Abstract
Accurate identification of mycobacterial species and subspecies is essential to evaluate their significance and to perform epidemiological studies. The subspecies of Mycobacterium avium have different attributes but coincide in their zoonotic potential. Our knowledge about M. avium subsp. silvaticum is limited, since its identification is uncertain. Mycobacterium avium subsp. avium and M. avium subsp. silvaticum can be discriminated from each other based only on phenotypic characteristics, as they have almost identical genome sequences. Here we describe the development of a diagnostic method which enables the molecular identification of M. avium subsp. silvaticum and discrimination from M. avium subsp. avium based on genomic differences in a duplex high-resolution melt and M. avium subsp. silvaticum-specific mismatch real-time PCR. The developed assay was tested on reference strains and 199 field isolates, which were analyzed by phenotypic methods previously. This assay not only identified all 63 M. avium subsp. silvaticum and 138 M. avium subsp. avium strains correctly but also enabled the detection of mixed M. avium subsp. avium-M. avium subsp. silvaticum cultures. This is the first time that such a large panel of strains has been analyzed, and we also report the first isolation of M. avium subsp. silvaticum from red fox, red deer, wild boar, cattle, and badger. This assay is reliable, rapid, simple, inexpensive, and robust. It eliminates the long-existing problem of ambiguous phenotypic identification and opens up the possibility for detailed and comprehensive strain studies.
INTRODUCTION
The species Mycobacterium avium is divided into three subspecies according to the currently valid taxonomical classification: Mycobacterium avium subsp. paratuberculosis, the etiological agent of Johne's disease; Mycobacterium avium subsp. avium, the pathogen of avian tuberculosis; and Mycobacterium avium subsp. silvaticum (1). In addition, the designation “Mycobacterium avium subsp. hominissuis” has been proposed for human/porcine-type M. avium isolates (2). M. avium subsp. silvaticum, previously called “wood pigeon Mycobacterium,” was assigned to the Mycobacterium avium complex (MAC) in 1990 (3). Besides wood pigeons, it was isolated from crane (3), penguin (4), roe deer (5), and hazel hen (5). The zoonotic potential of MAC members is renowned (6), but less is known about M. avium subsp. silvaticum; hence, the detection and study of this subspecies are hindered by its unreliable and ambiguous identification.
Accurate identification of mycobacterial species and subspecies is essential for evaluation of their significance, pathogenicity, and epidemiology. The four subspecies of M. avium differ greatly in their host range, growth potential, and environmental occurrence (4). Three subspecies can be easily differentiated by molecular biological methods thanks to distinct molecular differences. IS900 is the specific insertion sequence (IS) of M. avium subsp. paratuberculosis. “M. avium subsp. hominissuis” and M. avium subsp. avium both harbor IS1245 but can be discriminated with IS901 (absent from “M. avium subsp. hominissuis” and present in M. avium subsp. avium and M. avium subsp. silvaticum) (7). The identification of M. avium subsp. silvaticum is still based only on phenotypic characteristics, namely, the rough colony morphology and slow and mycobactin-dependent growth of M. avium subsp. silvaticum versus the smooth colony morphology, rapid growth, and mostly mycobactin-independent nature of M. avium subsp. avium (8). As phenotypic features can differ among isolates of the same subspecies (9), and mycobactin dependence can vanish in subcultures (1), misidentifications can occur, which urges the need for a reliable molecular biological identification method.
Attempts at molecular identification of MAC members were done by use of primary ISs (10). Moss and coworkers (11) assumed that IS902 would be a specific marker of M. avium subsp. silvaticum strains, but subsequently, it proved to be identical to IS901 (12). Other ISs, such as IS1311 (13) or IS1612 (9, 14), are also present in both M. avium subsp. avium and M. avium subsp. silvaticum, which hinder their differentiation by phenotypic characteristics further on.
Despite numerous other attempts, such as fatty acid composition studies (5), restriction fragment length polymorphism (RFLP) analysis (15, 16), large-sequence polymorphism analysis (17), multilocus sequence analysis (4), or multispacer sequence typing (18), some of which had promising results but were tested on only two to five isolates (4, 16, 17) or the reference strain alone (18), M. avium subsp. silvaticum generally remained undifferentiable from M. avium subsp. avium (19).
Before December 2013, only 19 partial sequences from M. avium subsp. silvaticum reference strain ATCC 49884T were available in GenBank, of which the 16S rRNA, 16S-23S rRNA intergenic spacer, hsp65, secA1, rpoB, ssrA, and tuf sequences were completely identical to the corresponding M. avium subsp. avium ones. Deposition of M. avium subsp. silvaticum reference strain ATCC 49884T whole-genome shotgun sequence data in GenBank enabled detailed comparative studies of the M. avium subsp. avium and M. avium subsp. silvaticum genomes (GenBank accession number AYOC00000000 and BioProject number PRJNA219418).
