Abstract
Cepacia syndrome (CS) is a fatal septic condition that develops in approximately 20% of cystic fibrosis (CF) patients chronically infected with the Burkholderia cepacia complex (Bcc). The most common causative agent is Burkholderia cenocepacia, a clinically dominant Bcc species that contains the globally distributed epidemic strain sequence type 32 (ST32). Using microarrays, we compared the transcriptomes of ST32 isolates from the bloodstream at the time of CS with their sputum counterparts recovered 1 to 2 months prior to the development of CS. Global gene expression profiles of blood isolates revealed greater activities of the virulence genes involved in the type III secretion system, the bacterial exopolysaccharide cepacian, and quorum sensing, while reduced expression was demonstrated for flagellar genes. Furthermore, a nonmotile phenotype (as evaluated by a swimming motility assay) was identified in blood isolates from 6 out of 8 patients with CS; this phenotype was traceable to 24 months prior to the onset of CS. Loss of motility was not observed in any of the 89 ST32 isolates recovered over the course of chronic infection from 17 patients without CS. In conclusion, the gene expression of Bcc bacteria disseminated during CS has been elucidated for the first time. This study demonstrated marked differences at the transcriptome level between isogenic ST32 isolates that are attributable to the stage and site of infection. The finding of a nonmotile B. cenocepacia isolate may serve as a warning sign for the development of CS in the near future.
INTRODUCTION
Patients with cystic fibrosis (CF) spend a lifetime at risk of contracting bacteria from the Burkholderia cepacia complex (Bcc), consisting of a group of 18 genetically closely related bacterial species (1). These microorganisms usually cause chronic respiratory infections in CF patients and result in little chance of treatment success due to their intrinsic resistance to most antimicrobials (2). Moreover, they pose a high risk for the development of a fatal clinical condition termed cepacia syndrome (CS). Because of this unfavorable outcome, Bcc species are considered particularly troublesome CF pathogens associated with not only increased morbidity but also increased mortality (3). Additionally, they spread relatively easily among CF patients and have resulted in several serious Bcc outbreaks in the past. A multicenter outbreak was reported for epidemic lineage ET12 (4), while in a local epidemic identified at the Prague CF Centre in the early 2000s, 30% of the CF population was found to be infected with a single Bcc strain. This strain was designated by multilocus sequence typing (MLST) (5) as sequence type 32 (ST32).
Cepacia syndrome (CS) is a worrying terminal phase of Bcc infection characterized by pulmonary exacerbation, high levels of inflammatory markers, new multifocal lung infiltrates visible by chest X-ray, and a positive blood culture for Bcc bacteria. CS is linked mostly to Burkholderia cenocepacia, a species that is clinically dominant among the Bcc members and that includes the infamous epidemic ET12 clone and ST32 strain. Nevertheless, the capacity to enter the bloodstream from the lung of a CF patient is not restricted to B. cenocepacia. Indeed, other Bcc species that have been implicated as the causative agents of sepsis are Burkholderia dolosa (6) and Burkholderia multivorans (7).
Empirical evidence indicates that approximately 20% of Bcc-infected patients die due to CS and that the preceding duration of the infection can vary from weeks (E. Tullis, unpublished data) to decades (8). Despite its clinical relevance, the mechanisms underlying the onset and development of CS remain unknown. We have been unable to pinpoint any apparent trigger or predict for which patients and under which circumstances the greatest risk of CS occurs.
To unravel the bacterial factors that are possibly associated with CS, we performed a microarray-based set of experiments to investigate the global gene expression changes characteristic of B. cenocepacia strain ST32. The bloodstream bacterial isolates were found to express higher levels of genes involved in the type III secretion system and the exopolysaccharide cepacian and suppress the activity of the flagellar system. Interestingly, changes in bacterial motility were almost exclusively observed in cases of ST32 infection that resulted in CS.
MATERIALS AND METHODS
Patients and bacterial isolates.
This retrospective study included 128 sputum and 9 blood isolates of B. cenocepacia ST32 recovered from 25 adult CF patients (8 of them presented with CS with blood culture positivity) (Table 1; see also Table S1 in the supplemental material). As part of our routine microbiological investigation of CF samples, each isolate was identified based on its growth on B. cepacia selective agar (BCSA; Oxoid) and confirmed by Bcc-specific PCR. Species and strain identifications were determined by MLST or ST32-specific PCR (9).
TABLE 1.
