Abstract
Acrophialophora fusispora is an emerging opportunistic fungus capable of causing human infections. The taxonomy of the genus is not yet resolved and, in order to facilitate identification of clinical specimens, we have studied a set of clinical and environmental Acrophialophora isolates by morphological and molecular analyses. This set included the available type strains of Acrophialophora species and similar fungi, some of which were considered by various authors to be synonyms of A. fusispora. Sequence analysis of the large subunit (LSU) and internal transcribed spacer (ITS) regions of the nuclear ribosomal DNA and a fragment of the β-tubulin (Tub) gene revealed that Acrophialophora belongs in the family Chaetomiaceae and comprises three different species, i.e., A. fusispora, Acrophialophora levis, and Acrophialophora seudatica; the latter was previously included in the genus Ampullifera. The most prevalent species among clinical isolates was A. levis (72.7%), followed by A. fusispora (27.3%), both of which were isolated mostly from respiratory specimens (72.7%), as well as subcutaneous and corneal tissue samples. In general, of the eight antifungal drugs tested, voriconazole had the greatest in vitro activity, while all other agents showed poor in vitro activity against these fungi.
INTRODUCTION
Acrophialophora is a thermotolerant soil fungus that is widely distributed in temperate and tropical regions. Given its capacity to produce large quantities of cellulases and xylanases, it is also commonly isolated as a decomposer of compost and other self-heating substrates (1, 2).
The genus Acrophialophora was erected by Edward (3) with a single species, Acrophialophora nainiana. This fungus forms grayish colonies with a black reverse with age. Microscopically, it produces darkly pigmented, straight, septate, unbranched, setae-like conidiophores with thick verrucose walls, which are fertile toward the apex, and flask-shaped, hyaline phialides grouped in verticils. Single flask-shaped phialides are also formed directly from the aerial hyphae. Acrophialophora was not fully accepted as a distinct genus, however, until the work of Samson and Mahmood (4), who, after studying a large set of isolates, demonstrated that the aforementioned morphological features were stable, which supported the differentiation of Acrophialophora from morphologically similar genera such as Paecilomyces and Masonia. Those authors accepted three species, based mainly on the size, pigmentation, and ornamentation of their conidia; these species were A. nainiana (4 to 10.5 by 2 to 5 μm, hyaline, and finely echinulate), Acrophialophora fusispora (5 to 12 by 3 to 6 μm, brown, and finely echinulate, forming spiral bands), which had been described earlier as Paecilomyces fusisporus (4), and Acrophialophora levis (4.5 to 8 by 2 to 3.5 μm, hyaline, and smooth to slightly roughened). However, while Ellis (5) regarded A. nainiana as conspecific with A. fusispora and the latter as the type species of the genus, Al-Mohsen et al. (1) considered the three species synonyms and conserved the single species name A. fusispora. In this wide concept of the species, other taxa were considered conspecific with A. fusispora, i.e., Masoniella indica and Ampullifera seudatica (4).
Acrophialophora fusispora is currently recognized as an emerging human opportunistic pathogen (6, 7), responsible for cases of keratitis (6, 8, 9), pulmonary colonization and infection (6, 10–12), and devastating cerebral infections requiring intensive antifungal therapy (1, 13–15). Antifungal susceptibility data for Acrophialophora are scarce and based mostly on a few clinical reports (1, 15).
The species delimitation for Acrophialophora, using a modern phylogenetic approach, has not been properly revised, and the taxonomic position and boundaries of the genus are unknown. Therefore, we carried out a phenotypic and molecular study with a set of clinical and environmental isolates, including all of the available type strains of the species historically included in the genus. In addition, in vitro antifungal susceptibility testing was performed with eight clinically available antifungal agents against these isolates.
MATERIALS AND METHODS
Fungal isolates and sequences.
A total of 39 isolates were included in this study, i.e., 32 from human clinical samples, 1 from an animal clinical sample, and 6 from environmental sources, including all of the available type strains of the genus (Table 1). Most of the clinical isolates were from the United States and were received by the Fungus Testing Laboratory at the University of Texas Health Science Center at San Antonio (UTHSCSA) from different parts of the country. In addition, 39 sequences retrieved from GenBank or the National Institute of Technology and Evaluation Biological Resource Center (NBRC) (Chiba, Japan) database were included in the phylogenetic analyses.
TABLE 1.
