Abstract
The recent development of the 1st WHO International Standard for human cytomegalovirus (CMV) and the introduction of commercially produced secondary standards have raised hopes of improved agreement among laboratories performing quantitative PCR for CMV. However, data to evaluate the trueness and uniformity of secondary standards and the consistency of results achieved when these materials are run on various assays are lacking. Three concentrations of each of the three commercially prepared secondary CMV standards were tested in quadruplicate by three real-time and two digital PCR methods. The mean results were compared in a pairwise fashion with nominal values provided by each manufacturer. The agreement of results among all methods for each sample and for like concentrations of each standard was also assessed. The relationship between the nominal values of standards and the measured values varied, depending upon the assay used and the manufacturer of the standards, with the degree of bias ranging from +0.6 to −1.0 log10 IU/ml. The mean digital PCR result differed significantly among the secondary standards, as did the results of the real-time PCRs, particularly when plotted against nominal log10 IU values. Commercially available quantitative secondary CMV standards produce variable results when tested by different real-time and digital PCR assays, with various magnitudes of bias compared to nominal values. These findings suggest that the use of such materials may not achieve the intended uniformity among laboratories measuring CMV viral load, as envisioned by adaptation of the WHO standard.
INTRODUCTION
Routine viral load measurements have become the standard of care for many patients, particularly those with severely compromised immune systems (1–5). However, despite their widespread clinical use, current testing strategies still have many limitations. Except for HIV and hepatitis B and C viruses, there has been little standardization of the testing process. Most methods are based on real-time PCR and have a high degree of result variability, particularly when testing among institutions is compared (6–9). The reasons for this variability are myriad. Real-time PCR is a dynamic process, with quantification based on normalization of the time to signal generation to a calibration curve that is in turn based on the use of calibration material with “known” values. Variations in any part of this complex procedure might theoretically affect result accuracy or precision. In fact, several factors have been shown to play a role (10); however, the most emphasis in the literature has been placed on the lack of universally accepted calibrators (11, 12). The lack of available international quantitative standards for many of the commonly tested viral analytes has led to the use of a wide variety of materials, intuitively reducing the agreement of results when common samples have been tested by different centers. It has been widely hoped that the development of such international reference material would help improve this situation. The agreement of quantitative values is important in ensuring the portability of patient results among institutions, as well as in data interpretation in the literature and in development of common breakpoints for therapeutic decision making.
Quantitative standards have recently been made available by the World Health Organization (WHO) for both cytomegalovirus (CMV) and Epstein-Barr virus (EBV) (13, 14). These are both biologic standards, with consensus international unit (IU) values assigned by international interlaboratory studies conducted for each standard. In the case of CMV, the Merlin strain of human CMV was grown in cell culture and provided in lyophilized vials to 32 laboratories running 53 assays. The mean reported values from all 53 assays were used to set the potency of the standard at 6.7 log10 IU/ml. A limited quantity of primary reference material was made available for purchase in October 2010, with subsequent generation of commercial secondary standards by various manufacturers; these may be purchased in larger volumes by end users. Alternatively, the manufacturers of FDA-cleared or CE-marked assays may directly normalize their results to the primary (WHO) material. One might guess that the introduction of such materials will generally result in a reduction in assay variability and a convergence of test results irrespective of the other elements of the assay design or use. However, the use of secondary standards, while necessary, introduces another potential source of variability. One must then ask if all such material uniformly contains the reported nominal concentrations of CMV and if the material from any given manufacturer behaves similarly, irrespective of the assay with which it is tested.
Although the CMV standard has been available for more than 3 years, few data that demonstrate the relative operating characteristics of secondary standards have as yet been generated. Here we tested the secondary standards from three manufacturers using two digital PCR systems and three real-time PCR methods. Digital PCR is a recently introduced method using limiting partition of endpoint PCRs to generate absolute quantification of the target (15–18). Now regarded as a reference standard by some (19), this methodology allows viral load testing without the use of calibration curves. A comparison of the results attained by testing secondary standards with both real-time and digital assays should help determine the uniformity of the viral concentrations among the commercial secondary standards and the consistency of the results among different methodologies.
MATERIALS AND METHODS
Experimental design.
Three concentrations of each of the three commercially available secondary CMV standards, normalized by their respective manufacturers to WHO international units (IU) were tested in four replicates by two droplet digital PCR systems and by three real-time PCR assays. The quantitative results for each method were compared both to each other in a pairwise manner and to nominal values provided by each manufacturer. The results of the real-time methods were also compared to those of the digital methods, with the latter serving as a reference standard.
