Abstract
In order to identify immunoreactive proteins that are usable for the immunological diagnosis of Babesia ovis infections, a phage lambda cDNA expression library was constructed and screened using parasite-specific immune serum. Immunoscreening resulted in the identification of a full-length cDNA clone encoding a secreted protein designated Babesia ovis secreted antigen 1 (BoSA1). The full-length BoSA1 cDNA contained a 1,137-bp open reading frame that encoded a protein of 378 amino acids, with a signal peptide and 2 internal repeat domains. The theoretical molecular mass of the mature protein was 42.5 kDa. Recombinant BoSA1 (rBoSA1) protein was expressed in Escherichia coli strain DH5α cells as a glutathione S-transferase (GST) fusion protein and was purified by affinity chromatography. Purified rBoSA1 was tested for reactivity with sera from animals experimentally or naturally infected with B. ovis, in an indirect enzyme-linked immunosorbent assay (ELISA). The results showed that specific antibodies against rBoSA1 were detectable on days 7 and 8 of the experimental infection and were maintained during the sampling period. Additionally, 38 field sera taken from sheep naturally infected with B. ovis gave strong positive reactions in the ELISA between day 20 and day 30 of treatment. As a result, the identified recombinant BoSA1 protein seems to be a promising diagnostic antigen that is usable for the development of serological assays for the diagnosis of ovine babesiosis. This is the first report on the molecular cloning, expression, and potential use of a recombinant antigen for the diagnosis of ovine babesiosis.
INTRODUCTION
Ovine babesiosis, caused by the tick-borne intraerythrocytic apicomplexan parasite Babesia ovis, is an acute disease characterized by clinical signs such as fever, hemolytic anemia, hemoglobinuria, and icterus in small ruminants. The disease is endemic in European, African, Asian, and Far Eastern countries (1–8). Babesia ovis is highly pathogenic and, in serious infections, causes pancytopenia. Untreated cases usually result in the death of sick animals, and even some treated animals may die as a result of heavy infection. Many recurrences also occur after the treatment period. Compensation for the abnormalities in the hematological profiles of acutely infected animals takes a long time after the treatment period (8). Currently, control of ovine babesiosis is based only on chemotherapy and limited tick control measures. In areas in which the status of B. ovis endemicity is unstable, an immunization program is needed (9). However, immunoprophylaxis is lacking for Babesia species in sheep.
Currently, the diagnosis of ovine babesiosis includes microscopic examination of Giemsa-stained thin blood smears. Although intraerythrocytic parasites are apparent in clinical cases, it is very difficult to detect the organisms by microscopy in chronic infections, because of the low levels of parasitemia (10–12). The analysis of blood smears usually depends on the experience of the observer. The morphological changes in the host cells and parasites, particularly in the later stages of acute infections, may result in inaccurate diagnoses. Serological tests and PCR assays are useful for detecting subclinical infections. Although serological testing has some limitations in confirming active or persistent disease and in distinguishing current and previous infections, circulating antibodies may be serologically detectable during chronic infections when the parasites are below the limit of PCR detection (10). Serological testing with the indirect fluorescent antibody test (IFAT) is the most commonly used method for diagnosing chronic B. ovis infections (8, 9, 13, 14). However, the IFAT requires experienced and trained personnel, is subjective in nature, makes results from different laboratories difficult to compare, and is time-consuming and exhausting in large-scale epidemiological studies. An enzyme-linked immunosorbent assay (ELISA) using synthetically derived Babesia bovis antigen has been used to detect anti-B. ovis antibodies in some research (15–17). However, immunoreactive recombinant proteins are widely used in serological assays for the diagnosis of equine, bovine, and canine babesiosis (18–21). There has been no report of the production of immunoreactive recombinant proteins of B. ovis. The development of diagnostic proteins usable for the diagnosis of ovine babesiosis is crucial for areas in which the disease is endemic. An optimized diagnostic test would benefit from the use of species-specific recombinant proteins. The purposes of the present study were to identify the immunoreactive proteins encoded by specific genes, through immunoscreening of a cDNA expression library, and to test the antigenic efficiency of those proteins in serological assays.
MATERIALS AND METHODS
Parasites and sera.
For construction of the cDNA expression library, a field strain of B. ovis was obtained from a natural case (50% parasitemia levels) located in an area in the central Anatolian region of Turkey in which ovine babesiosis is enzootic. The parasites were monitored by microscopic examination of Giemsa-stained thin blood smears, and results were verified by PCR analysis, as described previously (22). The serum samples used for the ELISA were available in our laboratories from previous studies (8, 14, 23) and were as follows: 15 negative samples, confirmed by microscopy and the IFAT, from experimentally infected lambs prior to infection (negative-control samples); 120 serum samples from the same experimentally infected lambs, at 6, 7, 8, 11, 21, 30, 45, and 75 days postinfection; and 76 serum samples from 38 naturally infected sheep, at the time of clinical infection and 20 to 30 days after treatment, in which the presence of the parasite was confirmed by microscopy.
Construction and immunoscreening of cDNA expression library.