Origin and GenBank accession numbers of the sequences of the Acrophialophora spp. included in this study
| Species and straina | Origin | GenBank accession no. |
||
|---|---|---|---|---|
| LSU | ITS | Tub | ||
| A. levis | ||||
| CBS 484.70 (type strain) | Germany, composted domestic waste | KM995840 | KM995878 | LN624419 |
| FMR 6662 = CBS 120407 | Spain, sputum | KM995841 | KM995879 | LN624420 |
| FMR 12780 | Spain, sputum | KM995842 | KM995880 | LN624421 |
| UTHSCSA DI-13-134 | USA, sputum | KM995843 | KM995881 | LN624422 |
| UTHSCSA DI-13-137 | USA, sputum | KM995844 | KM995882 | LN624423 |
| UTHSCSA DI-13-138 | USA, bronchoalveolar lavage fluid | KM995845 | KM995883 | LN624424 |
| UTHSCSA DI-13-139 | USA, bronchoalveolar lavage fluid | KM995846 | KM995884 | LN624425 |
| UTHSCSA DI-13-142 | USA, sputum | KM995847 | KM995885 | LN624426 |
| UTHSCSA DI-13-144 | USA, sputum | KM995848 | KM995886 | LN624427 |
| UTHSCSA DI-13-145 | USA, brain | KM995849 | KM995887 | LN624428 |
| UTHSCSA DI-13-146 | USA, bronchoalveolar lavage fluid | KM995850 | KM995888 | LN624429 |
| UTHSCSA DI-13-147 | USA, bronchoalveolar lavage fluid (canine) | KM995851 | KM995889 | LN624430 |
| UTHSCSA DI-13-148 | USA, lung, right upper lobe | KM995852 | KM995890 | LN624431 |
| UTHSCSA DI-13-150 | USA, sputum | KM995853 | KM995891 | LN624432 |
| UTHSCSA DI-13-151 | USA, sputum | KM995854 | KM995892 | LN624433 |
| UTHSCSA DI-13-152 | USA, leg tissue | KM995855 | KM995893 | LN624434 |
| UTHSCSA DI-13-153 | USA, tissue | KM995856 | KM995894 | LN624435 |
| UTHSCSA DI-13-154 | USA, bronchoalveolar lavage fluid | KM995857 | KM995895 | LN624436 |
| UTHSCSA DI-13-155 | USA, sputum | KM995858 | KM995896 | LN624437 |
| UTHSCSA DI-13-156 | USA, knee tissue | KM995859 | KM995897 | LN624438 |
| UTHSCSA DI-13-157 | USA, bronchoalveolar lavage fluid | KM995860 | KM995898 | LN624439 |
| UTHSCSA DI-13-158 | USA, sputum | KM995861 | KM995899 | LN624440 |
| UTHSCSA DI-13-159 | USA, bronchoalveolar lavage fluid | KM995862 | KM995900 | LN624441 |
| UTHSCSA DI-13-162 | USA, bronchoalveolar lavage fluid | KM995863 | KM995901 | LN624442 |
| UTHSCSA DI-13-163 | USA, bronchoalveolar lavage fluid | KM995864 | KM995902 | LN624443 |
| A. fusispora | ||||
| CBS 100.60 (A. nainiana type strain) | India, farm soil | KM995865 | KM995903 | LN624444 |
| CBS 149.64 (M. indica type strain) | India, forest soil | KM995866 | KM995904 | LN624445 |
| CBS 380.55 (P. fusisporus type strain) | India, forest soil | KM995867 | KM995905 | LN624446 |
| FMR 6258 = CBS 120406 | India, soil | KM995868 | KM995906 | LN624447 |
| FMR 8888 = CBS 120409 | India, cornea | KM995869 | KM995907 | LN624448 |
| UTHSCSA DI-13-135 | USA, left sphenoid sinus | KM995870 | KM995908 | LN624449 |
| UTHSCSA DI-13-136 | USA, brain abscess | KM995871 | KM995909 | LN624450 |
| UTHSCSA DI-13-140 | USA, bronchoalveolar lavage fluid | KM995872 | KM995910 | LN624451 |
| UTHSCSA DI-13-141 | USA, sputum | KM995873 | KM995911 | LN624452 |
| UTHSCSA DI-13-143 | USA, chest mass | KM995874 | KM995912 | LN624453 |
| UTHSCSA DI-13-149 | USA, cornea | KM995875 | KM995913 | LN624454 |
| UTHSCSA DI-13-160 | USA, bronchoalveolar lavage fluid | KM995876 | KM995914 | LN624455 |
| UTHSCSA DI-13-161 | USA, sputum | KM995877 | KM995915 | LN624456 |
| A. seudatica | ||||
| CBS 916.79 (Ampullifera seudatica type strain) | India, soil | LN736031 | LN736030 | LN736032 |
CBS, Fungal Biodiversity Centre (Utrecht, The Netherlands) culture collection; FMR, Facultat de Medicina, Universitat Rovira i Virgili (Reus, Spain); UTHSCSA, Fungus Testing Laboratory at the University of Texas Health Science Center at San Antonio (San Antonio, TX).
Phenotypic studies.
The isolates were grown on malt extract agar (MEA) (30 g of malt extract, 5 g of peptone, 15 g of agar, and 1 liter of distilled water) and oatmeal agar (OA) (30 g of filtered oat flakes, 20 g of agar, and 1 liter of distilled water). Colony features and growth rates were determined at 7 and 14 days of incubation at different temperatures (5, 15, 25, 35, 37, 40, 45, 50, and 52°C). Microscopic features were examined after 14 days of incubation at 25°C on both media, in wet mounts with 85% lactic acid, using light microscopy. All isolates were identified based on the features described by Edward (3), Samson and Mahmood (4), and Ellis (5). Photomicrographs were obtained with a Zeiss Axio-Imager M1 light microscope, using phase-contrast and Nomarski differential interference optics.
DNA extraction, amplification, and sequencing.
FastPrep kits (MP Biomedicals, Santa Ana, CA) were used to extract total genomic DNA from fungal mycelia harvested from colonies grown on potato dextrose agar (PDA) for 7 days at 25°C, according to the manufacturer's protocol. DNA was quantified using a Nanodrop 3000 apparatus (Thermo Scientific, Madrid, Spain).
Three nuclear DNA targets were amplified by PCR and sequenced using the following primer pairs: ITS4 and ITS5 (16) for a region spanning internal transcribed spacer 1 (ITS1) and ITS2 and the 5.8S gene of the ribosomal DNA (rDNA), LR0R and LR5 (17, 18) for a fraction of the 5′ end of the large subunit (LSU) gene of the rDNA, and BT2a and BT2b (19) for a fragment of the β-tubulin (Tub) gene. The amplified products were purified with the Diffinity RapidTip purification system (Sigma-Aldrich, St. Louis, MO) and stored at −20°C until sequencing.
Sequencing was performed in both directions, with the same primer pair as used for amplification, at Macrogen Europe (Macrogen Inc., Amsterdam, The Netherlands). The consensus sequences were obtained using SeqMan software (version 7.0.0; DNAStar Lasergene, Madison, WI).
