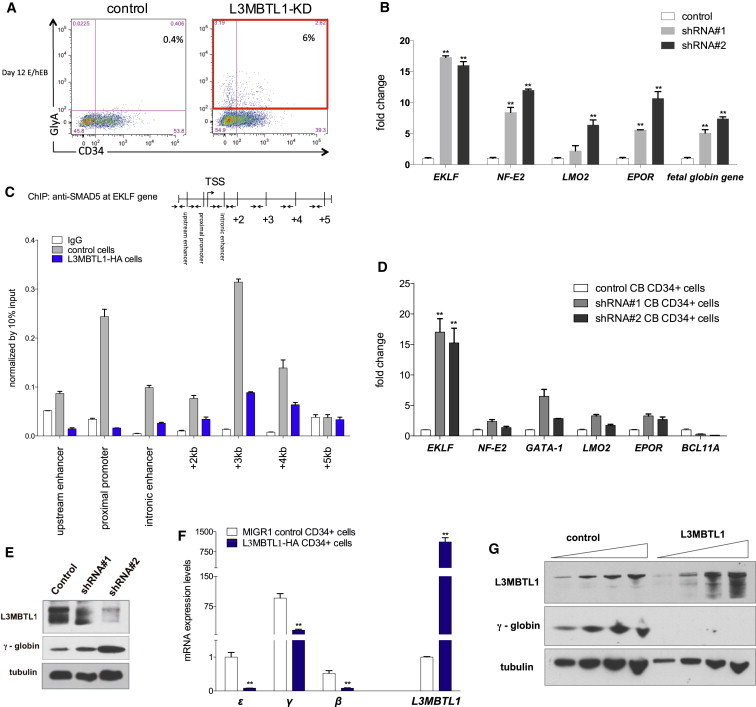
Regulation of SMAD5 by L3MBTL1 Occurs in Erythroid Cells
(A) KD of L3MBTL1 induces early expression of the erythroid-specific marker GlyA in iPSC-derived CD34+ cells. Representative flow cytometry analysis of iPSCs after 2 days in Epo-induced culture.
(B) Expression levels of EKLF, NF-E2, LMO2, EPOR, and fetal globin gene were assessed in iPSC-derived erythroid progeny by qRT-PCR. Data were normalized by GAPDH expression. The data represent the mean ± SD of the three independent experiments. ∗∗p < 0.01 by Student’s t test.
(C) K562 erythroleukemia cells were retrovirally transduced to express L3MBTL1-HA (blue bars) or empty MIGR1 control (gray bars), crosslinked with 1% formaldehyde, and immunoprecipitated with an anti-SMAD5 antibody or IgG antibody. Primers covering the SMAD-binding motifs (Adelman et al., 2002) across the upstream enhancer, the proximal promoter, and the intronic enhancer of the EKLF were utilized. Data were normalized by 10% input. The data represent the mean ± SD of the three independent experiments.
(D) EKLF, NF-E2, GATA-1, LMO2, EPOR, and BCL11A expression levels were evaluated by qRT-PCR in L3MBTL1-KD CB CD34+ cells after 7 days of erythroid-supporting culture (100 ng/ml SCF and 6 U/ml EPO) compared to controls. GAPDH served as a housekeeping gene control. The data represent the mean ± SD of the three independent experiments. ∗∗p < 0.01 by Student’s t test.
(E) Fetal globin levels were evaluated in L3MBTL1-KD CB CD34+ cells by western blot. Tubulin served as the loading control.
(F) Overexpression of L3MBTL1 decreases globin gene mRNA levels in cord blood CD34+ cells, as shown by qRT-PCR. Cells were cultured for 3 days in liquid culture that supports erythroid differentiation. GAPDH served as the housekeeping gene. The data represent the mean ± SD of the three independent experiments. ∗∗p < 0.01 by Student’s t test.
(G) Overexpression of L3MBTL1 dramatically decreases protein expression levels of gamma globin in K562 cells, as shown by western blot assay. Tubulin served as the loading control. See also Figure S4.
