Summary
Colinear HOX expression during hindbrain and spinal cord development diversifies and assigns regional neural phenotypes to discrete rhombomeric and vertebral domains. Despite the precision of HOX patterning in vivo, in vitro approaches for differentiating human pluripotent stem cells (hPSCs) to posterior neural fates coarsely pattern HOX expression thereby generating cultures broadly specified to hindbrain or spinal cord regions. Here, we demonstrate that successive activation of fibroblast growth factor, Wnt/β-catenin, and growth differentiation factor signaling during hPSC differentiation generates stable, homogenous SOX2+/Brachyury+ neuromesoderm that exhibits progressive, full colinear HOX activation over 7 days. Switching to retinoic acid treatment at any point during this process halts colinear HOX activation and transitions the neuromesoderm into SOX2+/PAX6+ neuroectoderm with predictable, discrete HOX gene/protein profiles that can be further differentiated into region-specific cells, e.g., motor neurons. This fully defined approach significantly expands capabilities to derive regional neural phenotypes from diverse hindbrain and spinal cord domains.
Graphical Abstract
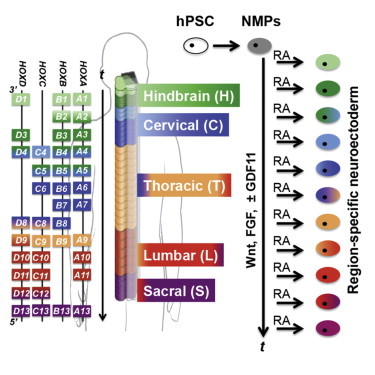
Highlights
-
•
Deterministic HOX expression in hPSC-derived neuromesoderm progenitors (NMPs)
-
•
Wnt/β-catenin, FGF, and GDF signaling regulate HOX activation in NMPs
-
•
Retinoic acid (RA) transitions NMPs to neuroectoderm and halts HOX activation
-
•
Neural cells can be patterned to any rostrocaudal hindbrain or spinal cord domain
Previous attempts to generate posterior CNS tissues from human pluripotent stem cells (hPSCs) yielded stochastic or heterogeneous rostrocaudal regional identity defined by HOX expression. Here, Ashton and colleagues demonstrate deterministic patterning of neural progenitors to defined domains throughout the hindbrain and spinal cord using a chemically defined system and temporal modulation of FGF, Wnt/β-catenin, GDF, and retinoic acid signaling.
Introduction
The human genome contains 39 HOX genes divided among four clusters and classified into 13 paralogous groups based on sequence homology (Figure 1A). During embryonic stages of body axis elongation, newly formed tissues express HOX genes in sequential, contiguous domains consistent with their order in each cluster, i.e., colinearly. This phenomenon is evolutionarily conserved in bilaterian species and spatially assigns body segment-specific differentiation trajectories to axial progenitors of all three germ layers (Lewis, 1978). During formation of the posterior CNS, progenitors proximal to the node progressively transition from a 3′ to 5′ HOX expression profile as the primitive streak regresses (Iimura and Pourquié, 2006). This process produces nested and overlapping axial domains of HOX expression within the neuroepithelium of hindbrain rhombomeric (HOX1-5) and spinal cord cervical (HOX5-9), thoracic (HOX9-10), and lumbosacral (HOX10-13) vertebral segments (Figure 1A) (Philippidou and Dasen, 2013). The spatial variation of HOX expression along the rostrocaudal (R/C) axis of the posterior CNS diversifies the fates of neuroepithelial progeny and precisely restricts the development of specific neural subtypes to discrete axial positions (Philippidou and Dasen, 2013).
Figure 1.
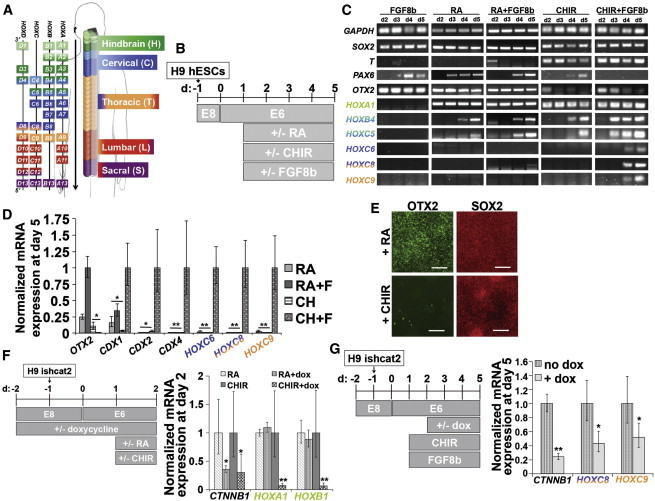
Wnt/β-Catenin and FGF Signaling Synergistically Coordinate HOX Activation during hPSC Differentiation
(A) HOX paralog expression in the hindbrain and spinal cord color-coded to the region where its expression is first detected (modified from a previous report; Philippidou and Dasen, 2013).
(B) Timeline of H9 hESC differentiation corresponding to (C)–(E).
(C) RT-PCR analysis of cultures at days 2–5.
(D) qPCR analysis at day 5. ∗p < 0.005; ∗∗p < 0.001.
(E) OTX2 and SOX2 expression at day 5 after RA or CHIR treatment. Scale bars, 100 μm. Adjacent images are the same field.
(F) qPCR analysis at day 2 using the H9 ishcat2 line with or without doxycycline treatment. ∗p < 0.02; ∗∗p < 0.005.
(G) qPCR analysis at day 5 using the H9 ishcat2 line with or without doxycycline treatment. ∗p < 0.02; ∗∗p < 0.0001.
All qPCR data are presented as mean ± SD calculated from independent biological triplicates. H9 data are normalized to the condition yielding maximum expression and ishcat2 data are normalized to each doxycycline-free condition. Statistical significance was calculated using the Student’s unpaired t test.
Retinoic acid (RA), wingless-type MMTV integration site protein family (Wnt), fibroblast growth factor (FGF), and growth differentiation factor (GDF) signaling intricately regulate HOX expression during posterior CNS development (Liu et al., 2001; Nordström et al., 2006). Yet, it remains controversial how these factors should be applied to human pluripotent stem cells (hPSCs) to recapitulate full colinear HOX activation and enable deterministic patterning of diverse R/C HOX profiles, i.e., regional hindbrain or spinal cord phenotypes, during neural differentiation. RA is used ubiquitously to caudalize hPSC-derived neuroectoderm because its secretion by paraxial somitic mesoderm is believed to convert 3′ HOX genes in hindbrain and spinal cord neural tissues from an epigenetically repressed to a transcriptionally active state (Gould et al., 1998; Mazzoni et al., 2013; Bel-Vialar et al., 2002). However, RA activates HOX1-5 chromatin domains in a saltatory (Mazzoni et al., 2013) versus colinear manner, which patterns mouse (mPSC) and human PSC-derived neuroectoderm with a broad caudal hindbrain thru cervical spinal cord identity. This is evidenced by demonstrations that RA-mediated caudalization generates HOXB4+ (Li et al., 2008), HOXC4+/HOXA5+/HOXC6−/HOXC8− (Mazzoni et al., 2013), HOXC5+/HOXC6+ (Peljto et al., 2010; Wichterle et al., 2002), or HOXC6+/HOXC8+ (Lee et al., 2007; Li et al., 2005) motor neurons (MNs), or HOXB4+ astrocytes (Krencik et al., 2011). Alternatively, Wnt and FGF treatments in the absence of RA can caudalize neurally differentiating mPSCs by inducing saltatory activation of HOX1-9 paralogs to coarsely pattern a heterogeneous mixture of HOXC6+ cervical and HOXD9+ thoracic spinal tissues, which can be further caudalized to also contain HOXD10+ lumbar tissues upon GDF11 supplementation (Mazzoni et al., 2013; Peljto et al., 2010). Also, a recent report demonstrated that manipulation of Wnt and RA concentrations could yield MNs possessing hindbrain or rostral spinal phenotypes but more caudal and further partitioning of regional HOX identity was not achieved (Maury et al., 2014). Overall, an approach for hPSC neural differentiation that recapitulates the precision, extent, and predictability of colinear HOX patterning observed in vivo remains elusive. Its development would enable unprecedented access to diverse regional phenotypes that populate posterior CNS tissues and generation of human disease models containing regional neural phenotypes with differential susceptibility to neurodegenerative disorders (Brockington et al., 2013; Kaplan et al., 2014; Sandoe and Eggan, 2013).
