Abstract
Human respiratory syncytial virus (hRSV) and human metapneumovirus (hMPV) share virologic and epidemiologic features and cause clinically similar respiratory illness predominantly in young children. In a previous study of acute febrile respiratory illness in Bangladesh, we tested paired serum specimens from 852 children presenting fever and cough for diagnostic increases in titers of antibody to hRSV and hMPV by enzyme immunoassay (EIA). Unexpectedly, of 93 serum pairs that showed a ≥4-fold increase in titers of antibody to hRSV, 24 (25.8%) showed a concurrent increase in titers of antibody to hMPV; of 91 pairs showing an increase to hMPV, 13 (14.3%) showed a concurrent increase to hRSV. We speculated that common antigens shared by these viruses explain this finding. Since the nucleocapsid (N) proteins of these viruses show the greatest sequence homology, we tested hyperimmune antisera prepared for each virus against baculovirus-expressed recombinant N (recN) proteins for potential cross-reactivity. The antisera were reciprocally reactive with both proteins. To localize common antigenic regions, we first expressed the carboxy domain of the hMPV N protein that was the most highly conserved region within the hRSV N protein. Although reciprocally reactive with antisera by Western blotting, this truncated protein did not react with hMPV IgG-positive human sera by EIA. Using 5 synthetic peptides that spanned the amino-terminal portion of the hMPV N protein, we identified a single peptide that was cross-reactive with human sera positive for either virus. Antiserum prepared for this peptide was reactive with recN proteins of both viruses, indicating that a common immunoreactive site exists in this region.
INTRODUCTION
Human respiratory syncytial virus (hRSV) and human metapneumovirus (hMPV) are negative single-stranded, enveloped RNA viruses that are coclassified within the Pneumovirinae subfamily of the Paramyxoviridae. hRSV is the leading cause of severe lower respiratory tract infections in infants and young children and has been associated with community-acquired pneumonia in adults, with the highest mortality rates found among the hospitalized elderly and immunocompromised (1, 2, 3). hMPV was identified first in 2001 by van den Hoogen et al. (4) and subsequently has been shown to have clinical and epidemiological features similar to but distinct from those of hRSV (5, 6). Most hMPV infections occur within the first few years of life, usually later than those for hRSV, and cause 5 to 10% of respiratory infections in children requiring hospitalization, second only to hRSV (7). Both viruses are ubiquitous globally, and recent studies have documented a high prevalence of infection among children in less developed countries (8, 9). Clinical outcomes similar to those for hRSV have been described for hMPV infections in the frail elderly (10).
The RNA genomes of hRSV and hMPV are complexed with nucleocapsid (N) protein that together serve as the templates for genome replication and transcription (11). The N protein is the most abundant viral protein and elicits a strong and long-lasting humoral immune response following acute infection (12, 13). Recombinant nucleocapsid (recN) protein has been used successfully to replace whole virus antigen in diagnostic serological assays for hRSV and hMPV by us and others (14, 15).
In a previous serologic study to assess the etiology of febrile respiratory illness in Bangladeshi children, we observed an unusually high number of increases titers of antibody to hRSV and hMPV (unpublished data). Although coinfections with hRSV and hMPV could explain this observation, coinfections could not be confirmed by reverse transcription-PCR (RT-PCR) of coincidently collected respiratory specimens from these children, and only limited cocirculation of these viruses was observed during the study period (9). An alternative hypothesis was that these heterotypic responses were due to hitherto-unreported shared antigens between these viruses. In this study, we identified a common immunoreactive site in the amino acid terminus of the N proteins of hRSV and hMPV and discuss the implications of this finding.
MATERIALS AND METHODS
Human serum specimens.
As part of a population-based respiratory disease surveillance study in Dhaka, Bangladesh, conducted by the International Center for Diarrheal Disease Research, Bangladesh (ICDDR, B), paired acute- and convalescent-phase serum specimens were collected from children <5 years of age presenting with acute respiratory tract illness and/or fever from April 2004 to Feb 2006. Serum pairs obtained from 852 children were sent to the Centers for Disease Control and Prevention (CDC; Atlanta, Georgia, USA) for serologic testing for respiratory pathogens (8). This study was reviewed and approved by the Research Review and Ethical Review Committees of ICDDR, B and the CDC Institutional Review Board. Written informed consent was obtained from the primary adult caretaker for all children from whom samples were collected.
Expression of the carboxy-terminal domain of hMPV N protein.
Methods used for expression, affinity purification, and Western blot analysis of the carboxy-terminal fragment of the hMPV N protein (designated recΔN) were as previously described for the full-length recN proteins of hRSV and hMPV (15). In that study, we found no difference in the performance of indirect IgG enzyme immunoassays (EIAs) with human sera using antigen derived from either hMPV subgroup A (CAN97-83) or subgroup B (CAN98-75) viruses, and results obtained using full-length CAN97-83 recN protein were highly correlated with whole-virus lysate antigen. Therefore, for our cross-reactivity studies, we chose the carboxy-terminal region of hMPV N protein from strain CAN97-83 (accession number AY297749.1), which showed the highest degree of amino acid homology with hRSV N protein, for expression in a baculovirus system (Fig. 1). Genomic RNA was extracted from CAN97-83 and amplified by RT-PCR using forward (5′-CACCCAAAATCAGAGGCCTTCAGCACCAG-3′) and reverse (5′-TTACTCATAATCATTTTGACTGTC-3′) primers to cover the region encoding amino acids (aa) 148 to 394 of hMPV N protein. The amplified gene fragment was cloned into entry vector pENTR/D-TOPO using the Baculovirus Expression System with Gateway technology (Life Technologies). A clone was selected, and sequence was confirmed and subcloned into Gateway destination vector pDEST10. This expression clone then was transformed into competent bacterial artificial chromosome (BAC) Escherichia coli DH10 cells. The resulting recombinant bacmid DNA that contained the His-tagged recΔN gene fragment was isolated and transfected into Spodoptera frugiperda clone 9 (Sf9; CRL 1711; ATCC) cells. The cells were grown and maintained in suspension at 27°C using serum-free medium Sf-900 II supplemented with antibiotics (penicillin, 10,000 U/ml; streptomycin, 10,000/ml) (GIBCO, Life Technologies). To obtain high-titer recombinant baculovirus expressing the hMPV recΔN protein, SF9 suspension cultures containing 2 × 106 cells/ml were infected with 1 PFU/cell of virus and harvested when 50% of the cells showed cytopathic effect.
