Abstract
Access to genotyping assays to determine successful antiretroviral treatment (ART) is limited in resource-constrained settings by high cost, suggesting the need for a cost-effective and simplified method to identify HIV-1 drug resistance (HIVDR) mutations. In this study, an amplification refractory mutation system (ARMS)-PCR assay was developed and used to investigate the most frequent HIVDR mutations affecting first-line ART in settings where WHO ART guidelines are applied. Seventy-five HIV-positive (HIV+) samples from Cameroon were used to assess the performance of this assay. Sequencing of HIV-1 reverse transcriptase was simultaneously performed for comparison, and discordant samples were tested with a Trugene HIV-1 genotyping kit. The ARMS-PCR assay was able to detect M184V, T215Y/F, K103N, and Y181C mutations with sensitivities of 96.8%, 85.7%, 91.3%, and 70%, respectively, and specificities of 90.6%, 95%, 100%, 96.9%, respectively, compared with data on sequencing. The results indicated the highest positive predictive value for K103N (100%) and the highest negative predictive value for M184V (97.5%). ARMS-PCR's limits of detection for mutations M184V, T215Y/F, K103N, and Y181C were <75 copies/ml, 143 copies/ml, 143 copies/ml, and 836 copies/ml, respectively. ARMS-PCR efficiently identified mutations in individuals harboring different HIV-1 clades (CRF02_AG and non-CRF02_AG). In addition, this approach was more cost-effective than other genotyping assays. The high throughput, the cost-effectiveness, and the simplicity of the ARMS-PCR assay make it a suitable tool to monitor HIVDR patterns in resource-constrained settings with broad HIV-1 genetic diversity.
INTRODUCTION
In recent years, significant progress has been made in the treatment of HIV-infected patients. Gains in antiretroviral treatment have been achieved in low- and middle-income countries with more than 9.7 million people living with HIV/AIDS, 7.2 million of whom lived in sub-Saharan Africa as of the end of 2012 (35). This scaling up of antiretroviral therapy in resource-constrained settings, where the logistics of medication disbursement and patient follow-up are not always optimal, results in the emergence and transmission of HIV drug-resistant variants, thus providing obstacles to successful antiretroviral treatment (ART) programs (1). The monitoring of HIV drug-resistant mutations in developed countries is an important aspect of clinical management of HIV infection. In these developed countries, sequencing-based drug resistance assays are used to select the most appropriate regimens when initiating or switching ART (2, 3). In resource-constrained settings, due to high cost, only a limited number of HIV drug resistance mutation tests are performed for patient management. In such countries, the majority of tests are used for research purposes or for surveillance of HIV-1 drug resistance (HIVDR) mutations recommended by WHO to update antiretroviral therapy policies (4, 5).
Resource-constrained settings use the WHO public health approach for the treatment of HIV-infected patients. This approach recommends the standardized and simplified treatment protocol consisting of two nucleoside reverse transcriptase (RT) inhibitors (NRTI) plus one non-NRTI (NNRTI) for first-line therapy. Treatment initiation or switching is based on clinical parameters and CD4 count because viral load (VL) assessment is not feasible for the majority of patients (6, 7). In addition to these challenges, the low genetic barrier of commonly used antiretrovirals (ARV) for first-line treatment further increases the risks of HIVDR mutations. The most prevalent mutations that cause intermediate- to high-level resistance to this first-line regimen in countries such as those in the central African region, including Cameroon, Central African Republic, Gabon, etc., that use the WHO approach are M184V (37% to 90%), T215Y/F (11% to 47.2%), K103N (14% to 44%), and Y181C (9.4% to 19.8%) (8–13). These can restrict treatment options and increase cost by requiring new and more expensive ARV regimens (36).
The detection of HIVDR mutations essentially depends on genotyping assays. However, their use is limited in resource-constrained settings because these assays require sophisticated equipment such as a genetic analyzer, costly reagents, and highly sophisticated manpower. Some laboratories in developing countries have developed and validated highly sensitive in-house HIV-1 genotyping assays comparable to FDA-approved assays to reduce the cost and to overcome subtype specificities (1, 14, 15). Despite these efforts, these in-house genotyping assays still remain unaffordable to the majority of patients in need.
Point mutation assays have been developed as alternatives for genotyping assays. These assays are allele specific and include mutant-allele-specific amplification (MASA), PCR amplification of specific alleles (PASA), and amplification refractory mutation system (ARMS)-PCR (12, 13, 16–18). These methods utilize the difference in extension efficiency between primers with matched and mismatched 3′ bases. Product detection and identification are the most variable steps that contribute to the throughput capacity of each particular method. The drawback with some of these assays is the detection method, such as fluorescence resonance energy transfer, fluorescence polarization, luminescence, or absorbance (19–21), requiring expensive equipment and reagents which still have limited use in resource-constrained settings. Thus, we have developed and evaluated the use of a simplified ARMS-PCR to detect the most frequent HIV-1 drug resistance mutations in individuals infected with strains of broad HIV-1 genetic diversity.
MATERIALS AND METHODS
Ethical considerations.
This study was approved by the Institutional Ethical Review Board of New York University School of Medicine, New York, USA, and by the Institutional Review Board of Cameroon's Ministry of Public Health. Written informed consent was obtained from all the study participants.
Characteristics of the study patients.
The samples analyzed in this study are part of an ongoing project in Cameroon aimed at analyzing viral evolution and humoral immune response to dual HIV-1 infection. Plasma samples were included from 75 HIV-1-infected patients in the study whose demographics and clinical characteristics are described in Table 1. Forty-eight (64%) of them were women, and the median age was 38 years (interquartile range [IQR], 33.5 to 45). Thirty-three (44%) patients were ART naive, 39 (52%) were receiving first-line ART, and three (4%) were receiving second-line ART. The median times on ART were 15 months (IQR, 11 to 26.5) for subjects on first-line ART and 32 months (IQR, 26 to 38) for patients receiving first-line ART. The median viral loads (VL) were 9,373 copies/ml for naive patients (IQR, 2,878 to 18,762), 17,314 (IQR, 7,031 to 34,927) for patients on first-line ART, and 14,535 (IQR, 8,890.5 to 16,394) for patients on second-line ART. Among the patients receiving first-line ART, combinations of zidovudine (AZT) plus lamivudine (3TC) plus nevirapine (NVP) and efavirenz (EFV) (NVP/EFV), tenofovir (TDF) plus 3TC plus NVP/EFV, stavudine (d4T) plus 3TC plus NVP, and 3TC plus NVP were taken by 28%, 18.6%, 2.6% and 2.6%, respectively. Among the patients receiving second-line ART, TDF plus 3TC plus lopinavir/ritonavir (LPV/r) and TDF plus 3TC plus ATV were taken by 2.6% and 1.3%, respectively.
TABLE 1.