Recent progress in understanding how the posterior CNS develops led us to consider other methods for in vitro HOX patterning. New evidence suggest that bipotent SOX2+/Brachyury(T)+ neuromesodermal progenitors (NMPs) residing in a stem zone proximal to the primitive streak’s regressing node are the cell source for both the posterior neural tube and flanking paraxial mesoderm (Delfino-Machín et al., 2005; Takemoto et al., 2011; Tzouanacou et al., 2009). These NMPs are maintained by synergistic Wnt/β-catenin and FGF signaling from the node and primitive streak that induces SOX2 expression via activation of enhancer N1 (Takemoto et al., 2006, 2011). FGF signaling from the node also helps maintain the NMP state by repressing the expression of the neuroectoderm fate determinant PAX6 (Bertrand et al., 2000; Mathis et al., 2001). As the node regresses, NMPs rostral to the stem zone experience a decline in FGF signaling, which initiates paraxial mesoderm differentiation (i.e., somite formation). Then, the somites secrete RA back to the presumptive neural tube, which induces PAX6 expression to generate definitive neuroectoderm of the posterior CNS (Diez del Corral et al., 2003; Molotkova et al., 2005). The discovery that Wnt/FGF signaling induces NMPs that progressively caudalize HOX expression and undergo RA-induced neuroectoderm differentiation to form the hindbrain and spinal cord suggests an alternative approach for patterning HOX expression in vitro. Guoti et al. recently had partial success recapitulating the HOX-expressing NMP state from hPSCs in vitro, but the NMP phenotype could only be maintained for 3 days preventing acquisition of more caudal HOX genes/proteins before exhibiting a mesodermal shift (Gouti et al., 2014). Also, they were unable to partition HOX patterning within hindbrain and spinal cord regions. Therefore, we proceeded to test whether (1) Wnt/β-catenin and FGF signaling could differentiate hPSCs to stable NMPs to permit full, colinear activation of HOX genes; (2) RA exposure could differentiate NMPs to neuroectoderm; (3) this approach to deriving human neuroectoderm could enable enhanced control of HOX patterning.
Using a chemically defined, monolayer neural differentiation system (Lippmann et al., 2014), we determined that by temporally modulating Wnt/β-catenin and FGF signaling, hPSCs could be differentiated into pure SOX2+/Brachyury+ NMPs that were stable for 7 days. Over this period, the NMPs exhibited colinear activation of hindbrain, cervical, and thoracic HOX genes plus lumbosacral HOX genes with GDF11 supplementation. At any point during NMP propagation, a transition to RA was necessary and sufficient to induce PAX6+/SOX2+ neuroectodermal differentiation, precisely arrest progressive HOX activation, and thereby generate discrete HOX protein profiles indicative of distinct R/C domains. Overall, we demonstrate the ability to deterministically pattern neuroectoderm to diverse R/C domains by simply varying the amount of time NMPs spend under Wnt/β-catenin, FGF, and GDF signaling prior to RA exposure. Furthermore, we demonstrate that neuroectoderm of differing HOX expression profiles produce region-specific hindbrain, cervical, thoracic, and lumbar MN populations. Therefore, this temporal biphasic approach could be used to produce regional neural phenotypes from diverse hindbrain and spinal cord domains.
Results
Regulation of HOX and CDX Expression by RA, Wnt/β-Catenin, and FGF Signaling
We used the chemically defined “E6” method to investigate the effects of Wnt, FGF, RA, and GDF11 during hPSC neural differentiation because it lacks the use of small molecule inhibitors that could influence fate choices and does not form non-neural germ layers that could initiate unknown signaling crosstalk (Lippmann et al., 2014). We first differentiated H9 human embryonic stem cells (hESCs) in E6 medium for 24 hr and then added RA, CHIR99021 (CHIR; a small molecule Wnt/β-catenin agonist), FGF8b, or their relevant combinations and monitored HOX expression by RT- and quantitative PCR (qPCR) over 4 days (Figures 1B–1D). The addition of RA or CHIR, but not FGF8b, induced temporal, progressive expression of HOXA1-HOXC5 (Figure 1C). However, CHIR/FGF8b, but not RA/FGF8b, accelerated the progression of HOX activation inducing colinear activation of HOXC6, HOXC8, and HOXC9 at levels ranging from 39- to 3,400-fold higher than the other treatments (Figures 1C and 1D). Moreover, the caudalizing effect of CHIR/FGF8b was not impeded by the addition of HX531, an RXR inhibitor, and CHIR but not RA treatment was able to reduce gene and protein expression of the midbrain/forebrain marker OTX2 (Figures 1C–1E; Figure S1). These results indicate that while RA can induce some degree of posterior identity indicated by HOX1-5 expression, CHIR/FGF8b induces rapid patterning of differentiating hESCs to an OTX2− posterior phenotype with caudal HOX1-9 expression in an RA-independent manner. This is in agreement with other neural differentiation studies (Gouti et al., 2014; Mazzoni et al., 2013)
Additionally, CHIR/FGF8b treatment yielded substantially elevated expression of CDX genes, which are the upstream regulators of HOX genes (Nordström et al., 2006): compared to RA, CHIR/FGF8b upregulated CDX1 by >6-fold, CDX2 by >1,800-fold, and CDX4 by >330-fold (Figure 1D). To confirm that CHIR-mediated HOX induction requires β-catenin signaling, we utilized the H9 ishcat2 line, which harbors a doxycycline-inducible β-catenin small hairpin RNA (shRNA) cassette (Lian et al., 2012). Addition of doxycycline prior to CHIR treatment reduced β-catenin (CTNNB1) expression by 3-fold and decreased HOXA1 and HOXB1 expression by 14- and 15-fold, respectively (Figure 1F). In contrast, while doxycycline treatment also reduced β-catenin by 3-fold in RA-treated cells, it did not reduce HOXA1 or HOXB1 expression, suggesting RA-mediated induction of HOX transcription is β-catenin independent (Figure 1F). To verify sustained β-catenin signaling promotes progressive HOX activation, we induced HOX expression with CHIR/FGF8b for 1 day and continued the treatment for 3 days with and without doxycycline. Doxycycline treatment reduced CTNNB1 by 4-fold, HOXC8 by 2-fold, and HOXC9 by 2-fold, indicating that sustained β-catenin signaling contributes to progressive HOX activation (Figure 1G).
Derivation of Stable NMPs Exhibiting Full Colinear HOX Activation
Gouti et al. recently demonstrated that simultaneous induction of Wnt/β-catenin and FGF signaling can differentiate hPSCs to SOX2+/Brachyury(T)+ NMPs, but NMPs persisted for only 3 days before shifting to a mesodermal fate under their treatment regimen (Gouti et al., 2014). We observed a similar trend with simultaneous CHIR and FGF8b treatment inducing uniform Brachyury but also causing a sharp decrease in SOX2 expression (23% ± 0% SOX2+), indicating a mesodermal fate shift (Figure 2A, Route 1). Conversely, pre-treatment with FGF8b prior to CHIR (pre-FGF8b/CHIR) yielded uniform expression of both Brachyury and SOX2 that could be maintained (75%–100% SOX2+/Brachyury+) for 7 days (Figure 2A, Route 2, and Figure 2B). Thus, FGF signaling upstream of Wnt/β-catenin signaling effectively induces a stable NMP identity during hPSC differentiation.
Figure 2.
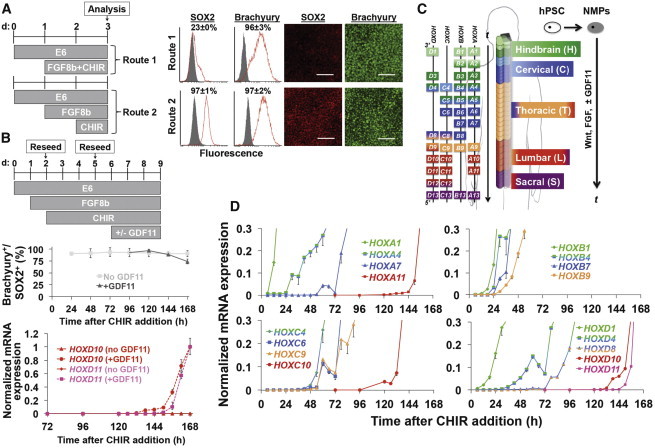
Colinear HOX Expression in hPSC-Derived NMPs
(A) Analysis of the NMP phenotype by flow cytometry and immunocytochemistry. Gray histogram, immunoglobulin G (IgG) control; red histogram, label of interest. Scale bars, 100 μm.