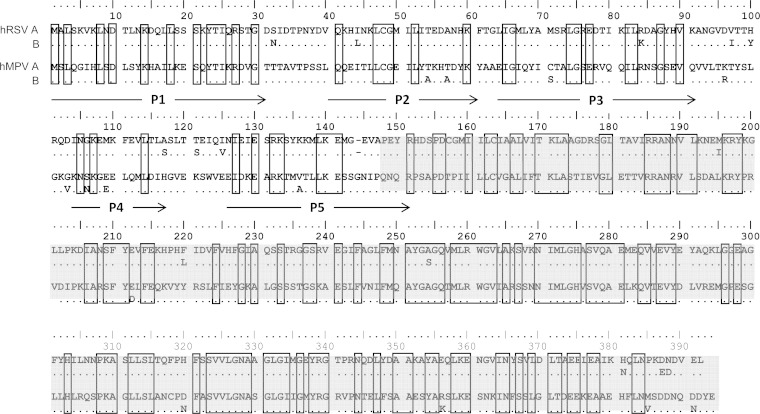
FIG 1.
Alignment of N proteins of representative hRSV and hMPV strains: hRSV subgroup A (A2; accession number M11486.1), hRSV subgroup B (CH-18537; accession number D00736.1), hMPV subgroup A (CAN97-83; accession number AY297749.1), and hMPV subgroup B (CAN98-75; accession number AY297748.1). Location of the amino-terminal peptides P1 to P5 (denoted by arrows) and expressed carboxyl domain (aa 148 to 394) of the hMPV N protein (highlighted in gray) used in this study.
Lysates of SF9 cells expressing recΔN protein and uninfected SF9 cells were run on 10% SDS-PAGE gels, and bands were analyzed by Western blotting with mouse anti-His tag antisera (Santa Cruz Biotechnology, Dallas, TX) and affinity-purified rabbit hyperimmune antiserum prepared for hRSV strain A2 (EMD Millipore) and mouse hyperimmune antiserum for hMPV strain CAN97-83 (CDC Scientific Resources Program). hMPV recΔN protein was reacted against human serum specimens by indirect IgG EIA as previously described for recN (15).
Amino-terminal peptides from hMPV N protein.
Five peptides (P1 to P5) covering conserved regions between the hRSV and hMPV N proteins were designed to the amino terminus of the hMPV N protein (strain CAN97-83) by following recommendations of the commercial vender (LifeTein, South Plainfield, NJ) (Fig. 1). Peptide-based seroassays were modified from recN protein bead-based assays developed on the MAGPIX platform (Luminex, Austin, TX) and described in detail elsewhere (15). One μg of each peptide was coupled to 2.5 × 106 MagPlex microspheres (Luminex), and the beads were combined to permit simultaneous testing. Reporter fluorescence of the peptide-coupled beads was expressed as the mean fluorescence intensity of at least 50 beads per well. Rabbit hyperimmune antisera was raised against peptide P1 (aa 1 to 31; MSLQGIHLSDLSYKHAILKESQYTIKRDVGT-Cys) that contained an added carboxy-terminal cysteine residue (Cys) to enable affinity purification using keyhole limpet hemocyanin (Lifetein).
RESULTS
Serological studies.
In a previous serologic study of 852 children with acute febrile respiratory illness using whole-virus-lysate antigen-based EIAs, 93 (10.9%) and 91 (10.7%) showed diagnostic increases (≥4-fold) in IgG titers of antibody to hRSV and hMPV, respectively (unpublished data). Unexpectedly, of those with diagnostic increases in levels of antibody to hRSV, 24 (25.8%) showed concurrent (≥4-fold and <4-fold) increases in levels of antibody to hMPV, and of those with diagnostic increases in levels of antibody to hMPV, 13 (14.3%) showed increased response to hRSV. In a subsequent study to evaluate the utility of expressed recN proteins as a substitute for whole-virus-lysate antigen, we randomly selected 87 serum pairs from this collection showing increases in levels of antibody to hRSV, hMPV, or both for comparison (15). Results obtained with whole-virus-lysate and recN EIAs were highly concordant, with 10 serum pairs showing clear diagnostic increases in IgG antibodies to both recN proteins. However, when we reviewed RT-PCR data from respiratory swab specimens available from some of these children, 3 were confirmed RT-PCR positive for hRSV, 4 were positive for hMPV, but none were positive for both viruses (9). In every case, the dominant seroresponse corresponded to the virus identified by RT-PCR.
Immunotypic cross-reactions between hRSV and hMPV full-length recN proteins.