With the advent of the high-resolution melt (HRM) technique, single base changes (single nucleotide polymorphisms [SNPs]), which cause only subtle changes in melting temperature (Tm), became sufficient for accurate species/subspecies/genotype/serovar differentiation (20).
The aim of our study was to develop a diagnostic method based on genomic differences between M. avium subsp. avium and M. avium subsp. silvaticum that enables omission of phenotypic identification and offers fast and reliable differentiation of these subspecies from one another for diagnostic laboratories.
MATERIALS AND METHODS
Sequence analysis and primer design.
We initially designed primers for IS1613 (GenBank accession number AJ011837.1) and IS1612 and then performed GenBank BLAST homology searches (http://www.ncbi.nlm.nih.gov/blast) of the nonredundant nucleotide collection (nr/nt) and whole-genome shotgun contig (wgs) databases. At the time of our first search, only 36 M. avium subsp. silvaticum nucleotide sequences were found in the nr/nt database, and 3 M. avium subsp. avium whole genomes were unassembled in the wgs database. Available sequences of M. avium subsp. silvaticum and M. avium subsp. avium were aligned with DNASTAR SeqMan Pro software (Lasergene 12; DNASTAR Inc., Madison, WI, USA), and the corresponding sequences were checked for differences.
As the contigs of the complete genome sequence of M. avium subsp. silvaticum reference strain ATCC 49884T (GenBank accession number AYOC00000000 and BioProject number PRJNA219418) were deposited in GenBank, a new search was conducted. The 808 M. avium subsp. silvaticum contigs were aligned with 258 contigs of M. avium subsp. avium ATCC 25291T (GenBank accession number ACFI00000000 and BioProject number PRJNA30909), 886 contigs of M. avium subsp. avium 10-9275, 577 contigs of M. avium subsp. avium 11-4751 (GenBank accession numbers AYOB00000000 and AYNY00000000 and BioProject numbers PRJNA216926 and PRJNA216924), 772 contigs of M. avium subsp. avium Env77, and 1,201 contigs of M. avium subsp. avium DT78 (21). Based on preliminary sequence and alignment studies, out of 2,800 nucleotide position differences, 50 potential sequence variations, such as single or multiple gaps, SNPs, or multiple mismatching (MM) base pairs, were selected. For 12 such sequence sections (including est, aspB, and pepB gene variable positions [4]), MM and HRM primer pairs were designed with DNASTAR Primer Design software (see Data Set S2 in the supplemental material). Primers were checked theoretically by BLAST homology searches in order to ensure specificity.
Test strain collection and DNA extraction.
The test strain collection used contained the M. avium subsp. avium ATCC 25291T, M. avium subsp. silvaticum ATCC 49884T, M. avium subsp. paratuberculosis ATCC 19851T, and Mycobacterium bovis AN5T reference strains and 202 field isolates. Field isolates were collected and typed at the Bacteriology Laboratory of the Veterinary Diagnostic Directorate (National Food Chain Safety Office [NFCSO]) in Budapest, Hungary, during routine Mycobacterium culture for samples from different wild and domestic mammals and birds. All strains were tested to determine whether they belonged to the MAC and whether they harbored or lacked the insertion sequences IS901, IS1245, and IS900, according to methods described previously by Wilton and Cousins (22), Álvarez et al. (23), and Castellanos et al. (24), respectively. M. avium subsp. silvaticum strains were identified phenotypically by their rough colony morphology, mycobactin dependence, slow growth, and inability to grow on egg medium. The 137 M. avium subsp. avium strains came from 14 cattle, 47 swine, 49 wild boars, 2 red deer, 8 red foxes, 7 chickens, 1 duck, 1 pigeon, 1 turkey, 1 long-eared owl, 1 tragopan, 1 tauraco, 1 monitor lizard, 1 pochard, 1 mallard, and 1 wigeon. The 62 M. avium subsp. silvaticum strains (see Data Set S1 in the supplemental material) were isolated from wild boars (44 isolates), red foxes (2), red deer (10), badger (1), and cattle (5). Isolates of Mycobacterium intracellulare (1 isolate), “M. avium subsp. hominissuis” (1 isolate), and M. saskatchewanense (1 isolate) were additionally sequenced to ensure correct identification.
Strains were grown on Lowenstein-Jensen and Middlebrook 7H11 (with mycobactin J) slants. The DNA was extracted by sonication at 80°C for 15 min at 80 Hz and boiling at 95°C for 10 min. Nucleic acid concentrations were determined with a NanoDrop 2000c spectrophotometer (Thermo Scientific, Waltham, MA, USA).
PCR settings and data analysis.
After conventional PCR amplification with reference strains, MM primers were optimized for real-time PCR by using a Rotor Gene 6000 real-time PCR machine and further tested on the field isolates.