Total RNA was extracted from B. ovis-infected sheep erythrocytes by using TRI Reagent BD (Sigma-Aldrich), according to the manufacturer's instructions. Isolation of poly(A) mRNA was performed with Oligotex-dT 30 latex beads (TaKaRa, Japan). Using 5 μg of mRNA, a cDNA library was constructed with a ZAP-cDNA synthesis kit using a Uni-ZAP XR vector and a ZAP-cDNA Gigapack Gold III packaging extract (Stratagene).
The cDNA library was screened with immune serum with a high titer of anti-B. ovis IgG antibodies (1:5,120), which was identified by the IFAT and was obtained from sheep naturally infected with B. ovis. Phage plates that had been incubated at 42°C for 4 h were overlaid with nitrocellulose transfer membranes (Protran; Whatman) that had been previously dried after being soaked in 10 mM isopropyl-β-d-1-thiogalactopyranoside (IPTG) for 3 min and were incubated at 37°C for 4 h. After being washed 3 times with Tris-buffered saline (TBS) with 0.05% Tween 20 (TBST) for 5 min at 4°C on a shaker, the membranes were soaked in blocking buffer (TBS containing 1% bovine serum albumin) overnight at 4°C on a shaker and then were treated with the primary antibody (B. ovis-specific sheep antiserum, diluted 1:100 in blocking buffer) for 2 h at room temperature (RT). After another four washes with TBST, the membranes were treated with the secondary antibody (alkaline phosphatase conjugate of rabbit anti-sheep IgG [Bethyl], diluted 1:4,000 in blocking buffer) for 1 h at RT. After being washed four times in TBST and then once again in TBS, the membranes were soaked in the dark for 3 to 5 min with freshly prepared substrate, i.e., 0.3% (vol/vol) 5-bromo-4-chloro-3-indolylphosphate (BCIP) and nitroblue tetrazolium (NBT) (Roche, Switzerland), in a color development solution (10 ml of 1 M Tris-HCl [pH 9.5], 2 ml of 5 M NaCl, and 0.5 ml of 1 M MgCl2 in 100 ml of distilled water) for color detection. Positive dots on the membranes, which were dried at RT after being rinsed with TBS, indicated positive clones on the plates. All procedures were repeated in the second and third immunoscreening steps, in order to isolate single clones. Following the identification of single positive clones, in vivo excision was performed to convert the lambda vectors into phagemid vectors.
Sequence analysis of BoSA1 cDNA.
The purified positive phage clones were excised in vivo using the ExAssist helper phage with the Escherichia coli SOLR strain, according to the single-clone excision protocol (Stratagene). The SOLR cell-based phagemid clones containing cDNA inserts were cultivated in Luria broth (LB) medium with ampicillin for 8 to 10 h at 37°C, and then DNA was extracted using a NucleoSpin plasmid QuickPure kit (TaKaRa, Japan). The recombinant plasmid clones were sequenced in an automated sequencer (ABI Prism 310 genetic analyzer) and amplified in a thermal cycler by using M13 forward and reverse primers. The amplified PCR products were subjected to 1.5% agarose gel electrophoresis.
Cloning, expression, and purification of rBoSA1 and preparation of anti-rBoSA1 mouse antibodies.
Using forward (5′-GCGGATCCCTTGGCGGAGACGAGGAT-3′; underlined bases indicate a BamHI site) and reverse (5′-GCCTCGAGTAATGGCATCGGGCAAG-3′; underlined bases indicate a XhoI site) primers designed to contain restriction enzyme sites in the expression vector pGEX-4T3 (Amersham Pharmacia Biotech), Babesia ovis secreted antigen 1 (BoSA1) cDNA was amplified by PCR. The BoSA1 cDNA fragments and pGEX-4T3 expression vectors were digested with the aforementioned restriction enzymes, run on a 1.5% agarose gel, and purified with a gel extraction kit (Qiagen, Germany). The purified BoSA1 cDNA fragments were ligated into the arms of the expression vector pGEX-4T3 using a DNA ligation kit (TaKaRa, Japan). A ligation reaction was carried out overnight at 16°C. The resulting plasmid, termed pGEX-4T3/BoSA1, was subsequently used for expression of the glutathione S-transferase (GST) fusion protein in E. coli DH5α. The GST-tagged recombinant BoSA1 (rBoSA1) fusion protein was purified by affinity chromatography according to the manufacturer's instructions, using glutathione-Sepharose 4B beads (Sigma), and was analyzed on a 12% SDS-PAGE gel.
Six female ICR mice (CLEA, Japan), 6 weeks of age, were intraperitoneally immunized with 100 μg of purified rBoSA1 emulsified with an equal volume of complete Freund's adjuvant (Difco Laboratories). Two additional immunizations, with 50 μg of the rBoSA1 antigen plus incomplete Freund's adjuvant, were administered at 2-week intervals. The mice were bled 14 days after the last immunization, and serum samples were stored at −30°C. The antibodies to purified rBoSA1 raised in ICR mice were used to identify native BoSA1 in the protein complex extracted from infected red blood cells (RBCs) and to determine the location of the native BoSA1 protein in the confocal laser microscopic observations.