Molecular identification and phylogenetic analysis.
In order to assess the taxonomic position of the genus Acrophialophora, a first phylogenetic analysis was carried out using partial LSU sequences of the available type strains of Acrophialophora species complemented with 15 sequences retrieved from public databases, selected on the basis of BLAST homology searches and representing 8 different genera from the families Chaetomiaceae and Sordariaceae, of the subclass Sordariomycetidae. A second phylogenetic analysis directed to assessing the species distribution of Acrophialophora was conducted using partial LSU, ITS, and Tub sequences and included all of the type strains of Acrophialophora species, the type strains of the putative synonyms Ampullifera seudatica and M. indica, and several clinical and environmental isolates morphologically identified as Acrophialophora spp. Multiple sequence alignments were made for each individual locus using Mega version 6.06 (20), with the ClustalW function, and were manually refined when necessary. The nucleotide substitution models for each data set (GTR+G+I for LSU, JC+G for ITS, and T92+G for Tub) were calculated using the Find best DNA/protein model tool in Mega 6.06. In order to compare the concordance of the different loci, individual phylogenetic analyses were carried out using the maximum likelihood (ML) algorithm in Mega; the resulting trees were compared visually using a 70% bootstrap cutoff and were complemented with the partition homogeneity test, carried out as implemented in PAUP* software (version 4.0b10; Sinauer, Sunderland, MA). Since no incongruence was found (P = 0.180), the three genes were combined into a single data set. The combined phylogenetic analyses were made using maximum likelihood (ML) and Bayesian inference (BI) methods in Mega and MrBayes (version 3.1.2) (21), respectively. For the ML analysis, nearest-neighbor interchange (NNI) was used as the heuristic method for tree inference. Support for the internal branches was assessed by a search of 1,000 bootstrapped sets of data. A bootstrap support (bs) value of ≥70 was considered significant. For BI analysis, two simultaneous runs of 3,000,000 generations were performed, and samples were stored every 100 generations. The 50% majority-rule consensus tree and posterior probability (pp) values were calculated after the first 25% of the samples were discarded. A pp value of ≥0.95 was considered significant.
Antifungal susceptibility testing.
Antifungal susceptibility testing was performed according to the methods in the CLSI M38-A2 standard (22). The antifungal drugs tested were amphotericin B (AMB), voriconazole (VRC), itraconazole (ITC), posaconazole (PSC), terbinafine (TRB), anidulafungin (AFG), caspofungin (CFG), and micafungin (MFG). The minimal effective concentration (MEC), defined as the lowest concentration that resulted in short, stubby, abnormally branched hyphae, was determined at 24 h for the echinocandins, and the MIC was determined at 48 h for the remaining drugs. The MIC was defined as the lowest concentration exhibiting 100% visual inhibition of growth for AMB, VRC, ITC, and PSC and 80% reduction in growth for TRB. Geometric mean (GM) MICs were compared using the Mann-Whitney test in GraphPad Prism for Windows (version 6; GraphPad Software, La Jolla CA).
Nucleotide sequence accession numbers.
Sequences newly generated in this study were deposited in GenBank under accession numbers KM995840 to KM995877 and LN736031 (LSU), KM995878 to KM995915 and LN736030 (ITS), and LN624419 to LN624456 and LN736032 (Tub) (Table 1).
RESULTS
Figure 1 shows the results of the analysis of the LSU sequences (431 bp) of Acrophialophora species and related fungi. The type strain of A. nainiana (CBS 100.60) clustered with the type strains of A. levis (CBS 484.70) and A. fusispora (CBS 380.55), being included in a fully supported clade containing several members of the family Chaetomiaceae of the order Sordariales. The species closest to Acrophialophora were members of Achaetomium, Botryotrichum, Chaetomidium, and Chaetomium. The latter genus was also found to be polyphyletic.
FIG 1.
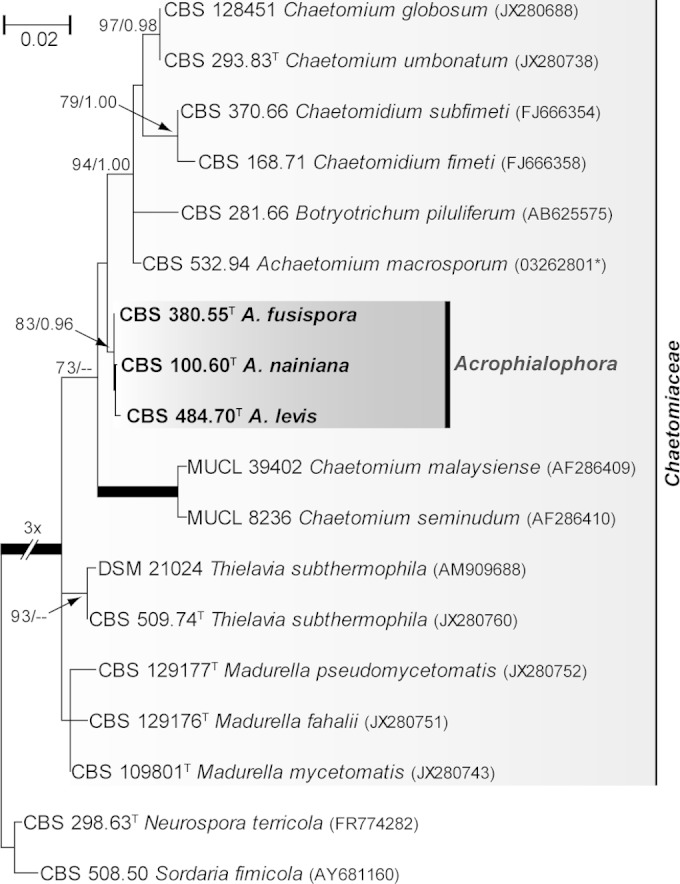
Maximum likelihood (ML) tree constructed with partial LSU sequences (431 bp) of Acrophialophora spp. and members of Chaetomiaceae. Branch lengths are proportional to the phylogenetic distance. ML bs and Bayesian pp values over 70% and 0.95, respectively, are shown on the nodes. Thickened branches indicate full statistical support. GenBank accession numbers are shown in parentheses. *, sequence retrieved from the National Institute of Technology and Evaluation Biological Resource Center (NBRC) (Chiba, Japan). The tree is rooted with Neurospora terricola and Sordaria fimicola. T, type strain; CBS, Fungal Biodiversity Centre (Utrecht, The Netherlands) culture collection; DSM, Leibniz Institute DSMZ-German Collection of Microorganisms and Cell Cultures (Braunschweig, Germany); MUCL, Mycothèque de l′Université Catholique de Louvain (Louvain-la-Neuve, Belgium).