HRM primer pairs were also tested and optimized with reference strains. Amplicons were sequenced by using an ABI Prism 3400 DNA sequencer. HRM primers 5′-CGGCGATCGGAATGGAAATA-3′ and 5′-CGGAACCCTGGTCAAGAT-3′ and M. avium subsp. silvaticum-specific MM primers 5′-TTCCTGGCCTGCTTCGACC-3′ and 5′-GTTGACCACCACGGCATTCC-3′ were chosen and used in the PCR master mix at a final concentration of 0.6 μM each. The PCR mixture was further composed of 0.1 μl (5 U/μl) GoTaq G2 Flexi DNA polymerase (Promega, Madison, WI, USA), 0.5 μl of 10 mM deoxynucleoside triphosphate mix (Fermentas, Burlington, Ontario, Canada), 5 μl of 10× PCR buffer, 2.2 μl of a 25 mM MgCl2 solution, 1.25 μl of 20× EvaGreen green fluorescent nucleic acid dye (Biotium, CA, USA), and PCR-grade H2O to a final volume of 25 μl with 20 ng template DNA per reaction mixture, under the following PCR conditions: denaturation at 95°C for 10 min followed by 40 cycles at 95°C for 20 s, 62°C for 40 s, and 72°C for 30 s. Melt curve conditions were 85°C to 99°C at a ramp rate of 0.5°C/s. HRM analysis was performed with temperatures from 87°C to 99°C with increases of 0.02°C. Data processing was performed with RotorGene Q 2.2.3 software (Qiagen, Venlo, Netherlands) based on the HRM plot and normalized graph.
To evaluate the reliability of identification of M. avium subsp. avium and M. avium subsp. silvaticum genotypes by HRM analysis, an independent-sample t test was used.
Specificity was tested with reference strains M. bovis AN5T, M. avium subsp. avium ATCC 25291T, M. avium subsp. silvaticum ATCC 49884T, and M. avium subsp. paratuberculosis ATCC 19851T and field isolates of M. intracellulare, “M. avium subsp. hominissuis,” and M. saskatchewanense.
For input template range determination and mixed-culture testing, reference strains M. avium subsp. silvaticum ATCC 49884T and M. avium subsp. avium ATCC 25291T were used.
Nucleotide sequence accession numbers.
The sequences of the reference strain and one field isolate obtained by using the MM primers were deposited in GenBank under accession numbers KP792235 and KP792233. The sequences of the reference strains and one field isolate obtained by using the HRM primers were deposited in GenBank under accession numbers KP792236, KP792232, and KP792234.
RESULTS
The primers designed for IS1613 gave no amplification products with either M. avium subsp. avium or M. avium subsp. silvaticum, while IS1612 primers generated amplicons with both M. avium subsp. avium and M. avium subsp. silvaticum reference strains.
Our first attempt at discrimination based on the sequence differences of the few available sequences at that time was unsuccessful, as the seemingly adequate sequence variations proved to be identical in our sequencing tests.
In our second attempt, of the 10 MM primers, only 1 was suitable. It was designed to contain a C/G difference in the aspB genes of the M. avium subsp. silvaticum and M. avium subsp. avium genomes. PCR identified the M. avium subsp. silvaticum reference strain correctly, gave a 369-bp-long amplification product with a Tm of 94.9°C, and was specific for M. avium subsp. silvaticum, as no products appeared with M. bovis AN5T, M. avium subsp. avium ATCC 25291T, M. avium subsp. paratuberculosis ATCC 19851T, M. intracellulare, “M. avium subsp. hominissuis,” or M. saskatchewanense. The sequenced products from the M. avium subsp. silvaticum reference strain and 10 M. avium subsp. silvaticum field isolates confirmed the presence of the sequence variation (see Data Set S2 in the supplemental material).
Of the 13 HRM primers, 4 were applicable. The most robustly working primer pair with an appropriate Tm was designed to contain a TT/CG difference in a putative membrane protein gene. The PCR products of the M. avium subsp. avium and M. avium subsp. silvaticum reference strains had unique melting-curve profiles in the normalized graph, with a temperature shift of 0.7°C (Fig. 1). The sequenced HRM products from both reference strains and 10 M. avium subsp. silvaticum field isolates confirmed the presence of the sequence variations (see Data Set S2 in the supplemental material). By testing mixed M. avium subsp. avium-M. avium subsp. silvaticum cultures, an altered midlevel melting-curve shape (Fig. 1) was detected.
FIG 1.