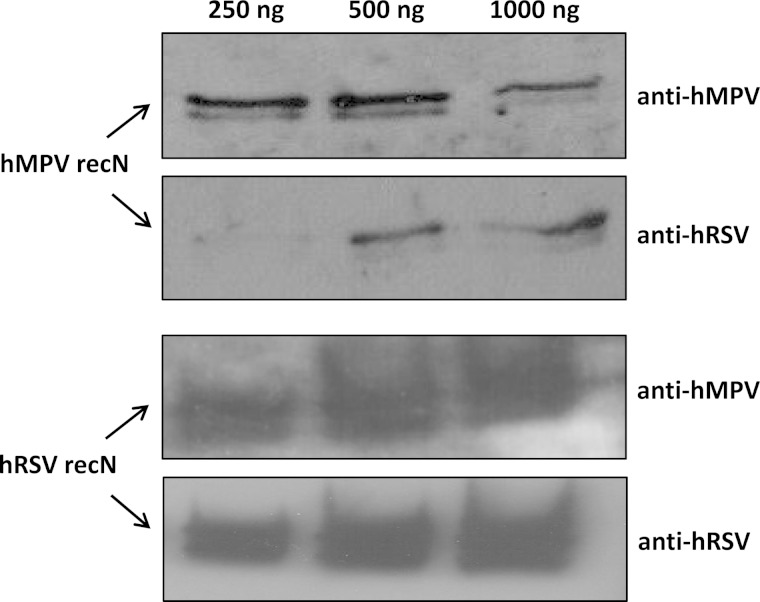
We speculated that serologic cross-reactions between the hRSV and hMPV N proteins explain the dual increases in antibody levels seen with some human serum pairs. To investigate this possibility, hyperimmune antisera were prepared for hRSV and hMPV and reacted with both full-length recN proteins and whole virus lysate by Western blotting. Both recN proteins (Fig. 2) and whole virus lysate were reciprocally reactive with antisera prepared against each virus. This finding suggested that the viruses share common antigenic determinants in the N protein. Notably, blots of hMPV recN with hMPV antisera revealed an additional smaller band which may represent the internal initiation of translation within the hMPV N gene, as described for the avian metapneumovirus nucleocapsid gene (16).
FIG 2.
Western blot analysis of different concentrations of purified full-length hRSV and hMPV recN proteins reciprocally reacted with affinity-purified goat and rabbit hyperimmune antisera prepared for hRSV (strain A2) and hMPV (strain CAN97-83), respectively.
Evaluation of the carboxy-terminal domain of hMPV N protein.
In an attempt to localize common immunoreactive regions within the hRSV and hMPV N proteins, we first investigated the carboxy-terminal portion of the hMPV N protein that showed the greatest sequence homology with hRSV. The carboxy domain of N proteins of all nonsegmented, negative-stranded RNA viruses have been reported to contain three relatively conserved domains and predicted secondary structure (17, 18). We expressed the carboxy-terminal truncated fragment of the hMPV N protein (recΔN; aa 148 to 394) as a His-tagged fusion protein in a baculovirus expression system and verified expression with His tag antisera and reactivity with hMPV hyperimmune sera by Western blotting (Fig. 3). The immunoreactivity of the hMPV recΔN protein also was evaluated by indirect IgG EIA with human sera containing high-titer IgG antibodies to hMPV, using full-length recN protein as a positive control. In contrast to hMPV recN, which showed strong reactivity with the human sera by EIA, recΔN was not reactive (data not shown).
FIG 3.
Western blot analysis of expressed carboxyl domain of the hMPV N protein (recΔN) and uninfected SF9 cell lysates reacted with mouse anti-His tag antisera and affinity-purified rabbit hyperimmune antisera prepared for hMPV (strain CAN97-83).
Evaluation of amino-terminal region of hMPV N protein using synthetic peptides.
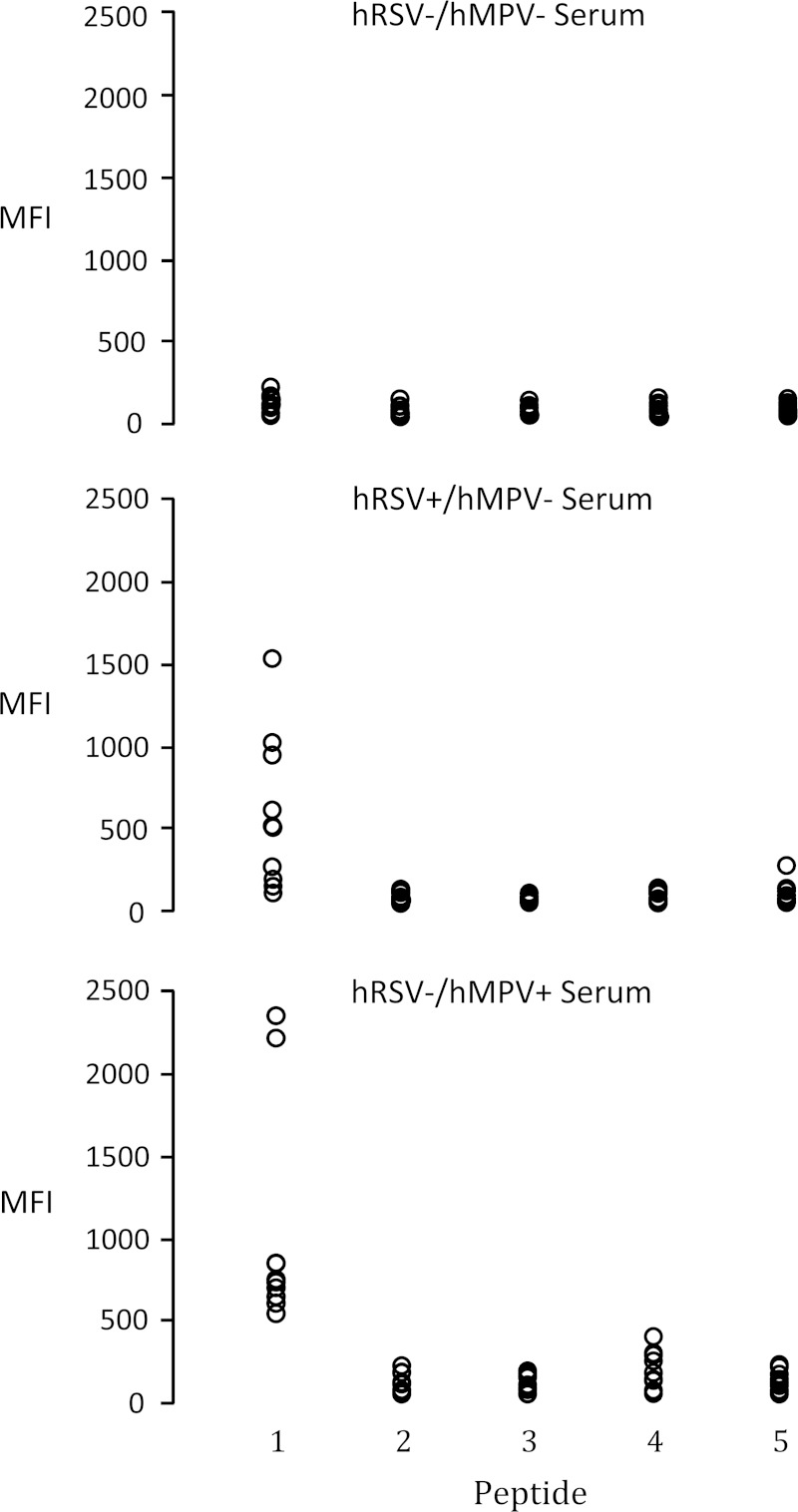
We next examined the amino-terminal region of hMPV N protein (aa 1 to 151) for possible common antigenic sites and designed 5 peptides (P1 to P5) covering the most conserved subregions between hRSV N and hMPV N (Fig. 1). The reactivity of these peptides with human serum specimens was tested using a modified Luminex MAGPIX seroassay previously developed for recN proteins (15). Each peptide was separately coupled to MagPlex microspheres (Luminex) that then were combined and tested against three groups of 10 human serum specimens for virus-specific IgG reactivity: (i) specimens lacking detectable antibodies to hMPV and hRSV by whole-virus-lysate and recN EIAs (hRSV−/hMPV−); (ii) specimens with stable levels of antibody to hRSV and negative for hMPV antibodies (hRSV+/hMPV−); and (iii) specimens with stable levels of antibody to hMPV and negative