SDS-PAGE and Western blotting.
Identification of the native BoSA1 protein in the extracts prepared from erythrocytes and plasma from B. ovis-infected and noninfected blood samples was performed using SDS-PAGE and Western blot analysis. In short, red blood cells (RBCs) were harvested with low-speed centrifugation and washed three times with phosphate-buffered saline (PBS). The pellets were suspended in an equal volume of PBS containing 0.5% saponin and then were incubated at 37°C for 10 min. The lysate, which was transferred into 1.5-ml tubes, was centrifuged at 10,000 rpm for 10 min. The resulting pellets were suspended in 50 μl of PBS, sonicated, and precipitated with acetone at −20°C for 15 min. The plasma proteins were precipitated with the same volume of saturated ammonium sulfate. After centrifugation at 15,000 rpm for 10 min, the precipitated proteins were dissolved in sample buffer containing 5% 2-mercaptoethanol, and the mixture was heated at 100°C for 10 min. The samples were subjected to electrophoresis on a 12% polyacrylamide gel and transferred to a nitrocellulose membrane. The membrane was soaked in blocking buffer (5% skim milk in PBS) for 1 h at room temperature, on a shaker, and incubated with anti-rBoSA1 antibody (1:100 dilution in blocking buffer) for 1 h at 37°C. After the membrane was washed with PBS containing 0.5% Tween 20 (PBST), it was incubated with peroxidase-conjugated goat anti-mouse IgG (1:5,000) for 1 h at 37°C. After the final wash, the membrane was reacted with the chromogenic substrate 3,3′-diaminobenzidine tetrahydrochloride (DAB) and H2O2, for detection of BoSA1 protein.
Confocal laser microscopy.
Thin blood smear films prepared from infected blood were fixed with methanol for 30 min and incubated with anti-rBoSA1 serum (1:100 dilution in PBS with 4% fetal bovine serum) for 2 h at 37°C. The slides were washed with PBST four times for 5 min, on a shaker, and incubated with Alexa Fluor 488-conjugated goat anti-mouse IgG (Molecular Probes) (1:200 dilution in PBS with 4% fetal bovine serum) for 1 h at 37°C. After four washes with PBST, the slides were incubated with 6.25 mg/ml propidium iodide (PI) (Wako, Japan) and 50 mg/ml RNase A (Qiagen, Germany) for 30 min at 37°C, washed with PBST, and mounted in 50% glycerol-PBS for confocal laser microscopic observations.
Enzyme-linked immunosorbent assay.
The specific antibody responses against rBoSA1 were analyzed in sera from B. ovis-infected animals by ELISA using the rBoSA1 protein. The purified rBoSA1-fused GST and control GST proteins were diluted with coating buffer (0.05 M carbonate-bicarbonate buffer [pH 9.6]) at a concentration of 2 μg/ml. Each well of the 96-well microtiter plates (Nunc, Denmark) was coated with 100 μl of protein overnight at 4°C. The plates were blocked with 5% skim milk in PBS for 1 h at 37°C and washed with PBS with 0.05% Tween 20 (PBST). The primary antibody (ovine serum positive for anti-B. ovis antibodies, at a dilution of 1:100) and the secondary antibody (peroxidase-conjugated anti-sheep IgG [Sigma] at a dilution of 1:4,000) were used. 3,3′-Diaminobenzidine tetrahydrochloride (Sigma) was used as the substrate. Absorbance was measured at 415 nm. The cutoff value was defined as the mean value plus 3 standard deviations of the mean optical density (OD) of 25 lamb serum samples seronegative for anti-B. ovis antibodies by the IFAT.
RESULTS
Identification of cDNA encoding BoSA1.
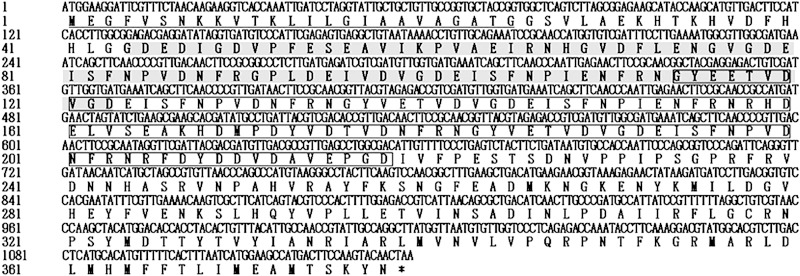
A total of 22 immunoreactive recombinant phage plaques were identified in the first immunoscreening of the amplified cDNA expression library (at 2 × 109 PFU/ml). These plaques were further purified in the second and third immunoscreenings. Fifty-four homogeneous plaques expressing the antigens were selected and excised to a pBluescript SK(−) phagemid. Sequencing analysis of the phagemid inserts showed that the sequences of the isolated cDNAs were similar to each other, with different base pair lengths. Only one of the inserts of phage plaques contained a full open reading frame (ORF) with a 3′ untranslated region (UTR) and a 5′ UTR. The cDNA encoding BoSA1, which contains an ORF of 1,137 nucleotides encoding a protein of 378 amino acids, with a signal peptide and 2 internal repeat domains, was selected for further molecular characterization (Fig. 1). A search of the protein database of NCBI with protein BLAST analysis, using the amino acid sequence of BoSA1, showed a low level of identity (28%) to the sequence of the hypothetical protein of Babesia bovis (GenBank accession no. XP_001609244). Furthermore, the amino acid sequences of BoSA1 were searched in the Eukaryotic Pathogen Genome Resources database (EuPathDB) among Amoebozoa, Apicomplexa, Diplomonadida, Kinetoplastida, Microsporidia, and Trichomonadida (http://eupathdb.org/eupathdb). The results indicate that the closest homologue of BoSA1 is the same Babesia bovis hypothetical protein mentioned above (GenBank accession no. XP_001609244).