Demographics and characteristics of the HIV-1-infected individuals studieda
| Characteristic | Values |
||
|---|---|---|---|
| Female | Male | Total | |
| Demographics | |||
| No. (%) | 48 (64) | 27 (36) | 75 (100) |
| Mean age (yrs) (IQR) | 37.5 (33.25–45) | 38 (34–45) | 38 (33.5–45) |
| VL [no. of copies/ml (%)] | |||
| VL < 75 | 4 (8.3) | 1 (3.7) | 5 (6.6) |
| 75 < VL < 5,000 | 17 (35.4) | 5 (18.5) | 22 (29.3) |
| VL > 5,000 | 27 (56.2) | 21 (77.7) | 48 (64) |
| ART (no. [%]) | |||
| Naïve | 22 (45.8) | 11 (40.7) | 33 (44) |
| First-line therapy | |||
| AZT + 3TC + NVP/EFV | 13 (27) | 8 (29.6) | 21 (28) |
| TDF + 3TC + NVP/EFV | 9 (18.7) | 5 (18.5) | 14 (18.6) |
| 3TC + NVP | 1 (2) | 1 (3.7) | 2 (2.6) |
| d4T + NVP | 1 (2) | 1 (3.7) | 2 (2.6) |
| Second-line therapy | |||
| TDF + 3TC + ATV | 1 (2) | 0 (0) | 1 (1.3) |
| TDF + 3TC + LPV/r | 1 (2) | 1 (3.7) | 2 (2.6) |
| Duration on ART (no. of mos (IQR) | |||
| First-line therapy | 15.5 (10.5–24.75) | 15 (10.5–27.25) | 15 (11–26.25) |
| Second-line therapy | 32 (20–44) | 36 (36–36) | 32 (26–38) |
ART, antiretroviral treatment; IQR, interquartile; AZT, zidovudine; 3TC, lamivudine; TDF, tenofovir; d4T, stavudine; NVP, nevirapine; EFV, efavirenz; ATV, atazanavir; LPV/r, lopinavir boosted with ritonavir. Viral load (VL) < 75 indicates successful ART; 75 < VL < 5,000 indicates ART failure that can still be managed with increased adherence; VL > 5,000 necessitates therapy change.
Viral load determination.
The plasma HIV-1 load was determined by the use of a Versant HIV RNA 3.0 assay (bDNA, Siemens, IL) as recommended by the manufacturer using the same plasma specimen used for ARMS-PCR and the sequencing of reverse transcriptase.
Selection of HIV drug resistance mutations for ARMS-PCR assay.
HIV drug resistance mutations were selected based on their prevalence, their effect on the first-line regimen recommended by WHO for resource-constrained settings, and the Stanford HIV drug resistance database mutation scoring system (http://hivdb.stanford.edu/DR/asi/releaseNotes/index.html). Using the aforementioned criteria, M184V and T215Y/F were selected as nucleoside reverse transcriptase inhibitor (NRTI) resistance mutations, while K103N and Y181C were selected as non-NRTI (NNRTI). M184V is the most common (37% to 90%) of the NRTI resistance mutations causing high-level resistance to lamivudine (3TC) and emtricitabine (FTC). T215Y/F is the most common (11% to 47.2%) thymidine analog mutation (TAM) that causes intermediate-level resistance to zidovudine (AZT) and stavudine (d4T) and with the potential of causing high level resistance to AZT and d4T when occurring with other TAMs (M41L, D67N, K70R, L210W, and K219QE). K103N is one of the most common (14% to 44%) NNRTI resistance mutations causing high-level resistance to nevirapine (NVP) and efavirenz (EFV). Y181C is a common (9.4% to 19.8%) NNRTI resistance mutation that reduces >50-fold the susceptibility to NVP (http://hivdb.stanford.edu/). The prevalence of these mutations ranges from 8.2% in naive patients to 90% in patients failing a first-line ART (8, 22, 23). The drugs affected (FTC, 3TC, AZT, ABC, EFV, and NVP) are among those most commonly used in the regimens recommended by WHO guidelines for first-line treatment in resource-constrained countries, especially in sub-Saharan Africa (6).
RNA extraction and RT-PCR.
Virus in 500 μl of plasma was concentrated by ultracentrifugation at 14,000 × g for 1 h at 4°C prior to RNA extraction. After removal of 360 μl of supernatant, the virus pellet was resuspended by vortex mixing and viral RNA was extracted from plasma using a QIAamp viral RNA minikit (Qiagen, Inc., Valencia, CA) according to the manufacturer's instructions. A total of 2.5 μl of RNA extract was used for the amplification of 1,750 bp of the pol region using a Superscript One-Step reverse transcription PCR (RT-PCR) system with platinum Taq (Invitrogen, Carlsbad, CA), following the manufacturer's instructions. For RT-PCR, forward primer RTPOL1F (5′-GTATTAGTAGGACCTACACCTGTC-3′; HXB2 locations 2475 to 2498) and reverse primer RTPOL1R (5′-ACCTTCCTGATTCCATTACTGAC-3′; HXB2 locations 4203 to 4225) were used. The cycling conditions included an initial cDNA synthesis step for 30 min at 50°C, followed by denaturation for 2 min at 94°C and then 35 cycles at 94, 50, and 72°C for 60, 60, and 90 s, respectively, and a final extension step at 72°C for 7 min.
Control.
The specificity of the ARMS-PCR assay was assessed using an HIV-1 subtype B molecular clone containing several NRTI- and NNRTI-associated mutations. This clone (AIDS reagent program catalog no. 12229; GenBank accession number JQ814884) was obtained from Robert Shafer through the NIH AIDS Research and Reference Reagent Program, Division of AIDS, NIAID, NIH.
ARMS-PCR procedure.
HIV-1-specific primers were manually designed after compiling the different HIV-1 pure and recombinant subtypes that are most prevalent in central Africa from the HIV sequence database (http://www.hiv.lanl.gov). For each mutation tested, two reverse primers were designed such that the sequences (wild-type [Wt] and mutant [Mut] variants) at their 3′-terminal nucleotide ends differed. Additional deliberate mismatches were introduced within the last four nucleotides at the 3′ terminus to increase the specificity of the assay using principles established for ARMS-PCR primer design (17, 24). The forward primers were designed following the classical rules and so as to obtain a short (200- to 600-bp) amplification product. Primers were manufactured by Integrated DNA Technologies (Coralville, IA) with the details provided (see Table S1 in the supplemental material).
An amplified pol gene fragment from RT-PCR was used as the template to detect wild-type and mutant sequences at different codons (M184V, T 215Y/F, K103N, and Y181C) by ARMS-PCR. Two reactions (wild type and mutant) were performed for each mutation with the respective specific primers using Platinum PCR High Fidelity SuperMix (Invitrogen, Carlsbad, CA). The cycling conditions included an initial denaturation of 2 min at 94°C, followed by 35 cycles at 94, 55, and 72°C, respectively. Ten microliters of ARMS-PCR products was visualized on a 2% agarose gel after ethidium bromide staining, and pictures were taken with inverted gel images using a ChemiDoc MP system from Bio-Rad. The two amplified products from each sample were run on the 2% agarose gel next to one another. The ARMS-PCR result was recorded as Wt or Mut depending on the amplicon band detected on the agarose gel picture. Only one band was expected for patients harboring the wild-type variant, while two bands of the same molecular size were expected for patients harboring a mutant variant due to the presence of mixed (wild-type and mutant) viral populations. ARMS-PCR results were compared to the sequencing data, the latter being the gold standard. The specificity of the ARMS-PCR primers for detection of M184V, T215Y/F, K103N, and Y181C mutations was assessed using the subtype B clone obtained from NIH. Only mutant amplicon bands for mutations M184V, T215Y/F, and K103N and a wild-type amplicon band for mutation Y181C were expected on the agarose gel.
Sequencing.