Figure 2 shows the results of the phylogenetic analysis of the species of the genus Acrophialophora using concatenated LSU, ITS, and Tub sequences. The final alignment consisted of 1,804 bp (LSU, 843 bp; ITS, 502 bp; Tub, 459 bp) of 41 isolates, i.e., one from an environmental source, 33 from clinical origins, and the type strains of A. fusispora, A. levis, and A. nainiana and the putative synonyms Ampullifera seudatica and M. indica. Chaetomium globosum and Chaetomium angustispirale were used to root the tree. The tree showed two fully supported main clades, one of which included the type strain of A. levis and the other that of A. fusispora. The latter clustered in a fully supported clade with the type strains of A. nainiana and M. indica, which demonstrated them to be conspecific, since their sequences were practically identical. The type strain of Ampullifera seudatica formed a single lineage, basal and distant to the A. fusispora clade (98.5% sequence similarity with A. fusispora in the combined analysis), and is here considered a different species of the genus named Acrophialophora seudatica.
FIG 2.
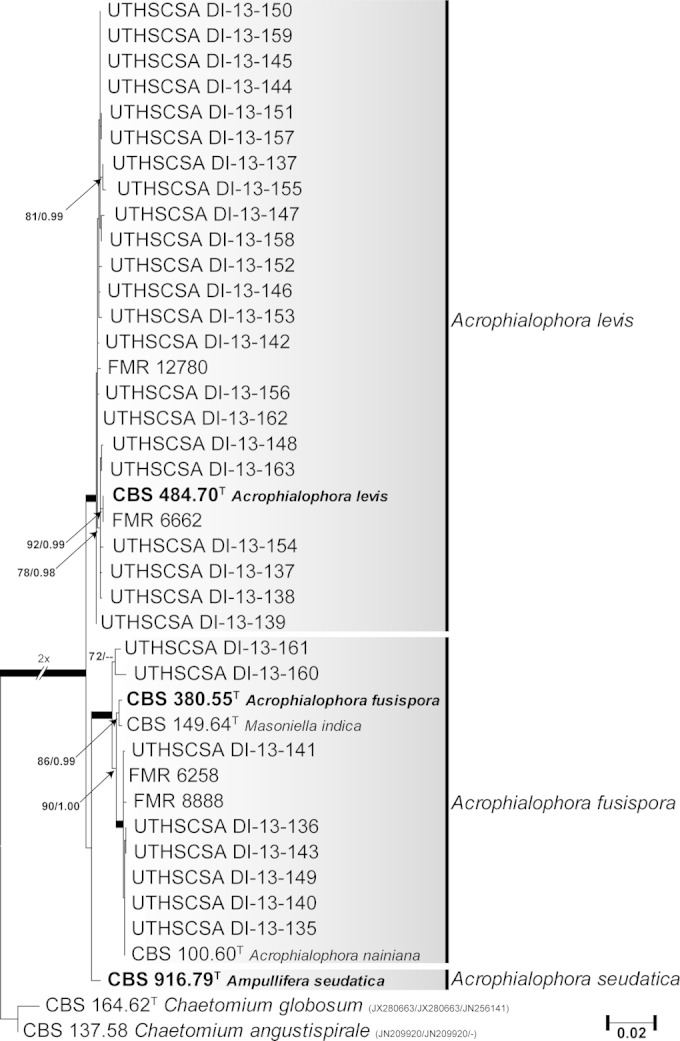
Maximum likelihood (ML) tree constructed with combined LSU (843 bp), ITS (502 bp), and Tub (459 bp) sequences of Acrophialophora clinical and environmental isolates. ML bs and Bayesian pp values are shown on the nodes. Thickened branches indicate full statistical support. The tree is rooted with Chaetomium globosum and Chaetomium angustispirale. GenBank accession numbers for LSU, ITS, and Tub sequences are shown in parentheses. T, type strain; CBS, Fungal Biodiversity Centre (Utrecht, The Netherlands) culture collection; FMR, Facultat de Medicina, Universitat Rovira i Virgili (Reus, Spain); UTHSCSA, Fungus Testing Laboratory at the University of Texas Health Science Center at San Antonio (San Antonio, TX).