for hRSV antibodies (hRSV−/hMPV+). Of the 5 peptides tested, only P1, which shows 38.7% amino acid identity to both hRSV subgroups, was reactive with serum specimens from both hRSV−/hMPV+ and hRSV+/hMPV− sample groups, with no reactivity detected with antibody-negative sera (Fig. 4). P1 reactivity was evaluated further with 10 serum pairs from children with acute virus infections showing diagnostic increases in titers of antibody to either hRSV or hMPV (Fig. 5). Increases in concentrations of antibody P1 were detected with most paired sera and were confirmed on repeat testing, indicating that this region of the N protein (aa 1 to 31) contains an immunoreactive site common to both viruses.
FIG 4.
Reactivity of peptides P1 to P5 with human sera. Peptides individually coupled to Luminex beads were reacted with three groups of 10 selected human serum specimens containing high stable levels of IgG antibodies to hRSV (hRSV+/hMPV−) and hMPV (hRSV−/hMPV+) and no detectable antibodies to either virus (hRSV−/hMPV−) by whole-virus-lysate EIAs and tested by MAGPIX immunoassay. The reporter fluorescence of the peptide-coupled beads was expressed as mean fluorescence intensity (MFI).
FIG 5.
Reactivity of peptide P1 with human sera. P1-coupled Luminex beads were reacted with 10 selected acute- and convalescent-phase serum pairs showing diagnostic increases (≥4-fold) in IgG antibodies to hRSV (6 pairs) or hMPV (4 pairs) by whole-virus-lysate EIAs and tested by MAGPIX immunoassay. Reporter fluorescence of the P1-coupled beads was expressed as mean fluorescence intensity (MFI). MFI ratios, calculated by dividing the convalescent-phase serum MFI by the MFI of acute-phase serum, show the relative magnitude of antibody responses.
Evaluation of hyperimmune antisera raised against peptide P1.
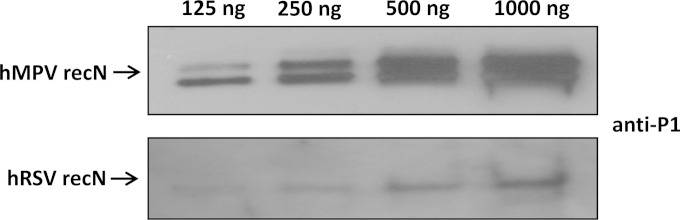
To affirm that P1 contained an immunoreactive site common to both hRSV and hMPV, we generated hyperimmune animal antisera against P1 and performed Western blotting against full-length recN proteins. As expected, the antisera prepared to P1 recognized both hMPV and hRSV recN proteins by Western blotting (Fig. 6).
FIG 6.
Western blot analysis of different concentrations of purified full-length hRSV and hMPV recN proteins reacted with affinity-purified rabbit hyperimmune antisera prepared for peptide P1.
DISCUSSION
During a serologic study of acute respiratory infections among young children in Dhaka, Bangladesh, we observed an unexpectedly high proportion of dual increases in titers of antibodies to hRSV and hMPV. Although coincident infections could explain this observation, there was minimal overlap in the temporal circulation of these viruses in this community, and RT-PCR testing of respiratory specimens from these children detected one or neither virus but never both (9). Further study revealed that full-length recN proteins were recognized reciprocally by hyperimmune antisera prepared for each virus, and a common immunoreactive site located at the amino terminus of hMPV N (aa 1 to 31) was identified using synthetic peptides. Our findings are consistent with those of previous studies that separately identified a linear epitope within the amino terminus of hMPV (19, 20) and hRSV (21), but this is the first study to demonstrate heterologous antibody responses in both immunized animals and naturally acquired human infections.