Two microliters of RT-PCR product was used for nested PCR using RTPOL2F as the forward primer (5′-TAAAGCCAGGAATGGATGGCCC-3′; HXB2 locations 2584 to 2605) and RTPOL2R as the reverse primer (5′-CCTCCAATCCCTTTGTGTGCTG-3′; HXB2 locations 4159 to 4180). The PCR cycling conditions were an initial denaturation for 2 min at 94°C, followed by 35 cycles at 94, 50, and 72°C for 60, 60, and 90 s, respectively, and a final extension step of 7 min at 72°C. Direct sequencing of the 5′ and 3′ ends of the purified PCR amplicons was performed with the two nested RTPOLF2 and RTPOLR2 primers by Microgen (New York, NY), using ABI BigDye Terminator cycle sequencing chemistry and an ABI 3730XL DNA analyzer.
Phylogenetic analysis and drug resistance genotyping.
All sequences were automatically aligned with reference sequences of all known HIV-1 group M subtypes and circulating recombinant forms (CRFs) from the Los Alamos HIV sequence database using CLUSTAL X with minor manual adjustments. Phylogenetic analyses were conducted using the MEGA version 3.1 software package (25) with pairwise evolutionary distances estimated by using Kimura's two-parameter method. Phylogenetic trees were constructed by the neighbor-joining method, and the reliability of the topologies was estimated by performing bootstrap analysis. Clustering of sequences with bootstrap values of more than 70% was considered significant for subtyping.
The reverse transcriptase DNA sequences amplified from patients were analyzed for potential drug resistance mutations using the Stanford University HIV database genotypic-resistance interpretation algorithm (http://hivdb.stanford.edu/index.html). Mutations in the study sequences were defined as differences from the consensus B reference sequence and were further characterized as NRTI resistance mutations or NNRTI resistance mutations.
Trugene HIV-1 genotyping assay.
Ten patients with concordant sequencing and ARMS-PCR results and 10 patients with discordant sequencing and ARMS-PCR results and with various viral loads and different HIV-1 clades were genotyped using a Trugene HIV-1 genotyping kit (Siemens, IL), an FDA-approved assay for the detection of HIV-1 drug resistance; the test was performed as recommended by the manufacturer.
ARMS-PCR performance analysis.
Using sequencing as the gold standard, the sensitivity, specificity, and positive predictive and negative predictive values were calculated from the following formulas: sensitivity = true positives/(true positives plus false negatives) × 100 (with true positives being the number of samples positive on ARMS-PCR and sequencing [gold standard] and false negatives being the number of samples negative on ARMS-PCR but positive on sequencing); specificity = true negatives/(true negatives plus false positives) × 100 (with true negatives being the number of samples negative on ARMS-PCR and sequencing [gold standard] and false positives being the number of samples positive on ARMS-PCR and negative on sequencing [gold standard]); positive predictive value (PPV) = true positive/(true positive plus false positive) × 100; and negative predictive value (NPV) = true negative/(true negative plus false negative) × 100.
RESULTS
Use of ARMS-PCR to detect the M184V, T215Y/F, K103N, and Y181C mutations in plasma samples.
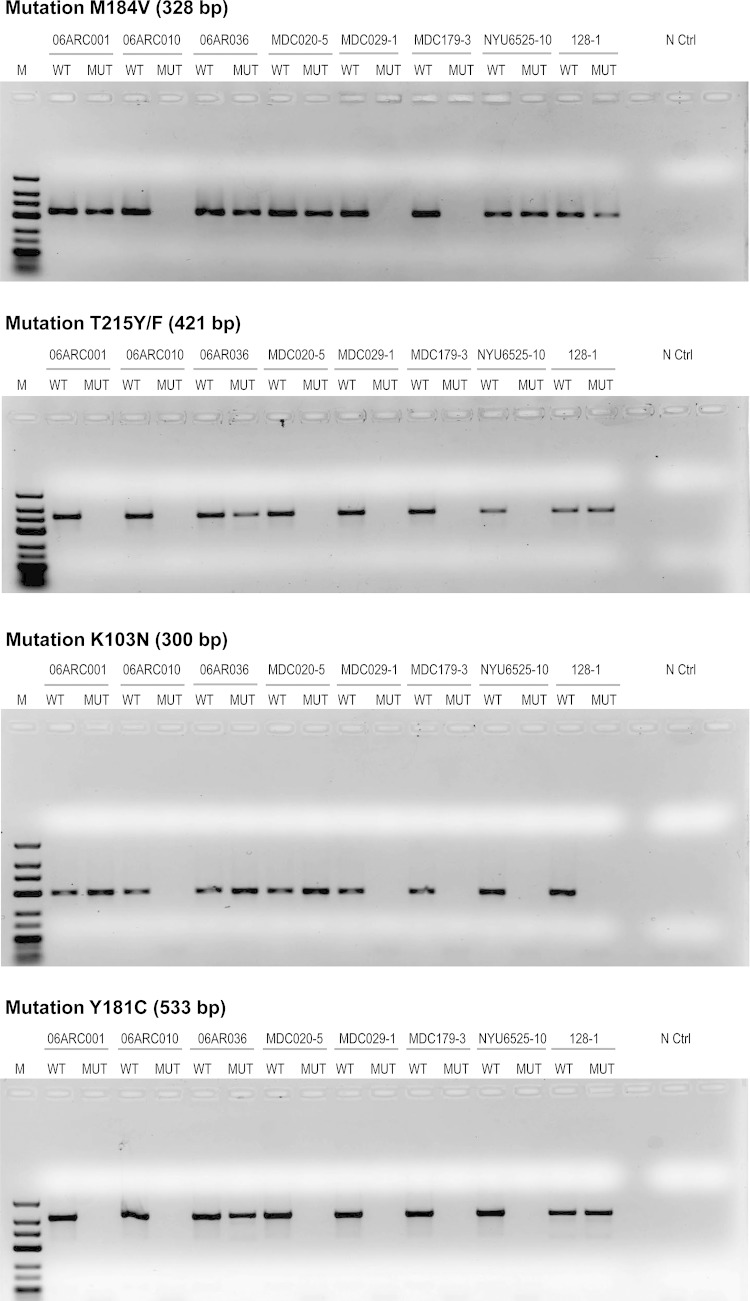
In these studies, we first ensured that the ARMS-PCR was able to correctly detect the different HIVDR mutations in the subtype B molecular clone which we obtained from the NIH reagent program. These mutations included K103N, M184V, Y181C, and T215Y/F and were correctly detected as shown in the gel picture presented in Fig. S1 in the supplemental material. Representative gel pictures revealing the detection of the different HIVDR mutations among eight HIV-1 patients infected with different HIV-1 subtypes, on different treatment regimens, and with various viral loads studied are shown in Fig. 1, to illustrate that this assay is able to detect the various HIVDR mutations studied. These HIVDR mutations as well as the wild-type amino acids identified by ARMS-PCR shown in the gel pictures in Fig. 1 were also confirmed by sequencing as shown in Fig. 2.
FIG 1.
HIV-1 drug resistance mutations detected by ARMS-PCR in plasma samples from HIV-1-infected individuals. These pictures represent 2% agarose gels stained with ethidium bromide, showing the results from eight individuals infected with different HIV-1 subtypes on different ART regimens and harboring different viral loads. M, molecular marker (low-range DNA ladder; Invitrogen). WT, wild type; MUT, mutant; N Ctrl, negative control. The presence of a band in the MUT lane indicates the presence of the mutation in the sample. Pictures were taken using an inverted gel image and a ChemiDoc imaging system (Bio-Rad). The viral loads in each sample were 3,135 copies/ml (06ARC001), 14,715 copies/ml (06ARC010), 8,221 copies/ml (06ARC036), 9,572 copies/ml (MDC020-5), 36,988 copies/ml (MDC029-1), 11,272 copies/ml (MDC179-3), 3,246 copies/ml (128-01), and 17,876 copies/ml (NYU6525-10). The infecting subtypes were CRF02_AG (06ARC001, 06ARC010, 06ARC036, and MDC020-5), D (MDC029-1), F2 (MDC179-3), CRF11_cpx (NYU6525-10), and CRF2201A1 (128-01).