CMV standards.
Commercially available secondary, whole viral CMV standards that were normalized to the 1st WHO International Standard (IU) for CMV were purchased from AcroMetrix (CMVtc panel; AcroMetrix Inc., Benicia, CA), ZeptoMetrix (NATtrol cytomegalovirus linearity panel; ZeptoMetrix Corporation, Buffalo, NY), and SeraCare Life Sciences (CMV DNA AccuSpan linearity panel; SeraCare Life Sciences, Inc., Milford, MA) and used in the study. Serial 10-fold dilutions of each secondary standard were performed in an AcroMetrix EDTA plasma dilution matrix, resulting in final concentrations of 300 to 30,000 IU/ml (AcroMetrix standards) and 500 to 50,000 IU/ml (both ZeptoMetrix and SeraCare standards). Both AcroMetrix and SeraCare used the Cobas AmpliPrep/Cobas TaqMan CMV test (Roche Molecular Diagnostics, Pleasanton, CA) as their reference method for determining nominal values in their respective secondary standard, while ZeptoMetrix used an in-house PCR assay normalized to quantified CMV, commercially produced by Advanced Biotechnologies, Inc. (Columbia, MD).
Internal controls specific to each assay were added, and DNA from 200 μl of the CMV standard was extracted and then eluted into 60 μl using the Qiagen EZ1 DSP virus kit on the Qiagen EZ1 advanced XL system (Qiagen, Inc.). Eight aliquots of 200 μl from each concentration were processed, and extracts were pooled, aliquoted, and stored at −20°C until molecular analysis. For the Abbott RealTime assay, 800 μl of CMV standard and template internal control was extracted using the m2000 platform (Abbott Molecular Inc., Des Plaines, IL).
ddPCR.
The QX100 Droplet Digital PCR (ddPCR) system (Bio-Rad, Pleasanton, CA) was used with RealStar (RS) CMV analyte- specific reagents (ASR) (Altona Diagnostics). The ddPCR reaction mixture consisted of 10 μl of a 2× ddPCR master mix (Bio-Rad), 0.7 μl each of RS-ASR CMV-Prm and RS-ASR CMV-Prb (Altona), and 5 μl of nucleic acid solution in a final volume of 20 μl. The entire reaction mixture was used to produce the droplets on the droplet generator (Bio-Rad). After processing, the droplets were transferred to a 96-well PCR plate (Eppendorf, Germany) and amplified on a T100 thermal cycler (Bio-Rad) beginning at 95°C for 10 min, followed by 40 cycles of 94°C for 30 s and 58°C for 60 s and 1 cycle of 98°C for 10 min, ending at 12°C. The plate was read on the droplet reader (Bio-Rad) at a rate of 32 wells per hour. ddPCR data were analyzed with QuantaSoft analysis software (Bio-Rad), and the results were generated in copies/μl of PCR.
The RainDrop digital PCR system (RainDance Technologies, Billerica, MA) consists of two parts, the RainDrop source (droplet generator) and sense (reader/counter) instruments. The ddPCR reaction mixture consisted of 30 μl of RealStar general purpose reagent (RS-GPR) (Altona), 1.67 μl each of RS-ASR CMV-Prm and RS-ASR CMV-Prb (Altona), and 10 μl of nucleic acid solution in a final volume of 50 μl. The entire reaction mixture was transferred to one of the 8 wells on a source chip (RainDance Technologies). The loaded source chip and an 8-tube strip (0.2 ml) were inserted into the RainDance source instrument for droplet generation. After processing, droplets in the tube strip were amplified on a GeneTouch thermal cycler (Bior Technology, Hangzhou, China): 1 cycle at 95°C for 10 min, followed by 45 cycles of 95°C for 15 s and 58°C for 60 s, and 1 cycle of 98°C for 10 min, ending at 15°C. After amplification, the 8-tube strip and a sense chip (RainDance Technologies) were inserted into the sense instrument. The latter instrument identifies and counts droplets at a rate of 8 samples per 5 h. The run data were analyzed with RainDrop Analyst software, and the results were generated in copies/PCR.
Quantitative real-time PCR. (i) Altona RealStar CMV.