B. cenocepacia ST32 isolates used in this study from patients with CS
| Patient with CS | Sitea and collection date (mo/yr) for sequential isolate no.: |
||||||||
|---|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | |
| A | Sputum 1/2002 | Sputum 1/2010 | Blood 3/2010 | ||||||
| B | Sputum 3/2003 | Sputum 10/2008 | Sputum 1/2009 | Sputum 10/2009 | Sputum 2/2010 | Sputum 7/2010 | Sputum 10/2010 | Blood 11/2010 | |
| C | Sputum 3/2002 | Sputum 1/2009 | Sputum 11/2009 | Sputum 2/2010 | Sputum 12/2011 | Blood 2/2012 | |||
| D | Sputum 1/2002 | Sputum 12/2006 | Sputum 04/2007 | Sputum 1/2009 | Sputum 10/2009 | Sputum 2/2010 | Blood 4/2010 | Blood 4/2010 | |
| E | Sputum 1/2004 | Sputum 9/2011 | Sputum 4/2012 | Sputum 4/2013 | Sputum 10/2013 | Blood 10/2013 | |||
| F | Sputum 11/2009 | Sputum 2/2011 | Blood 3/2011 | ||||||
| G | Sputum 3/2002 | Sputum 10/2009 | Sputum 2/2010 | Sputum 8/2010 | Blood 11/2010 | ||||
| H | Sputum 3/2006 | Sputum 5/2006 | Sputum 5/2007 | Sputum 1/2008 | Sputum 10/2009 | Sputum 5/2010 | Blood 11/2010 | Sputum 12/2010 | Sputum 4/2011 |
Isolates were recovered from a sputum sample (“sputum”) or blood culture (“blood”).
Global gene expression analyses were performed on two pairs of ST32 isolates from two patients (patient A, a female who died after 16 years of Bcc infection, and patient B, a male who died after 12 years of Bcc infection). One isolate per pair was recovered from blood at the time of CS (isolates A3 and B8 [Table 1]), while the other (isolates A2 and B7) was isolated from sputum 43 and 28 days earlier, respectively.
Microarrays. (i) Experimental design.
We worked with two-color custom 4x44K microarrays (ArrayExpress A-MEXP-1613; Agilent Technologies) designed for B. cenocepacia strain J2315 (10) and completed with unique features of CF strain AU1054 (11). The experimental layout and raw data were stored in ArrayExpress E-MTAB-3133. Briefly, we ran 36 microarrays, including 9 arrays for each of the 4 isolates tested. A Cy3 layer of the microarrays was occupied with Cy3-labeled J2315 DNA as a reference as described in reference 12 and used for robust signal normalization across all of the arrays.
(ii) CF sputum and serum samples.
Each type of clinical specimen used for the production of incubation medium was obtained from adult CF patients who provided informed consent. Sputum samples were immediately transferred to a −20°C freezer; whole-blood samples were left up to 5 days at 4°C. Separated serum was kept at −20°C.
(iii) Growth conditions.
Every isolate was subjected in triplicate to the following incubation conditions: (i) 10% (wt/vol) pooled CF sputum diluted in a basal salts medium (BSM) (13) (growth condition “sputum”), (ii) heat-inactivated (by exposure to 56°C for 30 min) pooled CF serum (growth condition “serum”), and (iii) BSM (“control”). For incubation in CF sputum, we followed a previously published protocol (13) with several modifications: (i) pooling of sputa or sera from 5 patients, (ii) total length of incubation set to 270 min, and (iii) processing after the cool down in liquid nitrogen, followed by centrifugation (5 min at 160 × g, 4°C), pellet vortexing with 1 ml of TRIzol (Ambion), and incubation at an ambient temperature for 5 min. Next, 0.2 ml of chloroform was added, the samples were incubated for 3 min and centrifuged for 15 min (160 × g, 4°C), and the upper water phase was transferred to 500 μl of ice-cooled 70% ethanol.
(iv) RNA processing.
RNA was extracted with the RiboPure-Bacteria kit (Ambion), incubated with DNase I (Ambion) for 1 h at 37°C, concentrated with 7.5 M lithium chloride (Ambion), and purified with the MICROBEnrich kit (Ambion) according to the manufacturer's instructions. The quality of the resulting mRNA was determined using total RNA Nano chips on a Bioanalyzer 2100 (Agilent). To label the samples with fluorescent dye, we applied the SuperScript Indirect cDNA labeling system (Invitrogen) and Cy5 or Cy3 dye (GE Healthcare). The hybridization conditions and the washing and scanning of the microarrays were performed as previously described (13).
(v) Analysis.
To preprocess the microarray data, we imported the data into R (www.R-project.org) using the function read.maimages from the package limma (14). Data were background corrected, normalized within arrays by loess, and normalized between arrays using the Gquantile method. Normalized red-channel data were analyzed with GeneSpring GX v.12 (Agilent) as single-color microarray data. Genes with statistically significant alterations in expression were identified by applying a 2-fold change filter and an adjusted P value of 0.05 (moderated t test, Benjamini Hochberg false-discovery rate [FDR]).