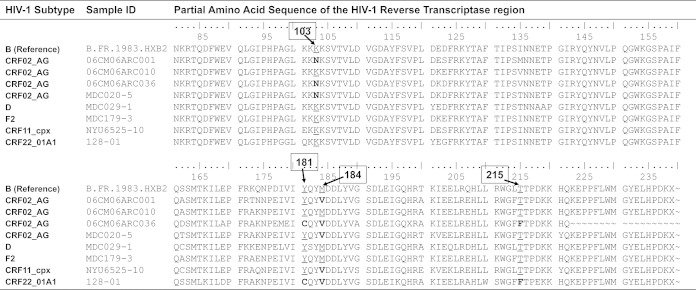
FIG 2.
Reverse transcriptase sequences of HIV-1 in plasma samples from eight patients indicating wild-type or HIV drug resistance mutations. These same samples were also analyzed by ARMS-PCR, and the results are shown in Fig. 1. The positions of the amino acids are indicated above the sequences in boxes. The HIV drug resistance mutations analyzed included M184V, K103N, Y181C, and T215Y/F. The presence of a wild-type amino acid is underlined, and the presence of a mutant amino acid is indicated in bold.
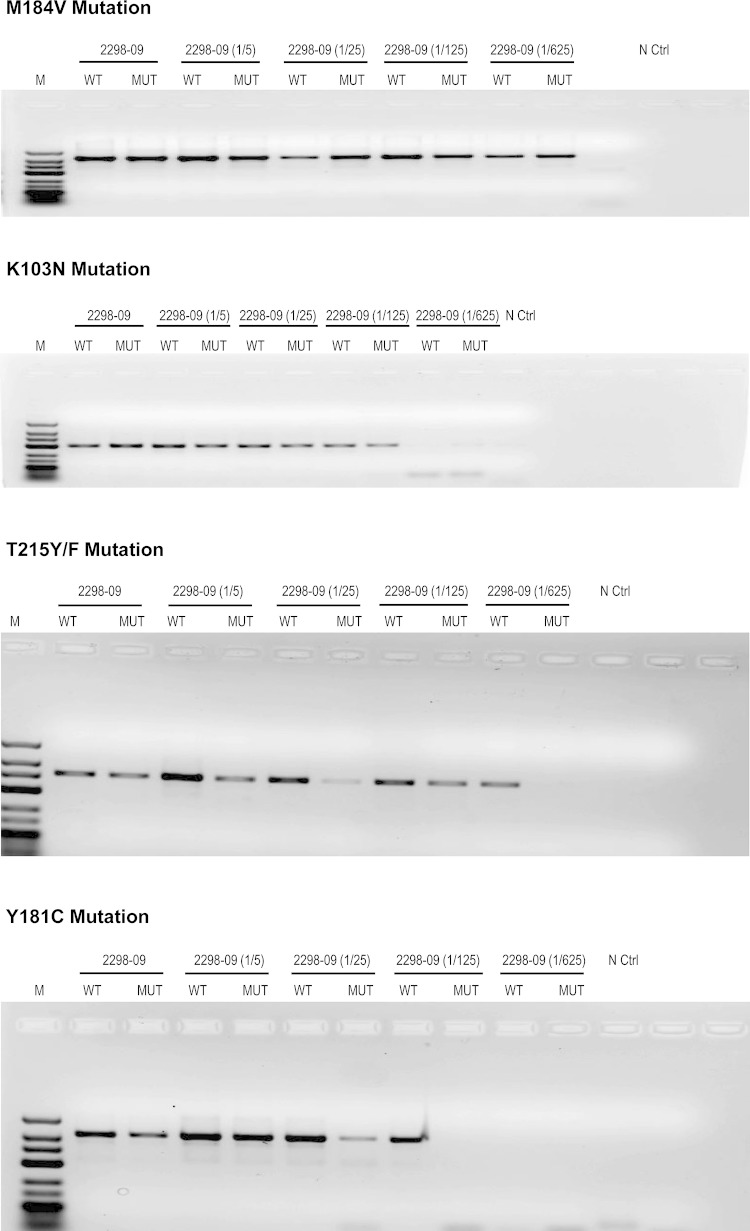
We also evaluated the ARMS-PCR assay to detect HIVDR mutations in samples with various viral loads as well as with different HIV-1 subtype variants. Thus, we tested the limit of detection of HIVDR mutations using a plasma sample from a patient with a high viral load (18,253 copies/ml) infected with a CRF02_AG variant. The plasma sample was serially diluted at 1/5, 1/25, 1/125, and 1/625 in RPMI medium, and the measured viral load corresponded to 4,977, 836, 143, and <75 copies/ml, respectively. All four HIVDR mutations, M184V, T215Y/F, K103N, and Y181C, were detected in samples corresponding to a viral load of ≥836 copies/ml. Furthermore, T215Y/F and K103N could also be detected at 143 copies/ml, and M184V was also detected at a viral load of as low as <75 copies/ml (Fig. 3). This analysis confirms detection of HIVDR mutations with low viral loads, confirming the high sensitivity of the ARMS-PCR assay.
FIG 3.
Detection of HIV-1 drug resistance mutations by ARMS-PCR in samples with different viral loads. Sample 2298-09 (viral load, 18,253 copies/ml) was serially diluted 1/5, 1/25, 1/125, and 1/625, corresponding to 4,977, 836, 143, and <75 copies/ml, respectively. The four HIVDR mutations, M184V, T215Y/F, K103N, and Y181C, were detected in samples corresponding to a viral load of ≥836 copies/ml. Furthermore, T215Y/F and K103N were detected at 143 copies/ml and M184V was detected at a viral load of <75 copies/ml.
The 75 HIV-1-infected patients studied harbored viral loads that ranged between <75 and 400,927 copies/ml. We noted that ARMS-PCR was able to detect mutations in patients with low viral loads and in those with high viral loads. For example, in patient 06ARC001, with a low viral load of 3,135 copies/ml, the M184V and K103N mutations were detected; in patient 06ARC036, with a moderate viral load of 8,221 copies/ml, mutations M184V, T215Y/F, K103N, and Y181C were detected; and in patient NYU6525-10, with a high viral load of 17,876 copies/ml, mutation M184V was detected (Fig. 1). These results suggest that these mutations can be detected in plasma samples with low to high viral loads.
While the CRF02_AG recombinant variant is the predominant strain infecting individuals in Cameroon, several other subtypes and recombinants also cause infections. Thus, we examined whether the ARMS-PCR assay was able to detect mutations in CRF02_AG variants as well as in non-CRF02_AG variants. As shown in Fig. 1 with the eight patient samples, infections due to CRF02_AG were present in patients 06ARC001, 06ARC010, 06ARC036, and MDC020-5, and non-CRF02_AG infections were present in patients MDC029-1 (subtype D), MDC179-3 (subtype F2), NYU6525-10 (subtype CRF11_cpx), and 128-01 (subtype CRF22_01A1). Using the samples from these patients, we were able to detect mutations in these different subtype infections. For example, M184V mutation was detected in patients 06ARC001 (CRF02_AG), NYU6525-10 (CRF11_cpx), and 128-01 (CRF22_01A1); mutation T215Y/F was detected in patients 06ARC036 (CRF02_AG) and 128-01 (CRF22_01A1); and mutation Y181C was detected in patients 06ARC036 (CRF02_AG) and 128-01 (CRF22_01A1). The results of these analyses were confirmed by sequencing of the reverse transcriptase gene (Fig. 2). Overall, HIV-1 drug resistance mutations were found in patients infected with CRF02_AG subtypes as well as in those with non-CRF02_AG infections; the M184V mutation was the most prevalent in both groups, and none of the mutations was found exclusively in one of the groups (see Table S2 in the supplemental material).