(B) Optimized NMP propagation scheme. The addition of CHIR is denoted as t = 0 hr in all panels. Purity of NMPs was assessed by flow cytometry in the presence or absence of GDF11 (data are presented as mean ± SD). qPCR analysis of HOXD10 and HOXD11 was conducted in the presence or absence of GDF11 (expression normalized to the time point of maximum expression).
(C) Schematic for HOX induction in NMPs. “t” indicates time under Wnt, FGF, and GDF signaling.
(D) qPCR analysis of colinear HOX expression normalized to the time point of maximum expression. GDF11 was included according to (B).
qPCR data in all panels are presented as mean ± SD calculated from technical duplicates. Full profiles for all analyzed genes can be found in Figure S2A. For all flow cytometry data, a minimum of two biological replicates was used to calculate mean ± SD.
Next, we investigated whether colinear HOX expression was evident within our NMP cultures. Analogous to their in vivo counterparts, these cultures exhibited colinear activation of HOX genes over the 7-day period (Figures 2B–2D; Figure S2). In our system, 3′ HOX transcripts remained expressed even after activation of caudal paralogs and fluctuated within an ∼5-fold range (Figure S2A), also agreeing with Gouti et al. (2014). Supplementation with GDF11, a transforming growth factor β (TGF-β) family member expressed at later stages of NMP propagation in vivo (Liu, 2006; Liu et al., 2001), was necessary for robust activation of lumbosacral HOX paralogs (Philippidou and Dasen, 2013) (Figures 2B–2D; Figure S2A) but did not significantly repress transcription of rostral HOX genes (Figure S2B) or disrupt the NMP state (Figure 2B). Moreover, premature GDF11 treatment (initiated after 1 day of FGF8b/CHIR) could induce HOXD10 but at >100-fold lower expression compared to GDF11 treatment after 4 days of FGF8b/CHIR, indicating robust lumbosacral patterning requires prior activation of rostral HOX genes (Figure S2C). We were able to detect colinear transcription of 33 of the 39 HOX genes in hPSC-derived NMP cultures, excluding HOXB3, HOXC12, and the HOX13 paralogs, over a time span consistent with posterior neural tube formation according to the Carnegie stages of human development (Figure S2A). Moreover, temporal HOX activation was also observed in NMPs derived from IMR90-4 iPSCs (Figure S2D), indicating these mechanisms translate to other hPSC lines. While we believe the lack of HOXB3 detection was due to experimental or qPCR reagent error, our extensive literature search was unable to find definitive documentation of HOXC12 or HOX13 paralog expression in the posterior CNS (Dasen et al., 2005).
RA Converts NMPs to Neuroectoderm
In vivo, regression of the stem zone along the primitive streak yields a rostral decline in FGF signaling that coincides with RA secretion from newly formed somites and PAX6 expression in the newly formed neural tube (Diez del Corral et al., 2003). This suggests that RA could induce a NMP-to-neuroectoderm fate switch. To test this in vitro, we exposed hPSC-derived NMPs to E6 medium containing RA, CHIR, or neither for 2 days. Cultures exposed to only E6 medium maintained SOX2 and Brachyury expression but gained minimal PAX6 (96% ± 4% SOX2+, 15% ± 8% PAX6+, 78% ± 16% Brachyury+), while exposure to only CHIR induced a mesodermal fate shift as evidenced by SOX2 downregulation, no PAX6 induction, and uniform maintenance of Brachyury expression (50% ± 8% SOX2+, 96% ± 1% Brachyury+) (Figure 3A). However, when exposed to RA, the cells gained PAX6 expression along with SOX2 maintenance (98% ± 4% SOX2+, 97% ± 3% PAX6+) and Brachyury downregulation (43% ± 4%) indicating a neuroectodermal fate shift (Figure 3A). Moreover, transitioning to RA at any point during NMP propagation generated a highly pure (>83%) PAX6+/SOX2+ neuroepithelial culture, i.e., polarized N-cadherin, within 4 days (Figures 3B–3D). If GDF11 was added to facilitate lumbosacral patterning, PAX6 expression in response to RA was decreased in a dose-dependent manner (Figures S3A–S3C). The ability of GDF11 to control R/C patterning is mediated by signaling via ALK5 (Andersson et al., 2006) and activation of the SMAD2/3 complex (Liu, 2006). GDF11 has also been shown to activate SMAD1/5/8 in vitro (Liu, 2006), which contributes to dorsal patterning (Tozer et al., 2013). Since dorsomorphin selectively inhibits ALK2, ALK3, and ALK6, which blocks SMAD1/5/8 signaling (Yu et al., 2008), we hypothesized the addition of dorsomorphin should prevent acquisition of a dorsal phenotype without affecting R/C patterning. Indeed, the addition of dorsomorphin with GDF11 and throughout RA treatment was sufficient to recover PAX6 expression (83% ± 4% PAX6+; Figure S3D). Thus, highly pure neuroectoderm cultures could be obtained from NMPs at any point during colinear HOX activation.
Figure 3.
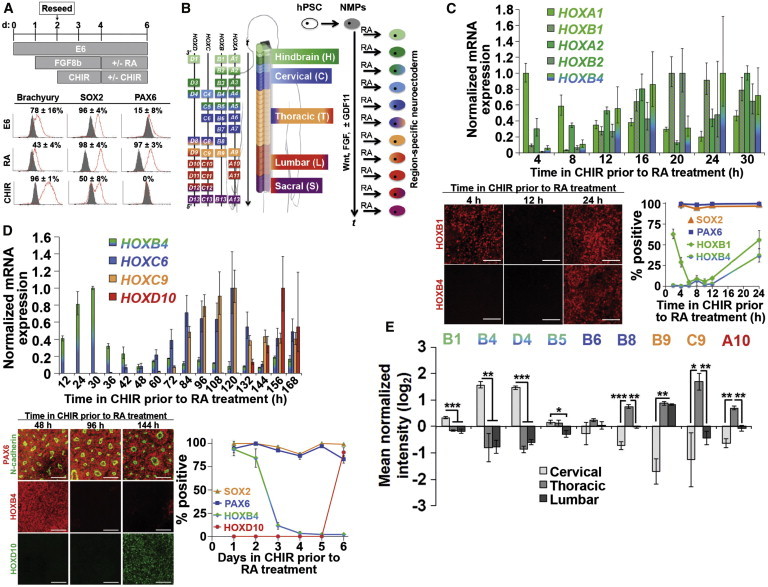
RA Induces a Neuroectodermal Fate and Halts Colinear HOX Activation
(A) NMPs exposed to RA or CHIR acquire a neuroectodermal or mesodermal fate as assessed by flow cytometry at day 6 (gray histogram, IgG control; red histogram, antigen of interest).
(B) Schematic for transition from NMPs to neuroectoderm by RA treatment. “t” indicates time under Wnt, FGF, and GDF signaling prior to RA treatment, which halts HOX progression to yield a defined rostrocaudal identity.
(C and D) Representative hindbrain (C) and spinal cord (D) cultures assessed by qPCR, immunocytochemistry, and flow cytometry. For all plots, flow cytometry data are presented as mean ± SD from biological duplicates, and qPCR data are mean ± SD from technical duplicates normalized to the time point of maximum expression for each gene. For hindbrain cultures, HOXB1 and HOXB4 were quantified by immunocytochemistry relative to DAPI+ nuclei (technical triplicates per time point, minimum 10,000 cells counted, and relative HOX expression patterns were qualitatively verified across biological duplicates), while SOX2 and PAX6 were quantified by flow cytometry. For spinal cord cultures, HOXD10 was quantified by immunocytochemistry relative to DAPI+ nuclei (technical duplicates, >2,000 cells counted), while SOX2, PAX6, and HOXB4 were quantified by flow cytometry. Scale bars, 100 μm. For (D), dorsomorphin was included with GDF11 and throughout RA treatment (further details in Figure S3).