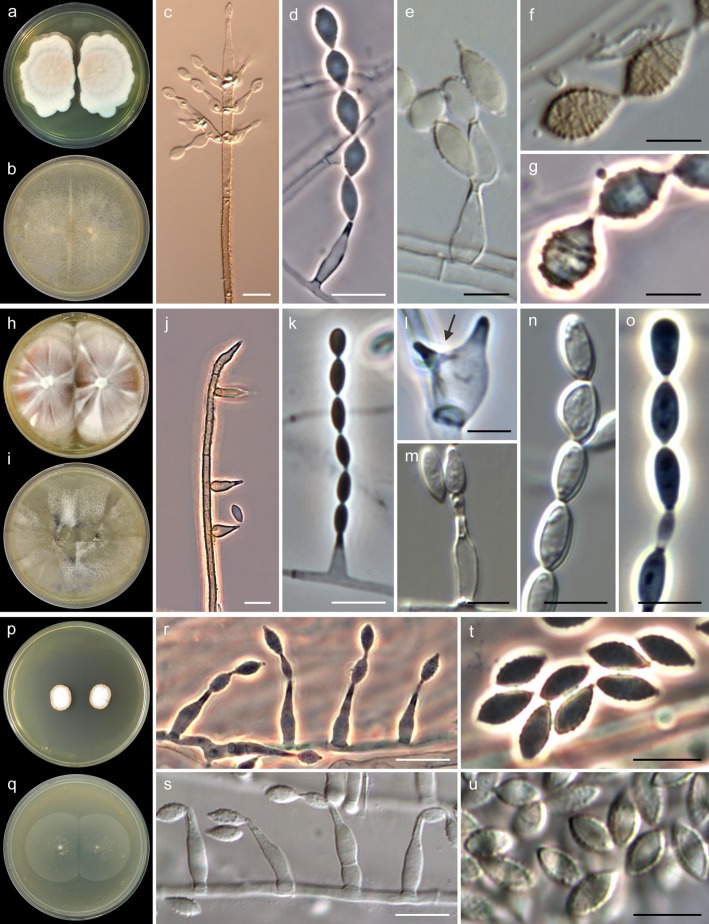
The isolates identified as A. fusispora and A. levis showed similar macroscopic features on all media tested. The colonies on MEA (Fig. 3a and h) ranged from 30 to 60 mm in diameter after 14 days at 25°C and were flat to slightly umbonate, at first white but soon becoming pale yellow to brownish gray, velvety to felty, with irregular margins and a yellow, brown, or black reverse. Acrophialophora seudatica grew more slowly (20 to 25 mm in diameter in 14 days), and its colonies were flat, at first white but rapidly turning pale orange, velvety, with a pale orange reverse (Fig. 3p). The optimal temperature for growth was between 35 and 40°C, with a minimum of 15°C and a maximum of 50°C, for all of the species. Microscopically, the isolates of A. fusispora were characterized by the abundant production of flask-shaped phialides and polyphialides measuring 5 to 19 by 1.5 to 5 μm (Fig. 3d and e), swollen at the base and tapering abruptly to a narrow neck, mostly formed directly from the aerial hyphae or in the apex of well-differentiated conidiophores, which were erect, unbranched, dark-colored, and with a spiny to warted wall surface (Fig. 3c). Conidia were produced on basipetal chains and were one-celled, subhyaline to brownish, ovoid to fusiform, finely echinulate or forming spiral bands, and measuring 5 to 12 by 2 to 5 μm (Fig. 3d to g). The isolates of A. levis also produced abundant flask-shaped phialides and frequently polyphialides, measuring 4 to 13 by 1.5 to 5 μm (Fig. 3i and m), and hyaline to subhyaline ellipsoid to cylindrical conidia, smooth to finely echinulate and measuring 4 to 9 by 2 to 6 μm (Fig. 3m to o). The isolate of A. seudatica exhibited flask-shaped phialides measuring 8 to 22 by 2.5 to 4.5 μm, with long necks (Fig. 3r and s), and ovoid to fusiform conidia measuring 6 to 8 by 3 to 4 μm (Fig. 3t and u), with thick and finely verruculose walls, subhyaline or turning pale yellow when mature. This isolate was unable to produce the typical pigmented conidiophores of Acrophialophora. Table 2 summarizes the key morphological features that distinguish the three Acrophialophora species.
FIG 3.
Key morphological features of Acrophialophora fusispora (a to g), A. levis (h to o), and A. seudatica (p to u). (a, h, and p) Colonies on MEA after 14 days at 25°C. (b, i, and q) Colonies on OA after 14 days at 25°C. (c and j) Conidiophores. (d, e, k, l, m, r, and s) Phialides (polyphialides indicated with an arrow). (f, g, n, o, t, and u) Conidia. White bars, 10 μm; black bars, 5 μm.
TABLE 2.
Key differential features of the three species of Acrophialophora
| Characteristica | A. fusispora | A. levis | A. seudatica |
|---|---|---|---|
| Colony diameter (mm) | 30–50 | 35–60 | 20–25 |
| Colony color | White, pale yellow, or gray | White, pale yellow, or gray | White to pale orange |
| Colony texture | Velvety to felty | Velvety to felty | Velvety |
| Phialide size (μm) | 5–19 by 1.5–5 | 4–13 by 1.5–5 | 8–22 by 2.5–4.5 |
| Conidial size (μm) | 5–12 by 2–5 | 4–9 by 2–6 | 6–8 by 3–4 |
| Conidial shape | Ovoid to fusiform | Ellipsoid to cylindrical | Ovoid to fusiform |
| Conidial ornamentation | Finely echinulate to spiral sculpted | Smooth to finely echinulate | Finely verruculose |
| Conidial color | Subhyaline to brown | Hyaline to subhyaline | Subhyaline to pale yellow |
Characteristics were determined after growth on MEA for 14 days at 25°C.