DNA extraction for the samples was performed on the Qiagen EZ1 advanced XL system as described above. The amplification reaction mixture consisted of 18 μl of a master mix of RS-GPR (Altona), 1 μl each of RS-ASR CMV-Prm and RS-ASR CMV-Prb (Altona Diagnostics), and 10 μl of nucleic acid solution in a final volume of 30 μl. The amplification was carried out on an ABI Prism 7500 SDS instrument (Applied Biosystems) under the thermal profile of 1 cycle of 95°C for 10 min, followed by 40 cycles of 94°C for 15 s and 58°C for 60 s, and ending at 1 cycle of 58°C for 10 min. A set of four-member quantification standards ranging from 1 × 10 to 1 × 104 IU/μl that were calibrated against the 1st WHO International Standard for human cytomegalovirus for nucleic acid amplification techniques was purchased from Altona Diagnostics and used to generate a calibration curve for the assay.
(ii) Qiagen artus CMV.
DNA extraction for the samples was performed on the Qiagen EZ1 advanced XL system as described above. The controls (two positive controls [low and high] and a negative control) (AcroMetrix Inc.) were also subjected to DNA extraction and included in each PCR run. Following DNA extraction, the artus CMV real-time PCR was performed using a Qiagen artus CMV kit (Qiagen, Inc., Valencia, CA) on the Abbott m2000rt instrument with the Qiagen recommended PCR setup and cycling conditions (artus CMV RG PCR kit handbook, January 2011 version; Qiagen). Four external quantitation standards supplied by Qiagen (CMV RG QS 1 to 4) were used to generate a calibration curve in order to estimate the CMV DNA concentrations in samples. These standards were provided by the manufacturer with nominal concentrations given only in copies/ml; IU/ml data were not available. Therefore, all results from the Qiagen assay are given in copies/ml.
(iii) Abbott real-time CMV.
Quantitation of CMV in plasma was carried out on the m2000 platform (Abbott Molecular Inc., Des Plaines, IL) that includes the m2000sp instrument for automated extraction of DNA and the m2000rt instrument for real-time PCR. DNA extraction was performed from 800 μl of plasma using the Abbott mSample preparation system DNA kit on the m2000sp instrument and eluted in a volume of 70 μl. An internal control was added to the lysis buffer prior to the extraction. The automated PCR setup was performed on the m2000sp instrument, and the PCR plate was loaded on the m2000rt instrument for quantitation of CMV DNA. Two positive controls (low and high) and a negative control (AcroMetrix Inc.) were included in each run. The AcroMetrix CMVtc panel run in duplicate was used to establish the calibration curve and calculate the CMV DNA concentrations in samples.
Statistical analysis.
Continuous variables were summarized by their means and standard deviations. Bias was estimated by the difference between the measured and nominal values. Analysis of covariance (ANCOVA) was first applied to examine whether the two digital assays (Bio-Rad and RainDance) yielded similar measures when each CMV standard was used. If the measures were found similar, an average of digital PCR values would be taken to represent the digital measure, which would be adopted in subsequent analyses. ANCOVA was also conducted to check whether each digital assay had similar measured values across the three CMV standards. Linear regressions were applied to examine the quantitative correlations of digital or real-time PCR measures against nominal values, as well as real-time PCR measures against average digital PCR values. Comparisons were also made by hypothesis tests against the line of identity based on linear regressions to check whether measures were close to nominal values and average digital PCR values. All statistical analyses were performed using SAS (SAS Institute, Cary, NC) with Windows (version 9.3).
RESULTS
Three concentrations of each of the three commercially prepared secondary CMV IU standards (AcroMetrix, ZeptoMetrix, and SeraCare) were tested in quadruplicate by three real-time and two digital PCR methods. The mean results for each concentration of each standard, stratified by method, are presented as the means of log10-transformed concentrations in Tables 1 to 3. Tables S1 to S3 in the supplemental material demonstrate the difference (bias) between mean measured results and the nominal values provided by each manufacturer for each concentration of standard. The overall bias is shown in Fig. 1. This represents the mean bias for each assay, and each quantitative standard (compared to nominal values), showing more than a log unit spread in some cases. Note that in many cases, the lowest concentration of the standard was either not detected or detected in some replicates.
TABLE 1.