(E) Mass spectrometry comparison of HOX profiles in cervical, thoracic, and lumbar neuroectoderm cultures. Cervical differentiation: 1 day FGF8b, 2 days FGF8b/CHIR, 4 days RA; thoracic differentiation: 1 day FGF8b, 6 days FGF8b/CHIR, 4 days RA; lumbar differentiation: 1 day FGF8b, 4 days FGF8b/CHIR, 2 days FGF8b/CHIR/GDF11, 4 days RA. Biological triplicates (neuroepithelium isolated individually from three separate culture wells) were used to calculate statistical significance between cervical, thoracic, and lumbar samples.
Data are presented as mean ± SD and statistical significance was calculated using the Student’s unpaired t test. ∗p < 0.01; ∗∗p < 0.002; ∗∗∗p < 0.0001.
Deterministic Hox Patterning Can Be Achieved Using a Temporal Transition between Wnt/FGF/GDF and RA Treatment
Because RA production in vivo and treatment in vitro coincides with neural differentiation, we and others (Diez del Corral and Storey, 2004) posited that it might also serve as a signal to arrest colinear HOX activation. We therefore tested the ability of RA to generate neuroectoderm with discrete, region-specific HOX expression profiles in accordance with the status of colinear HOX activation observed in the NMP state (Figure 3B). Also, we assessed the expression of HOX transcription factors (TFs) that would demarcate distinct hindbrain and spinal cord domains (Dasen et al., 2003, 2005). First, we evaluated our ability to pattern neuroectoderm to hindbrain domains using a 0- to 30-hr FGF8b/CHIR NMP propagation period prior to 4 days of RA treatment (Figures 3B and 3C). qPCR analysis of the resulting neuroectoderm cultures revealed elevated expression of HOXA1 and HOXA2 prior to HOXB1, HOXB2, and HOXB4. Immunocytochemical analysis revealed that early cultures were HOXB1+/HOXB4− (2 hr FGF8b/CHIR, 4 days RA; 63% ± 6% HOXB1+) but rapidly lost HOXB1 expression if derived after only an additional 4 hr of NMP propagation (6 hr FGF8b/CHIR, 4 days RA; 5% ± 2% HOXB1+) (Figure 3C). If neuroectoderm was derived after 24 hr of NMP propagation, it reacquired HOXB1 and now also expressed HOXB4 (24 hr FGF8b/CHIR, 4 days RA; 63% ± 5% HOXB1+ and 37% ± 8% HOXB4+) (Figure 3C). This mirrors HOX TF expression during development of the fourth, fifth, and sixth through eighth rhombomeres, respectively, indicating that transitioning from Wnt/FGF to RA at the 2- to 4-hr time point generates HOXB1+ neuroectoderm that could potentially produce facial nerve MNs whereas transitioning at 24 hr generates HOXB4+ neuroectoderm of the caudal hindbrain (Bell et al., 1999; Philippidou and Dasen, 2013; Bel-Vialar et al., 2002). Interestingly, the high percentage of HOXB1+ cells at the 2 hr mark coincided with low HOXB1 expression (∼10% of the maximum observed expression at 20 hr) suggesting that while transcript levels are useful for identifying relative HOX gene expression, they may not be directly instructive for predicting HOX TF profiles that arise due to cross-repressive interactions (Bell et al., 1999; Dasen et al., 2003, 2005). Also, the observed abrupt transitions in neuroectoderm HOX TF expression suggest fairly uniform progressive HOX activation within the NMPs (Figure 3C).
Next, we performed a similar Wnt/FGF/GDF-to-RA transition time-course analysis focusing on later time points that would generate neuroectoderm patterned to spinal cord domains (Figures 3B and 3D). qPCR analysis again demonstrated that RA treatment could halt the induction of 5′ paralogs that would otherwise have become expressed in continuous Wnt/FGF/GDF11 treatment. Using both immunocytochemistry and flow cytometry, we observed uniform HOXB4 expression in neuroectodermal cultures derived from NMPs transitioned to RA after 24–48 hr of propagation, correlating to a rostral cervical spinal cord domain (Figures 3B and 3D). HOXB4 expression dropped sharply (from 84% ± 10% down to 12% ± 4% HOXB4+ cells) when the transition to RA occurred at 72 hr (Figure 3D), which coincided with the initiation of thoracic HOXC9 expression, a known repressor of HOX4 paralog expression (Jung et al., 2010). Furthermore, HOXD10 was only detected in neuroectodermal cultures when RA was added after ∼144 hr of NMP propagation (Figures 3B and 3D; Figure S3E), while HOXB4 remained repressed. Additional insights into the diversity of HOX expression in neuroectoderm patterned to cervical, thoracic, and lumbar spinal cord domains were obtained using quantitative mass spectrometry (Figure 3E; Table S1). As expected, HOXC9 was expressed in thoracic neuroectoderm but repressed in the HOXD10+ lumbar culture, and many (HOXB1, HOXB4, HOXD4, HOXB5, HOXB8, HOXC9, and HOXA10) but not all (HOXB9) detected HOX factors exhibited expression patterns indicative of similar cross-repressive interactions (Dasen et al., 2005) (Figure 3E). Thus, these results demonstrate region-specific patterning of hPSC-derived neuroectoderm in the spinal cord, and similar to the hindbrain neuroectoderm cultures, repression of HOX proteins occurred even though their HOX transcripts remained expressed, possibly indicating cross-repressive interactions at this stage are occurring at the post-transcriptional stage (Yekta et al., 2004). Hence, our temporal biphasic approach for differentiating hPSCs to neuroectoderm through a stable NMP intermediate enables deterministic HOX patterning to impart diverse and discrete hindbrain and spinal cord regional identities (Figure 3B).
Neuroectoderm with Region-Specific HOX Profiles Generate Regional MN Populations
We used MN differentiation to evaluate whether the regional specification imparted by our HOX patterning approach yielded characteristic differences in the neuroectoderm progeny. First, we verified that MNs derived from neuroectoderm of discrete hindbrain domains likewise exhibited distinct HOX signatures. Hindbrain neuromesoderm was propagated for 4, 12, and 24 hr before conversion to ventralized neuroectoderm using RA, sonic hedgehog (SHH) and purmorphamine (PM, a sonic hedgehog agonist) exposure and further differentiation to MNs using media containing RA and neurotrophic factors (NTFs). MN differentiation was verified by the presence of cells co-expressing βIII-tubulin/ISL1 and SMI-32 reactive non-phosphorylated neurofilament heavy chain with ISL1 or synapsin (Amoroso et al., 2013; Li et al., 2005) (Figure 4A). Immunocytochemical analysis of HB9+ MNs generated from 4 hr neuromesoderm revealed no co-expression of HOXB1 or HOXB4. However, HB9+ MNs generated from 12 hr neuromesoderm predominantly co-expressed HOXB1 but not HOXB4, and those generated from 24 hr neuromesoderm predominantly co-expressed both HOXB1 and HOXB4 (Figure 4A). Also, within all cultures, many ISL1+ cells co-expressed PHOX2B indicative of hindbrain identity (Maury et al., 2014; Pattyn et al., 2000). Thus, MNs derived from hindbrain neuroectoderm with discrete Hox gene and protein profiles (Figure 3C) likewise express distinct HOX profiles, although the precise regional identity of these MNs based on their HOX signatures is currently unclear.
Figure 4.
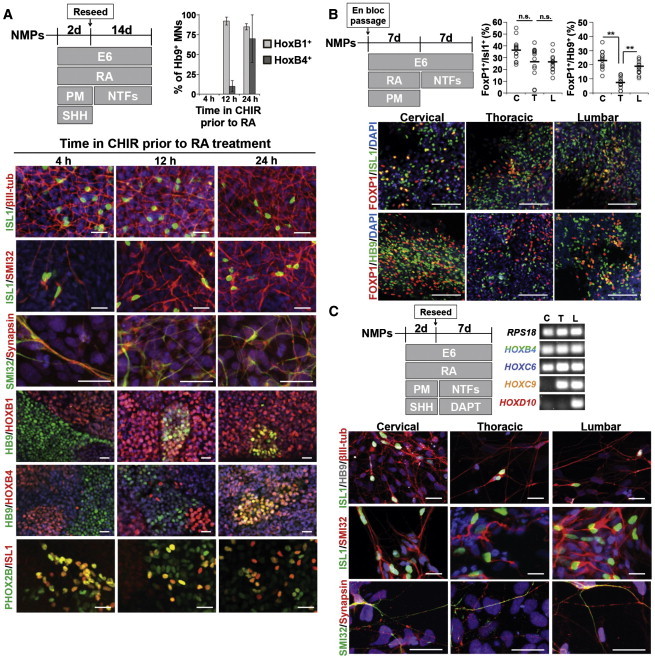
Derivation of Region-Specific MNs
(A) Neuronal maturation from various hindbrain locales. NMPs were propagated for 4, 12, or 24 hr before conversion to ventralized neuroectoderm and differentiation to neurons. DAPI (blue) is overlaid in most images. Scale bars, 20 μm. Quantified data are presented as mean ± SD (four technical replicates, >100 HB9+ cells counted per sample).