The majority of isolates from clinical sources belonged to A. levis (72.7%), while A. fusispora accounted for the remaining 34.3% of the isolates. The main source of isolates was the respiratory tract (72.7%), mostly from sputum and bronchoalveolar lavage (BAL) fluid specimens, followed by subcutaneous tissues (9.1%), brain tissue, and corneas (6.1% each). Other sites from which the fungi were cultured included the sphenoid sinus and a chest mass (3% each). No major differences regarding the origins of isolates were observed between A. levis and A. fusispora.
The antifungal susceptibility results for the isolates belonging to A. levis and A. fusispora are shown in Table 3. Overall, the highest MIC values were observed for AMB, with geometric mean (GM) MIC and MIC90 values of 5.66 μg/ml and 16 μg/ml, respectively. The azole drugs exhibited the best in vitro activity, with VRC being the most potent, with overall GM MIC and MIC90 values of 0.17 μg/ml and 0.25 μg/ml, respectively, followed by PSC and ITC. The echinocandins exhibited poor in vitro activity, with AFG showing the lowest GM MEC and MEC90 values (1.86 μg/ml and 4 μg/ml, respectively). TRB showed GM MIC and MIC90 values of 0.51 μg/ml and 1 μg/ml, respectively. Although the differences were subtle, the MICs for VRC, ITC, CFG, AFG, MFG, and TRB were significantly lower against A. levis than A. fusispora (P < 0.0001).
TABLE 3.
Results of in vitro antifungal susceptibility testing for the 33 clinical isolates of Acrophialophora spp. included in the study
| Species and parameter | MIC or MEC (μg/ml)a |
|||||||
|---|---|---|---|---|---|---|---|---|
| AMB | VRC | PSC | ITC | CFGb | AFGb | MFGb | TRB | |
| A. levis (n = 24) | ||||||||
| GM | 6.77 | 0.16 | 0.50 | 1.15 | 3.58 | 1.65 | 2.64 | 0.42 |
| Range | 1–32 | 0.06–05 | 0.25–1 | 0.5–4 | 0.25–32 | 0.25–8 | 0.25–32 | 0.125–4 |
| MIC90 | 32 | 0.25 | 1 | 2 | 16 | 4 | 32 | 1 |
| A. fusispora (n = 9) | ||||||||
| GM | 4.22 | 0.18 | 0.50 | 0.85 | 11.02 | 2.61 | 6.46 | 0.75 |
| Range | 2–32 | 0.125–0.25 | 0.25–1 | 0.125–1 | 4–32 | 2–8 | 0.125–32 | 0.5–1 |
| MIC90 | 16 | 0.25 | 1 | 1 | 16 | 4 | 32 | 1 |
| Overall (n = 33) | ||||||||
| GM | 5.66 | 0.17 | 0.49 | 1 | 4.98 | 1.86 | 3.21 | 0.51 |
| Range | 1–32 | 0.06–05 | 0.25–1 | 0.125–4 | 0.25–32 | 0.25–8 | 0.125–32 | 0.125–4 |
| MIC90 | 16 | 0.25 | 1 | 1 | 16 | 4 | 32 | 1 |
AMB, amphotericin B; VRC, voriconazole; PSC, posaconazole; ITC, itraconazole; CFG, caspofungin; AFG, anidulafungin; MFG, micafungin; TRB, terbinafine.
This column contains MEC data.
TAXONOMY
According to the results of our phylogenetic and morphological analyses, the following new combination is proposed: Acrophialophora seudatica (Subrahm.) Sandoval-Denis, Gené & Guarro comb. nov., Mycobank accession number MB811225. Basionym: Ampullifera seudatica Subrahmanyam, Nova Hedwigia 31:159 (1979).
DISCUSSION
To our knowledge, this is the first study involving molecular assessment of the fungal genus Acrophialophora, a rare opportunistic human and animal pathogen. It also includes the largest set of clinical isolates of the species studied to date. The taxonomy of the genus has been revised and the spectrum of species associated with human disease determined.
According to our results, the genus Acrophialophora, belonging to the sordariomycetous family Chaetomiaceae, comprises three species, i.e., A. fusispora, A. levis, and A. seudatica. This family includes mostly soilborne cellulose decomposers but also thermotolerant opportunistic pathogens, including neurotropic species such as Achaetomium strumarium and Chaetomium atrobrunneum (23, 24). Although historically the family Chaetomiaceae encompassed mainly ascosporulating fungi, Acrophialophora is not the first genus of the family showing strictly asexual reproduction. Recently, de Hoog et al. (24) demonstrated that agents of black-grain mycetomas such as Madurella species, which fail to produce fertile sexual morphs, also belong to Chaetomiaceae. According to our LSU phylogeny (Fig. 1), members of the genera Acrophialophora, Chaetomidium, and Thielavia nested unambiguously in highly supported terminal clades, while the positions of the genera Achaetomium, Botryotrichum, Chaetomium, and Madurella are unclear, with the genus Chaetomium forming two polyphyletic clades. The classifications of the latter genus, however, have been shown to differ significantly when molecular and conventional approaches are compared (24).
Because the morphological features used to distinguish the three Acrophialophora species recognized by Samson and Mahmood (4) tended to overlap, Al-Mohsen et al. (1) considered them morphological variations of a single species, A. fusispora. However, our phylogenetic analysis of the different type strains does not support this conclusion. Our results confirm that A. fusispora, A. nainiana, and M. indica are conspecific, while A. levis and A. seudatica are two different species. In contrast to the observations of Al-Mohsen et al. (1), our phylogenetic results indicate that subtle morphological findings for these fungi, such as conidial size, shape, color, and ornamentation, show consistent differences to distinguish A. fusispora, A. levis, and A. seudatica (Table 2). The presence (mainly on OA) of erect pigmented conidiophores is typical of cultures of the two clinically relevant species of Acrophialophora, i.e., A. fusispora and A. levis, and can be important for the initial generic diagnosis. However, these pigmented conidiophores are absent in A. seudatica. This species was originally described as having simple, hyaline, straight conidiophores (25), a feature that is confirmed here. However, A. seudatica is known only from its type specimen isolated from a soil sample from India. Therefore, confirmation of the presence or absence of the typical conidiophores of Acrophialophora will be possible only by studying more isolates of this rare species.