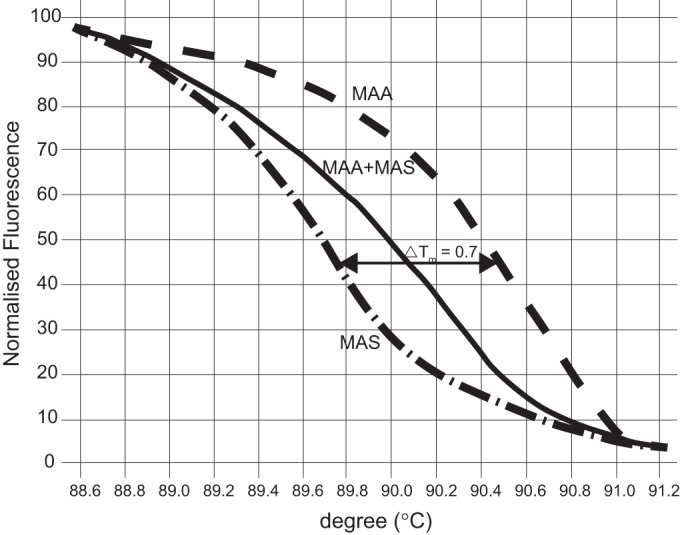
Normalized HRM graph of Mycobacterium avium subsp. avium (MAA) and Mycobacterium avium subsp. silvaticum (MAS). The normalized graph displays the unique melting-curve profiles of M. avium subsp. avium and M. avium subsp. silvaticum with a temperature shift of 0.7°C and contains the altered midlevel melting curve shape of a mixed M. avium subsp. avium-M. avium subsp. silvaticum sample.
The two systems were combined, optimized to run in a single tube as a duplex HRM and M. avium subsp. silvaticum-specific real-time PCR (Fig. 2; see also Data Set S3 in the supplemental material), and tested on 199 field isolates, which were all members of the MAC, IS900 negative, and IS901 and IS1245 positive.
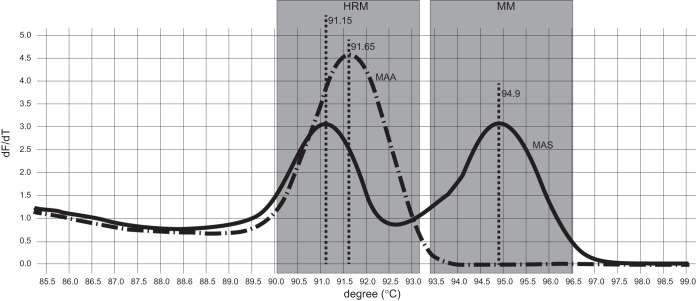
FIG 2.
Melting-curve graph for Mycobacterium avium subsp. silvaticum (MAS) identification by duplex real-time PCR. The graph displays the melting curves of reference strains Mycobacterium avium subsp. avium (MAA) ATCC 25291 and Mycobacterium avium subsp. silvaticum ATCC 49884 in the developed duplex HRM and M. avium subsp. silvaticum-specific real-time PCR assay. The M. avium subsp. silvaticum-specific MM amplification product has a Tm of 94.9°C, while the obtained HRM products had Tms of 91.15°C and 91.65°C for the M. avium subsp. silvaticum and M. avium subsp. avium genotypes, respectively.
This method correctly identified the 199 field isolates. All 137 strains previously identified as M. avium subsp. avium had the M. avium subsp. avium-specific melting-curve profile with no amplification of the aspB gene product. The remaining 62 strains, phenotypically identified as M. avium subsp. silvaticum, yielded the M. avium subsp. silvaticum-specific PCR product with Tms of 94.65°C to 95.1°C and the M. avium subsp. silvaticum-specific melting-curve profile in the HRM analysis.
As samples of mixed M. avium subsp. avium-M. avium subsp. silvaticum cultures have an altered midlevel melting curve shape (Fig. 1), mixed-culture samples can also be distinguished with our assay.
The Tms and their standard deviations in the HRM assay were 91.17°C ± 0.13°C and 91.63°C ± 0.05°C for M. avium subsp. silvaticum and M. avium subsp. avium genotypes, respectively. The independent-sample t test confirmed that the results of the HRM analysis were statistically significant (P < 0.0001). The input DNA template range extended between 100 ng and 15pg.
Besides the developed molecular identification assay, we report the first isolation of M. avium subsp. silvaticum from wild boars, red foxes, red deer, badgers, and naturally infected cattle.
DISCUSSION
Since its first isolation, the existence of M. avium subsp. silvaticum as a distinct species or subspecies has been debated (25), as no molecular difference from M. avium subsp. avium could be traced. However, in 1990, it was classified as a M. avium subspecies, and its identification is still based on the detection of phenotypic characteristics, which can be subjective and may vary among laboratories, depending strongly on personal experience.
There is a significant lack of data on the pathogenicity and zoonotic potential of M. avium subsp. silvaticum. Apart from its first description as a pathogenic agent causing tuberculosis in wood pigeon (1), only a few experimental and hypothetical data are available in the literature (26).