Ten SMAD-binding motifs were identified within the upstream enhancer, proximal promoter, and intronic enhancer of EKLF using EBs (Adelman et al., 2002; Lohmann and Bieker, 2008); thus, we investigated whether L3MBTL1 regulates EKLF gene expression by SMAD5 in cells that are more mature than pluripotent stem cells. We found SMAD5 at most of these regions using ChIP assays in K562 erythroleukemia cells (Figure 4C, gray bars), and by retrovirally overexpressing L3MBTL1-HA in K562 cells we found impaired recruitment of SMAD5 to these EKLF regulatory regions (Figure 4C, blue bars). Furthermore, L3MBTL1 overexpression decreased SMAD5 expression in K562 cells (Figure S4D).
When we examined primary cord blood CD34+ cells, we found primarily increased expression of EKLF among several erythroid-specific genes following KD of L3MBTL1 (Figure 4D). We also found increased globin gene expression in the L3MBTL1-KD CD34+ cells (Figures 4E and S4E) and decreased globin expression following overexpression of L3MBTL1 (Figure 4F). Similar observations were made in K562 cells (Figures 4G, S4F, and S4G), where we also found an increase in EKLF expression following L3MBTL1-KD (Figure S4H).
Overall, these data indicate that the regulation of SMAD targets by L3MBTL1 occurs in mature erythroid cells as well as pluripotent stem cells; this effect further promotes erythroid differentiation.
Discussion
This study shows that loss of the Polycomb protein L3MBTL1 primes human iPSCs for hematopoietic differentiation and enhances commitment toward the erythroid lineage.
Epigenetic Priming of Hematopoietic Lineage
In many biological systems, epigenetic regulation is important for modulating the expression of critical developmental genes (Talbert and Henikoff, 2010). For example, in hESCs, epigenetic priming of genomic regulatory regions is an important mechanism underlying the early specification of future developmental fates. During the erythroid differentiation of HSPCs, many genes involved in development and differentiation lose their H3K27me3 mark, and a fraction of them become enriched with H3K4me1 and H3K9me1 marks, suggesting that these genes are poised for activation during the differentiation process (Bernstein et al., 2006; Cui et al., 2009). Several epigenetic regulatory mechanisms control gene induction and repression during erythroid development (Hattangadi et al., 2011). The enhancer-mediated recruitment of JMJD3 leads to exclusion of the repressive Polycomb repressive complex 2 (PRC2) from the poised alpha-globin promoter in erythroid cells (Vernimmen et al., 2011). The L3MBTL1-KD undifferentiated iPSCs display a GEP that predicts the precocious generation of hemogenic precursors and an expansion of induced hematopoietic cells under culture conditions that support hematopoietic cell specification. These changes in gene expression include upregulation of BMP/SMAD targets and downregulation of inhibitors of this signaling pathway, which may mediate, at least in part, a trophectoderm versus epiblast decision, as well as a mesoderm versus ectoderm decision, and finally a non-neural ectoderm versus neural ectoderm decision. Accordingly, the KD iPSCs cannot efficiently generate neural progenitors under conditions that allow activation of the BMP/SMAD-signaling pathway. The impaired neural development that we observed in L3MBTL1-KD cells supports the observation that activation of BMP4 signaling is inhibitory to the development of non-hematopoietic lineages (Dee et al., 2007). Consistent with our findings, using two inhibitors of SMAD signaling, Noggin and SB431542, one can induce the rapid and nearly complete conversion of >80% of hESCs to neural cells (Chambers et al., 2009). This epigenetic silencing mechanism involving L3MBTL1 also may underlie previous findings in the L3MBTL1-KD hESCs, which are able to generate neural progenitors when cultured with SB431542 and dorsomorphin, strong inhibitors of the SMAD-signaling pathways, and spontaneously differentiate toward the trophoblast lineage (mirroring the BMP4-treated hESCs) (Hoya-Arias et al., 2011).
It is possible that a feedback regulatory mechanism of L3MBTL1 occurs upon the addition of extrinsic BMP4, as downregulation of L3MBTL1 expression occurs during mesodermal specification (Figure 1A). This could mean not only that mesodermal differentiation requires silencing of L3MBTL1, but also that BMP4 regulates L3MBTL1 expression, especially given that changes in culture conditions (days 6–10) restore the initial levels of L3MBTL1 expression.