Detection of M184V/T215Y/F (NRTI) and K103N/Y181C (NNRTI) drug resistance mutations using the ARMS-PCR assay.
After establishing that ARMS-PCR could correctly detect HIVDR mutations, we then used the assay to study the mutations in the 75 study subjects, including those who were ART naive (n = 33) and those on ART (n = 42). The combinations of HIVDR mutations are shown in Table 2.
TABLE 2.
Drug resistance mutations among HIV-1-infected individuals determined by sequencing and ARMS-PCRa
| ARV class | Mutation(s) | No. (%) of patients |
|||||||
|---|---|---|---|---|---|---|---|---|---|
| ART-naïve (n = 33) |
First-line ART (n = 39) |
Second-line ART (n = 3) |
Overall (n = 75) |
||||||
| Sequencing (%) | ARMS-PCR (%) | Sequencing (%) | ARMS-PCR (%) | Sequencing (%) | ARMS-PCR (%) | Sequencing (%) | ARMS-PCR (%) | ||
| NRTI | M184V | 3 (9) | 4 (12) | 26 (66.6) | 29 (74.3) | 3 (100) | 3 (100) | 32 (42.6) | 36 (48) |
| T215Y/F | 1 (3) | 3 (9) | 10 (25.6) | 9 (23) | 3 (100) | 3 (100) | 14 (18.6) | 15 (20) | |
| M184V or T215Y/F | 3 (9) | 6 (18) | 26 (66.6) | 29 (74.3) | 3 (100) | 3 (100) | 32 (42.6) | 38 (50.6) | |
| M184V + T215Y/F | 1 (3) | 1 (3) | 10 (25.6) | 9 (23) | 3 (100) | 3 (100) | 14 (18.6) | 13 (17.3) | |
| NNRTI | K103N | 5 (15) | 5 (15) | 17 (43.5) | 16 (41) | 1 (33.3) | 1 (33.3) | 23 (30.6) | 22 (29.3) |
| Y181C | 0 (0) | 0 (0) | 9 (23) | 8 (20.5) | 1 (33.3) | 1 (33.3) | 10 (13.3) | 9 (12) | |
| K103N or Y181C | 5 (15) | 5 (15) | 22 (56.4) | 19 (48.7) | 2 (66.6) | 2 (66.6) | 29 (38.6) | 26 (34.6) | |
| K103N + Y181C | 0 (0) | 0 (0) | 5 (12.8) | 4 (10.2) | 0 (0) | 0 (0) | 5 (6.6) | 4 (5.3) | |
ARV, antiretroviral; ART, antiretroviral treatment; NRTI, nucleoside reverse transcriptase inhibitor; NNRTI, non-NRTI. Percentages in each column were calculated based on the number of individuals in the respective ART group (ART-naive, first-line ART, or second-line ART).
Detection of HIVDR mutations in ART-naive patients.
Of the 33 ART-naive patients studied, 6 (18.1%) harbored viruses with M184V or T215Y/F mutations, and 1 (3%) concomitantly had both mutations. Four (12%) patients had the M184V mutation, whereas 3 (9%) patients had the T215Y/F mutation. Five (15.1%) ART-naive patients had the K103N mutation, and none of them had the Y181C mutation.
Detection of HIVDR mutations in patients on first-line ART.
Among the 39 patients on first-line ART, 29 (74.3%) harbored viruses with M184V or T215Y/F mutations, and 9 (23%) concomitantly had both mutations. Twenty-nine (74.3%) patients had the M184V mutation, whereas 9 (23%) had the T215Y/F mutation. Nineteen (48.7%) patients harbored viruses with K103N or Y181C mutations, and 4 (10.2%) concomitantly had both mutations. Sixteen (41%) patients had the K103N mutation, whereas 9 (23%) patients had the Y181C mutation.
Detection of HIVDR mutations in patients on second-line ART.
All three patients harbored viruses with the two NRTI (M184V and T215Y/F) mutations tested. Of the NNRTI mutations, one patient harbored virus with the K103N mutation and another patient had virus with the Y181C mutation.
Detection of M184V/T215Y/F (NRTI) and K103N/Y181C (NNRTI) drug resistance mutations using sequencing.
The HIVDR mutations identified by sequence analysis are shown in Table 2. The results reveal that among the 33 ART-naive patients studied, 3 (9%) harbored viruses with M184V or T215Y/F mutation, and 1 (3%) concomitantly had both mutations. Three patients (9%) had the M184V mutation, whereas 1 (3%) had the T215Y/F mutation. Five (15.1%) patients had the K103N mutation, and no ART-naive patient was identified with a Y181C mutation.
Among the 39 patients on first-line ART, 26 (66.6%) harbored viruses with M184V or T215Y/F; 10 (25.6%) concomitantly had both mutations. Twenty-six (66.6%) patients had the M184V mutation, whereas 10 (25.6%) had the T215Y/F mutation. Twenty-two (56.4%) patients harbored viruses with K103N or Y181C; among those patients, 5 (12.8%) concomitantly had both mutations. Seventeen (43.5%) patients had the K103N mutation, whereas 9 (23%) had the Y181C mutation.
The results of sequence analysis of all the three patients on second-line ART were identical to those of ARMS-PCR in detecting the two NRTI (M184V and T215Y/F) mutations tested. Furthermore, the detection of the NNRTI mutation in only one patient who harbored virus with the K103N mutation and another patient that had virus with the Y181C mutation was also similarly revealed by the ARMS-PCR assay.
Performance of the ARMS-PCR assay versus sequencing and a Trugene HIV-1 genotyping kit.
The mutation rates obtained with ARMS-PCR and sequencing were comparable. Of 75 patients, 63 had similar results for all the mutations tested using ARMS-PCR and sequencing. The highest number of false positives (n = 4) was obtained for the M184V mutation, whereas the highest number of false negatives (n = 3) was obtained for the Y181C mutation as shown in Table 3.
TABLE 3.
Detection of HIVDR mutations in corresponding plasma samples by ARMS-PCR compared with sequence analysisa
| Mutation | No. of samples |
Sensitivity (%) | Specificity (%) | PPV (%) | NPV (%) | |||
|---|---|---|---|---|---|---|---|---|
| POS ARMS-PCR | POS sequencing | False-POS ARMS-PCR | False-NEG ARMS-PCR | |||||
| M184V | 35 | 32 | 4 | 1 | 96.8 | 90.6 | 88.5 | 97.5 |
| T215Y/F | 15 | 14 | 3 | 2 | 85.7 | 95 | 80 | 96.6 |
| K103N | 21 | 23 | 0 | 2 | 91.3 | 100 | 100 | 96.2 |
| Y181C | 9 | 10 | 2 | 3 | 70 | 96.9 | 77.7 | 95.4 |
POS, positive; NEG, negative; PPV, positive predictive value; NPV; negative predictive value. Sensitivity, specificity, PPV, and NPV of ARMS-PCR were calculated using sequencing as the gold standard.