(B) Evaluation of FOXP1 columnar identity in ISL1+ and HB9+ MNs. Cervical differentiation (C): 1 day FGF8b, 2 days FGF8b/CHIR; thoracic differentiation (T): 1 day FGF8b, 5 days FGF8b/CHIR; lumbar differentiation (L): 1 day FGF8b, 4 days FGF8b/CHIR, 2 days FGF8b/CHIR/GDF11; prior to en bloc passage and further differentiation as indicated. NTFs, neurotrophic factors (described in Experimental Procedures). For plotted data, open circles are percentages from individual fields and bars indicate the mean. Statistical significance was calculated using the Student’s unpaired t test (three to five fields were counted across three biological replicates for each condition, minimum 2,500 cells counted). n.s., p > 0.02; ∗∗p < 0.000001. Scale bars, 100 μm.
(C) Neuronal maturation from cervical, thoracic, and lumbar patterned neuroectoderm corresponding to the NMP state described in (B).
DAPI is overlaid in all images. Scale bars, 20 μm. RT-PCR (50 cycles) demonstrates maintenance of regional identity.
In the developed spinal cord, MNs reside in distinct motor columns: the lateral (LMC) and medial motor columns (MMC) at cervical and lumbar levels, and the preganglionic (PGC), MMC, and hypaxial motor column (HMC) at thoracic levels (Philippidou and Dasen, 2013). MMC MNs exclusively express LHX3 along with HB9 and ISL1, while LMC MNs express LHX1 and FOXP1 with either HB9 or ISL1 depending on their further subdivision into medial and lateral domains (Thaler et al., 1999; Tsuchida et al., 1994). PGC MNs are FOXP1+/ISL1+/HB9−, while HMC MNs are LHX3−/FOXP1− but express both HB9 and ISL1 (Dasen et al., 2008; William et al., 2003). Thus, quantitative immunocytochemistry can be used to observe whether cervical, thoracic, and lumbar neuroectoderm could be differentiated into MN precursors populations exhibiting relative FOXP1/HB9 co-expression patterns characteristic of their R/C domain in vivo, i.e., the presence of FOXP1+/HB9+ cells in cervical and lumbar but not thoracic cultures (Dasen et al., 2008).
Regional spinal MN precursors were derived by exposing appropriately patterned NMPs to RA and PM for 7 days prior to an additional week of differentiation in neurotrophic factors (Li et al., 2005) (Figure 4B). We observed no significant differences in the numbers of FOXP1+/ISL1+ MN precursors (37% ± 9%, 27% ± 13%, and 26% ± 8%, respectively, of the total ISL1+ population). However, FOXP1+/HB9+ MN precursors were prevalent in cervical and lumbar cultures (23% ± 6% and 18% ± 4% of the total HB9+ population) but scarce in thoracic cultures (8% ± 4%; p < 0.000001, Figure 4B). We initially expected that no FOXP1+/HB9+ MN precursors would be present in the thoracic cultures, but PGC MNs proceed through an HB9+ state before its expression is rapidly lost (William et al., 2003). Therefore, the small subset of FOXP1+/HB9+ cells in our thoracic cultures likely represents immature PGC MNs. The significant difference in the propensity of cervical and lumbar versus thoracic neuroectoderm to generate FOXP1+/HB9+ MN precursors characteristic of the LMC demonstrates proper regional specification as a result of our deterministic HOX patterning approach.
Furthermore, we investigated whether accelerated MN differentiation induced by treating ventralized cervical (HOXC6+/HOXC9−/HOXD10−), thoracic (HOXC6+/HOXC9+/HOXD10−), and lumbar (HOXC6+/HOXC9+/HOXD10+) (Figure 3D) neuroectoderm with the Notch signaling inhibitor DAPT (Maury et al., 2014), which yielded >92% NeuN+ neuronal cultures within 7 days (Figures 4C and S4A), altered the regional specification imposed at the neuroectoderm stage. Each differentiated culture contained MNs co-expressing βIII-tubulin with ISL1 or HB9 and SMI-32 with ISL1 or synapsin (Figure 4C). RT-PCR analysis revealed that each culture’s regional spinal cord identity patterned at the neuroectodermal state was preserved throughout accelerated neuronal differentiation (Figure 4C). Also, electrophysiological analysis of these cervical, thoracic, and lumbar cultures after additional weeks of maturation determined that each region-specific culture possessed neurons exhibiting mature action potentials (Figures S4B and S4C). Hence, our approach to deterministic patterning of HOX expression should be broadly applicable to deriving functional and region-specific hindbrain and spinal cord tissues from hPSCs.
Discussion
Regulation of HOX Expression during Posterior Neural Differentiation of hPSCs
Our results provide unique evidence that HOX expression during posterior neural differentiation of hPSCs can be precisely regulated by temporal control of two opposing mechanisms. Wnt/β-catenin, FGF, and GDF signaling synergistically control colinear HOX activation while maintaining an NMP state, and RA arrests HOX activation to yield a fixed R/C position and transitions the cultures to definitive neuroectoderm. This approach should enable the derivation of neuroectoderm from any R/C position spanning the hindbrain to lumbosacral spinal cord in a predictable manner.
Specifically, we have demonstrated the ability to generate neuroectoderm with HOX profiles corresponding to specific rhombomeric segments and cervical, thoracic, and lumbosacral vertebral segments. Our findings are distinct from other PSC neural differentiation approaches that unpredictably pattern cells to primarily caudal hindbrain thru cervical HOX profiles (Amoroso et al., 2013; Lee et al., 2007; Li et al., 2005) or to heterogeneous mixtures of cervical, thoracic, and lumbar tissues (Patani et al., 2011; Peljto et al., 2010). Given the data presented in our manuscript, it is perhaps surprising that RA-only patterning approaches can yield HOX6-8 expression, but this may be inherent to the choice of differentiation methods. Whereas we utilized a chemically defined monolayer system to screen HOX patterning cues, most other studies have used embryoid body (EB) cultures for posterior neural differentiation. EB differentiation has been shown to activate endogenous Wnt signaling (Li et al., 2009), but cells in outer layers of EBs are exposed to different concentrations of soluble media cues than those on the inside of EBs due to diffusional limitations (Van Winkle et al., 2012). Therefore, we suspect in these EB-based systems that competition between endogenous Wnt (activating HOX progression) and exogenously added RA (inducing HOX arrest and neural differentiation) could cause heterogeneous R/C patterning, especially when considering relative heterogeneity in EB sizes and shapes. In contrast, exposure to soluble factors in our monolayer differentiation system should be more uniform, possibly enabling our enhanced HOX patterning precision.
We also provide evidence that Wnt/β-catenin and FGF signaling alone can induce substantial expression of CDX genes, which are core regulators of HOX expression in vivo, similar to results from tissue explant and other PSC differentiation studies (Gouti et al., 2014; Maury et al., 2014; Mazzoni et al., 2013; Nordström et al., 2006). Thus, RA and Wnt/FGF signaling may regulate different epigenetic states, which could potentially explain discrepancies between reports of coarse, saltatory HOX activation in vitro by RA alone (Mazzoni et al., 2013) versus the induction of organized, colinear HOX activation in vivo by the temporal integration of multiple signaling pathways. Moreover, though previous studies have suggested RA serves as a “rostralizing” factor (Philippidou and Dasen, 2013), our treatment of thoracic or lumbosacral NMPs with RA did not generate rostralized neuroectoderm (Figure 3D); as such, we postulate these previous studies were actually observing premature termination of HOX progression. From our results, we posit that HOX patterning may involve a temporal, biphasic mechanism where Wnt/FGF primarily controls colinear HOX activation and RA controls its termination. This is different than the widely cited gradient mechanism that supposes RA controls 3′ HOX patterning and Wnt/FGF controls 5′ HOX patterning (Philippidou and Dasen, 2013). Our patterning mechanism resonates with a progress/stem zone model that has been heavily scrutinized for limb development (Towers and Tickle, 2009). However, our in vitro approach does not allow us to draw direct conclusions about the role each of these factors plays in HOX regulation in vivo. We are instead hopeful that our patterning approach will enable detailed studies of HOX regulation in vitro, which could then be used to conduct more informative studies of HOX patterning in vivo. In addition, since much of the “Hoxasome” is still poorly understood (Mann et al., 2009), the scalability of our chemically defined, deterministic HOX patterning protocol will be invaluable to elucidating the molecular mechanisms underlying Hox regulation of downstream neural cell fates.