An enhanced differentiation toward other non-hematopoietic mesodermal lineages, or non-neural ectoderm lineages, cannot be excluded in the L3MBTL1-KD cells. Indeed, while GSEA primarily supported our findings (Figure S3B), we also found significant enrichment of gene sets related to targets of other PcG proteins in ESCs, such as SUZ12 (Pasini et al., 2007). These epigenetic signatures regulate establishment of expression programs for ESC differentiation. In fact, given the diverse signaling pathways that govern hematopoietic cell development, it is likely that additional pathways contribute to hematopoiesis at later stages of differentiation.
Transcriptional Networks and Signaling Pathways in Lineage Specification
Developmental decisions require the integration of a variety of signaling pathways, and transitions between poised and active regulatory states are closely integrated with various developmental signals. There appears to be a close relationship between core lineage regulators and signaling pathway outputs. Hence, the genomic binding of transcriptional effectors of TGFβ, BMP, and WNT pathways not only follows the binding of master regulators of lineage identity, but can be broadly redirected by the overexpression of a regulatory or lineage-specifying factor for another cell type (Mullen et al., 2011; Trompouki et al., 2011).
Our study shows that modulation of L3MBTL1 expression directs lineage-specific gene expression by regulating the expression of SMAD5 and its recruitment to critical regulatory regions, such as the erythroid transcription factor EKLF, allowing it to regulate globin gene expression.
Combinatorial crosstalk among transcription factors is essential to direct the lineage specification of HSPCs. In agreement with larger maps of combinatorial interactions among transcription factors (Ravasi et al., 2010), our work implies that lineage specification is not completely determined by tissue-specific transcription factors (such as EKLF), but can be modulated by crosstalk with transcription factors that are broadly expressed (like SMAD5). A similar effect is seen in the regulation of monocyte differentiation by SMAD3 (generally expressed) and FLI1 (whose expression is more restricted primarily to macrophage-related tissues, such as spleen and lymph nodes) (Ravasi et al., 2010).
We identified SMAD5 as a target of L3MBTL1 and showed that L3MBTL1 represses its ability to drive target gene expression. KD of SMAD5 impairs maintenance of the erythroid program (Fuchs et al., 2002; Liu et al., 2003; McReynolds et al., 2007), whereas upregulation of EKLF, a crucial erythroid-specific transcription factor (Alhashem et al., 2011; Siatecka and Bieker, 2011), occurs in response to BMP4 treatment in EBs (Adelman et al., 2002; Lohmann and Bieker, 2008). Here we demonstrated the recruitment of SMAD5 to these binding sites in definitive hematopoietic cells and the regulation of EKLF expression and function via L3MBTL1.
As a readout of EKLF expression and function, we evaluated globin gene expression in definitive hematopoietic cells, which was inversely correlated with L3MBTL1 expression. Increased understanding of how genetic and epigenetic programs regulate different stages of red blood cell (RBC) development may enable large-scale production of functional RBCs from ESCs and iPSC-derived cells. Hopefully, a deeper understanding of the mechanisms regulating erythropoiesis will result in the development of drugs that are useful in treating patients with erythropoietin-resistant anemias, including bone marrow failure disorders.
Experimental Procedures
Generation and hematopoietic differentiation of human iPSCs were performed according to previous protocols (Papapetrou et al., 2011).
Neural Differentiation of EBs
Stem cell colonies were dissociated into single cells using accutase and cultured in suspension, without any differentiation factors, for 7 to 8 days until they formed three-dimensional multicellular aggregates. Once EBs formed, they were landed on PO/Laminin/Fibronectin-coated plates and further differentiated in N2 media supplemented with BDNF and ascorbic acid. Immunocytochemical and flow cytometry analyses were performed after an additional 7–14 days in culture.
Purification and in vitro primary culture of human cord blood CD34+ cells were performed according to previous protocols (Perna et al., 2010).
Generation of Lenti- and Retroviruses and Cell Cultures
Lenti- and retroviral vectors were produced by transfection of 293T and Phoenix A cells as previously described (Perna et al., 2010). Detailed description of the constructs was provided previously (Kalakonda et al., 2008; Perna et al., 2010). K562 cells were cultured in RPMI medium supplemented with 10% fetal bovine serum.