MAC members can interact in tuberculin skin tests, thus hampering tuberculosis diagnoses. M. avium subsp. avium and “M. avium subsp. hominissuis” can cause severe conditions in immunocompromised people, and M. avium subsp. paratuberculosis is associated with human Crohn's disease (1). Contrary to the thoroughly studied subspecies M. avium subsp. avium, “M. avium subsp. hominissuis,” and M. avium subsp. paratuberculosis, the ecology and zoonotic potential of M. avium subsp. silvaticum are barely known. Moss et al. (27) and McFadden et al. (28) reported the identification of M. avium subsp. silvaticum in human samples, but the almost identical M. avium subsp. avium genome sequence makes their results dubious. However, the significance of this less characterized fourth member of the MAC should not be dismissed without further clarification.
Numerous attempts to differentiate M. avium subsp. silvaticum strains from M. avium subsp. avium have been undertaken. Most recently, Chiers and coworkers designed a PCR-RFLP method that targeted the 85B antigen gene (29). Among others, it was tested on M. avium subsp. silvaticum ATCC 49884T and M. avium subsp. avium ITG75/219, but not one M. avium subsp. avium reference strain was included. By retesting the protocol with M. avium subsp. avium ATCC 25291T and M. avium subsp. silvaticum ATCC 49884T, contrary to their results, we obtained identical RFLP profiles (data not shown) for these subspecies. Our results call the applicability of this method and also the identification of the horse isolate into question.
IS1613 is supposed to be a M. avium subsp. avium-specific IS (GenBank accession number AJ011837) (10), but by searching the GenBank wgs database for the 1,734-bp-long sequence, we found only 99% (1,468/1,474) sequence identity with the M. avium subsp. avium 2285(S)gma2285S.contig.5 wgs sequence (GenBank accession number JAOD01000006.1). Furthermore, IS1613 could not be aligned with either the M. avium subsp. avium ATCC 25291T or the M. avium subsp. silvaticum ATCC 49884T wgs database sequence, which supports our negative results.
The results of our IS1612 PCR are consistent with those reported by Bull et al. (9) but cast doubt on the results of Orrú et al. (14). When we tested their primers, we obtained identical amplification products from both the M. avium subsp. avium and M. avium subsp. silvaticum reference strains. Although IS1612 is not present in M. avium subsp. paratuberculosis, it cannot be used to reliably identify M. avium subsp. silvaticum, as it is also present in M. avium subsp. avium strains.
The results of our first attempt at sequence variation-based differentiation of M. avium subsp. silvaticum from M. avium subsp. avium raise questions regarding sequences deposited in GenBank. Higgins et al. (30) reported sequence differences in the same gene of the M. avium subsp. silvaticum reference strain, presumably as a result of a sequencing failure. Partial and sometimes even faulty sequence data are not a suitable basis for the design of identification assays.
In this study, a duplex HRM and M. avium subsp. silvaticum-specific real-time PCR was developed to discriminate thus-far-undifferentiable M. avium subsp. silvaticum and M. avium subsp. avium strains by molecular analysis reliably and easily.
Until the discovery and development of HRM analysis, the SNPs observed in the est, aspB, and pepB genes (4) and at several other points of the genome could have been tested only by sequencing, which is time-consuming and expensive for routine laboratory diagnostic purposes.
As HRM analysis does not require probes, labeled primers, or any post-PCR processing, it is a rapid and cost-effective identification method.
The sequence difference in the putative membrane protein gene targeted by the chosen HRM primer was not described previously. The MM primer targeted the C/G SNP in the aspB gene, as described previously by Turenne et al. (4). The est and pepB gene differences (described in that same study) failed to bring the expected results.
Due to practical considerations, the two systems were combined in a duplex real-time PCR for routine use on both solid and liquid culture isolates in high-throughput laboratories.
Parallel gene scanning strengthens the trustworthiness of the method applied, and as amplification is simultaneous in a single closed tube, its implementation is easy to perform.
The stable Tm differences of the HRM curves and the low standard deviation of the Tm of the M. avium subsp. silvaticum-specific amplification products contribute to the robustness of the system.
The fact that our assay identified all 199 tested field isolates correctly makes it an accurate and reliable method for the identification of M. avium subsp. silvaticum and the discrimination of M. avium subsp. silvaticum from M. avium subsp. avium.
HRM analysis usually requires preamplification template concentration normalization. The wide input DNA range of this system is a great advantage in rendering DNA quantification redundant, thus making our assay flexible and ready to use for routine diagnostic laboratories.
As M. avium subsp. silvaticum is usually first identified in liquid cultures, and mixed cultures with other mycobacterial strains can occur, besides M. avium subsp. silvaticum identification, the ability of our assay to detect mixed M. avium subsp. avium-M. avium subsp. silvaticum cultures enhances its importance.
This is the first time that such a large panel of strains has been analyzed. In previous studies, at most 5 strains were tested, and in some cases, even the same few strains were used across various studies (31). The numerous field isolates better reflect environmental circumstances and are able to reveal potential strain differences.