Swimming motility assay.
To measure bacterial swimming activity, we used Luria-Bertani (LB) broth plates containing 0.3% (wt/vol) Bacto agar (Sigma-Aldrich). We inoculated 2 μl of bacteria grown in LB broth (optical density at 260 nm [OD600], 1) into the central part of the soft agar by piercing the agar surface and letting the bacteria grow at 37°C for up to 120 h. The extent of motility was determined by measuring the diameter of the bacterial halos. For the purpose of this study, we defined a considerable decrease in motility as a first occurrence of nonmotility or decrease in motility by a minimum of 40 mm over the last 12 months.
Quantitative PCR.
Changes in gene expression were examined for 6 selected genes in 21 longitudinal isolates collected from 8 patients with CS. The minimal time interval between the collections of two isolates from a single patient was set to 1 month.
All isolates were cultivated in BSM and their RNAs were extracted as described above. cDNA was synthesized using the QuantiTect reverse transcription kit (Qiagen), and the PCR was performed using the QuantiTect SYBR green PCR kit (Qiagen). The recA gene was used as a reference based on its stable and unchanged expression across the conditions tested by microarrays.
RESULTS AND DISCUSSION
Microarray analysis.
To compare transcriptomes, we chose a hybridization-based approach that has been frequently applied in Bcc research (15). Because we utilized a microarray designed for B. cenocepacia strain J2315 (a member of ET12), we first evaluated its suitability for the ST32 strain. Based on preliminary results from ST32 genome sequencing (provided by M. Kolar and H. Strnad from IMG, unpublished data), the J2315 chip possessed 6,755 probes with close matches to ST32 sequences. Of these, 5,436 represented putative coding sequences (CDS), and 1,049 were localized within intergenic regions. Presuming that the ST32 strain has a total of 7,150 CDS (based on results of in silico comparative analysis of the ST32 sequence with the whole genomes of the J2315 [10], HI2424, and AU1054 [11] strains), the microarray used in this study enabled the detection of approximately 76% of all ST32 CDS. This calculation revealed an apparent limitation of the microarray-generated data in that one-quarter of the ST32-specific genes would be absent from the array. However, in this study, we aimed to identify key virulence factors; most of these genes are likely common between the genomes of the highly problematic B. cenocepacia clinical strains and, thus, are detectable using this array. Furthermore, analogous data-mining approaches for strains other than J2315 were proven to be valid in several other studies (performed for the H111 [16] and IST439 [17] strains).
A total of 36 arrays were performed to capture expression profiles from two sputum and two blood culture isolates that were each incubated in triplicate in (i) sputum, (ii) serum, and (iii) control medium. The four isolates formed two pairs that had been recovered from two adult CF patients. While the first isolates per pair were obtained from sputum samples (isolates A2 and B7 [Table 1]), the remaining two isolates (isolates A3 and B8) came from blood cultures at the time of CS.
Our experimental layout enabled the comparison of two different test conditions and the identification of changes in gene expression as consequences of either bacterial exposure to laboratory-defined growth conditions (sputum or serum) or the site of infection from which they were recovered (lung or bloodstream). The number of genes with altered expression levels for either of the two conditions is summarized in Tables 2 and 3; these tables also propose various meaningful subvariants of the analyses for each condition (i.e., merged or broken down to individual patients).
TABLE 2.
Summary of microarray data analyses applied and the number of genes with altered expression when focused on growth conditiona
| Parameter | No. of altered genes | No. of upregulated genes | No. of down-regulated genes |
|---|---|---|---|
| Growth condition sputum | |||
| Isolates obtained from blood culture | |||
| A3 and B8 isolates together | 30 | 22 | 8 |
| A3 only | 61 | 37 | 24 |
| B8 only | 163 | 93 | 70 |
| Isolates obtained from sputum culture | |||
| A2 and B7 isolates together | 208 | 89 | 119 |
| A2 only | 262 | 141 | 121 |
| B7 only | 359 | 153 | 206 |
| Isolates obtained from both blood and sputum | |||
| A3, B8, A2, and B7 | 157 | 70 | 87 |
| Growth condition serum | |||
| Isolates obtained from blood culture | |||
| A3 and B8 isolates together | 169 | 90 | 79 |
| A3 only | 67 | 33 | 34 |
| B8 only | 304 | 137 | 167 |
| Isolates obtained from sputum culture | |||
| A2 and B7 isolates together | 374 | 177 | 197 |
| A2 only | 488 | 200 | 288 |
| B7 only | 381 | 186 | 195 |
| Isolates obtained from both blood and sputum | |||
| A3, B8, A2, and B7 | 265 | 134 | 131 |
Gene changes observed while the same isolate was incubated in sputum or serum compared with the control medium.