FACS Analysis and Sorting
Transduced CB CD34+ cells were sorted for green fluorescence (GFP) and CD34 expression after staining with an APC-conjugated anti-CD34 antibody (BD340667, Becton Dickinson), using a fluorescence-activated cell sorting (FACS) Vantage cell sorter. Cells were stained with the following antibodies: CD34-APC (BD340667, BD), CD34-PE (BD348057, BD), CD71-APC (BD561940, BD), Glycophorin A-PE (MHGLA04, Invitrogen), CD31-PE (BD555446, BD), CD34-PEcy7 (BD560710, BD), and CD45-APC (MHCD4505, Invitrogen), and analyzed by FACS. Neural progenitors were stained with Annexin V (BD556421, BD) and Ki-67 (BD558615, BD) antibodies upon fixation. Isotype-matched antibodies were utilized as negative controls.
RNA Extraction and Quantitative Real-Time PCR
For quantitative real-time PCR, total RNA was isolated from 2 × 105 cells using the RNeasy mini kit (QIAGEN), and then subjected to reverse transcription with random hexamers (SuperScript III kit, Invitrogen). Real-time PCR reactions were performed using an ABI 7500 sequence detection system. A list of PCR primers is available upon request.
Antibodies
The following antibodies were used for western blot assays: the affinity purified anti-L3MBTL1 antibody (described in Kalakonda et al., 2008), pSMAD1/5 (9516), SMAD1 (6944P), SMAD5 (9517S), pSMAD2 (3108S), SMAD2 (5339P) (Cell Signaling Technology), HHEX (Chemicon 10071), EKLF (Abcam 49158), and HA (sc-805).
IF
Cultures were fixed in 4% paraformaldehyde and processed for immunocytochemistry. The following primary antibodies were used: Tuj1 (Covance MRB435P-100), Nestin (Neuromics MO15012), and TAL1 (Abcam 119754).
ChIP Assays
Approximately 4 × 106 cells were used per ChIP reaction (per antibody) after crosslinking with 1% formaldehyde for 10 min at room temperature. ChIP was performed according to the previously reported methodology (Zhao et al., 2008). The associated DNA after purification was subjected to quantitative real-time PCR to detect specific DNA sequences. Quantitative results are represented as percentages to 10% DNA input. The primer sequences are available upon request.
GEP and GSEA
RNA was extracted and hybridized to Affymetrix Human U133 plus 2.0 arrays, scanned, and subjected to quality control parameters. The GSEA (Subramanian et al., 2005) was performed on 4,722 curated pathway gene sets from the Broad Molecular Signature Database (http://www.broadinstitute.org/gsea/msigdb/).
Acknowledgments
This work was supported by an American Society of Hematology Scholar Award and Memorial Sloan Kettering Cancer Center Clinical Scholars Biomedical Research Fellowship (Dana Foundation) to F.P., National Cancer Institute R01 CA-102202 to S.D.N. and Gene Repression Fund to S.D.N. and R.H.-A., and National Institute of Diabetes and Digestive and Kidney Diseases K99DK087923 to E.P.P.
Footnotes
This is an open access article under the CC BY-NC-ND license (http://creativecommons.org/licenses/by-nc-nd/4.0/).
Contributor Information
Fabiana Perna, Email: pernaf@mskcc.org.
Stephen D. Nimer, Email: snimer@med.miami.edu.
Accession Numbers
Data have been deposited in the NCBI Gene Expression Omnibus under accession number GSE65438.