FIG 1.
Nucleotide and predicted amino acid sequences for cDNA encoding BoSA1. The underlined region indicates the predicted signal peptide. The gray and boxed regions indicate 2 internal repeat domains.
Expression, purification, and reactivity of recombinant BoSA1.
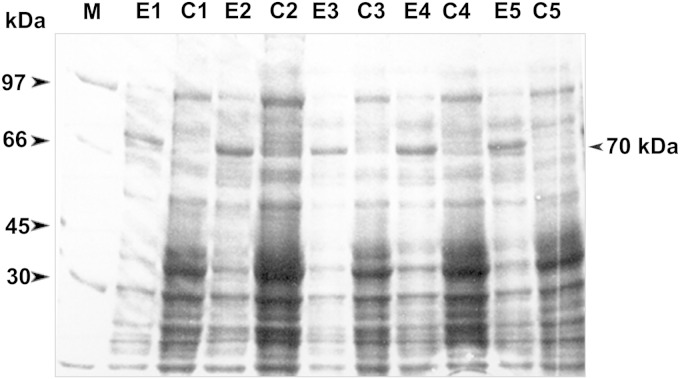
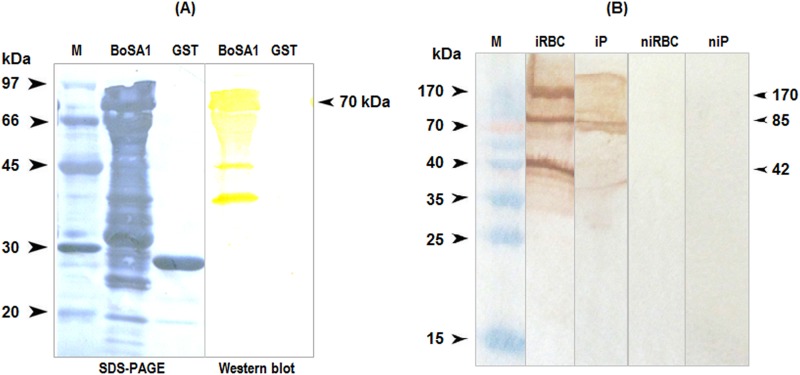
The BoSA1 cDNA insert cloned in the bacterial expression vector pGEX-4T3 was expressed in E. coli DH5α as a GST fusion protein, with an approximate molecular mass of 70 kDa (Fig. 2). Immune sera from lambs that had been experimentally infected with B. ovis recognized the purified rBoSA1-GST fusion protein but not the GST protein in Western blots (Fig. 3A).
FIG 2.
SDS-PAGE analysis of the recombinant BoSA1 protein expressed in E. coli DH5α. Lane M, protein ladder (kDa). The predicted rBoSA1 protein band expressed in cells is indicated. The figure includes five randomly selected clones of E. coli DH5α cells transformed with recombinant plasmid pGEX-4T3/BoSA1. Lanes E1, E2, E3, E4, and E5, E. coli DH5α cells treated with IPTG. Lanes C1, C2, C3, C4, and C5, uninduced control E. coli DH5α cells.
FIG 3.
Western blot analysis of recombinant and native BoSA1 proteins. (A) Western blots showing the reactivity of anti-B. ovis antibodies with rBoSA1. The anti-B. ovis antibodies reacted with several components of rBoSA1 between ∼70 and ∼40 kDa but not with the GST protein. (B) Western blots showing the reactivity of anti-rBoSA1 mouse antibodies with native BoSA1 proteins in the erythrocytes and plasma of B. ovis-infected blood. Lane M, protein ladder; lane iRBC, infected erythrocyte lysate extracted with saponin; lane iP, infected plasma proteins precipitated with saturated ammonium sulfate; lane niRBC, noninfected erythrocyte lysate extracted with saponin; lane niP, noninfected plasma proteins precipitated with saturated ammonium sulfate. Comparisons with molecular size markers indicated estimated molecular masses consistent with the monomeric, dimeric, and tetrameric forms of native BoSA1.