TABLE 3.
Summary of microarray data analyses applied and the number of genes with altered expression when focused on the site of infectiona
| Parameter | No. of altered genes | No. of upregulated genes | No. of down-regulated genes |
|---|---|---|---|
| Incubated in sputum | |||
| Isolates of patients A and B together (A3 and B8 vs A2 and B7) | 41 | 31 | 10 |
| Isolates of patient A only (A3 vs A2) | 210 | 113 | 97 |
| Isolates of patient B only (B8 vs B7) | 139 | 90 | 49 |
| Incubated in serum | |||
| Isolates of patients A and B together (A3 and B8 vs A2 and B7) | 20 | 13 | 7 |
| Isolates of patient A only (A3 vs A2) | 145 | 99 | 46 |
| Isolates of patient B only (B8 vs B7) | 90 | 47 | 53 |
| Incubated in control medium | |||
| Isolates of patients A and B together (A3 and B8 vs A2 and B7) | 57 | 24 | 33 |
| Isolates of patient A only (A3 vs A2) | 34 | 28 | 6 |
| Isolates of patient B only (B8 vs B7) | 188 | 92 | 96 |
| Irrespective of culture medium | |||
| Isolates of patients A and B together (A3 and B8 vs A2 and B7) | 60 | 41 | 19 |
| Isolates of patient A only (A3 vs A2) | 95 | 65 | 30 |
| Isolates of patient B only (B8 vs B7) | 129 | 63 | 66 |
Gene changes observed between blood and sputum isolates.
With respect to the aim of the study, we focused on the biological origin of the isolate to pinpoint differences in gene activities of the ST32 isolate found in blood (expression changes inherent to CS) as opposed to the ST32 isolate isolated from a CF patient's lung (changes typical of localized infection). Specifically, we performed a subanalysis comparing 18 technical replicates of blood isolates (a combined set of two isolates in growth medium sputum, serum, and control; each in triplicate) with 18 technical replicates of sputum isolates (a subanalysis represented in the “Irrespective of culture medium” rows of Table 3). This approach filtered out any possible effects of the growth media that mimicked in vivo conditions and revealed 60 genes with minimal 2-fold changes and P values lower than 0.05 in their expression (Table 4).
TABLE 4.
Genes resulting from comparison of blood isolates with sputum isolatesa
| Gene | Annotation or putative gene function | Fold change |
|---|---|---|
| Metal ion transport or metabolism | ||
| BCAM2233 | Putative pyochelin biosynthetic protein PchC | 2.7 ↓ |
| BCAM0810 | Putative aromatic oxygenase | 2.5 ↑ |
| BCAM0811 | Putative aromatic oxygenase | 3.1 ↑ |
| Carbohydrate transport and metabolism | ||
| BCAL2793 | Major facilitator superfamily protein | 3.5 ↓ |
| BCAL2472 | Alpha, alpha-trehalose-phosphate synthase, otsA | 2.1 ↑ |
| BCAM0783 | Major facilitator superfamily protein | 3.4 ↑ |
| Cell envelope biogenesis, membrane proteins | ||
| BCAM0081 | Putative outer membrane protein-OmpW family | 2.4 ↓ |
| BCAL1391 | Cellulose biosynthesis protein | 2.1 ↓ |
| BCAL1395 | Cellulose synthase catalytic subunit, bscA | 2.9 ↓ |
| BCAL1396 | Cellulose synthase operon protein YhjU | 2.2 ↓ |
| BCAM0805 | Muconate cycloisomerase I 1, catB1 | 2.9 ↑ |
| BCAM1004 | GDP-mannose 4,6-dehydratase, gca | 2.6 ↑ |
| BCAM1008 | Glycosyltransferase | 2.9 ↑ |
| BCAM1010 | Putative UTP-glucose-1-phosphate uridylyltransferase, gtaB | 3.5 ↑ |
| Lipid transport and metabolism | ||
| BCAM0080 | AMP-binding domain-containing protein | 2.7 ↓ |
| BCAM1005 | Putative acyltransferase | 3.2 ↑ |
| Energy production | ||
| BCAL2795 | Aldehyde dehydrogenase family protein | 2.1 ↑ |
| BCAL3184 | Homogentisate 1,2-dioxygenase, hmgA | 3.2 ↑ |
| BCAL3187 | FADb-dependent oxidoreductase | 2.4 ↑ |