Supplemental Information
References
- Adelman C.A., Chattopadhyay S., Bieker J.J. The BMP/BMPR/Smad pathway directs expression of the erythroid-specific EKLF and GATA1 transcription factors during embryoid body differentiation in serum-free media. Development. 2002;129:539–549. doi: 10.1242/dev.129.2.539. [DOI] [PubMed] [Google Scholar]
- Alhashem Y.N., Vinjamur D.S., Basu M., Klingmüller U., Gaensler K.M., Lloyd J.A. Transcription factors KLF1 and KLF2 positively regulate embryonic and fetal beta-globin genes through direct promoter binding. J. Biol. Chem. 2011;286:24819–24827. doi: 10.1074/jbc.M111.247536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aziz A., Baxter E.J., Edwards C., Cheong C.Y., Ito M., Bench A., Kelley R., Silber Y., Beer P.A., Chng K. Cooperativity of imprinted genes inactivated by acquired chromosome 20q deletions. J. Clin. Invest. 2013;123:2169–2182. doi: 10.1172/JCI66113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bench A.J., Nacheva E.P., Hood T.L., Holden J.L., French L., Swanton S., Champion K.M., Li J., Whittaker P., Stavrides G., UK Cancer Cytogenetics Group (UKCCG) Chromosome 20 deletions in myeloid malignancies: reduction of the common deleted region, generation of a PAC/BAC contig and identification of candidate genes. Oncogene. 2000;19:3902–3913. doi: 10.1038/sj.onc.1203728. [DOI] [PubMed] [Google Scholar]
- Bench A.J., Li J., Huntly B.J., Delabesse E., Fourouclas N., Hunt A.R., Deloukas P., Green A.R. Characterization of the imprinted polycomb gene L3MBTL, a candidate 20q tumour suppressor gene, in patients with myeloid malignancies. Br. J. Haematol. 2004;127:509–518. doi: 10.1111/j.1365-2141.2004.05278.x. [DOI] [PubMed] [Google Scholar]
- Bernardo A.S., Faial T., Gardner L., Niakan K.K., Ortmann D., Senner C.E., Callery E.M., Trotter M.W., Hemberger M., Smith J.C. BRACHYURY and CDX2 mediate BMP-induced differentiation of human and mouse pluripotent stem cells into embryonic and extraembryonic lineages. Cell Stem Cell. 2011;9:144–155. doi: 10.1016/j.stem.2011.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein B.E., Mikkelsen T.S., Xie X., Kamal M., Huebert D.J., Cuff J., Fry B., Meissner A., Wernig M., Plath K. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell. 2006;125:315–326. doi: 10.1016/j.cell.2006.02.041. [DOI] [PubMed] [Google Scholar]
- Boccuni P., MacGrogan D., Scandura J.M., Nimer S.D. The human L(3)MBT polycomb group protein is a transcriptional repressor and interacts physically and functionally with TEL (ETV6) J. Biol. Chem. 2003;278:15412–15420. doi: 10.1074/jbc.M300592200. [DOI] [PubMed] [Google Scholar]
- Chambers S.M., Fasano C.A., Papapetrou E.P., Tomishima M., Sadelain M., Studer L. Highly efficient neural conversion of human ES and iPS cells by dual inhibition of SMAD signaling. Nat. Biotechnol. 2009;27:275–280. doi: 10.1038/nbt.1529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui K., Zang C., Roh T.Y., Schones D.E., Childs R.W., Peng W., Zhao K. Chromatin signatures in multipotent human hematopoietic stem cells indicate the fate of bivalent genes during differentiation. Cell Stem Cell. 2009;4:80–93. doi: 10.1016/j.stem.2008.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dee C.T., Gibson A., Rengifo A., Sun S.K., Patient R.K., Scotting P.J. A change in response to Bmp signalling precedes ectodermal fate choice. Int. J. Dev. Biol. 2007;51:79–84. doi: 10.1387/ijdb.062204cd. [DOI] [PubMed] [Google Scholar]
- Detmer K., Walker A.N. Bone morphogenetic proteins act synergistically with haematopoietic cytokines in the differentiation of haematopoietic progenitors. Cytokine. 2002;17:36–42. doi: 10.1006/cyto.2001.0984. [DOI] [PubMed] [Google Scholar]