Implications for Regenerative Therapies and Disease Modeling
Recent studies of cell replacement therapy in the anterior CNS using hPSC-derived progenitors have demonstrated that the implanted cells must possess a regional phenotype that mimics endogenous tissues in order to effectively engraft and correct neural deficits (Kriks et al., 2011; Ma et al., 2012). Since HOX expression patterns are crucial determinants of cellular phenotype, organization, and neural circuit integration in the developing hindbrain and spinal cord (Philippidou and Dasen, 2013), our patterning approach could serve as the basis for generating a spectrum of posterior neural progeny with highly specific regional identities that may aid regenerative therapy efforts. Also, regional MN phenotypes exhibit differential susceptibility to several neurodegenerative disorders, including amyotrophic lateral sclerosis (ALS) and spinal muscular atrophy (SMA) (Brockington et al., 2013; Kaplan et al., 2014). Our deterministic HOX patterning approach will facilitate elucidation of the molecular basis underlying differential susceptibility of iPSC-derived MNs with the same disease-causing mutations but different regional phenotypes, including previously inaccessible hindbrain phenotypes. The ability to execute such experimental paradigms should significantly enhance the utility of iPSC-derived MN disease models to identify relevant pathways of neurodegenerative disease induction and progression (Chen et al., 2014; Kiskinis et al., 2014).
Experimental Procedures
Induction and Propagation of NMPs and Differentiation to Neural Fates
H9 hESCs (Thomson et al., 1998) (passage 25–45), H9 ishcat2 hESCs (Lian et al., 2012) (passage 33–43), and IMR90-4 iPSCs (Yu et al., 2007) (passage 31–40) were maintained in E8 medium on Matrigel (BD Biosciences) as previously described (Lippmann et al., 2014). 2 μg/ml doxycycline (Sigma) was used for β-catenin knockdown in the ishcat2 line. To initiate differentiation, hPSCs were passaged with accutase (Life Technologies) onto vitronectin (VTN-NC)-coated (Chen et al., 2011) plates at a density of 1 × 105 cells/cm2 with 10 μM ROCK inhibitor (Y27632; R&D Systems) as previously described (Lippmann et al., 2014). The following day, cells were changed to E6 medium and then changed to E6 medium containing 200 ng/ml FGF8b 24 hr later (PeproTech). After another day, the cells were washed once with 2 ml PBS, accutased for 2 min, and removed from the surface by gentle pipetting. After collection by centrifugation, cells were gently resuspended in E6 medium containing 200 ng/ml FGF8b and CHIR99021 (CHIR, 3 μM for H9 hESCs and 2 μM for IMR90-4 iPSCs; R&D Systems) and re-seeded on VTN-NC-coated plates at a density of 1.5 × 105 cells/cm2. 10 μM Y27632 was included during the re-seeding process, and the medium was not changed again until 48 hr after passaging. If extended NMP propagation was required, this passaging process was repeated on day 3 of CHIR treatment (re-seed density of 1.2 × 105 cells/cm2), and 50 ng/ml GDF11 (PeproTech) was added on day 4 of CHIR treatment to initiate lumbosacral patterning. For some experiments, 1 μM dorsomorphin (R&D Systems) was included with GDF11. At any point during CHIR treatment, cells were changed to E6 medium containing 1 μM retinoic acid (RA; Sigma) to elicit neural induction and arrest HOX progression. For assessment of FOXP1 expression in MN precursors, NMPs were passaged en bloc by scraping and re-seeded in VTN-NC-coated chamber slides at a 1:200 ratio in E6 medium containing 1 μM RA and 100 nM purmorphamine (PM; Cayman Chemicals) for 7 days, followed by an additional 7-day treatment with 10 ng/ml brain-derived neurotrophic factor (BDNF; PeproTech), 10 ng/ml glial-derived neurotrophic factor (GDNF; PeproTech), and cAMP (1 μM; Sigma). For assessment of HOX identity in MNs, NMPs were neuralized and ventralized with 1 μM RA, 2 μM PM, and 1 μg/ml sonic hedgehog (SHH; R&D Systems) for 2 days and then passaged en bloc or with accutase and re-seeded in VTN-NC-coated chamber slides at 2 × 104 cells/cm2 in E6 medium containing RA, 10 μM Y27632, 10 ng/ml BDNF, 10 ng/ml GDNF, and 1 μM cAMP for 7–14 days (Y27632 was removed after the first 3 days). 10 nM RA was used for cultures with hindbrain HOX signatures, and 1 μM RA was used for cultures with spinal cord HOX signatures. 1 μM DAPT (Cayman Chemicals) was included for cultures with spinal cord identity. For extended neuronal differentiation to conduct electrophysiology studies, NMPs were neuralized and ventralized with 1 μM RA, 2 μM PM, and 1 μg/ml SHH, passaged 1:100 onto Matrigel-coated glass coverslips, and cultured for 7 days in E6 medium containing 1 μM RA, 100 nM PM, 100 ng/ml SHH, and 1 μM DAPT. The cultures were then matured in E6 medium containing 10 ng/ml BDNF, 10 ng/ml GDNF, and 1 μM cAMP for at least 2 months prior to electrophysiology measurements. The cells were supplemented with 1 μg/ml mouse laminin (Invitrogen) once per week to help maintain attachment.
Author Contributions
E.S.L. and R.S.A. conceived the study. E.S.L. and M.C.E.-S. performed all cell-culture experiments and related analyses. C.E.W. and J.J.C. designed the mass spectrometry proteomic analysis and C.E.W. conducted these experiments. D.A.R. and E.R.C. conducted and supervised the electrophysiology experiments, respectively. All authors contributed to manuscript preparation.
Acknowledgments
We would like to thank Dr. James Thomson and Nicholas Propson for providing the vitronectin peptide, Dr. Sean Palecek for providing the H9 ishcat2 line, and Dr. John Yin for use of the Biorad CFX96 qPCR machine. The PAX6, HOXB4, HB9, and ISL1 antibodies used in this study were obtained from the Developmental Studies Hybridoma Bank developed under the auspices of the NICHD and maintained by the University of Iowa. The HOXD10 antibody was obtained as a pre-release reagent from R&D Systems. This work was supported by funding from the Wisconsin Institutes for Discovery (R.S.A.), a Draper Technology Innovation Fund Award from the Wisconsin Alumni Research Foundation (R.S.A.), and NIH grants R21 NS082618 (R.S.A.), R01 GM080148 (J.J.C.) and R01 MH061876 (E.R.C.). E.S.L. was supported by a postdoctoral fellowship from the University of Wisconsin Stem Cell and Regenerative Medicine Center (SCRMC) and an Individual Postdoctoral National Research Service Award (NRSA) from the NIH (1F32NS083291-01A1). D.A.R. was supported by a National Science Foundation (NSF) GRSP fellowship (DGE-1256259) and the Neuroscience Training Program (T32-GM007507). E.R.C. is an Investigator of the Howard Hughes Medical Institute. R.S.A. holds an Innovation in Regulatory Science Award for the Burroughs Wellcome Fund.
Footnotes
This is an open access article under the CC BY-NC-ND license (http://creativecommons.org/licenses/by-nc-nd/4.0/).