hRSV and hMPV are each comprised of two major antigenic subgroups, which are distinguishable genetically and immunologically by neutralization with animal hyperimmune antisera and reactivity with monoclonal antibodies (22, 23). However, these antigenic differences are associated primarily with the highly variable F and G envelope glycoproteins and are less pronounced with the immunodominant N proteins that show >95% amino acid sequence identity between subgroup members (24, 25). hRSV and hMPV N proteins share up to 44% amino acid identity, with the greatest homology found in the carboxy domain, where we focused our efforts to identify cross-reactive sites (17, 18). However, we were unsuccessful, because, unlike full-length hMPV recN, the truncated protein did not react with IgG-positive human sera by EIA. The hMPV recΔN protein was reciprocally recognized by both hMPV and hRSV antisera by Western blotting (data not shown), suggesting that a linear epitope(s) common to both viruses is present in this region. In fact, other studies have identified multiple linear epitopes within the carboxy domain of the hRSV N protein (21, 26, 27), and their potential common antigenicity with hMPV should be investigated. One possible explanation for the loss of immunoreactivity of the hMPV recΔN protein with antibody-positive human sera by EIA is that N protein antigenicity is conformation dependent, and the truncation of the amino-terminal portion of the protein changes its natural configuration, resulting in the loss of conformational epitopes or sequestering of linear epitopes. The existence of conformational epitopes on the hMPV N protein was demonstrated by Petraitytė-Burneikienė et al. (20), who showed that monoclonal antibodies directed against these epitopes reacted with native viral nucleocapsid but not with its truncated form. Murray et al. (21) showed that linear epitopes in the carboxyl terminus of the hRSV N protein are blocked on binding to phosphoprotein during formation of the viral RNA-dependent RNA polymerase complex.
Given the possibility that the amino terminus of the hMPV and hRSV N protein contained common antigenic sites, we revisited a study by Alvarez et al. (19), who identified a peptide (designated N protein10-29) common to hMPV and group C avian metapneumovirus (AMPV) that computational analysis predicted to be highly antigenic. Rabbit hyperimmune antiserum prepared for this peptide was shown by these authors to react by both Western blotting and EIA with cultured virus lysate of hMPV (strain CAN98-75) and AMPV subtypes A to C, but not hRSV (strain A2). Their finding of a lack of cross-reactivity with hRSV was inconsistent with our results. However, close examination of the Western blot image presented in Fig. 2A of their manuscript shows a faint band of the expected size in lane 6, demonstrating possible reactivity between N protein10-29 antisera and hRSV A2 lysate. We suspect that the use of hRSV recN protein in our assays increased sensitivity over whole-virus antigen and may account for this difference in results.
Patterns of temporal circulation of respiratory viruses in a given community is governed by many factors, including climate, geography, population demographics and density, and nonspecific viral interference, among others (28, 29), and distinct recurring seasonal peaks have been described for hRSV and hMPV in the United States (30, 31), although substantial variation in individual years and communities can occur (32). Community circulation patterns also may be influenced by population immunological barriers (herd immunity) that can affect the timing and duration of virus activity. For example, two other members of the Paramyxoviridae family, human parainfluenza virus (hPIV) types 1 and 3, possess common antigens that show cross-reactions in serological assays (33). In our study, 50 of 86 (58.1%) children that showed diagnostic increases in titers of antibody to hPIV1 or hPIV3 had increases for both types, but no codetections occurred by RT-PCR in concurrently collected respiratory specimens (data not shown). Shared antigenicity and immunologic cross protection has been posited as one explanation for the distinctive seasonal peaks and apparent interactions seen in a national prospective study monitoring hPIV circulation over a 15-year period; hPIV1 peaks biennially in the fall of odd-numbered years, while hPIV3 peaks annually in late spring with increased activity occurring during years when hPIV1 is absent (34). Discontinuous circulation possibly related to shared antigenicity also was evident with hRSV and hMPV in the larger RT-PCR study of Bangladeshi children conducted from February 2004 through February 2008, of which our serological study was a subset (9). hRSV activity periods were well defined, with peaks in January 2006, July 2006, and October 2007. In contrast, these peaks were preceded by four episodes of hMPV peak activity in February 2005, November 2005, February 2007, and September 2007 that showed limited overlap with hRSV. In the longest consecutive study of hRSV and hMPV seasonality, Williams et al. (30) conducted an etiological assessment of acute upper respiratory infections in 1,532 children monitored over a 20-year period from January 1982 through December 2001 in Nashville, TN. The authors found that hMPV infections occurred during the late winter and early spring months, overlapping the winter peak of hRSV, but with peak frequency occurring approximately 2 months after the hRSV peak. A similar pattern is found in multiple studies involving different populations over different time periods from varied geographic regions, including Argentina (35), Canada (36), China (37), England (38), Japan (39), and the United States (40, 41), including Alaska (42).
It is interesting to speculate on the possible effect of prior infection with one virus on the clinical outcome of subsequent infection with the other. For example, multiple infections with hRSV and hMPV occur throughout life, but repeat infections with the same virus generally are much less severe (53), likely due to prior immunologic priming. Therefore, the presence of common antigenic determinants within the hRSV and hMPV N protein may have implications for cross-protection from natural virus infections and may help inform vaccine development and evaluation strategies. Following hRSV or hMPV infection, serum-neutralizing antibodies are produced, principally against the F and G envelope glycoproteins, and have been shown to be protective (43, 44). Both hRSV and hMPV F and G proteins share limited sequence identity, and antisera generated against either virus have not been shown to be cross-neutralizing (45). Although it is well known that hRSV serum-neutralizing antibodies are protective, inducing adequate neutralizing antibodies at the mucosal surface by vaccination has been unsuccessful (46). It has been suggested that hRSV vaccines that induce antibody and CD8+ T cells are more effective (47). Notably, the hRSV N protein was the major target for memory CD8+ T-cell responses (48, 49, 50), and multiple human cytotoxic T lymphocyte epitopes have been identified within the hRSV N protein, including an epitope located at amino acid positions 16 to 24 within the P1 peptide (50, 51).