Mean results achieved with each quantitative PCR method using the highest concentration of standard from each manufacturera
| Standard | Mean (SD) (log10 IU/ml) for: |
||||
|---|---|---|---|---|---|
| Bio-Rad | RainDance | Altona | Abbott | Qiagen | |
| AcroMetrix | 3.93 (0.07) | 3.84 (0.07) | 4.65 (0.08) | 4.27 (0.06) | 4.14 (0.02) |
| SeraCare | 4.92 (0.01) | 5.07 (0.41) | 5.40 (0.06) | 4.55 (0.14) | 4.99 (0.07) |
| ZeptoMetrix | 3.66 (0.06) | 3.63 (0.22) | 3.95 (0.12) | 3.81 (0.11) | 3.71 (0.002) |
The highest concentrations were 4.48 log10 IU/ml for AcroMetrix and 4.70 log10 IU/ml for ZeptoMetrix and SeraCare. Qiagen was compared against 4.71 log10 copies/ml for AcroMetrix, 4.00 log10 copies/ml for ZeptoMetrix, and 4.70 log10 copies/ml for SeraCare.
TABLE 2.
Mean results achieved with each quantitative PCR method using the second highest concentration of standard from each manufacturera
| Standard | Mean (SD) (log10 IU/ml) for: |
||||
|---|---|---|---|---|---|
| Bio-Rad | RainDance | Altona | Abbott | Qiagen | |
| AcroMetrix | 3.13 (0.16) | 2.95 (0.10) | 3.63 (0.13) | 3.39 (0.12) | 3.08 (0.03) |
| SeraCare | 3.96 (0.06) | 3.77 (0.08) | 4.13 (0.13) | 3.78 (0.09) | 3.89 (0.04) |
| ZeptoMetrix | 2.65 (0.15) | 2.52 (0.24) | 3.10 (0.04) | 2.77 (0.17) | 2.63 (0.10) |
The second highest concentrations were 3.48 log10 IU/ml for AcroMetrix and 3.70 log10 IU/ml for ZeptoMetrix and SeraCare. Qiagen was compared against 3.71 log10 copies/ml for AcroMetrix, 3.00 log10 copies/ml for ZeptoMetrix, and 3.70 log10 copies/ml SeraCare.
TABLE 3.
Mean results achieved with each quantitative PCR method using the lowest concentration of standard from each manufacturera
| Standard | Mean (SD) (log10 IU/ml) for: |
||||
|---|---|---|---|---|---|
| Bio-Rad | RainDanceb | Altona | Abbott | Qiagen | |
| AcroMetrix | 1.96 (0.02)c | 1.88 (0.35)c | 2.43 (0.40) | 2.44 (0.29) | 2.26 (0.04) |
| SeraCare | 3.01 (0.20) | 2.87 (0.37) | 3.50 (0.17) | 2.56 (0.16) | 2.94 (0.14) |
| ZeptoMetrix | 2.09 (0.20)c | 1.74 (0.37)c | 1.72 (0.31) | 1.65 (0.24)c | |
The lowest concentrations were 2.48 log10 IU/ml for AcroMetrix and 2.70 log10 IU/ml for ZeptoMetrix and SeraCare. Qiagen was compared against 2.71 log10 copies/ml for AcroMetrix, 2.00 log10 copies/ml for ZeptoMetrix, and 2.70 log10 copies/ml SeraCare.
No replicates were detected for ZeptoMetrix from RainDance.
n = <4 replicates detected and used.
FIG 1.
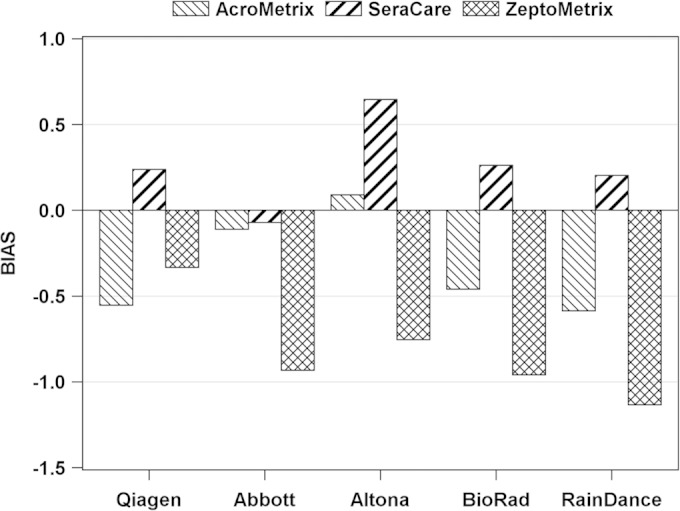
Overall bias between mean measured results and the nominal values provided by each manufacturer.