| Regulatory proteins | ||
| BCAL0534 | Two-component regulatory system, response regulator protein | 2.1 ↓ |
| BCAM2329 | GntR family regulatory protein | 2.0 ↓ |
| BCAL3205a | Putative phage-related regulator | 2.1 ↓ |
| BCAM1290 | RpiR-family transcriptional regulator | 2.2 ↓ |
| BCAM0835 | AraC family regulatory protein | 3.7 ↓ |
| BCAM0784 | LysR family regulatory protein | 2.0 ↑ |
| Quorum sensing | ||
| BCAM1870 | N-Acyl-homoserine lactone synthase CepI | 3.9 ↑ |
| BCAM1871 | Hypothetical protein | 4.5 ↑ |
| Secretion | ||
| BCAM2041 | Yop virulence translocation protein R | 4.2 ↑ |
| BCAM2042 | Lytic transglycosylase | 4.1 ↑ |
| BCAM2043_J_0 | Putative uncharacterized protein | 2.4 ↑ |
| BCAM2044 | Asparagine synthetase | 3.0 ↑ |
| BCAM2046 | Type III secretion protein SctT | 2.6 ↑ |
| BCAM2048 | Type III secretion apparatus H+-transporting two-sector ATPase | 4.9 ↑ |
| BCAM2049 | BcscL, type III secretion apparatus protein | 4.9 ↑ |
| BCAM2050 | BcscK, putative uncharacterized protein | 7.3 ↑ |
| BCAM2051 | Type III secretion apparatus lipoprotein | 3.8 ↑ |
| BCAM2052 | Uncharacterized protein | 3.4 ↑ |
| BCAM2053 | Uncharacterized protein | 6.9 ↑ |
| BCAM2055 | Type III secretion outer membrane pore | 4.0 ↑ |
| Exopolysaccharide | ||
| BCAM0859 | Capsular exopolysaccharide family | 2.8 ↑ |
| BCAM0860 | Glycosyl transferase | 4.7 ↑ |
| BCAM0861 | Glycosyltransferase-like protein | 3.1 ↑ |
| BCAM0863 | Glycosyltransferase | 3.0 ↑ |
| BCAM0864 | Glycosyltransferase | 2.0 ↑ |
| BCAM0856_J_0 | Sugar transferase | 2.1 ↑ |
| BCAM0856_J_1 | Sugar transferase | 4.9 ↑ |
| Motility and adherence | ||
| BCAL1677 | Type 1 fimbrial protein | 3.4 ↓ |
| Biofilm | ||
| BCAM0184 | Fucose-binding lectin II, photopexin A | 3.3 ↑ |
| Other proteins | ||
| BCAL2025 | Hypothetical protein | 2.0 ↓ |
| BCAL2793 | Major facilitator superfamily protein | 3.5 ↓ |
| BCAL3207 | Hypothetical protein | 2.5 ↓ |
| BCAM0414 | Hypothetical protein | 2.3 ↓ |
| BCAM2685 | Hypothetical protein | 2.6 ↓ |
| BCAL3006 | Cold shock-like protein | 2.2 ↑ |
| BCAL3186 | Hypothetical protein | 2.4 ↑ |
Irrespective of culture medium used, defined in Table 3.
FAD, flavin adenine dinucleotide.
Changes in expression of virulence factors.
Blood isolates were characterized by increased expression of the gene clusters for the type III secretion system (T3SS) and the exopolysaccharide cepacian. They also overexpressed the quorum-sensing (QS) gene cepI. Interestingly, the activities of genes for the cepacian and other surface polysaccharides were most noticeable during growth in sputum (Table 2, “Growth condition sputum” rows), while T3SS activities were most pronounced in the serum environment (Table 2, “Growth condition serum” rows). Genes for flagellar assembly and siderophore pyochelin emerged from the list of downregulated genetic features. Indeed, clusters of flagellar genes (see Table S2 in the supplemental material) were found to exhibit significantly decreased expressions only in blood isolate A3 but not in B8 (discussed below).
Confirmatory quantitative PCR for the T3SS and cepacian genes provided results consistent with the microarray data for isolates of all but one patient with CS (see Table S3 in the supplemental material). Variability in the quantitative PCR results for the QS genes and nonuniformity across the isolates likely reflected a role for QS as a global regulator whose expression is very sensitive to conditions outside the cell (15, 18). Nevertheless, the expression of QS genes in isolates from patients A and B matched the microarray data.