The test strains originated from 17 different species, which underlines the ubiquitous nature and wide host range of M. avium subsp. avium. To our knowledge, isolation of M. avium subsp. silvaticum from wild boars, red foxes, red deer, badgers, and naturally infected cattle has not been reported previously. The infected cattle gave positive reactions by the intradermal tuberculin skin test. However, the sample size from wild animals was limited; macroscopic lesions were found in 35% (20/57) of samples, of which 65% (13/20) resembled tuberculotic lesions in their microscopic structure.
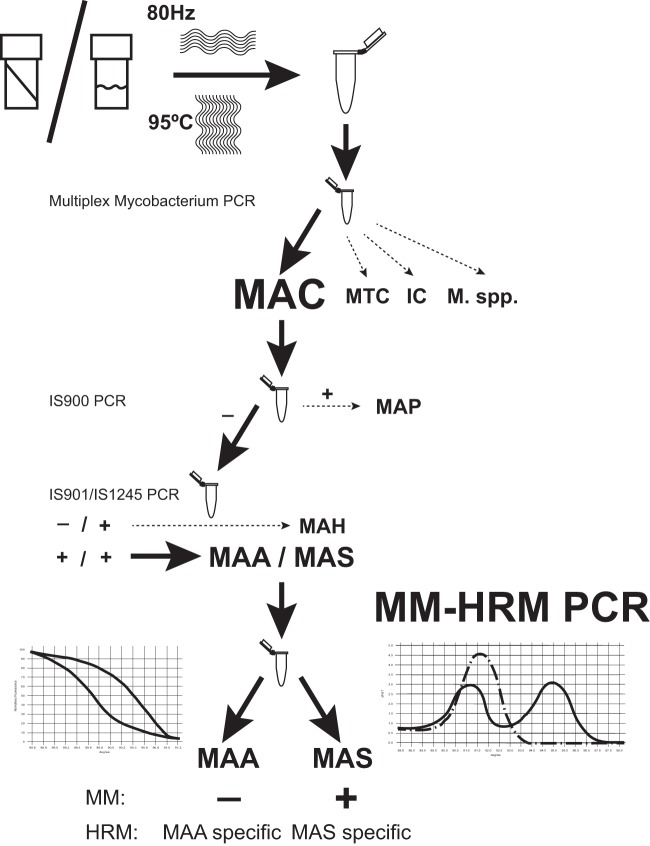
Considering the results of specificity testing, it can be stated that the specificity of the assay is high, but we recommend previous MAC identification and IS900, IS901, and IS1245 characterization steps for M. avium subsp. paratuberculosis and “M. avium subsp. hominissuis” discrimination (Fig. 3).
FIG 3.
Flow chart for the recommended mycobacterial strain characterization with special emphasis on Mycobacterium avium subspecies identification. The steps of the potential Mycobacterium avium complex subspecies-level identification method are presented in the flow chart. Abbreviations: MTC, Mycobacterium tuberculosis complex; MAC, Mycobacterium avium complex; MI, Mycobacterium intracellulare; M., Mycobacterium; MAP, Mycobacterium avium subsp. paratuberculosis; MAH, “Mycobacterium avium subsp. hominissuis”; MAA, Mycobacterium avium subsp. avium; MAS, Mycobacterium avium subsp. silvaticum.
Altogether, a reliable, rapid, and robust M. avium subsp. silvaticum identification method was developed, which eliminates the long-existing problem of the ambiguous phenotypic M. avium subsp. silvaticum identification and opens up the possibility of detailed and comprehensive M. avium subsp. silvaticum strain studies.
Supplementary Material
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JCM.03556-14.