Ten patients with concordant results and 10 patients with discordant results between sequencing and ARMS-PCR were tested with the Trugene HIV-1 genotyping assay, and the results obtained were 100% concordant with the sequencing results. The sensitivity, specificity, PPV, and NPV of ARMS-PCR assay were calculated for each mutation, and results are presented in Table 3. The highest sensitivity (96.8%) and the highest NPV (97.5%) were for the detection of the M184V mutation, while the highest specificity (100%) and the highest PPV (100%) were for the detection of K103N mutation.
Concordance of prevalence rates of mutations based on ARMS-PCR and sequencing.
The overall prevalence rates using ARMS-PCR were 48%, 20%, 29.3%, and 12% for the M184V, T215Y/F, K103N, and Y181C mutations, respectively, whereas the prevalence rates using sequencing were 42.6%, 18.6%, 30.6%, and 13.3% for the M184V, T215Y/F, K103N, and Y181C mutations, respectively (see Fig. S2 in the supplemental material). The ability of ARMS-PCR to detect the various HIVDR mutations at the individual level and at the population level in analysis of all 75 samples was comparable to that of sequence analysis (see Fig. S2).
HIV-1 subtypes infecting study subjects.
Phylogenetic analysis of the reverse transcriptase gene of studied patients revealed a predominance of CRF02_AG and several other variants. These HIV-1 subtypes included CRF02_AG (70.6%), CRF11_cpx (9.3%), CRF22_01A1 (8%), F2 (4%), D (4%), A1 (1.3%), G (1.3%), and CRF18_cpx (1.3%). The distribution of the infecting subtypes among treatment-naive patients was similar to that among those on treatment (data not shown).
DISCUSSION
The scaling-up of antiretroviral treatment (ART) in resource-constrained settings without adequate virological follow-up raises concerns with respect to ART program sustainability. Sequencing technology is not available for all patients in need, and because of financial constraints, it is not affordable where it is available; therefore, providing an alternative cost-effective and less laborious assay to identify HIVDR mutations will be a great asset. In this study, an ARMS-PCR assay was developed and used to investigate the presence of the most common HIVDR mutations (M184V, T215Y/F, K103N, and Y181C) affecting the first-line ARV regimen. The results of the ARMS-PCR were highly concordant with those of sequencing in identifying all four mutations studied in comparisons of the results determined for each sample in both assays for each mutation (see Fig. S2a in the supplemental material) or in comparisons of all the samples at the population level (see Fig. S2b). In particular, the assay was most sensitive in identifying the M184V and K103N mutations, with sensitivities of 96.8% and 91.3%, respectively. The specificities of the M184V and K103N mutations were also high (88.5% and 100%, respectively). HIVDR drug resistance mutations were found in patients infected with CRF02_AG subtypes and non-CRF02_AG at comparable rates, and the M184V mutation was the most prevalent in both groups.
Taken together, among all the mutations studied, a few samples (n = 9) revealed discordant results for the different mutations; some were identified by ARMS-PCR as positive results whereas they were negative on sequence analysis. Also, a few (n = 8) were negative on ARMS-PCR but were positive on sequence analysis (Table 3). The few false positives identified could have been due to the fact that, like many point mutation assays, the ARMS-PCR is more sensitive than sequencing in detecting mutations in mixed HIV-1 genotypic populations (26). The few results that were false negatives could have been due to unusual polymorphisms. These false negatives and positives indicate the need to further study the variables that account for them to improve the assay. These also indicate the need to randomly select specimens and subject the results to quality control in other reference laboratories, especially in conducting population-based surveillance in public health programs prior to introduction of antiretroviral drugs. Nonetheless, the ARMS-PCR assay still revealed high performance in identifying HIVDR compared with sequence analysis and has several advantages over other alternative approaches that have been developed. For example, most of the point mutation assays use mass spectrometry, fluorescence, or luminescence to identify the allele-specific PCR products (4, 16); this implies the use of expensive equipment and reagents. Furthermore, these assays are designed to be performed in 96-well plates requiring many samples to perform the test, while ARMS-PCR can be performed even with only one sample. The use of ARMS-PCR reduces the time to results, taking an average of 8 h. The high throughput of this assay also reduces the cost of supplies and of reagents. These advantages make ARMS-PCR cost-effective and affordable for resource-constrained settings.
The results of our newly developed ARMS-PCR assay, which were confirmed by sequence analysis and the use of a Trugene HIV-1 genotyping kit, now allow us to describe the HIVDR mutations identified in our study subjects. The samples from the study subjects were obtained during the period 2006 to 2014. This study revealed a high rate of the first-line drug resistance mutations studied in ART-naive patients (n = 33) and in patients on ART (n = 42). Among the ART-naive patients, 18% and 15% harbored viruses with NRTI-associated mutations (M184V and T215Y/F) and viruses with NNRTI-associated mutations (K103N and Y181C), respectively. These prevalence rates are higher than the rates seen in earlier studies in Cameroon, which reported transmitted drug resistance rates in Cameroon ranging from 1.9% to 12.3% (1, 11, 23) in ART-naive patients. This increasing rate of transmitted HIVDR mutation is consistent with the increase in the availability of ARV in Cameroon over time. A meta-analysis of data from many low- and middle-income countries, including African countries with ART programs similar to Cameroon's, has also shown an increase of transmitted drug resistance mutations (27). Similarly, reports in developed countries with long ART experience have shown an increase in transmitted drug resistance mutations to up to 25% (9, 28, 29). The most prevalent mutation was K103N, with a 15% rate. This could be explained by the wide use of nevirapine in first-line combinations and prophylaxis for prevention of mother-to-child transmission of HIV-1; therefore, the K103N mutation is more likely to develop and spread in the population (30).
Treated patients exhibited high rates of the HIVDR mutations tested: M184V (76.1%), T215Y/F (28.5%), K103N (40.4%), and Y181C (21.4%). These findings are consistent with previous studies that have reported similar rates of acquired drug resistance mutations in Cameroon (8, 23) and other African countries with similar ART programs (10, 31, 32). The most prevalent mutations identified were M184V and K103N. The high frequency of these mutations in patients on ART is related to the wide use of 3TC- and NVP/EFV-based first-line combinations, representing 94.8% (37/39) of drug used by patients on first-line ART in our study; therefore, these mutations are more likely to develop in this population. This high rate of HIVDR mutations is the result of a median time of only 15 months for the patients that were on first-line ART and of 32 months for patients on second-line ART.
The mutations identified in this study confer intermediate- to high-level resistance to all the ARVs (3TC, FTC, AZT, NVP, and EFV) used in resource-constrained settings for first-line treatment except TDF. Patients with these mutations require a switch to second-line treatment only after a median time of 15 months on first-line treatment in settings where the drug options for therapeutic switch are already limited, thus requiring new and more expensive ARV regimens (36). The high rate of transmitted and acquired HIVDR mutations observed in this study underlines the necessity to improve efforts for closer monitoring of viral load in HIV-infected patients and the necessity to make drug resistance testing assays more accessible and more affordable. This will guide physicians in the choice of the most appropriate therapy for treatment initiation and a therapy switch in a timely manner to sustain ART programs in resource-constrained settings. It should be noted that better outcomes are expected in patients who failed first-line therapy only when they switch ART within 8 weeks of failure (33).
Because of financial constraints, evaluations of ARV treatment programs in poor settings are usually done in a limited number of patients and in only a few sites (5, 34). The ARMS-PCR assay would be an advantageous tool to evaluate current antiretroviral programs in larger populations and in more sites, to better assess the need for the introduction of new ARVs. In fact, the ARMS-PCR assay costs only $10 to cover the reagent cost for one mutation ($40 for four mutations) compared to sequencing and commercial genotyping assays that cost $120 to $280 in resource-constrained settings (28). The use of this assay will facilitate surveillance studies and will improve ART programs, thereby reducing the risk of transmitted and acquired drug resistance mutations and improving the monitoring and care of HIV-1-infected patients.