In Western blotting of lysates of B. ovis-infected and normal sheep erythrocytes, the anti-rBoSA1 mouse antibodies reacted with the three major >40-kDa components of the native proteins in infected erythrocytes but not in normal sheep erythrocyte lysates. These multiple bands were the expected molecular masses for the monomeric, dimeric, and tetrameric forms of native BoSA1. The Western blots of the proteins precipitated with saturated ammonium sulfate from the infected plasma samples also showed that the two major native protein bands of >70 kDa reacted with the anti-rBoSA1 mouse antibodies (Fig. 3B).
Localization of the BoSA1 protein in parasites.
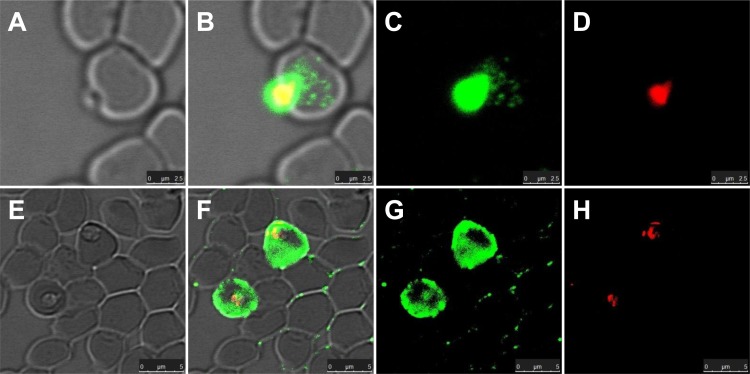
To determine the locations of the BoSA1 protein in B. ovis-infected erythrocytes, B. ovis merozoites were examined by the IFAT with a confocal laser microscope. The specific fluorescence was localized both in and on the B. ovis merozoites and inside infected erythrocytes (Fig. 4B, C, F, and G).
FIG 4.
Localization of native BoSA1 proteins recognized by anti-rBoSA1 mouse antibodies in confocal laser micrographs. The images were derived from two sections. (A and E) Phase-contrast images of B. ovis merozoites. (B and F) Overlaid images of fluorescent reactivity and red PI staining on the phase-contrast images of the parasites. (C and G) Immunofluorescence images of parasite-specific antigens. (D and H) Propidium iodide staining of B. ovis merozoite nuclei. Bars = 5 µm.
Diagnostic potential of the rBoSA1 protein.
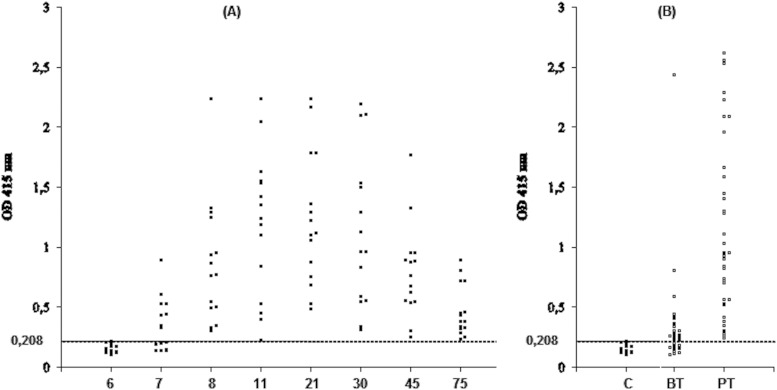
As shown in Fig. 5, the anti-B. ovis serum antibodies from 15 lambs experimentally infected with B. ovis showed strong reactivity with the rBoSA1 antigen in an indirect ELISA, at OD values above 0.208, for 8 sera on day 7 and for all sera on day 8 after the experimental infection. The cutoff value was calculated as 0.208 by using the following formula: arithmetic mean plus 3 times the standard deviation of the OD values for the 25 negative sheep sera. The antibody titers remained high during the 75 days of the postinfection period for the experimentally infected lambs (Fig. 5A). All of the sera were negative on day 6 of infection and on the prior days (data not shown). For the field serum samples taken from sheep naturally infected with B. ovis, 26 of the 38 sera yielded positive reactions during the time of clinical infection prior to treatment, and all of the 38 sera had strong positivity in the ELISA between 20 and 30 days posttreatment (Fig. 5B).
FIG 5.
Scatter diagrams of absorbance values measured in ELISAs with the recombinant BoSA1 protein. (A) Sera from 15 lambs experimentally infected with B. ovis, assayed on days 6, 7, 8, 11, 21, 30, 45, and 75 postinfection. The negative results obtained prior to infection and until day 6 of the experimental infection are not shown in the diagram. (B) Sera from 38 sheep naturally infected with B. ovis. Each dot above a horizontal line indicates an ELISA-positive sample. BT, before treatment; PT, 20 to 30 days posttreatment; C, negative-control samples.