REFERENCES
- 1.Pavlik I, Falkinham JO III, Kazda J. 2009. Potentially pathogenic mycobacteria, p 31–40. In Kazda J, Pavlik I, Falkinham JO III, Hruska K (ed), The ecology of mycobacteria: impact on animal's and human's health. Springer, New York, NY. [Google Scholar]
- 2.Mijs W, de Haas P, Rossau R, Van der Laan T, Rigouts L, Portaels F, van Soolingen D. 2002. Molecular evidence to support a proposal to reserve the designation Mycobacterium avium subsp. avium for bird-type isolates and ‘M. avium subsp hominissuis’ for the human/porcine type of M. avium. Int J Syst Evol Microbiol 52:1505–1518. doi: 10.1099/ijs.0.02037-0. [DOI] [PubMed] [Google Scholar]
- 3.Thorel MF, Krichevsky M, Lévy-Frébault VV. 1990. Numerical taxonomy of mycobactin-dependent mycobacteria, emended description of Mycobacterium avium, and description of Mycobacterium avium subsp. avium subsp. nov., Mycobacterium avium subsp. paratuberculosis subsp. nov., and Mycobacterium avium subsp. silvaticum subsp. nov. Int J Syst Bacteriol 40:254–260. [DOI] [PubMed] [Google Scholar]
- 4.Turenne CY, Collins DM, Alexander DC, Behr MA. 2008. Mycobacterium avium subsp. paratuberculosis and M. avium subsp. avium are independently evolved pathogenic clones of a much broader group of M. avium organisms. J Bacteriol 190:2479–2487. doi: 10.1128/JB.01691-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Saxegaard F, Baess I. 1988. Relationship between Mycobacterium avium, Mycobacterium paratuberculosis and “wood pigeon mycobacteria” determinations by DNA-DNA hybridization. APMIS 96:37–42. doi: 10.1111/j.1699-0463.1988.tb05265.x. [DOI] [PubMed] [Google Scholar]
- 6.Kiehn TE, Edwards FF, Brannon P, Tsang AY, Maio M, Gold JW, Whimbey E, Wong B, McClatchy JK, Armstrong D. 1985. Infections caused by Mycobacterium avium complex in immunocompromised patients: diagnosis by blood culture and fecal examination, antimicrobial susceptibility tests, and morphological and seroagglutination characteristics. J Clin Microbiol 21:168–173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Moravkova M, Hlozek P, Beran V, Pavlik I, Preziuso S, Cuteri V, Bartos M. 2008. Strategy for the detection and differentiation of Mycobacterium avium species in isolates and heavily infected tissues. Res Vet Sci 85:257–264. doi: 10.1016/j.rvsc.2007.10.006. [DOI] [PubMed] [Google Scholar]
- 8.Castellanos E, Aranaz A, De Buck J. 2010. PCR amplification and high-resolution melting curve analysis as a rapid diagnostic method for genotyping members of the Mycobacterium avium-intracellulare complex. Clin Microbiol Infect 16:1658–1662. doi: 10.1111/j.1469-0691.2010.03198.x. [DOI] [PubMed] [Google Scholar]
- 9.Bull TJ, Sheridan JM, Martin H, Sumar N, Tizard M, Hermon-Taylor J. 2000. Further studies on the GS element. A novel mycobacterial insertion sequence (IS1612) inserted into an acetylase gene (mpa) in Mycobacterium avium subsp. silvaticum but not in Mycobacterium avium subsp. paratuberculosis. Vet Microbiol 77:453–463. doi: 10.1016/S0378-1135(00)00330-8. [DOI] [PubMed] [Google Scholar]
- 10.Legrand E, Sola C, Rastogi N. 1999. Le complexe Mycobacterium avium-intracellulare: marqueurs phénotypiques et génotypiques et les bases moléculaires de la transmission inter-espèces. Manuscript 2155/RIP 7. 3e Colloque du Réseau International des Instituts Pasteur et Instituts Associés, 14 to 15 October 1999. Institut Pasteur de Paris, Paris, France: http://www.pathexo.fr/documents/articles-bull/T93-3-2155-RIP7.pdf. [Google Scholar]
- 11.Moss MT, Malik ZP, Tizard MLV, Green EP, Sanderson JD, Hermon-Taylor J. 1992. IS902, an insertion element of the chronic-enteritis-causing Mycobacterium avium subsp. silvaticum. J Gen Microbiol 138:139–145. doi: 10.1099/00221287-138-1-139. [DOI] [PubMed] [Google Scholar]
- 12.Rindi L, Garzelli C. 2014. Genetic diversity and phylogeny of Mycobacterium avium. Infect Genet Evol 21:375–383. doi: 10.1016/j.meegid.2013.12.007. [DOI] [PubMed] [Google Scholar]
- 13.Shin SJ, Lee BS, Koh W-J, Manning EJB, Anklam K, Sreevatsan S, Lambrecht RS, Collins MT. 2010. Efficient differentiation of Mycobacterium avium complex species and subspecies by use of five-target multiplex PCR. J Clin Microbiol 48:4057–4062. doi: 10.1128/JCM.00904-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Orrú G, Meloni M, Spissu F, Isola D, Palmieri G, Melis E, Besharati E, Liciardi M. 2007. Rilevamento di Mycobacterium avium subsp. silvaticum in campioni clinici di ovino mediante PCR real time. Large Anim Rev 13:13–17. [Google Scholar]