Supplementary Material
ACKNOWLEDGMENTS
This study was supported by the National Institutes of Health (NIH) grant AI083142 from the National Institute of Allergy and Infectious Diseases (NIAID), grant CA153726 from the National Cancer Institute (NCI), and grant TW009604 from the Fogarty International Center.
We thank the Cameroon Ministry of Public Health for supporting our studies. We also thank Charles Kouanfack and Josephine Meli for their assistance in patient recruitment.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JCM.00114-15.
REFERENCES
- 1.Aghokeng AF, Kouanfack C, Laurent C, Ebong E, Atem-Tambe A, Butel C, Montavon C, Mpoudi-Ngole E, Delaporte E, Peeters M. 2011. Scale-up of antiretroviral treatment in sub-Saharan Africa is accompanied by increasing HIV-1 drug resistance mutations in drug-naive patients. AIDS 25:2183–2188. doi: 10.1097/QAD.0b013e32834bbbe9. [DOI] [PubMed] [Google Scholar]
- 2.Hirsch MS, Gunthard HF, Schapiro JM, Brun-Vezinet F, Clotet B, Hammer SM, Johnson VA, Kuritzkes DR, Mellors JW, Pillay D, Yeni PG, Jacobsen DM, Richman DD. 2008. Antiretroviral drug resistance testing in adult HIV-1 infection: 2008 recommendations of an International AIDS Society-USA panel. Clin Infect Dis 47:266–285. doi: 10.1086/589297. [DOI] [PubMed] [Google Scholar]
- 3.Little SJ, Frost SD, Wong JK, Smith DM, Pond SL, Ignacio CC, Parkin NT, Petropoulos CJ, Richman DD. 2008. Persistence of transmitted drug resistance among subjects with primary human immunodeficiency virus infection. J Virol 82:5510–5518. doi: 10.1128/JVI.02579-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bennett DE, Bertagnolio S, Sutherland D, Gilks CF. 2008. The World Health Organization's global strategy for prevention and assessment of HIV drug resistance. Antivir Ther 13(Suppl 2):S1–S13. [PubMed] [Google Scholar]
- 5.Billong SC, Fokam J, Aghokeng AF, Milenge P, Kembou E, Abessouguie I, Meva'a-Onglene FB, Bissek AC, Colizzi V, Mpoudi EN, Elat JB, Shiro KS. 2013. Population-based monitoring of emerging HIV-1 drug resistance on antiretroviral therapy and associated factors in a sentinel site in Cameroon: low levels of resistance but poor programmatic performance. PLoS One 8:e72680. doi: 10.1371/journal.pone.0072680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gilks CF, Crowley S, Ekpini R, Gove S, Perriens J, Souteyrand Y, Sutherland D, Vitoria M, Guerma T, De Cock K. 2006. The WHO public-health approach to antiretroviral treatment against HIV in resource-limited settings. Lancet 368:505–510. doi: 10.1016/S0140-6736(06)69158-7. [DOI] [PubMed] [Google Scholar]
- 7.Kouanfack C, Laborde-Balen G, Aghokeng AF, Bourgeois A, Dontsop M, Mben JM, Kaze S, Mpoudi-Ngole E, Koulla-Shiro S, Delaporte E, Laurent C. 2010. WHO clinical criteria-based initiation of antiretroviral therapy: lessons from rural district hospitals in Cameroon with regard to 2009 revised WHO recommendations. Trop Med Int Health 15:580–583. [DOI] [PubMed] [Google Scholar]
- 8.Aghokeng AF, Kouanfack C, Eymard-Duvernay S, Butel C, Edoul GE, Laurent C, Koulla-Shiro S, Delaporte E, Mpoudi-Ngole E, Peeters M. 2013. Virological outcome and patterns of HIV-1 drug resistance in patients with 36 months' antiretroviral therapy experience in Cameroon. J Int AIDS Soc 16:18004. doi: 10.7448/IAS.16.1.18004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Descamps D, Chaix ML, Montes B, Pakianather S, Charpentier C, Storto A, Barin F, Dos Santos G, Krivine A, Delaugerre C, Izopet J, Marcelin AG, Maillard A, Morand-Joubert L, Pallier C, Plantier JC, Tamalet C, Cottalorda J, Desbois D, Calvez V, Brun-Vezinet F, Masquelier B, Costagliola D. 2010. Increasing prevalence of transmitted drug resistance mutations and non-B subtype circulation in antiretroviral-naive chronically HIV-infected patients from 2001 to 2006/2007 in France. J Antimicrob Chemother 65:2620–2627. doi: 10.1093/jac/dkq380. [DOI] [PubMed] [Google Scholar]
- 10.Djoko CF, Wolfe ND, Vidal N, Tamoufe U, Montavon C, LeBreton M, Pike BL, Fair J, Mbacham WF, Benito A, Rimoin AW, Saylors K, Mpoudi-Ngole E, Grillo MP, Peeters M. 2010. HIV type 1 pol gene diversity and genotypic antiretroviral drug resistance mutations in Malabo, Equatorial Guinea. AIDS Res Hum Retroviruses 26:1027–1031. doi: 10.1089/aid.2010.0046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ndembi N, Abraha A, Pilch H, Ichimura H, Mbanya D, Kaptue L, Salata R, Arts EJ. 2008. Molecular characterization of human immunodeficiency virus type 1 (HIV-1) and HIV-2 in Yaounde, Cameroon: evidence of major drug resistance mutations in newly diagnosed patients infected with subtypes other than subtype B. J Clin Microbiol 46:177–184. doi: 10.1128/JCM.00428-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Okayama H, Curiel DT, Brantly ML, Holmes MD, Crystal RG. 1989. Rapid, nonradioactive detection of mutations in the human genome by allele-specific amplification. J Lab Clin Med 114:105–113. [PubMed] [Google Scholar]
- 13.Richter S, Seth A. 1998. One step direct detection of recurrent mutations in the breast cancer susceptibility gene, BRCA1. Int J Oncol 12:1263–1267. [DOI] [PubMed] [Google Scholar]
- 14.Fokam J, Salpini R, Santoro MM, Cento V, D'Arrigo R, Gori C, Perno CF, Colizzi V, Nanfack A, Gwom LC, Cappelli G, Takou D. 2011. Performance evaluation of an in-house human immunodeficiency virus type-1 protease-reverse transcriptase genotyping assay in Cameroon. Arch Virol 156:1235–1243. doi: 10.1007/s00705-011-0982-3. [DOI] [PubMed] [Google Scholar]
- 15.Saravanan S, Vidya M, Balakrishnan P, Kumarasamy N, Solomon SS, Solomon S, Kantor R, Katzenstein D, Ramratnam B, Mayer KH. 2009. Evaluation of two human immunodeficiency virus-1 genotyping systems: ViroSeq 2.0 and an in-house method. J Virol Methods 159:211–216. doi: 10.1016/j.jviromet.2009.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hayashi N, Ito I, Yanagisawa A, Kato Y, Nakamori S, Imaoka S, Watanabe H, Ogawa M, Nakamura Y. 1995. Genetic diagnosis of lymph-node metastasis in colorectal cancer. Lancet 345:1257–1259. doi: 10.1016/S0140-6736(95)90922-2. [DOI] [PubMed] [Google Scholar]