It is important to note that heterotypic antibody responses were not detected in most children that showed diagnostic increases in antibodies to hRSV or hMPV, and of those children with increases in titers of antibody to both viruses, most exhibited a strong and weak response, with the former being attributable to the infecting virus. Nevertheless, our findings suggest that there will be some diagnostic uncertainty when using native or recombinant N protein-based assays for serological assessment of acute infection with these viruses. That some children demonstrate heterotypic antibody responses and others do not may reflect differences in their history with either virus; with hPIVs, co-antibody titer rises are seen more often following reinfection with a heterotypic virus (52). However, our study examined only immunoglobulin subclass G in serum and did not evaluate virus-specific IgM or IgA antibody responses in serum or respiratory mucosa. Moreover, our study was restricted to available serum specimens from young Bangladeshi children; therefore, our findings may not be generalizable due to unanticipated differences associated with this geographically and demographically distinct population. Further studies are warranted to establish the full antigenic relatedness of hRSV and hMPV using well-characterized sera from different population groups.
ACKNOWLEDGMENT
The findings and conclusions in this report are those of the authors and do not necessarily represent the official position of the Centers for Disease Control and Prevention.
REFERENCES
- 1.Meng J, Stobart CC, Hotard AL, Moore ML. 2014. An overview of respiratory syncytial virus. PLoS Pathog 10(4):e1004016. doi: 10.1371/journal.ppat.1004016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Thompson WW, Shay DK, Weintraub E, Brammer L, Cox N, Anderson LJ, Fukuda K. 2003. Mortality associated with influenza and respiratory syncytial virus in the United States. JAMA 289:179–186. doi: 10.1001/jama.289.2.179. [DOI] [PubMed] [Google Scholar]
- 3.Falsey AR, Hennessey PA, Formica MA, Cox C, Walsh EE. 2005. Respiratory syncytial virus infection in elderly and high-risk adults. N Engl J Med 352:1749–1759. doi: 10.1056/NEJMoa043951. [DOI] [PubMed] [Google Scholar]
- 4.van den Hoogen BG, de Jong JC, Groen J, Kuiken T, de Groot R, Fouchier RA. 2001. A newly discovered human pneumovirus isolated from young children with respiratory tract disease. Nat Med 7:719–724. doi: 10.1038/89098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wilkesmann A, Schildgen O, Eis-Hubinger AM, Geikowski T, Glatzel T, Lentze MJ. 2006. Human metapneumovirus infections cause similar symptoms and clinical severity as respiratory syncytial virus infections. Eur J Pediatr 165:467–475. doi: 10.1007/s00431-006-0105-4. [DOI] [PubMed] [Google Scholar]
- 6.Papenburg J, Boivin G. 2010. The distinguishing features of human metapneumovirus and respiratory syncytial virus. Rev Med Virol 20:245–260. doi: 10.1002/rmv.651. [DOI] [PubMed] [Google Scholar]
- 7.Edwards KM, Zhu Y, Griffin MR, Weinberg GA, Hall CB, Szilagyi PG, Staat MA, Iwane M, Prill MM, Williams JV. 2013. New vaccine surveillance network: burden of human metapneumovirus infection in young children. N Engl J Med 368:633–643. doi: 10.1056/NEJMoa1204630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brooks WA, Erdman D, Terebuh P. 2007. Human metapneumovirus infection among children, Bangladesh. Emerg Infect Dis 13:1611–1613. doi: 10.3201/eid1310.070337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Stockman LJ, Brooks WA, Streatfield PK, Rahman M, Goswami D, Nahar K, Rahman MZ, Luby SP, Anderson LJ. 2013. Challenges to evaluating respiratory syncytial virus mortality in Bangladesh, 2004-2008. PLoS One 8:e53857. doi: 10.1371/journal.pone.0053857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Falsey AR, Erdman D, Anderson LJ, Walsh EE. 2003. Human metapneumovirus infections in young and elderly adults. J Infect Dis 187:785–790. doi: 10.1086/367901. [DOI] [PubMed] [Google Scholar]
- 11.Bhella D, Ralph A, Murphy LB, Yeo RP. 2002. Significant differences in nucleocapsid morphology within the Paramyxoviridae. J Gen Virol 83:1831–1839. [DOI] [PubMed] [Google Scholar]
- 12.Buraphacheep W, Britt WJ, Sullender WM. 1997. Detection of antibodies to respiratory syncytial virus attachment and nucleocapsid proteins with recombinant baculovirus-expressed antigens. J Clin Microbiol 35:354–357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.O'Shaughnessy L, Carr M, Crowley B, Carberry S, Doyle S. 2011. Recombinant expression and immunological characterisation of proteins derived from human metapneumovirus. J Clin Virol 52:236–243. doi: 10.1016/j.jcv.2011.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liu L, Bastien N, Sidaway F, Chan E, Li Y. 2007. Seroprevalence of human metapneumovirus (hMPV) in the Canadian province of Saskatchewan analyzed by a recombinant nucleocapsid protein-based enzyme-linked immunosorbent assay. J Med Virol 79:308–313. doi: 10.1002/jmv.20799. [DOI] [PubMed] [Google Scholar]
- 15.Zhang Y, Brooks WA, Goswami D, Rahman M, Luby SP, Erdman DD. 2014. A duplex recombinant viral nucleoprotein microbead immunoassay for simultaneous detection of seroresponses to human respiratory syncytial virus and metapneumovirus infections. J Virol Methods 206:55–62. doi: 10.1016/j.jviromet.2014.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tedcastle AB, Fenwick F, Ingram RE, King BJ, Robinson MJ, Toms GL. 2012. The characterization of monoclonal antibodies to human metapneumovirus and the detection of multiple forms of the virus nucleoprotein and phosphoprotein. J Med Virol 84:1061–1070. doi: 10.1002/jmv.23298. [DOI] [PubMed] [Google Scholar]