Supplemental Information
References
- Amoroso M.W., Croft G.F., Williams D.J., O’Keeffe S., Carrasco M.A., Davis A.R., Roybon L., Oakley D.H., Maniatis T., Henderson C.E., Wichterle H. Accelerated high-yield generation of limb-innervating motor neurons from human stem cells. J. Neurosci. 2013;33:574–586. doi: 10.1523/JNEUROSCI.0906-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersson O., Reissmann E., Ibáñez C.F. Growth differentiation factor 11 signals through the transforming growth factor-β receptor ALK5 to regionalize the anterior-posterior axis. EMBO Rep. 2006;7:831–837. doi: 10.1038/sj.embor.7400752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bel-Vialar S., Itasaki N., Krumlauf R. Initiating Hox gene expression: in the early chick neural tube differential sensitivity to FGF and RA signaling subdivides the HoxB genes in two distinct groups. Development. 2002;129:5103–5115. doi: 10.1242/dev.129.22.5103. [DOI] [PubMed] [Google Scholar]
- Bell E., Wingate R.J.T., Lumsden A. Homeotic transformation of rhombomere identity after localized Hoxb1 misexpression. Science. 1999;284:2168–2171. doi: 10.1126/science.284.5423.2168. [DOI] [PubMed] [Google Scholar]
- Bertrand N., Médevielle F., Pituello F. FGF signalling controls the timing of Pax6 activation in the neural tube. Development. 2000;127:4837–4843. doi: 10.1242/dev.127.22.4837. [DOI] [PubMed] [Google Scholar]
- Brockington A., Ning K., Heath P.R., Wood E., Kirby J., Fusi N., Lawrence N., Wharton S.B., Ince P.G., Shaw P.J. Unravelling the enigma of selective vulnerability in neurodegeneration: motor neurons resistant to degeneration in ALS show distinct gene expression characteristics and decreased susceptibility to excitotoxicity. Acta Neuropathol. 2013;125:95–109. doi: 10.1007/s00401-012-1058-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen G., Gulbranson D.R., Hou Z., Bolin J.M., Ruotti V., Probasco M.D., Smuga-Otto K., Howden S.E., Diol N.R., Propson N.E. Chemically defined conditions for human iPSC derivation and culture. Nat. Methods. 2011;8:424–429. doi: 10.1038/nmeth.1593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H., Qian K., Du Z., Cao J., Petersen A., Liu H., Blackbourn L.W., 4th, Huang C.-L., Errigo A., Yin Y. Modeling ALS with iPSCs reveals that mutant SOD1 misregulates neurofilament balance in motor neurons. Cell Stem Cell. 2014;14:796–809. doi: 10.1016/j.stem.2014.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dasen J.S., Liu J.-P., Jessell T.M. Motor neuron columnar fate imposed by sequential phases of Hox-c activity. Nature. 2003;425:926–933. doi: 10.1038/nature02051. [DOI] [PubMed] [Google Scholar]
- Dasen J.S., Tice B.C., Brenner-Morton S., Jessell T.M. A Hox regulatory network establishes motor neuron pool identity and target-muscle connectivity. Cell. 2005;123:477–491. doi: 10.1016/j.cell.2005.09.009. [DOI] [PubMed] [Google Scholar]
- Dasen J.S., De Camilli A., Wang B., Tucker P.W., Jessell T.M. Hox repertoires for motor neuron diversity and connectivity gated by a single accessory factor, FoxP1. Cell. 2008;134:304–316. doi: 10.1016/j.cell.2008.06.019. [DOI] [PubMed] [Google Scholar]
- Delfino-Machín M., Lunn J.S., Breitkreuz D.N., Akai J., Storey K.G. Specification and maintenance of the spinal cord stem zone. Development. 2005;132:4273–4283. doi: 10.1242/dev.02009. [DOI] [PubMed] [Google Scholar]
- Diez del Corral R., Storey K.G. Opposing FGF and retinoid pathways: a signalling switch that controls differentiation and patterning onset in the extending vertebrate body axis. BioEssays. 2004;26:857–869. doi: 10.1002/bies.20080. [DOI] [PubMed] [Google Scholar]
- Diez del Corral R., Olivera-Martinez I., Goriely A., Gale E., Maden M., Storey K. Opposing FGF and retinoid pathways control ventral neural pattern, neuronal differentiation, and segmentation during body axis extension. Neuron. 2003;40:65–79. doi: 10.1016/s0896-6273(03)00565-8. [DOI] [PubMed] [Google Scholar]
- Gould A., Itasaki N., Krumlauf R. Initiation of rhombomeric Hoxb4 expression requires induction by somites and a retinoid pathway. Neuron. 1998;21:39–51. doi: 10.1016/s0896-6273(00)80513-9. [DOI] [PubMed] [Google Scholar]
- Gouti M., Tsakiridis A., Wymeersch F.J., Huang Y., Kleinjung J., Wilson V., Briscoe J. In vitro generation of neuromesodermal progenitors reveals distinct roles for wnt signalling in the specification of spinal cord and paraxial mesoderm identity. PLoS Biol. 2014;12:e1001937. doi: 10.1371/journal.pbio.1001937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iimura T., Pourquié O. Collinear activation of Hoxb genes during gastrulation is linked to mesoderm cell ingression. Nature. 2006;442:568–571. doi: 10.1038/nature04838. [DOI] [PubMed] [Google Scholar]
- Jung H., Lacombe J., Mazzoni E.O., Liem K.F., Jr., Grinstein J., Mahony S., Mukhopadhyay D., Gifford D.K., Young R.A., Anderson K.V. Global control of motor neuron topography mediated by the repressive actions of a single hox gene. Neuron. 2010;67:781–796. doi: 10.1016/j.neuron.2010.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaplan A., Spiller K.J., Towne C., Kanning K.C., Choe G.T., Geber A., Akay T., Aebischer P., Henderson C.E. Neuronal matrix metalloproteinase-9 is a determinant of selective neurodegeneration. Neuron. 2014;81:333–348. doi: 10.1016/j.neuron.2013.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiskinis E., Sandoe J., Williams L.A., Boulting G.L., Moccia R., Wainger B.J., Han S., Peng T., Thams S., Mikkilineni S. Pathways disrupted in human ALS motor neurons identified through genetic correction of mutant SOD1. Cell Stem Cell. 2014;14:781–795. doi: 10.1016/j.stem.2014.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krencik R., Weick J.P., Liu Y., Zhang Z.-J., Zhang S.-C. Specification of transplantable astroglial subtypes from human pluripotent stem cells. Nat. Biotechnol. 2011;29:528–534. doi: 10.1038/nbt.1877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kriks S., Shim J.-W., Piao J., Ganat Y.M., Wakeman D.R., Xie Z., Carrillo-Reid L., Auyeung G., Antonacci C., Buch A. Dopamine neurons derived from human ES cells efficiently engraft in animal models of Parkinson’s disease. Nature. 2011;480:547–551. doi: 10.1038/nature10648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee H., Shamy G.A., Elkabetz Y., Schofield C.M., Harrsion N.L., Panagiotakos G., Socci N.D., Tabar V., Studer L. Directed differentiation and transplantation of human embryonic stem cell-derived motoneurons. Stem Cells. 2007;25:1931–1939. doi: 10.1634/stemcells.2007-0097. [DOI] [PubMed] [Google Scholar]
- Lewis E.B. A gene complex controlling segmentation in Drosophila. Nature. 1978;276:565–570. doi: 10.1038/276565a0. [DOI] [PubMed] [Google Scholar]
- Li X.-J., Du Z.-W., Zarnowska E.D., Pankratz M., Hansen L.O., Pearce R.A., Zhang S.-C. Specification of motoneurons from human embryonic stem cells. Nat. Biotechnol. 2005;23:215–221. doi: 10.1038/nbt1063. [DOI] [PubMed] [Google Scholar]
- Li X.-J., Hu B.-Y., Jones S.A., Zhang Y.-S., Lavaute T., Du Z.-W., Zhang S.-C. Directed differentiation of ventral spinal progenitors and motor neurons from human embryonic stem cells by small molecules. Stem Cells. 2008;26:886–893. doi: 10.1634/stemcells.2007-0620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X.-J., Zhang X., Johnson M.A., Wang Z.-B., Lavaute T., Zhang S.-C. Coordination of sonic hedgehog and Wnt signaling determines ventral and dorsal telencephalic neuron types from human embryonic stem cells. Development. 2009;136:4055–4063. doi: 10.1242/dev.036624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lian X., Hsiao C., Wilson G., Zhu K., Hazeltine L.B., Azarin S.M., Raval K.K., Zhang J., Kamp T.J., Palecek S.P. Robust cardiomyocyte differentiation from human pluripotent stem cells via temporal modulation of canonical Wnt signaling. Proc. Natl. Acad. Sci. USA. 2012;109:E1848–E1857. doi: 10.1073/pnas.1200250109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lippmann E.S., Estevez-Silva M.C., Ashton R.S. Defined human pluripotent stem cell culture enables highly efficient neuroepithelium derivation without small molecule inhibitors. Stem Cells. 2014;32:1032–1042. doi: 10.1002/stem.1622. [DOI] [PubMed] [Google Scholar]