Compared against each other, the two ddPCR methods (Bio-Rad and RainDance) showed no significant difference in their quantitative relationships with nominal values for material from any of the three manufacturers using ANCOVA. This is also confirmed by Fig. 2 with plots of digital PCR results against nominal values, showing a similar pattern for Bio-Rad and RainDance. In both graphs, there is a clear difference in the amount of target present at a given nominal value among the secondary standard materials from each of the three manufacturers (P < 0.001 for Bio-Rad; P < 0.001 for RainDance).
FIG 2.
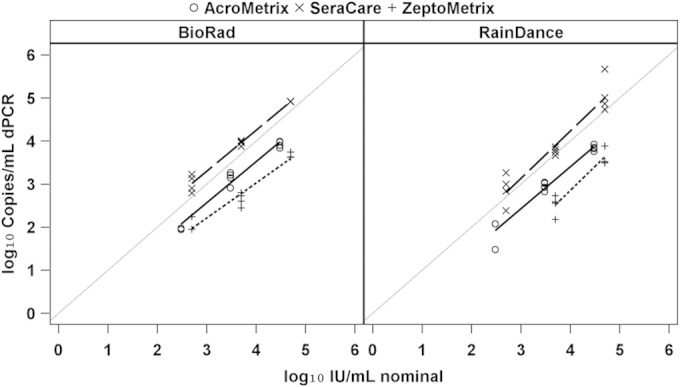
Regression analysis of measured values of ddPCR compared against nominal values provided by each manufacturer.
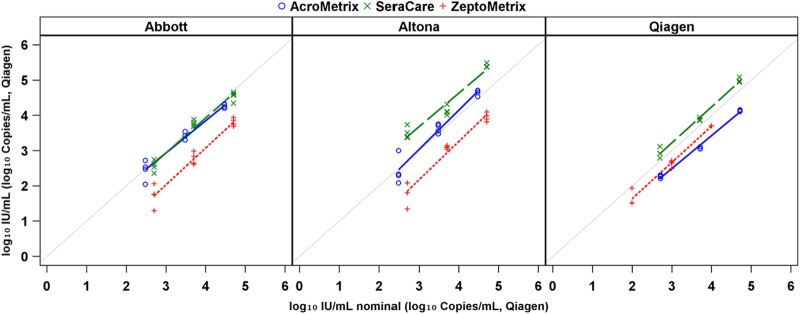
The differences in detected viral loads among like concentrations of secondary standards persisted when these materials were tested by real-time methods. The degree and significance of quantitative differences varied markedly based on the assay/standard combination and on whether real-time results were compared to nominal or digital values. The apparent difference between real-time results appeared to diminish when digital values were used (rather than nominal values provided by each manufacturer in IU). This is shown by a convergence of regression y intercepts (Fig. 3 and 4; see also Fig. S1 and Tables S4 and S5 in the supplemental material). If data are stratified by standard rather than by assay (see Fig. S1 and S2 and Tables S4 and S5 in the supplemental material), the convergence appears somewhat less marked, but generally the intercepts of regression lines still diverge from each other.
FIG 3.
Regression analysis of real-time PCR measures compared against nominal values stratified by assay.
FIG 4.
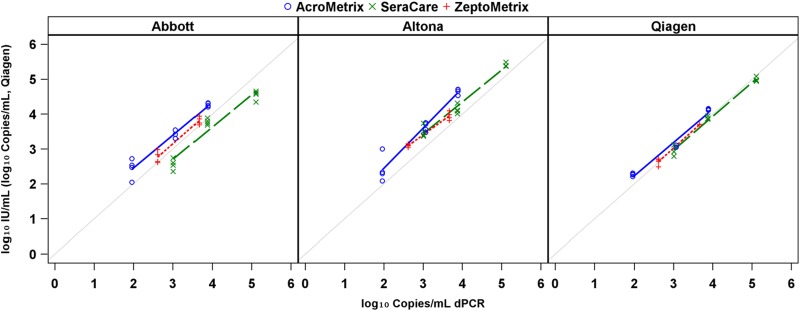
Regression analysis of real-time PCR measures compared against the average ddPCR measure stratified by assay.