DISCUSSION
Immunoreactive recombinant proteins generated from parasite genes are valuable as diagnostic antigens and vaccine candidates (24–29). Recombinant antigen-based serological assays have been widely and effectively used for the diagnosis of bovine, equine, and canine babesiosis (18–21). Although ovine babesiosis has a significant economic impact on the livestock industry, to date no study has been conducted on production of the recombinant proteins of B. ovis and their use as antigens in any serological test. The present study was performed in order to identify the genes encoding the immunoreactive proteins of B. ovis, to express the specific genes in an E. coli expression system, and to evaluate the diagnostic potential of these proteins in an enzyme-linked immunosorbent assay. We constructed a cDNA expression library from mRNA obtained from B. ovis merozoites, screened the library using immune serum from sheep naturally infected with B. ovis, and identified a secreted antigen designated Babesia ovis secreted antigen 1 (BoSA1).
In the phylum Apicomplexa, including intracellular protozoans such as Toxoplasma, Plasmodium, and Theileria, secretion of parasite proteins into the host cells is one of the methods the parasites use for interactions with the host immune system and for host cell modifications (30). In the present study, the BoSA1 protein, as detected by anti-rBoSA1 mouse antibodies in immunofluorescence assays, was identified both in and on B. ovis merozoites, which demonstrated that the protein was secreted exclusively into the cytoplasm of infected erythrocytes (Fig. 5). Considering the importance of secreted proteins as rich sources of new therapeutic agents, drug targets, diagnostic antigens, and vaccine candidates (31), studies should be performed to find and to characterize the functions of the BoSA1 protein in host interactions.
In order to test the diagnostic potential of the rBoSA1 protein in a serological assay, the protein was used as an antigen in an indirect ELISA. The results indicated that the ELISA system using the rBoSA1 protein as an antigen could detect specific anti-B. ovis antibodies in sera from B. ovis-infected animals. The specific antibodies used against rBoSA1 were detectable in the B. ovis-infected lamb sera from days 7 and 8 to day 75 of the experimental infection. Additionally, 26 (68.42%) of 38 field serum samples from sheep naturally infected with B. ovis were seropositive during the period of infection, and all 38 samples taken between day 20 and day 30 of the specific treatment showed strong reactivity in the ELISA. Our results indicate that the recombinant BoSA1 protein is useful for the diagnosis of B. ovis infections. However, further testing of serum samples for any potential cross-reactivity between the recombinant BoSA1 antigen and antibodies against other ovine infectious agents, such as Theileria spp., Anaplasma ovis, and Babesia species other than B. ovis, has significance for the development of serological assays for the detection of anti-B. ovis antibodies.
In the Western blots of infected RBC lysates, the anti-rBoSA1 antiserum recognized native BoSA1 with the expected molecular size of ∼42 kDa. Two additional bands, of ∼85 and ∼170 kDa, also reacted with the anti-rBoSA1 antibodies in Western blots; these bands corresponded to the sizes expected for the dimeric and tetrameric forms of BoSA1, respectively. Our results demonstrate that the rBoSA1 produced in E. coli is a monomeric protein and forms oligomers under physiological conditions. A recent study indicated that a family of membrane attack complex/perforin (MACPF) domain proteins of Babesia bovis are expressed and oligomerized to form large prepore complexes on the surfaces of target cells (32), and a dissertation showed that heat shock protein 20 (HSP20) of Babesia bovis can exist as an oligomer (33). Further studies with a large number of infected blood samples are required to determine whether native BoSA1 exists in monomeric, dimeric, and tetrameric forms in vivo. Purified rBoSA1 reacted with the immune sheep serum in Western blots, and no recognition was found with normal sheep serum. These results show the presence of specific antibodies against the expressed protein in this immune serum.
In conclusion, we performed immunoscreening of the cDNA expression library constructed from B. ovis mRNA by using anti-B. ovis serum. We identified and characterized the gene encoding a secreted protein designated B. ovis secreted antigen 1 (BoSA1), the cDNA of which was expressed in E. coli. In the ELISA, recombinant BoSA1 showed strong reactivity with anti-B. ovis antibodies. The specific anti-rBoSA1 antiserum recognized native parasite proteins in B. ovis-infected erythrocyte lysates as well as in the plasma from infected blood. Because of the importance of circulating antigens released into plasma by the parasites for the definition of current infections, future experiments will be performed in order to define their diagnostic efficiency in antigen-detecting assays.
ACKNOWLEDGMENT
This work was supported by the Scientific and Technological Research Council in Turkey (grant number 113O336).