The three species of Acrophialophora shared very similar LSU sequences (99.9%), but the large differences in their ITS and Tub sequences (<96.1% and <96.6% sequence similarity, respectively) show that both loci can discriminate between the three species, making them good candidates for barcoding targets in Acrophialophora.
Some authors stated that Acrophialophora infections may have been underdiagnosed due to the rarity of these fungi and the potential confusion with similar opportunistic molds, such as Lomentospora prolificans and Scopulariopsis chartarum (9, 13, 26, 27). Only six well-documented cases of human infections exist in the literature, most of which lacks molecular confirmation of the etiological agent. In only one case was the fungus confirmed as A. fusispora by sequencing of the ITS region (15). The rarity is also reflected by the scarcity of reference sequences for comparison in fungal databases, which do not include any type or correctly identified reference strains.
Since the synonymy of the species of Acrophialophora was formally proposed by Al-Mohsen et al. (1), A. fusispora has remained the only accepted species of the genus and, as such, has been cited as the causative agent in the reported clinical cases (1, 7, 14, 23). According to our results, however, A. levis seems to be the most common species isolated from human clinical samples. The identification of some of the isolates included in the study by Guarro et al. (6) was reassessed here by sequence comparison. One clinical isolate (FMR 8888, from a corneal infection) was confirmed as A. fusispora, while another (FMR 6662, isolated from sputum) was reidentified here as A. levis. The third clinical isolate included in that study (FMR 6404) was not available for analysis, and thus its final identification remains unknown.
Most published cases refer to pulmonary involvement, with or without systemic dissemination (6, 10–12). Similarly, the majority of our clinical isolates were obtained from respiratory specimens, with one-half of them being from BAL fluid samples. It was not possible, however, to distinguish between true infectious agents, colonizers, and environmental contaminants, given the nature of the samples and the absence of appropriate clinical or histopathological data. The second most common infection in clinical reports is keratitis (6, 9), while in our study it was soft tissue infection, particularly lower extremity tissue infection. Corneal and cerebral samples, in equal proportions, were the third most common sites of isolation. The lack of isolates from the central nervous system (CNS) does not allow us to confirm the potential neurotropism attributed to Acrophialophora (14).
The antifungal treatment of Acrophialophora infections has been hampered by the paucity of in vitro susceptibility data and the lack of specific treatment guidelines. The clinical cases have demonstrated variable results. Arthur et al. (9) reported a favorable outcome with the use of AMB and surgical debridement in a case of keratouveitis. In one report of a pulmonary infection, monotherapy with liposomal AMB (LAMB) was not effective, but the patient responded to combination therapy with LAMB and ITC (1). Guarro et al. (6) reported the use of VRC in two clinical cases, one a case of keratitis that responded favorably to the drug and one a pulmonary infection with a fatal outcome. In addition, Li et al. (15) described a negative outcome using VRC in a case of cerebral infection. The two latter cases, however, were in highly immunocompromised patients with systemic involvement. Our susceptibility results showed that, while AMB and the echinocandins have almost no activity against Acrophialophora species, VRC exhibits potent in vitro activity. This confirms the observations of Guarro et al. (6), suggesting that VRC may be a potential treatment option for Acrophialophora infections.
In conclusion, Acrophialophora includes three closely related species, A. fusispora, A. levis, and A. seudatica, that can be accurately identified on the basis of ITS or Tub sequencing and detailed morphological study. Acrophialophora levis appears to be the most frequent species in clinical samples. VRC shows potent in vitro activity against these fungi.
ACKNOWLEDGMENTS
We thank Dea Garcia-Hermoso (Institut Pasteur, Centre National de Réfeìrence Mycoses Invasives et Antifongiques, Paris, France) for her help in obtaining the clinical isolates included in this study.
This study was supported by the Spanish Ministerio de Economía y Competitividad (grant CGL 2011-27185).