DISCUSSION
Results here demonstrate the challenges in producing and implementing a common quantitative standard for CMV, particularly when one is faced with the myriad assays in current use. It has been hoped that the development of the 1st WHO International Standard for CMV might mitigate the variability among assay results previously demonstrated in the testing of common patient samples or proficiency testing materials. The application of this standard in a manner most likely to bring about consensus requires that the commercial secondary standards are in quantitative agreement with one another and with the primary WHO material. It also requires that such secondary standards behave similarly among different assays (similarly to the demonstration of commutability in the testing of patient samples) (20, 21). The data here suggest that these parameters have not been met. The use of digital PCR as a reference method demonstrated the lack of quantitative agreement among the three different secondary standards, while real-time methods showed disparate results for the same material tested with different assays.
Digital PCR offers numerous advantages when used as a quantitative reference standard. By design, it is a more absolute method of quantification, obviating the need for calibration curves and demonstrating reduced susceptibility to common sources of variability, such as amplification inhibition, seen in real-time methods. The use of two digital methods, as done here, added further robustness to the analysis. Both methods showed close quantitative agreement, despite marked disagreement among real-time assays, with the mean values achieved from both systems used for comparative purposes. Although proposed as a primary means of viral load testing, digital PCR may play a critical role in the evaluation of quantitative calibrators to be used for real-time methods. This might be in the comparison of different manufacturers' materials or in the lot-to-lot assessment of either primary or secondary standards. Here, the digital PCR values differed among all three secondary standards, despite the fact that the nominal values for two of those three standards were initially assigned using the same methodology. None of the manufacturers used digital methods in the assignment of nominal values. We have observed in our study that agreement across real-time assays seemingly converges compared against digital values. Although the explanation is unclear, this finding may support the role of digital PCR in serving as a valuable reference measure when the performance of secondary standards is characterized. The lack of uniformity in how nominal values are assigned may hinder agreement between standards and ultimately reduce agreement when clinical samples are tested. However, this may not represent the entire explanation for such disagreement.
While this study suggests disparity of content and performance among currently available quantitative CMV standards, it does not directly demonstrate the impact of such differences on testing of clinical patient samples. The use of common secondary standards in some cases has been shown to increase agreement among laboratories using various quantitative assays (6, 14, 20). However, noncommutable standards may actually decrease interlaboratory/interassay agreement (22). It is clear that in some cases, some secondary standards showed similar results across different assays, and some showed improved agreement compared with the digital results rather than with the nominal values of secondary standards. This may suggest that the materials behave differently with the experimental real-time methods than with the methods originally used for nominal value assignment, which may in turn suggest that when tested with clinical samples they may lack sufficient commutability or they may need to be normalized to IU with an assay-specific correction factor. Future studies may help clarify the impact of recalibration on the final quantitative results. The evaluation here of the AcroMetrix standards (particularly when one is looking at bias against nominal values using the Abbott method) was somewhat limited by the fact that the Abbott real-time assay was calibrated with the same material. This was an unavoidable limitation, but mitigated to some extent by the fact that results of all real-time methods were also compared against those of ddPCR.
It has been demonstrated that use of common quantitative calibrators can improve agreement among viral load tests (6, 13). That other factors may also contribute to such variability does not reduce the need for addressing this issue. The availability of an international quantitative CMV standard represents the best hope to date of bringing uniformity to high-impact clinical testing, used for some of our most acutely ill patients. The disparity of composition and performance found here among commercial secondary CMV standards suggests reasons that we may still be falling short of our goals and but also helps point the way to achieving those goals in the future. The increasing use of reference methodology such as digital PCR, together with assay-specific evaluation of quantitative standards, including both commutability and conversion to IU should lead to improved agreement, portability, and comparability of viral load results, essential for improved patient care and advances in the determination of clinically relevant quantitative thresholds for therapy.
Supplementary Material
ACKNOWLEDGMENTS
This study was supported by ALSAC and Life Span/Tufts/Brown CFAR, P30AI042853.
We thank Tony Godfrey (Boston University, Boston, MA) for the use of the RainDance digital PCR equipment in his laboratory.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JCM.03375-14.