REFERENCES
- 1.Yeruham I, Hadani A, Galker F. 1998. Some epizootiological and clinical aspects of ovine babesiosis caused by Babesia ovis: a review. Vet Parasitol 74:153–163. doi: 10.1016/S0304-4017(97)00143-X. [DOI] [PubMed] [Google Scholar]
- 2.Kuttler KL. 1988. World-wide impact of babesiosis, p 1–22. In Ristic M. (ed), Babesiosis of domestic animals and man. CRC Press, Boca Raton, FL. [Google Scholar]
- 3.Razmi GR, Naghibi A, Aslani MR, Dastjerdi K, Hossieni H. 2003. An epidemiological study on Babesia infection in small ruminants in Mashhad suburb, Khorasan Province, Iran. Small Rumin Res 50:39–44. doi: 10.1016/S0921-4488(03)00107-X. [DOI] [Google Scholar]
- 4.Uilenberg G. 2006. Babesia: a historical overview. Vet Parasitol 138:3–10. doi: 10.1016/j.vetpar.2006.01.035. [DOI] [PubMed] [Google Scholar]
- 5.Ahmed JS, Luo J, Schnittger L, Seitzer U, Jongejan F, Yin H. 2006. Phylogenetic position of small-ruminant infecting piroplasms. Ann N Y Acad Sci 1081:498–504. doi: 10.1196/annals.1373.074. [DOI] [PubMed] [Google Scholar]
- 6.Ranjbar-Bahadori S, Eckert B, Omidian Z, Shirazi NS, Shayan P. 2012. Babesia ovis as the main causative agent of sheep babesiosis in Iran. Parasitol Res 110:1531–1536. doi: 10.1007/s00436-011-2658-z. [DOI] [PubMed] [Google Scholar]
- 7.Fakhar M, Hajihasani A, Maroufi S, Alizadeh H, Shirzad H, Piri F, Pagheh AS. 2012. An epidemiological survey on bovine and ovine babesiosis in Kurdistan Province, western Iran. Trop Anim Health Prod 44:319–322. doi: 10.1007/s11250-011-0023-y. [DOI] [PubMed] [Google Scholar]
- 8.Sevinc F, Sevinc M, Ekici OD, Yildiz R, Isik N, Aydogdu U. 2013. Babesia ovis infections: detailed clinical and laboratory observations in the pre- and post-treatment periods of 97 field cases. Vet Parasitol 191:35–43. doi: 10.1016/j.vetpar.2012.07.025. [DOI] [PubMed] [Google Scholar]
- 9.Ekici OD, Sevinc F, Isik N. 2012. Instability of ovine babesiosis in an endemic area in Turkey. Vet Parasitol 188:372–375. doi: 10.1016/j.vetpar.2012.04.001. [DOI] [PubMed] [Google Scholar]
- 10.Bose R, Jorgensen WK, Dalgliesh RJ, Friedhoff KT, de Vos AJ. 1995. Current state and future trends in the diagnosis of babesiosis. Vet Parasitol 57:61–74. doi: 10.1016/0304-4017(94)03111-9. [DOI] [PubMed] [Google Scholar]
- 11.Georges K, Loria GR, Riili S, Greco A, Caracappa S, Jongejan F, Sparagano O. 2001. Detection of haemoparasites in cattle by reverse line blot hybridisation with a note on the distribution of ticks in Sicily. Vet Parasitol 99:273–286. doi: 10.1016/S0304-4017(01)00488-5. [DOI] [PubMed] [Google Scholar]
- 12.Bock R, Jackson L, de Vos A, Jorgensen W. 2004. Babesiosis of cattle. Parasitology 129(Suppl):S247–S269. doi: 10.1017/S0031182004005190. [DOI] [PubMed] [Google Scholar]
- 13.Wright IG. 1990. Immunodiagnosis of and immunoprophylaxis against the haemoparasites Babesia sp. and Anaplasma sp. in domestic animals. Rev Sci Tech 9:345–356. [DOI] [PubMed] [Google Scholar]
- 14.Sevinc F, Turgut K, Sevinc M, Derinbay Ekici O, Coskun A, Koc Y, Erol M, Ica A. 2007. Therapeutic and prophylactic efficacy of imidocarb dipropionate on experimental Babesia ovis infection of lambs. Vet Parasitol 149:65–71. doi: 10.1016/j.vetpar.2007.07.014. [DOI] [PubMed] [Google Scholar]
- 15.Duzgun A, Wright IG, Waltisbuhl DJ, Gale KR, Goodger BV, Dargie JD, Alabay M, Cerci H. 1991. An ELISA for the diagnosis of Babesia ovis infection utilizing a synthetic, Babesia bovis-derived antigen. Vet Parasitol 39:225–231. doi: 10.1016/0304-4017(91)90039-X. [DOI] [PubMed] [Google Scholar]
- 16.Emre Z, Duzgun A, Iriadam M, Sert H. 2001. Seroprevalence of Babesia ovis in Awassi sheep in Urfa, Turkey. Turk J Vet Anim Sci 25:759–762. [Google Scholar]
- 17.Cicek H, Duzgun A, Emre Z, Karaer Z. 2004. Seroprevalence of Babesia ovis in sheep around Afyon. Turk J Vet Anim Sci 28:683–686. [Google Scholar]
- 18.Ikadai H, Osorio CR, Xuan X, Igarashi I, Kanemaru T, Nagasawa H, Fujisaki K, Suzuki N, Mikami T. 2000. Detection of Babesia caballi infection by enzyme-linked immunosorbent assay using recombinant 48-kDa merozoite rhoptry protein. Int J Parasitol 30:633–635. doi: 10.1016/S0020-7519(00)00008-4. [DOI] [PubMed] [Google Scholar]