- Fuchs O., Simakova O., Klener P., Cmejlova J., Zivny J., Zavadil J., Stopka T. Inhibition of Smad5 in human hematopoietic progenitors blocks erythroid differentiation induced by BMP4. Blood Cells Mol. Dis. 2002;28:221–233. doi: 10.1006/bcmd.2002.0487. [DOI] [PubMed] [Google Scholar]
- Gruber T.A., Larson Gedman A., Zhang J., Koss C.S., Marada S., Ta H.Q., Chen S.C., Su X., Ogden S.K., Dang J. An Inv(16)(p13.3q24.3)-encoded CBFA2T3-GLIS2 fusion protein defines an aggressive subtype of pediatric acute megakaryoblastic leukemia. Cancer Cell. 2012;22:683–697. doi: 10.1016/j.ccr.2012.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hattangadi S.M., Wong P., Zhang L., Flygare J., Lodish H.F. From stem cell to red cell: regulation of erythropoiesis at multiple levels by multiple proteins, RNAs, and chromatin modifications. Blood. 2011;118:6258–6268. doi: 10.1182/blood-2011-07-356006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong S.H., Lee J.H., Lee J.B., Ji J., Bhatia M. ID1 and ID3 represent conserved negative regulators of human embryonic and induced pluripotent stem cell hematopoiesis. J. Cell Sci. 2011;124:1445–1452. doi: 10.1242/jcs.077511. [DOI] [PubMed] [Google Scholar]
- Hoya-Arias R., Tomishima M., Perna F., Voza F., Nimer S.D. L3MBTL1 deficiency directs the differentiation of human embryonic stem cells toward trophectoderm. Stem Cells Dev. 2011;20:1889–1900. doi: 10.1089/scd.2010.0437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalakonda N., Fischle W., Boccuni P., Gurvich N., Hoya-Arias R., Zhao X., Miyata Y., Macgrogan D., Zhang J., Sims J.K. Histone H4 lysine 20 monomethylation promotes transcriptional repression by L3MBTL1. Oncogene. 2008;27:4293–4304. doi: 10.1038/onc.2008.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawabata M., Imamura T., Miyazono K. Signal transduction by bone morphogenetic proteins. Cytokine Growth Factor Rev. 1998;9:49–61. doi: 10.1016/s1359-6101(97)00036-1. [DOI] [PubMed] [Google Scholar]
- Larsson J., Karlsson S. The role of Smad signaling in hematopoiesis. Oncogene. 2005;24:5676–5692. doi: 10.1038/sj.onc.1208920. [DOI] [PubMed] [Google Scholar]
- Lengerke C., Schmitt S., Bowman T.V., Jang I.H., Maouche-Chretien L., McKinney-Freeman S., Davidson A.J., Hammerschmidt M., Rentzsch F., Green J.B. BMP and Wnt specify hematopoietic fate by activation of the Cdx-Hox pathway. Cell Stem Cell. 2008;2:72–82. doi: 10.1016/j.stem.2007.10.022. [DOI] [PubMed] [Google Scholar]
- Lenox L.E., Perry J.M., Paulson R.F. BMP4 and Madh5 regulate the erythroid response to acute anemia. Blood. 2005;105:2741–2748. doi: 10.1182/blood-2004-02-0703. [DOI] [PubMed] [Google Scholar]
- Li H., Fischle W., Wang W., Duncan E.M., Liang L., Murakami-Ishibe S., Allis C.D., Patel D.J. Structural basis for lower lysine methylation state-specific readout by MBT repeats of L3MBTL1 and an engineered PHD finger. Mol. Cell. 2007;28:677–691. doi: 10.1016/j.molcel.2007.10.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu B., Sun Y., Jiang F., Zhang S., Wu Y., Lan Y., Yang X., Mao N. Disruption of Smad5 gene leads to enhanced proliferation of high-proliferative potential precursors during embryonic hematopoiesis. Blood. 2003;101:124–133. doi: 10.1182/blood-2002-02-0398. [DOI] [PubMed] [Google Scholar]
- Lohmann F., Bieker J.J. Activation of Eklf expression during hematopoiesis by Gata2 and Smad5 prior to erythroid commitment. Development. 2008;135:2071–2082. doi: 10.1242/dev.018200. [DOI] [PubMed] [Google Scholar]
- Massagué J., Chen Y.G. Controlling TGF-beta signaling. Genes Dev. 2000;14:627–644. [PubMed] [Google Scholar]