Virulence factors, such as motility, T3SS, and exopolysaccharides, are QS dependent, and their orchestrated regulation in Bcc bacteria was previously reported (19, 20). In the context of our test condition (the biological origin of the isolate), we presumed that their significant gene expression changes had important implications for survival in the bloodstream and contributed to bacterial cell defense against the host immune system. This notion was supported by studies on another CF respiratory pathogen, Pseudomonas aeruginosa, which highlighted the protective role of T3SS (21) and a nonmotile phenotype (22) against phagocytosis. Moreover, mucoidity associated with the production of cepacian was reported to be an important factor for B. cenocepacia resistance to phagocytosis (23).
Flagella and their link to CS.
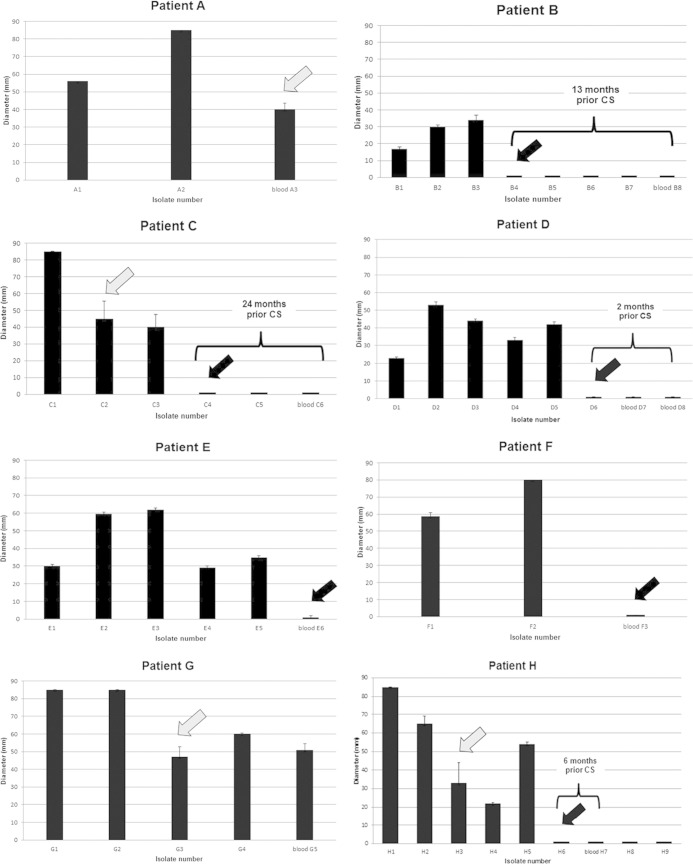
Our previous transcriptomic data from a CF sputum-based infection model indicated that the Bcc bacteria maintained their motility during chronic infection, in contrast to P. aeruginosa (13). Therefore, the downregulation of the flagellar genes in blood isolate A3 was an unexpected observation, but it was confirmed by a follow-up experiment in which a steep decrease in flagellum-mediated motility between A2 and A3 was verified using a swimming motility assay (Fig. 1).
FIG 1.
Swimming motility of isolates from patients A through H with CS. Detected changes in motility status due to nonmotility or a substantial decrease in motility are indicated with a black or gray arrow, respectively. The time period preceding CS is shown only for cases where a nonmotile sputum isolate was detected prior to CS.
Because the expression changes for flagellar genes were detectable only by microarray between the two isolates of patient A, we hypothesize that the functions of the flagella in patient B may have already been altered in earlier sputum isolates. Indeed, when we screened isolates preceding the most recent sputum isolate (B7), we discovered a nonmotile phenotype as early as 13 months prior to CS. Subsequently, the loss of motility was identified in longitudinal data from 6 of 8 patients who developed CS; isolates of the 2 remaining patients (A and G) presented a rapid decrease in their motility (>40-mm reductions of the growth zone; Fig. 1). The maximum time interval between the first detected nonmotile isolate and the last blood culture isolate was 24 months. In contrast, isolates from 17 patients who suffered from chronic ST32 infection without signs of CS (see Table S1 in the supplemental material) were all found to be motile (see Fig. S1; infection span, 4 to 12 years). A considerable and maintained decrease in motility was detected in 18% of the patients (patients L, U, and W).
These findings suggest a link between nonmotility (or a rapid change in B. cenocepacia ST32 motility) and CS (P = 0.0002, Fisher's exact test). Nonetheless, it is noteworthy that such data need to be considered strictly strain specific. A recent study on a large collection of various B. cenocepacia CF strains did not find any correlation between motility and clinical outcomes, despite the fact that 38% of the tested isolates were nonmotile (24).
We believe that a simple phenotypic test to investigate the swimming activity of ST32 isolates can serve as a useful tool for monitoring chronic infection and help identify patients at the greatest risk for developing CS, which is currently the case for patients L, U, and W, whose latest isolates expressed a marked decrease in motility (see Fig. S1 in the supplemental material).