- Liu J.-P. The function of growth/differentiation factor 11 (Gdf11) in rostrocaudal patterning of the developing spinal cord. Development. 2006;133:2865–2874. doi: 10.1242/dev.02478. [DOI] [PubMed] [Google Scholar]
- Liu J.-P., Laufer E., Jessell T.M. Assigning the positional identity of spinal motor neurons: rostrocaudal patterning of Hox-c expression by FGFs, Gdf11, and retinoids. Neuron. 2001;32:997–1012. doi: 10.1016/s0896-6273(01)00544-x. [DOI] [PubMed] [Google Scholar]
- Ma L., Hu B., Liu Y., Vermilyea S.C., Liu H., Gao L., Sun Y., Zhang X., Zhang S.-C. Human embryonic stem cell-derived GABA neurons correct locomotion deficits in quinolinic acid-lesioned mice. Cell Stem Cell. 2012;10:455–464. doi: 10.1016/j.stem.2012.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mann R.S., Lelli K.M., Joshi R. Hox specificity: unique roles for cofactors and collaborators. In: Pourquié O., editor. Chapter 3. Academic Press; 2009. pp. 63–101. (Current Topics in Developmental Biology). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathis L., Kulesa P.M., Fraser S.E. FGF receptor signalling is required to maintain neural progenitors during Hensen’s node progression. Nat. Cell Biol. 2001;3:559–566. doi: 10.1038/35078535. [DOI] [PubMed] [Google Scholar]
- Maury Y., Côme J., Piskorowski R.A., Salah-Mohellibi N., Chevaleyre V., Peschanski M., Martinat C., Nedelec S. Combinatorial analysis of developmental cues efficiently converts human pluripotent stem cells into multiple neuronal subtypes. Nat. Biotechnol. 2014 doi: 10.1038/nbt.3049. [DOI] [PubMed] [Google Scholar]
- Mazzoni E.O., Mahony S., Peljto M., Patel T., Thornton S.R., McCuine S., Reeder C., Boyer L.A., Young R.A., Gifford D.K., Wichterle H. Saltatory remodeling of Hox chromatin in response to rostrocaudal patterning signals. Nat. Neurosci. 2013;16:1191–1198. doi: 10.1038/nn.3490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molotkova N., Molotkov A., Sirbu I.O., Duester G. Requirement of mesodermal retinoic acid generated by Raldh2 for posterior neural transformation. Mech. Dev. 2005;122:145–155. doi: 10.1016/j.mod.2004.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nordström U., Maier E., Jessell T.M., Edlund T. An early role for WNT signaling in specifying neural patterns of Cdx and Hox gene expression and motor neuron subtype identity. PLoS Biol. 2006;4:e252. doi: 10.1371/journal.pbio.0040252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patani R., Hollins A.J., Wishart T.M., Puddifoot C.A., Álvarez S., de Lera A.R., Wyllie D.J.A., Compston D.A., Pedersen R.A., Gillingwater T.H. Retinoid-independent motor neurogenesis from human embryonic stem cells reveals a medial columnar ground state. Nat. Commun. 2011;2:214. doi: 10.1038/ncomms1216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pattyn A., Hirsch M., Goridis C., Brunet J.F. Control of hindbrain motor neuron differentiation by the homeobox gene Phox2b. Development. 2000;127:1349–1358. doi: 10.1242/dev.127.7.1349. [DOI] [PubMed] [Google Scholar]
- Peljto M., Dasen J.S., Mazzoni E.O., Jessell T.M., Wichterle H. Functional diversity of ESC-derived motor neuron subtypes revealed through intraspinal transplantation. Cell Stem Cell. 2010;7:355–366. doi: 10.1016/j.stem.2010.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Philippidou P., Dasen J.S. Hox genes: choreographers in neural development, architects of circuit organization. Neuron. 2013;80:12–34. doi: 10.1016/j.neuron.2013.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandoe J., Eggan K. Opportunities and challenges of pluripotent stem cell neurodegenerative disease models. Nat. Neurosci. 2013;16:780–789. doi: 10.1038/nn.3425. [DOI] [PubMed] [Google Scholar]
- Takemoto T., Uchikawa M., Kamachi Y., Kondoh H. Convergence of Wnt and FGF signals in the genesis of posterior neural plate through activation of the Sox2 enhancer N-1. Development. 2006;133:297–306. doi: 10.1242/dev.02196. [DOI] [PubMed] [Google Scholar]
- Takemoto T., Uchikawa M., Yoshida M., Bell D.M., Lovell-Badge R., Papaioannou V.E., Kondoh H. Tbx6-dependent Sox2 regulation determines neural or mesodermal fate in axial stem cells. Nature. 2011;470:394–398. doi: 10.1038/nature09729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thaler J., Harrison K., Sharma K., Lettieri K., Kehrl J., Pfaff S.L. Active suppression of interneuron programs within developing motor neurons revealed by analysis of homeodomain factor HB9. Neuron. 1999;23:675–687. doi: 10.1016/s0896-6273(01)80027-1. [DOI] [PubMed] [Google Scholar]
- Thomson J.A., Itskovitz-Eldor J., Shapiro S.S., Waknitz M.A., Swiergiel J.J., Marshall V.S., Jones J.M. Embryonic stem cell lines derived from human blastocysts. Science. 1998;282:1145–1147. doi: 10.1126/science.282.5391.1145. [DOI] [PubMed] [Google Scholar]
- Towers M., Tickle C. Growing models of vertebrate limb development. Development. 2009;136:179–190. doi: 10.1242/dev.024158. [DOI] [PubMed] [Google Scholar]
- Tozer S., Le Dréau G., Marti E., Briscoe J. Temporal control of BMP signalling determines neuronal subtype identity in the dorsal neural tube. Development. 2013;140:1467–1474. doi: 10.1242/dev.090118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuchida T., Ensini M., Morton S.B., Baldassare M., Edlund T., Jessell T.M., Pfaff S.L. Topographic organization of embryonic motor neurons defined by expression of LIM homeobox genes. Cell. 1994;79:957–970. doi: 10.1016/0092-8674(94)90027-2. [DOI] [PubMed] [Google Scholar]
- Tzouanacou E., Wegener A., Wymeersch F.J., Wilson V., Nicolas J.-F. Redefining the progression of lineage segregations during mammalian embryogenesis by clonal analysis. Dev. Cell. 2009;17:365–376. doi: 10.1016/j.devcel.2009.08.002. [DOI] [PubMed] [Google Scholar]
- Van Winkle A.P., Gates I.D., Kallos M.S. Mass transfer limitations in embryoid bodies during human embryonic stem cell differentiation. Cells Tissues Organs (Print) 2012;196:34–47. doi: 10.1159/000330691. [DOI] [PubMed] [Google Scholar]
- Wichterle H., Lieberam I., Porter J.A., Jessell T.M. Directed differentiation of embryonic stem cells into motor neurons. Cell. 2002;110:385–397. doi: 10.1016/s0092-8674(02)00835-8. [DOI] [PubMed] [Google Scholar]
- William C.M., Tanabe Y., Jessell T.M. Regulation of motor neuron subtype identity by repressor activity of Mnx class homeodomain proteins. Development. 2003;130:1523–1536. doi: 10.1242/dev.00358. [DOI] [PubMed] [Google Scholar]
- Yekta S., Shih I.H., Bartel D.P. MicroRNA-directed cleavage of HOXB8 mRNA. Science. 2004;304:594–596. doi: 10.1126/science.1097434. [DOI] [PubMed] [Google Scholar]
- Yu J., Vodyanik M.A., Smuga-Otto K., Antosiewicz-Bourget J., Frane J.L., Tian S., Nie J., Jonsdottir G.A., Ruotti V., Stewart R. Induced pluripotent stem cell lines derived from human somatic cells. Science. 2007;318:1917–1920. doi: 10.1126/science.1151526. [DOI] [PubMed] [Google Scholar]
- Yu P.B., Hong C.C., Sachidanandan C., Babitt J.L., Deng D.Y., Hoyng S.A., Lin H.Y., Bloch K.D., Peterson R.T. Dorsomorphin inhibits BMP signals required for embryogenesis and iron metabolism. Nat. Chem. Biol. 2008;4:33–41. doi: 10.1038/nchembio.2007.54. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