- McReynolds L.J., Gupta S., Figueroa M.E., Mullins M.C., Evans T. Smad1 and Smad5 differentially regulate embryonic hematopoiesis. Blood. 2007;110:3881–3890. doi: 10.1182/blood-2007-04-085753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullen A.C., Orlando D.A., Newman J.J., Lovén J., Kumar R.M., Bilodeau S., Reddy J., Guenther M.G., DeKoter R.P., Young R.A. Master transcription factors determine cell-type-specific responses to TGF-β signaling. Cell. 2011;147:565–576. doi: 10.1016/j.cell.2011.08.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakao A., Afrakhte M., Morén A., Nakayama T., Christian J.L., Heuchel R., Itoh S., Kawabata M., Heldin N.E., Heldin C.H., ten Dijke P. Identification of Smad7, a TGFbeta-inducible antagonist of TGF-beta signalling. Nature. 1997;389:631–635. doi: 10.1038/39369. [DOI] [PubMed] [Google Scholar]
- Nostro M.C., Cheng X., Keller G.M., Gadue P. Wnt, activin, and BMP signaling regulate distinct stages in the developmental pathway from embryonic stem cells to blood. Cell Stem Cell. 2008;2:60–71. doi: 10.1016/j.stem.2007.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papapetrou E.P., Lee G., Malani N., Setty M., Riviere I., Tirunagari L.M., Kadota K., Roth S.L., Giardina P., Viale A. Genomic safe harbors permit high β-globin transgene expression in thalassemia induced pluripotent stem cells. Nat. Biotechnol. 2011;29:73–78. doi: 10.1038/nbt.1717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasini D., Bracken A.P., Hansen J.B., Capillo M., Helin K. The polycomb group protein Suz12 is required for embryonic stem cell differentiation. Mol. Cell. Biol. 2007;27:3769–3779. doi: 10.1128/MCB.01432-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perna F., Gurvich N., Hoya-Arias R., Abdel-Wahab O., Levine R.L., Asai T., Voza F., Menendez S., Wang L., Liu F. Depletion of L3MBTL1 promotes the erythroid differentiation of human hematopoietic progenitor cells: possible role in 20q- polycythemia vera. Blood. 2010;116:2812–2821. doi: 10.1182/blood-2010-02-270611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pimanda J.E., Donaldson I.J., de Bruijn M.F., Kinston S., Knezevic K., Huckle L., Piltz S., Landry J.R., Green A.R., Tannahill D., Göttgens B. The SCL transcriptional network and BMP signaling pathway interact to regulate RUNX1 activity. Proc. Natl. Acad. Sci. USA. 2007;104:840–845. doi: 10.1073/pnas.0607196104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravasi T., Suzuki H., Cannistraci C.V., Katayama S., Bajic V.B., Tan K., Akalin A., Schmeier S., Kanamori-Katayama M., Bertin N. An atlas of combinatorial transcriptional regulation in mouse and man. Cell. 2010;140:744–752. doi: 10.1016/j.cell.2010.01.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Real P.J., Ligero G., Ayllon V., Ramos-Mejia V., Bueno C., Gutierrez-Aranda I., Navarro-Montero O., Lako M., Menendez P. SCL/TAL1 regulates hematopoietic specification from human embryonic stem cells. Mol. Ther. 2012;20:1443–1453. doi: 10.1038/mt.2012.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmerer M., Evans T. Primitive erythropoiesis is regulated by Smad-dependent signaling in postgastrulation mesoderm. Blood. 2003;102:3196–3205. doi: 10.1182/blood-2003-04-1094. [DOI] [PubMed] [Google Scholar]
- Siatecka M., Bieker J.J. The multifunctional role of EKLF/KLF1 during erythropoiesis. Blood. 2011;118:2044–2054. doi: 10.1182/blood-2011-03-331371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian A., Tamayo P., Mootha V.K., Mukherjee S., Ebert B.L., Gillette M.A., Paulovich A., Pomeroy S.L., Golub T.R., Lander E.S., Mesirov J.P. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talbert P.B., Henikoff S. Histone variants—ancient wrap artists of the epigenome. Nat. Rev. Mol. Cell Biol. 2010;11:264–275. doi: 10.1038/nrm2861. [DOI] [PubMed] [Google Scholar]
- Trojer P., Li G., Sims R.J., 3rd, Vaquero A., Kalakonda N., Boccuni P., Lee D., Erdjument-Bromage H., Tempst P., Nimer S.D. L3MBTL1, a histone-methylation-dependent chromatin lock. Cell. 2007;129:915–928. doi: 10.1016/j.cell.2007.03.048. [DOI] [PubMed] [Google Scholar]