In conclusion, we performed a thorough transcriptomic analysis of B. cenocepacia isolates recovered at the terminal stage of chronic infection in CF patients. Bacterial isolates from the bloodstream exhibited upregulated gene expressions for T3SS and cepacians compared to isolates from sputum before the onset of CS and showed reduced expressions of flagellar genes; this loss of motility was maintained in sputum isolates up to 24 months prior to the development of CS.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by the Ministry of Education (grant LD11029), the Ministry of Health (grant NT12405-5 and DRO, Motol University Hospital grant 00064203), and the Grant Agency of Charles University (grant GAUK640214), Czech Republic, and received institutional support from the Academy of Sciences of the Czech Republic (grant no. RVO: 68378050 to M.K.).
We are most grateful to Hynek Strnad from IMG for providing us with the preliminary genome annotation of strain ST32.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JCM.03605-14.
REFERENCES
- 1.Peeters C, Zlosnik JE, Spilker T, Hird TJ, Lipuma JJ, Vandamme P. 2013. Burkholderia pseudomultivorans sp. nov., a novel Burkholderia cepacia complex species from human respiratory samples and the rhizosphere. Syst Appl Microbiol 36:483–489. doi: 10.1016/j.syapm.2013.06.003. [DOI] [PubMed] [Google Scholar]
- 2.Chernish RN, Aaron SD. 2003. Approach to resistant Gram-negative bacterial pulmonary infections in patients with cystic fibrosis. Curr Opin Pulm Med 9:509–515. doi: 10.1097/00063198-200311000-00011. [DOI] [PubMed] [Google Scholar]
- 3.Mahenthiralingam E, Urban TA, Goldberg JB. 2005. The multifarious, multireplicon Burkholderia cepacia complex. Nat Rev Microbiol 3:144–156. doi: 10.1038/nrmicro1085. [DOI] [PubMed] [Google Scholar]
- 4.Govan JR, Brown PH, Maddison J, Doherty CJ, Nelson JW, Dodd M, Greening AP, Webb AK. 1993. Evidence for transmission of Pseudomonas cepacia by social contact in cystic fibrosis. Lancet 342:15–19. doi: 10.1016/0140-6736(93)91881-L. [DOI] [PubMed] [Google Scholar]
- 5.Drevinek P, Vosahlikova S, Cinek O, Vavrova V, Bartosova J, Pohunek P, Mahenthiralingam E. 2005. Widespread clone of Burkholderia cenocepacia in cystic fibrosis patients in the Czech Republic. J Med Microbiol 54:655–659. doi: 10.1099/jmm.0.46025-0. [DOI] [PubMed] [Google Scholar]
- 6.Kalish LA, Waltz DA, Dovey M, Potter-Bynoe G, McAdam AJ, Lipuma JJ, Gerard C, Goldmann D. 2006. Impact of Burkholderia dolosa on lung function and survival in cystic fibrosis. Am J Respir Crit Care Med 173:421–425. doi: 10.1164/rccm.200503-344OC. [DOI] [PubMed] [Google Scholar]
- 7.Blackburn L, Brownlee K, Conway S, Denton M. 2004. “Cepacia syndrome” with Burkholderia multivorans, 9 years after initial colonization. J Cyst Fibros 3:133–134. doi: 10.1016/j.jcf.2004.03.007. [DOI] [PubMed] [Google Scholar]
- 8.Drevinek P, Mahenthiralingam E. 2010. Burkholderia cenocepacia in cystic fibrosis: epidemiology and molecular mechanisms of virulence. Clin Microbiol Infect 16:821–830. doi: 10.1111/j.1469-0691.2010.03237.x. [DOI] [PubMed] [Google Scholar]
- 9.Dedeckova K, Kalferstova L, Strnad H, Vavrova J, Drevinek P. 2013. Novel diagnostic PCR assay for Burkholderia cenocepacia epidemic strain ST32 and its utility in monitoring infection in cystic fibrosis patients. J Cyst Fibros 12:475–481. doi: 10.1016/j.jcf.2012.12.007. [DOI] [PubMed] [Google Scholar]
- 10.Holden MT, Seth-Smith HM, Crossman LC, Sebaihia M, Bentley SD, Cerdeno-Tarraga AM, Thomson NR, Bason N, Quail MA, Sharp S, Cherevach I, Churcher C, Goodhead I, Hauser H, Holroyd N, Mungall K, Scott P, Walker D, White B, Rose H, Iversen P, Mil-Homens D, Rocha EP, Fialho AM, Baldwin A, Dowson C, Barrell BG, Govan JR, Vandamme P, Hart CA, Mahenthiralingam E, Parkhill J. 2009. The genome of Burkholderia cenocepacia J2315, an epidemic pathogen of cystic fibrosis patients. J Bacteriol 191:261–277. doi: 10.1128/JB.01230-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.LiPuma JJ, Spilker T, Coenye T, Gonzalez CF. 2002. An epidemic Burkholderia cepacia complex strain identified in soil. Lancet 359:2002–2003. doi: 10.1016/S0140-6736(02)08836-0. [DOI] [PubMed] [Google Scholar]