- 17.Barr J, Chambers P, Pringle CR, Easton AJ. 1991. Sequence of the major nucleocapsid protein gene of pneumonia virus of mice: sequence comparisons suggest structural homology between nucleocapsid proteins of pneumoviruses, paramyxoviruses, rhabdoviruses and filoviruses. J Gen Virol 72:677–685. doi: 10.1099/0022-1317-72-3-677. [DOI] [PubMed] [Google Scholar]
- 18.Dar AM, Munir S, Goyal SM, Abrahamsen MS, Kapur V. 2001. Sequence analysis of the nucleocapsid and phosphoprotein genes of avian pneumoviruses circulating in the US. Virus Res 79:15–25. doi: 10.1016/S0168-1702(01)00276-3. [DOI] [PubMed] [Google Scholar]
- 19.Alvarez R, Jones LP, Seal BS, Kapczynski DR, Tripp RA. 2004. Serological cross-reactivity of members of the Metapneumovirus genus. Virus Res 105:67–73. doi: 10.1016/j.virusres.2004.04.019. [DOI] [PubMed] [Google Scholar]
- 20.Petraitytė-Burneikienė R, Nalivaiko K, Lasickienė R, Firantienė R, Emužytė R, Sasnauskas K, Zvirblienė A. 2011. Generation of recombinant metapneumovirus nucleocapsid protein as nucleocapsid-like particles and development of virus-specific monoclonal antibodies. Virus Res 161:131–139. doi: 10.1016/j.virusres.2011.07.009. [DOI] [PubMed] [Google Scholar]
- 21.Murray J, Loney C, Murphy LB, Graham S, Yeo RP. 2001. Characterization of monoclonal antibodies raised against recombinant respiratory syncytial virus nucleocapsid (N) protein: identification of a region in the carboxy terminus of N involved in the interaction with P protein. Virology 289:252–261. doi: 10.1006/viro.2001.1150. [DOI] [PubMed] [Google Scholar]
- 22.Anderson LJ, Hierholzer JC, Tsou C, Hendry RM, Fernie BF, Stone Y, McIntosh K. 1985. Antigenic characterization of respiratory syncytial virus strains with monoclonal antibodies. J Infect Dis 151:626–633. doi: 10.1093/infdis/151.4.626. [DOI] [PubMed] [Google Scholar]
- 23.van den Hoogen BG, Herfst S, Sprong L. 2004. Antigenic and genetic variability of human metapneumoviruses. Emerg Infect Dis 10:658–666. doi: 10.3201/eid1004.030393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bastien N, Normand S, Taylor T, Ward D, Peret TC, Boivin G, Anderson LJ, Li Y. 2003. Sequence analysis of the N, P, M and F genes of Canadian human metapneumovirus strains. Virus Res 93:51–62. doi: 10.1016/S0168-1702(03)00065-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Skiadopoulos MH, Biacchesi S, Buchholz UJ. 2004. The two major human metapneumovirus genetic lineages are highly related antigenically, and the fusion (F) protein is a major contributor to this antigenic relatedness. J Virol 78:6927–6937. doi: 10.1128/JVI.78.13.6927-6937.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Leonov SV, Waris M, Norrby E. 1995. Linear antigenic and immunogenic regions of human respiratory syncytial virus N protein. J Gen Virol 76(Part 2):357–364. doi: 10.1099/0022-1317-76-2-357. [DOI] [PubMed] [Google Scholar]
- 27.Li J, Ling R, Randhawa JS, Shaw K, Davis PJ, Juhasz K, Pringle CR, Easton AJ, Cavanagh D. 1996. Sequence of the nucleocapsid protein gene of subgroup A and B avian pneumoviruses. Virus Res 41:185–191. doi: 10.1016/0168-1702(96)01288-9. [DOI] [PubMed] [Google Scholar]
- 28.Shek LP, Lee BW. 2003. Epidemiology and seasonality of respiratory tract virus infections in the tropics. Paediatr Respir Rev 4:105–111. doi: 10.1016/S1526-0542(03)00024-1. [DOI] [PubMed] [Google Scholar]
- 29.Paynter S, Sly PD, Ware RS, Williams G, Weinstein P. 2014. The importance of the local environment in the transmission of respiratory syncytial virus. Sci Total Environ 493:521–525. doi: 10.1016/j.scitotenv.2014.06.021. [DOI] [PubMed] [Google Scholar]
- 30.Williams JV, Wang CK, Yang C-F, Tollefson SJ, House FS, Heck JM, Chu M, Brown JB, Lintao LD, Quinto JD, Chu D, Spaete RR, Edwards KM, Wright PF, Crowe JE Jr. 2006. The role of human metapneumovirus in upper respiratory tract infections in children: a 20-year experience. J Infect Dis 193:387–395. doi: 10.1086/499274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Centers for Disease Control and Prevention. 2013. Respiratory syncytial virus activity–United States, July 2011–January 2013. MMWR 62:141–144. [PMC free article] [PubMed] [Google Scholar]
- 32.Peret TC, Hall CB, Schnabel KC, Golub JA, Anderson LJ. 1998. Circulation patterns of genetically distinct group A and B strains of human respiratory syncytial virus in a community. J Gen Virol 79:2221–2229. [DOI] [PubMed] [Google Scholar]
- 33.Henrickson KJ. 2003. Parainfluenza viruses. Clin Microbiol Rev 16:242–264. doi: 10.1128/CMR.16.2.242-264.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fry AM, Curns AT, Harbour K, Hutwagner L, Holman RC, Anderson LJ. 2006. Seasonal trends of human parainfluenza viral infections: United States, 1990-2004. Clin Infect Dis 43:1016–1022. doi: 10.1086/507638. [DOI] [PubMed] [Google Scholar]
- 35.Marcone DN, Ellis A, Videla C, Ekstrom J, Ricarte C, Carballal G, Vidaurreta SM, Echavarría M. 2013. Viral etiology of acute respiratory infections in hospitalized and outpatient children in Buenos Aires, Argentina. Pediatr Infect Dis J 32:e105–10. doi: 10.1097/INF.0b013e31827cd06f. [DOI] [PubMed] [Google Scholar]