- 17.Newton CR, Graham A, Heptinstall LE, Powell SJ, Summers C, Kalsheker N, Smith JC, Markham AF. 1989. Analysis of any point mutation in DNA. The amplification refractory mutation system (ARMS). Nucleic Acids Res 17:2503–2516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sommer SS, Groszbach AR, Bottema CD. 1992. PCR amplification of specific alleles (PASA) is a general method for rapidly detecting known single-base changes. Biotechniques 12:82–87. [PubMed] [Google Scholar]
- 19.Bengra C, Mifflin TE, Khripin Y, Manunta P, Williams SM, Jose PA, Felder RA. 2002. Genotyping of essential hypertension single-nucleotide polymorphisms by a homogeneous PCR method with universal energy transfer primers. Clin Chem 48:2131–2140. [PubMed] [Google Scholar]
- 20.Gelsthorpe AR, Wells RS, Lowe AP, Tonks S, Bodmer JG, Bodmer WF. 1999. High-throughput class I HLA genotyping using fluorescence resonance energy transfer (FRET) probes and sequence-specific primer-polymerase chain reaction (SSP-PCR). Tissue Antigens 54:603–614. doi: 10.1034/j.1399-0039.1999.540611.x. [DOI] [PubMed] [Google Scholar]
- 21.Myakishev MV, Khripin Y, Hu S, Hamer DH. 2001. High-throughput SNP genotyping by allele-specific PCR with universal energy-transfer-labeled primers. Genome Res 11:163–169. doi: 10.1101/gr.157901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Burda ST, Viswanath R, Zhao J, Kinge T, Anyangwe C, Tinyami ET, Haldar B, Powell RL, Jarido V, Hewlett IK, Nyambi PN. 2010. HIV-1 reverse transcriptase drug-resistance mutations in chronically infected individuals receiving or naive to HAART in Cameroon. J Med Virol 82:187–196. doi: 10.1002/jmv.21677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ceccarelli L, Salpini R, Moudourou S, Cento V, Santoro MM, Fokam J, Takou D, Nanfack A, Dori L, Torimiro J, Sarmati L, Andreoni M, Perno CF, Colizzi V, Cappelli G. 2012. Characterization of drug resistance mutations in naive and ART-treated patients infected with HIV-1 in Yaounde, Cameroon. J Med Virol 84:721–727. doi: 10.1002/jmv.23244. [DOI] [PubMed] [Google Scholar]
- 24.Kwok S, Kellogg DE, McKinney N, Spasic D, Goda L, Levenson C, Sninsky JJ. 1990. Effects of primer-template mismatches on the polymerase chain reaction: human immunodeficiency virus type 1 model studies. Nucleic Acids Res 18:999–1005. doi: 10.1093/nar/18.4.999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kumar S, Tamura K, Nei M. 2004. MEGA3: Integrated software for Molecular Evolutionary Genetics Analysis and sequence alignment. Brief Bioinform 5:150–163. doi: 10.1093/bib/5.2.150. [DOI] [PubMed] [Google Scholar]
- 26.Van Laethem K, Van Vaerenbergh K, Schmit JC, Sprecher S, Hermans P, De Vroey V, Schuurman R, Harrer T, Witvrouw M, Van Wijngaerden E, Stuyver L, Van Ranst M, Desmyter J, De Clercq E, Vandamme AM. 1999. Phenotypic assays and sequencing are less sensitive than point mutation assays for detection of resistance in mixed HIV-1 genotypic populations. J Acquir Immune Defic Syndr 22:107–118. doi: 10.1097/00126334-199910010-00001. [DOI] [PubMed] [Google Scholar]
- 27.Gupta RK, Jordan MR, Sultan BJ, Hill A, Davis DH, Gregson J, Sawyer AW, Hamers RL, Ndembi N, Pillay D, Bertagnolio S. 2012. Global trends in antiretroviral resistance in treatment-naive individuals with HIV after rollout of antiretroviral treatment in resource-limited settings: a global collaborative study and meta-regression analysis. Lancet 380:1250–1258. doi: 10.1016/S0140-6736(12)61038-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Grant RM, Hecht FM, Warmerdam M, Liu L, Liegler T, Petropoulos CJ, Hellmann NS, Chesney M, Busch MP, Kahn JO. 2002. Time trends in primary HIV-1 drug resistance among recently infected persons. JAMA 288:181–188. doi: 10.1001/jama.288.2.181. [DOI] [PubMed] [Google Scholar]
- 29.Little SJ, Holte S, Routy JP, Daar ES, Markowitz M, Collier AC, Koup RA, Mellors JW, Connick E, Conway B, Kilby M, Wang L, Whitcomb JM, Hellmann NS, Richman DD. 2002. Antiretroviral-drug resistance among patients recently infected with HIV. N Engl J Med 347:385–394. doi: 10.1056/NEJMoa013552. [DOI] [PubMed] [Google Scholar]
- 30.Kurle SN, Gangakhedkar RR, Sen S, Hayatnagarkar SS, Tripathy SP, Paranjape RS. 2007. Emergence of NNRTI drug resistance mutations after single-dose nevirapine exposure in HIV type 1 subtype C-infected infants in India. AIDS Res Hum Retroviruses 23:682–685. doi: 10.1089/aid.2006.0167. [DOI] [PubMed] [Google Scholar]
- 31.Adje C, Cheingsong R, Roels TH, Maurice C, Djomand G, Verbiest W, Hertogs K, Larder B, Monga B, Peeters M, Eholie S, Bissagene E, Coulibaly M, Respess R, Wiktor SZ, Chorba T, Nkengasong JN. 2001. High prevalence of genotypic and phenotypic HIV-1 drug-resistant strains among patients receiving antiretroviral therapy in Abidjan, Cote d'Ivoire. J Acquir Immune Defic Syndr 26:501–506. [DOI] [PubMed] [Google Scholar]
- 32.Vergne L, Diagbouga S, Kouanfack C, Aghokeng A, Butel C, Laurent C, Noumssi N, Tardy M, Sawadogo A, Drabo J, Hien H, Zekeng L, Delaporte E, Peeters M. 2006. HIV-1 drug-resistance mutations among newly diagnosed patients before scaling-up programmes in Burkina Faso and Cameroon. Antivir Ther 11:575–579. [PubMed] [Google Scholar]
- 33.Li L, Eron JJ, Ribaudo H, Gulick RM, Johnson BA. 2012. Evaluating the effect of early versus late ARV regimen change if failure on an initial regimen: results from the AIDS Clinical Trials Group Study A5095. J Am Stat Assoc 107:542–554. doi: 10.1080/01621459.2011.646932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dzangare J, Gonese E, Mugurungi O, Shamu T, Apollo T, Bennett DE, Kelley KF, Jordan MR, Chakanyuka C, Cham F, Banda RM. 2012. Monitoring of early warning indicators for HIV drug resistance in antiretroviral therapy clinics in Zimbabwe. Clin Infect Dis 54(Suppl 4):S313–S316. doi: 10.1093/cid/cir1014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.World Health Organization. 2013. Global update on HIV treatment 2013: results, impact and opportunities. World Health Organization, Geneva, Switzerland: http://apps.who.int/iris/bitstream/10665/85326/1/9789241505734_eng.pdf. [Google Scholar]
- 36.World Health Organization. 2012. WHO HIV drug resistance report 2012. World Health Organization, Geneva, Switzerland: http://apps.who.int/iris/bitstream/10665/75183/1/9789241503938_eng.pdf. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.