- 12.Dobbin K, Shih JH, Simon R. 2003. Statistical design of reverse dye microarrays. Bioinformatics 19:803–810. doi: 10.1093/bioinformatics/btg076. [DOI] [PubMed] [Google Scholar]
- 13.Drevinek P, Holden MT, Ge Z, Jones AM, Ketchell I, Gill RT, Mahenthiralingam E. 2008. Gene expression changes linked to antimicrobial resistance, oxidative stress, iron depletion and retained motility are observed when Burkholderia cenocepacia grows in cystic fibrosis sputum. BMC Infect Dis 8:121. doi: 10.1186/1471-2334-8-121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Smyth G. 2005. Limma: linear models for microarray data, p 397–420. In Gentleman R, Carey V, Dudoit S, Irizarry R, Huber W (ed), Bioinformatics and computational biology solutions using R and Bioconductor. Springer, New York, NY. [Google Scholar]
- 15.Sass A, Drevinek P, Sokol PA. 2014. Comparative transcriptomics in Burkholderia, p 77–98. In Coenye T, Mahenthiralingam E (ed), Burkholderia: from genomes to function. Caister Academic Press, Norfolk, United Kingdom. [Google Scholar]
- 16.Agnoli K, Schwager S, Uehlinger S, Vergunst A, Viteri DF, Nguyen DT, Sokol PA, Carlier A, Eberl L. 2012. Exposing the third chromosome of Burkholderia cepacia complex strains as a virulence plasmid. Mol Microbiol 83:362–378. doi: 10.1111/j.1365-2958.2011.07937.x. [DOI] [PubMed] [Google Scholar]
- 17.Mira NP, Madeira A, Moreira AS, Coutinho CP, Sa-Correia I. 2011. Genomic expression analysis reveals strategies of Burkholderia cenocepacia to adapt to cystic fibrosis patients' airways and antimicrobial therapy. PLoS One 6:e28831. doi: 10.1371/journal.pone.0028831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.O'Grady EP, Sokol PA. 2011. Burkholderia cenocepacia differential gene expression during host-pathogen interactions and adaptation to the host environment. Front Cell Infect Microbiol 1:15. doi: 10.3389/fcimb.2011.00015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Loutet SA, Di Lorenzo F, Clarke C, Molinaro A, Valvano MA. 2011. Transcriptional responses of Burkholderia cenocepacia to polymyxin B in isogenic strains with diverse polymyxin B resistance phenotypes. BMC Genomics 12:472. doi: 10.1186/1471-2164-12-472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sass A, Marchbank A, Tullis E, Lipuma JJ, Mahenthiralingam E. 2011. Spontaneous and evolutionary changes in the antibiotic resistance of Burkholderia cenocepacia observed by global gene expression analysis. BMC Genomics 12:373. doi: 10.1186/1471-2164-12-373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Diaz MR, King JM, Yahr TL. 2011. Intrinsic and extrinsic regulation of type III secretion gene expression in Pseudomonas aeruginosa. Front Microbiol 2:89. doi: 10.3389/fmicb.2011.00089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Amiel E, Lovewell RR, O'Toole GA, Hogan DA, Berwin B. 2010. Pseudomonas aeruginosa evasion of phagocytosis is mediated by loss of swimming motility and is independent of flagellum expression. Infect Immun 78:2937–2945. doi: 10.1128/IAI.00144-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Conway BA, Chu KK, Bylund J, Altman E, Speert DP. 2004. Production of exopolysaccharide by Burkholderia cenocepacia results in altered cell-surface interactions and altered bacterial clearance in mice. J Infect Dis 190:957–966. doi: 10.1086/423141. [DOI] [PubMed] [Google Scholar]
- 24.Zlosnik JE, Mori PY, To D, Leung J, Hird TJ, Speert DP. 2014. Swimming motility in a longitudinal collection of clinical isolates of Burkholderia cepacia complex bacteria from people with cystic fibrosis. PLoS One 9:e106428. doi: 10.1371/journal.pone.0106428. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.