- 36.Robinson JL, Lee BE, Bastien N, Li Y. 2005. Seasonality and clinical features of human metapneumovirus infection in children in northern Alberta. J Med Virol 76:98–105. doi: 10.1002/jmv.20329. [DOI] [PubMed] [Google Scholar]
- 37.Feng L, Li Z, Zhao S, Nair H, Lai S, Xu W, Li M, Wu J, Ren L, Liu W, Yuan Z, Chen Y, Wang X, Zhao Z, Zhang H, Li F, Ye X, Li S, Feikin D, Yu H, Yang W. 2014. Viral etiologies of hospitalized acute lower respiratory infection patients in China, 2009-2013. PLoS One 9:e99419. doi: 10.1371/journal.pone.0099419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhao H, Green H, Lackenby A, Donati M, Ellis J, Thompson C, Bermingham A, Field J, Sebastianpillai P, Zambon M, Watson J, Pebody R. 2014. A new laboratory-based surveillance system (Respiratory DataMart System) for influenza and other respiratory viruses in England: results and experience from 2009 to 2012. Euro Surveill 19:20680. [DOI] [PubMed] [Google Scholar]
- 39.Mizuta K, Abiko C, Aoki Y, Ikeda T, Matsuzaki Y, Itagaki T, Katsushima F, Katsushima Y, Noda M, Kimura H, Ahiko T. 2013. Seasonal patterns of respiratory syncytial virus, influenza A virus, human metapneumovirus, and parainfluenza virus type 3 infections on the basis of virus isolation data between 2004 and 2011 in Yamagata, Japan. Jpn J Infect Dis 66:140–145. doi: 10.7883/yoken.66.140. [DOI] [PubMed] [Google Scholar]
- 40.Williams JV, Harris PA, Tollefson SJ, Halburnt-Rush LL, Pingsterhaus JM, Edwards KM, Wright PF, Crowe JE Jr. 2004. Human metapneumovirus and lower respiratory tract disease in otherwise healthy infants and children. N Engl J Med 350:443–450. doi: 10.1056/NEJMoa025472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Litwin CM, Bosley JG. 2014. Seasonality and prevalence of respiratory pathogens detected by multiplex PCR at a tertiary care medical center. Arch Virol 159:65–72. doi: 10.1007/s00705-013-1794-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Singleton RJ, Bulkow LR, Miernyk K, DeByle C, Pruitt L, Hummel KB, Bruden D, Englund JA, Anderson LJ, Lucher L, Holman RC, Hennessy TW. 2010. Viral respiratory infections in hospitalized and community control children in Alaska. J Med Virol 82:1282–1290. doi: 10.1002/jmv.21790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Connors M, Collins PL, Firestone CY, Murphy BR. 1991. Respiratory syncytial virus (RSV) F, G, M2 (22K), and N proteins each induce resistance to RSV challenge, but resistance induced by M2 and N proteins is relatively short-lived. J Virol 65:1634–1637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Skiadopoulos MH, Biacchesi S, Buchholz UJ. 2006. Individual contributions of the human metapneumovirus F, G, and SH surface glycoproteins to the induction of neutralizing antibodies and protective immunity. Virology 345:492–501. doi: 10.1016/j.virol.2005.10.016. [DOI] [PubMed] [Google Scholar]
- 45.Wyde PR, Chetty SN, Jewell AM, Boivin G, Piedra PA. 2003. Comparison of the inhibition of human metapneumovirus and respiratory syncytial virus by ribavirin and immune serum globulin in vitro. Antiviral Res 60:51–59. doi: 10.1016/S0166-3542(03)00153-0. [DOI] [PubMed] [Google Scholar]
- 46.de Swart RL, van den Hoogen BG, Kuiken T, Herfst S, van Amerongen G, Yüksel S, Sprong L, Osterhaus AD. 2007. Immunization of macaques with formalin-inactivated human metapneumovirus induces hypersensitivity to hMPV infection. Vaccine 25:8518–8528. doi: 10.1016/j.vaccine.2007.10.022. [DOI] [PubMed] [Google Scholar]
- 47.Graham BS. 2011. Biological challenges and technological opportunities for respiratory syncytial virus vaccine development. Immunol Rev 239:149–166. doi: 10.1111/j.1600-065X.2010.00972.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bangham CR, Openshaw PJ, Ball LA, King AM, Wertz GW, Askonas BA. 1986. Human and murine cytotoxic T cells specific to respiratory syncytial virus recognize the viral nucleoprotein (N), but not the major glycoprotein (G), expressed by vaccinia virus recombinants. J Immunol 137:3973–3977. [PubMed] [Google Scholar]
- 49.Goulder PJ, Lechner F, Klenerman P, McIntosh K, Walker BD. 2000. Characterization of a novel respiratory syncytial virus-specific human cytotoxic T-lymphocyte epitope. J Virol 74:7694–7697. doi: 10.1128/JVI.74.16.7694-7697.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Venter M, Rock M, Puren AJ, Tiemessen CT, Crowe JE. 2003. Respiratory syncytial virus nucleoprotein-specific cytotoxic T-cell epitopes in a South African population of diverse HLA types are conserved in circulating field strains. J Virol 77:7319–7329. doi: 10.1128/JVI.77.13.7319-7329.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Terrosi C, Di Genova G, Savellini GG, Correale P, Blardi P, Cusi MG. 2007. Immunological characterization of respiratory syncytial virus N protein epitopes recognized by human cytotoxic T lymphocytes. Viral Immunol 20:399–406. doi: 10.1089/vim.2007.0041. [DOI] [PubMed] [Google Scholar]
- 52.Julkunen I. 1984. Serological diagnosis of parainfluenza virus infections by enzyme immunoassay with special emphasis on purity of viral antigens. J Med Virol 14:177–187. doi: 10.1002/jmv.1890140212. [DOI] [PubMed] [Google Scholar]
- 53.Ohuma EO, Okiro EA, Ochola R, Sande CJ, Cane PA. 2012. The natural history of respiratory syncytial virus in a birth cohort: the influence of age and previous infection on reinfection and disease. Am J Epidemiol 176:794–802. doi: 10.1093/aje/kws257. [DOI] [PMC free article] [PubMed] [Google Scholar]