- Trompouki E., Bowman T.V., Lawton L.N., Fan Z.P., Wu D.C., DiBiase A., Martin C.S., Cech J.N., Sessa A.K., Leblanc J.L. Lineage regulators direct BMP and Wnt pathways to cell-specific programs during differentiation and regeneration. Cell. 2011;147:577–589. doi: 10.1016/j.cell.2011.09.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Galen P., Kreso A., Wienholds E., Laurenti E., Eppert K., Lechman E.R., Mbong N., Hermans K., Dobson S., April C. Reduced lymphoid lineage priming promotes human hematopoietic stem cell expansion. Cell Stem Cell. 2014;14:94–106. doi: 10.1016/j.stem.2013.11.021. [DOI] [PubMed] [Google Scholar]
- Vernimmen D., Lynch M.D., De Gobbi M., Garrick D., Sharpe J.A., Sloane-Stanley J.A., Smith A.J., Higgs D.R. Polycomb eviction as a new distant enhancer function. Genes Dev. 2011;25:1583–1588. doi: 10.1101/gad.16985411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W.K., Tereshko V., Boccuni P., MacGrogan D., Nimer S.D., Patel D.J. Malignant brain tumor repeats: a three-leaved propeller architecture with ligand/peptide binding pockets. Structure. 2003;11:775–789. doi: 10.1016/s0969-2126(03)00127-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L., Li L., Shojaei F., Levac K., Cerdan C., Menendez P., Martin T., Rouleau A., Bhatia M. Endothelial and hematopoietic cell fate of human embryonic stem cells originates from primitive endothelium with hemangioblastic properties. Immunity. 2004;21:31–41. doi: 10.1016/j.immuni.2004.06.006. [DOI] [PubMed] [Google Scholar]
- Wang Z., Oron E., Nelson B., Razis S., Ivanova N. Distinct lineage specification roles for NANOG, OCT4, and SOX2 in human embryonic stem cells. Cell Stem Cell. 2012;10:440–454. doi: 10.1016/j.stem.2012.02.016. [DOI] [PubMed] [Google Scholar]
- Wismar J., Löffler T., Habtemichael N., Vef O., Geissen M., Zirwes R., Altmeyer W., Sass H., Gateff E. The Drosophila melanogaster tumor suppressor gene lethal(3)malignant brain tumor encodes a proline-rich protein with a novel zinc finger. Mech. Dev. 1995;53:141–154. doi: 10.1016/0925-4773(95)00431-9. [DOI] [PubMed] [Google Scholar]
- Yeo C., Whitman M. Nodal signals to Smads through Cripto-dependent and Cripto-independent mechanisms. Mol. Cell. 2001;7:949–957. doi: 10.1016/s1097-2765(01)00249-0. [DOI] [PubMed] [Google Scholar]
- Zafonte B.T., Liu S., Lynch-Kattman M., Torregroza I., Benvenuto L., Kennedy M., Keller G., Evans T. Smad1 expands the hemangioblast population within a limited developmental window. Blood. 2007;109:516–523. doi: 10.1182/blood-2006-02-004564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C., Evans T. BMP-like signals are required after the midblastula transition for blood cell development. Dev. Genet. 1996;18:267–278. doi: 10.1002/(SICI)1520-6408(1996)18:3<267::AID-DVG7>3.0.CO;2-7. [DOI] [PubMed] [Google Scholar]
- Zhang W., Yatskievych T.A., Cao X., Antin P.B. Regulation of Hex gene expression by a Smads-dependent signaling pathway. J. Biol. Chem. 2002;277:45435–45441. doi: 10.1074/jbc.M208056200. [DOI] [PubMed] [Google Scholar]
- Zhang L., Prak L., Rayon-Estrada V., Thiru P., Flygare J., Lim B., Lodish H.F. ZFP36L2 is required for self-renewal of early burst-forming unit erythroid progenitors. Nature. 2013;499:92–96. doi: 10.1038/nature12215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao X., Jankovic V., Gural A., Huang G., Pardanani A., Menendez S., Zhang J., Dunne R., Xiao A., Erdjument-Bromage H. Methylation of RUNX1 by PRMT1 abrogates SIN3A binding and potentiates its transcriptional activity. Genes Dev. 2008;22:640–653. doi: 10.1101/gad.1632608. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