- 19.Boonchit S, Xuan X, Yokoyama N, Goff WL, Wagner G, Igarashi I. 2002. Evaluation of an enzyme-linked immunosorbent assay with recombinant rhoptry-associated protein 1 antigen against Babesia bovis for the detection of specific antibodies in cattle. J Clin Microbiol 40:3771–3775. doi: 10.1128/JCM.40.10.3771-3775.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Huang X, Xuan X, Yokoyama N, Xu L, Suzuki H, Sugimoto C, Nagasawa H, Fujisaki K, Igarashi I. 2003. High-level expression and purification of a truncated merozoite antigen-2 of Babesia equi in Escherichia coli and its potential for immunodiagnosis. J Clin Microbiol 41:1147–1151. doi: 10.1128/JCM.41.3.1147-1151.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhou J, Jia H, Nishikawa Y, Fujisaki K, Xuan X. 2007. Babesia gibsoni rhoptry-associated protein 1 and its potential use as a diagnostic antigen. Vet Parasitol 145:16–20. doi: 10.1016/j.vetpar.2006.10.022. [DOI] [PubMed] [Google Scholar]
- 22.Aktas M, Altay K, Dumanli N. 2005. Development of a polymerase chain reaction method for diagnosis of Babesia ovis infection in sheep and goats. Vet Parasitol 133:277–281. doi: 10.1016/j.vetpar.2005.05.057. [DOI] [PubMed] [Google Scholar]
- 23.Sevinc F, Sevinc M, Koc Y, Alkan F, Ekici OD, Yildiz R, Isik N, Aydogdu U. 2014. The effect of 12 successive blood passages on the virulence of Babesia ovis in splenectomized lambs: a preliminary study. Small Rumin Res 116:66–70. doi: 10.1016/j.smallrumres.2013.10.010. [DOI] [Google Scholar]
- 24.Kumar S, Malhotra DV, Nichani AK. 2002. Identification of immunoreactive polypeptides of Babesia equi parasite during immunization. Vet Parasitol 107:295–301. doi: 10.1016/S0304-4017(02)00161-9. [DOI] [PubMed] [Google Scholar]
- 25.Huang X, Xuan X, Yokoyama N, Katayama Y, Anzai T, Igarashi I. 2006. Evaluation of enzyme-linked immunosorbent assays with recombinant antigens for the serodiagnosis of equine Babesia infections. Vet Parasitol 140:158–161. doi: 10.1016/j.vetpar.2006.03.013. [DOI] [PubMed] [Google Scholar]
- 26.Terkawi MA, Jia H, Gabriel A, Goo YK, Nishikawa Y, Yokoyama N, Igarashi I, Fujisaki K, Xuan X. 2007. A shared antigen among Babesia species: ribosomal phosphoprotein P0 as a universal babesial vaccine candidate. Parasitol Res 102:35–40. doi: 10.1007/s00436-007-0718-1. [DOI] [PubMed] [Google Scholar]
- 27.Goo YK, Jia H, Aboge GO, Terkawi MA, Kuriki K, Nakamura C, Kumagai A, Zhou J, Lee EG, Ishikawa Y, Igarashi I, Fujisaki K, Xuan X. 2008. Babesia gibsoni: serodiagnosis of infection in dogs by an enzyme-linked immunosorbent assay with recombinant BgTRAP. Exp Parasitol 118:555–560. doi: 10.1016/j.exppara.2007.11.010. [DOI] [PubMed] [Google Scholar]
- 28.Guan GQ, Chauvin A, Rogniaux H, Luo JX, Yin H, Moreau E. 2010. Merozoite proteins from Babesia sp. BQ1 (Lintan) as potential antigens for serodiagnosis by ELISA. Parasitology 137:927–938. doi: 10.1017/S0031182009991922. [DOI] [PubMed] [Google Scholar]
- 29.Ooka H, Terkawi MA, Cao S, Aboge G, Goo YK, Luo Y, Li Y, Nishikawa Y, Igarashi I, Xuan X. 2012. Molecular and immunological characterization of a novel 32-kDa secreted protein of Babesia microti. J Parasitol 98:1045–1048. doi: 10.1645/GE-2999.1. [DOI] [PubMed] [Google Scholar]
- 30.Ravindran R, Boothroyd JC. 2008. Secretion of proteins into host cells by apicomplexan parasites. Traffic 9:647–656. doi: 10.1111/j.1600-0854.2008.00723.x. [DOI] [PubMed] [Google Scholar]
- 31.Bonin-Debs AL, Boche I, Gille H, Brinkmann U. 2004. Development of secreted proteins as biotherapeutic agents. Expert Opin Biol Ther 4:551–558. doi: 10.1517/14712598.4.4.551. [DOI] [PubMed] [Google Scholar]
- 32.Kafsack BFC, Carruthers VB. 2010. Apicomplexan perforin-like proteins. Commun Integr Biol 3:18–23. doi: 10.4161/cib.3.1.9794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Carson KH. 2006. Study and characterization of a novel small heat shock protein from Babesia. Ph.D. thesis Texas A&M University, College Station, TX. [Google Scholar]