REFERENCES
- 1.Al-Mohsen IZ, Sutton DA, Sigler L, Almodovar E, Mahgoub N, Frayha H, Al-Hajjar S, Rinaldi MG, Walsh TJ. 2000. Acrophialophora fusispora brain abscess in a child with acute lymphoblastic leukemia: review of cases and taxonomy. J Clin Microbiol 38:4569–4576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barros RR, Oliveira RA, Gottschalk LM, Bon EP. 2010. Production of cellulolytic enzymes by fungi Acrophialophora nainiana and Ceratocystis paradoxa using different carbon sources. Appl Biochem Biotechnol 161:448–454. doi: 10.1007/s12010-009-8894-3. [DOI] [PubMed] [Google Scholar]
- 3.Edward JC. 1959. A new genus of the Moniliaceae. Mycologia 51:781–786. doi: 10.2307/3755830. [DOI] [Google Scholar]
- 4.Samson RA, Mahmood T. 1970. The genus Acrophialophora (Fungi, Moniliales). Acta Bot Neerl 19:804–808. [Google Scholar]
- 5.Ellis MG. 1971. Dematiaceous Hyphomycetes, p 533–534. Commonwealth Mycological Institute, Kew, United Kingdom. [Google Scholar]
- 6.Guarro J, Mendiratta DK, De Sequeira H, Rodríguez V, Thamke D, Gomes AM, Shukla AK, Menezes F, Narang P, Roldão Vieira J, Gené J. 2007. Acrophialophora fusispora: an emerging agent of human mycoses: a report of 3 new clinical cases. Diagn Microbiol Infect Dis 59:85–88. doi: 10.1016/j.diagmicrobio.2007.04.001. [DOI] [PubMed] [Google Scholar]
- 7.de Hoog GS, Guarro J, Gené J, Figueras MJ. 2011. Atlas of clinical fungi. CD-ROM version 3.1. CBS-KNAW Fungal Biodiversity Centre, Utrecht, The Netherlands. [Google Scholar]
- 8.Shukla PK. 1983. Clinical and experimental keratitis caused by Colletotrichum state of Glomerella cingulata and Acrophialophora fusispora. Sabouraudia 21:137–147. doi: 10.1080/00362178385380211. [DOI] [PubMed] [Google Scholar]
- 9.Arthur S, Steed LL, Apple DJ, Peng Q, Howard G, Escobar-Gomez M. 2001. Scedosporium prolificans keratouveitis in association with a contact lens retained intraocularly over a long term. J Clin Microbiol 39:4579–4582. doi: 10.1128/JCM.39.12.4579-4582.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sutton DA, Sigler L, Kalassian KG, Fothergill AW, Rinaldi MG. 1997. Pulmonary Acrophialophora fusispora: case history, literature, review and mycology. Abstr 13th Int Soc Hum Anim Mycol Congr, abstr 363. [Google Scholar]
- 11.González-Escalada A, Del Palacio A, Calvo MT, Gené J, Guarro J. 2000. Two cases of scalp wound colonization and respiratory tract by mycelial fungi. Rev Iberoam Micol 17:149–151. (In Spanish.) [PubMed] [Google Scholar]
- 12.Cimon B, Challier S, Béguin H, Carrère J, Chabasse D, Bouchara JP. 2005. Airway colonization by Acrophialophora fusispora in patients with cystic fibrosis. J Clin Microbiol 43:1484–1487. doi: 10.1128/JCM.43.3.1484-1487.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Welsh RD, Ely RW. 1999. Scopulariopsis chartarum systemic mycosis in a dog. J Clin Microbiol 37:2102–2103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Revankar SG, Sutton DA. 2010. Melanized fungi in human disease. Clin Microbiol Rev 23:884–928. doi: 10.1128/CMR.00019-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li CW, Lee HC, Chang TC, Wan JY, Chen HM, Wu CJ, Lee NY, Chang CM, Lee CC, Ko WC. 2013. Acrophialophora fusispora brain abscess in a patient with acquired immunodeficiency syndrome: a case report and review of the literature. Diagn Microbiol Infect Dis 76:368–371. doi: 10.1016/j.diagmicrobio.2013.03.016. [DOI] [PubMed] [Google Scholar]
- 16.White TJ, Bruns T, Lee S, Taylor JW. 1990. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics, p 315–322. In Innis MA, Gelfand DH, Sninsky JJ, White TJ (ed), PCR protocols: a guide to methods and applications. Academic Press, New York, NY. [Google Scholar]
- 17.Vilgalys R, Hester M. 1990. Rapid genetic identification and mapping of enzymatically amplified ribosomal DNA from several Cryptococcus species. J Bacteriol 172:4238–4246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Vilgalys R, Sun BL. 1994. Ancient and recent patterns of geographic speciation in the oyster mushroom Pleurotus revealed by phylogenetic analysis of ribosomal DNA sequences. Proc Natl Acad Sci U S A 91:4599–4603. doi: 10.1073/pnas.91.10.4599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Glass NL, Donaldson GC. 1995. Development of primer sets designed for use with the PCR to amplify conserved genes from filamentous ascomycetes. Appl Environ Microbiol 61:1323–1330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. 2013. MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol Biol Evol 30:2725–2729. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Huelsenbeck JP, Ronquist F. 2001. MrBayes: Bayesian inference of phylogenetic trees. Bioinformatics 17:754–755. doi: 10.1093/bioinformatics/17.8.754. [DOI] [PubMed] [Google Scholar]
- 22.Clinical and Laboratory Standards Institute. 2008. Reference method for broth dilution antifungal susceptibility testing of filamentous fungi; approved standard—2nd ed. Document M38-A2 Clinical and Laboratory Standards Institute, Wayne, PA. [Google Scholar]
- 23.Sigler L. 2003. Miscellaneous opportunistic fungi: Microascaceae and other Ascomycetes, Hyphomycetes, Coelomycetes, and Basidiomycetes, p 637–676. In Howard DH. (ed), Pathogenic fungi in humans and animals, 2nd ed Marcel Dekker, New York, NY. [Google Scholar]
- 24.de Hoog GS, Ahmed SA, Najafzadeh MJ, Sutton DA, Keisari MS, Fahal AH, Eberhardt U, Verkleij GJ, Xin L, Stielow B, van de Sande WWJ. 2013. Phylogenetic findings suggest possible new habitat and routes of infection of human eumycetoma. PLoS Negl Trop Dis 7:e2229. doi: 10.1371/journal.pntd.0002229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Subrahmanyam A. 1979. Ampullifera seudatica A Subrahm sp. nov. Nova Hedwigia 31:159–161. [Google Scholar]
- 26.Guarro J, Gené J. 2002. Acrophialophora fusispora misidentified as Scedosporium prolificans: comment letter 1. J Clin Microbiol 40:3544–3545. doi: 10.1128/JCM.40.9.3544-3545.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sigler L, Sutton DA. 2002. Acrophialophora fusispora misidentified as Scedosporium prolificans: comment letter 2. J Clin Microbiol 40:3544–3545. doi: 10.1128/JCM.40.9.3544-3545.2002. [DOI] [PubMed] [Google Scholar]