- 15.van Soolingen D, Bauer J, Ritacco V, Leão SC, Pavlik I, Vincent V, Rastogi N, Gori A, Bodmer T, Garzelli C, Garcia MJ. 1998. IS1245 restriction fragment length polymorphism typing of Mycobacterium avium isolates: proposal for standardization. J Clin Microbiol 36:3051–3054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dvorska L, Bull TJ, Bartos M, Matlova L, Svastova P, Weston RT, Kintr J, Parmova I, van Soolingen D, Pavlik I. 2003. A standardised restriction fragment length polymorphism (RFLP) method for typing Mycobacterium avium isolates links IS901 with virulence for birds. J Microbiol Methods 55:11–27. doi: 10.1016/S0167-7012(03)00092-7. [DOI] [PubMed] [Google Scholar]
- 17.Paustian ML, Zhu X, Sreevatsan S, Robbe-Austerman S, Kapur V, Bannantine JP. 2008. Comparative genomic analysis of Mycobacterium avium subspecies obtained from multiple host species. BMC Genomics 9:135–150. doi: 10.1186/1471-2164-9-135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cayrou C, Turenne C, Behr MA, Drancourt M. 2010. Genotyping of Mycobacterium avium complex organisms using multispacer sequence typing. Microbiology 156:687–694. doi: 10.1099/mic.0.033522-0. [DOI] [PubMed] [Google Scholar]
- 19.Tran QT, Han XY. 2014. Subspecies identification and significance of 257 clinical strains of Mycobacterium avium. J Clin Microbiol 52:1201–1206. doi: 10.1128/JCM.03399-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zeinzinger J, Pietzka AT, Stöger A, Kornschober C, Kunert R, Allerberger F, Mach R, Ruppitsch W. 2012. One-step triplex high-resolution melting analysis for rapid identification and simultaneous subtyping of frequently isolated Salmonella serovars. Appl Environ Microbiol 78:3352–3360. doi: 10.1128/AEM.07668-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hsu CY, Wu CW, Talaat AM. 2011. Genome-wide sequence variation among Mycobacterium avium subspecies paratuberculosis isolates: a better understanding of Johne's disease transmission dynamics. Front Microbiol 2:236. doi: 10.3389/fmicb.2011.00236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wilton S, Cousins D. 1992. Detection and identification of multiple mycobacterial pathogens by DNA amplification in a single tube. Genome Res 1:269–273. doi: 10.1101/gr.1.4.269. [DOI] [PubMed] [Google Scholar]
- 23.Álvarez J, García IG, Aranaz A, Bezos J, Romero B, de Juan L, Mateos A, Gómez-Mampaso E, Domínguez L. 2008. Genetic diversity of Mycobacterium avium isolates recovered from clinical samples and from the environment: molecular characterization for diagnostic purposes. J Clin Microbiol 46:1246–1251. doi: 10.1128/JCM.01621-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Castellanos E, Aranaz A, de Juan L, Alvarez J, Rodrígez S, Romero B, Bezos J, Stevenson K, Mateos A, Domínguez L. 2009. Single nucleotide polymorphisms in the IS900 sequence of Mycobacterium avium subsp. paratuberculosis are strain type specific. J Clin Microbiol 47:2260–2264. doi: 10.1128/JCM.00544-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Turenne CY, Wallace R Jr, Behr MA. 2007. Mycobacterium avium in the postgenomic era. Clin Microbiol Rev 20:205–229. doi: 10.1128/CMR.00036-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Matthews PRJ, McDiarmid A. 1979. The production in bovine calves of a disease resembling paratuberculosis with a Mycobacterium sp. isolated from a woodpigeon (Columba palumbus L). Vet Rec 104:286. [DOI] [PubMed] [Google Scholar]
- 27.Moss MT, Sanderson JD, Tizard MLV, Hermon-Taylor J, el-Zaatari FA, Markesich DC, Graham DY. 1992. Polymerase chain reaction detection of Mycobacterium paratuberculosis and Mycobacterium avium subsp silvaticum in long term cultures from Crohn's disease and control tissues. Gut 33:1209–1213. doi: 10.1136/gut.33.9.1209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.McFadden J, Collins J, Beaman B, Arthur M, Gitnick G. 1992. Mycobacteria in Crohn's disease: DNA probes identify the wood pigeon strain of Mycobacterium avium and Mycobacterium paratuberculosis from human tissue. J Clin Microbiol 30:3070–3073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chiers K, Deschaght P, De Baere T, Dabrowski S, Kotlowski R, De Clercq D, Ducatelle R, Vaneechoutte M. 2012. Isolation and identification of Mycobacterium avium subspecies silvaticum from a horse. Comp Immunol Microbiol Infect Dis 35:303–307. doi: 10.1016/j.cimid.2012.01.011. [DOI] [PubMed] [Google Scholar]
- 30.Higgins J, Camp P, Farrell D, Bravo D, Pate M, Robbe-Austerman S. 2011. Identification of Mycobacterium spp. of veterinary importance using rpoB gene sequencing. BMC Vet Res 7:77. doi: 10.1186/1746-6148-7-77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Turenne CY, Semret M, Cousins DV, Collins DM, Behr MA. 2006. Sequencing of hsp65 distinguishes among subsets of the Mycobacterium avium complex. J Clin Microbiol 44:433–440. doi: 10.1128/JCM.44.2.433-440.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.