REFERENCES
- 1.Fan H, Gulley ML. 2001. Epstein-Barr viral load measurement as a marker of EBV-related diseases. Mol Diagn 6:279–289. doi: 10.1054/modi.2001.29161. [DOI] [PubMed] [Google Scholar]
- 2.Gärtner B, Preiksaitis JK. 2010. EBV viral load detection in clinical virology. J Clin Virol 48:82–90. doi: 10.1016/j.jcv.2010.03.016. [DOI] [PubMed] [Google Scholar]
- 3.Gerna G, Campanini G, Rognoni V, Marchi A, Rovida F, Piralla A, Percivalle E. 2008. Correlation of viral load as determined by real-time RT-PCR and clinical characteristics of respiratory syncytial virus lower respiratory tract infections in early infancy. J Clin Virol 41:45–48. doi: 10.1016/j.jcv.2007.10.018. [DOI] [PubMed] [Google Scholar]
- 4.Heim A, Ebnet C, Harste G, Pring-Akerblom P. 2003. Rapid and quantitative detection of human adenovirus DNA by real-time PCR. J Med Virol 70:228–239. doi: 10.1002/jmv.10382. [DOI] [PubMed] [Google Scholar]
- 5.Ljungman P. 2010. Molecular monitoring of viral infections after hematopoietic stem cell transplantation. Int J Hematol 91:596–601. doi: 10.1007/s12185-010-0570-4. [DOI] [PubMed] [Google Scholar]
- 6.Hayden RT, Hokanson KM, Pounds SB, Bankowski MJ, Belzer SW, Carr J, Diorio D, Forman MS, Joshi Y, Hillyard D, Hodinka RL, Nikiforova MN, Romain CA, Stevenson J, Valsamakis A, Balfour HH Jr, U.S. EBV Working Group. 2008. Multicenter comparison of different real-time PCR assays for quantitative detection of Epstein-Barr virus. J Clin Microbiol 46:157–163. doi: 10.1128/JCM.01252-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hirsch HH, Lautenschlager I, Pinsky BA, Cardenoso L, Aslam S, Cobb B, Vilchez RA, Valsamakis A. 2013. An international multicenter performance analysis of cytomegalovirus load tests. Clin Infect Dis 56:367–373. doi: 10.1093/cid/cis900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hoffman NG, Cook L, Atienza EE, Limaye AP, Jerome KR. 2008. Marked variability of BK virus load measurement using quantitative real-time PCR among commonly used assays. J Clin Microbiol 46:2671–2680. doi: 10.1128/JCM.00258-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pang XL, Fox JD, Fenton JM, Miller GG, Caliendo AM, Preiksaitis JK. 2009. Interlaboratory comparison of cytomegalovirus viral load assays. Am J Transplant 9:258–268. doi: 10.1111/j.1600-6143.2008.02513.x. [DOI] [PubMed] [Google Scholar]
- 10.Hayden RT, Yan X, Wick MT, Rodriguez AB, Xiong X, Ginocchio CC, Mitchell MJ, Caliendo AM. 2012. Factors contributing to variability of quantitative viral PCR results in proficiency testing samples: a multivariate analysis. J Clin Microbiol 50:337–345. doi: 10.1128/JCM.01287-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Holden MJ, Madej RM, Minor P, Kalman LV. 2011. Molecular diagnostics: harmonization through reference materials, documentary standards and proficiency testing. Expert Rev Mol Diagn 11:741–755. doi: 10.1586/erm.11.50. [DOI] [PubMed] [Google Scholar]
- 12.Madej RM, Davis J, Holden MJ, Kwang S, Labourier E, Schneider GJ. 2010. International standards and reference materials for quantitative molecular infectious disease testing. J Mol Diagn 12:133–143. doi: 10.2353/jmoldx.2010.090067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.National Institute for Biological Standards and Control. 2013. 1st WHO International Standard for Epstein-Barr virus for nucleic acid amplification techniques. NIBSC code 09/260, National Institute for Biological Standards and Control, Potters Bar, Hertfordshire, United Kingdom. [Google Scholar]
- 14.Fryer JF, Heath AB, Anderson R, Minor PD, Collaborative Study Group. 2010. Collaborative study to evaluate the proposed 1st WHO international standard for human cytomegalovirus (HCMV) for nucleic acid amplification (NAT)-based assays. National Institute for Biological Standards and Control, Potters Bar, Hertfordshire, United Kingdom. [Google Scholar]
- 15.Hindson BJ, Ness KD, Masquelier DA, Belgrader P, Heredia NJ, Makarewicz AJ, Bright IJ, Lucero MY, Hiddessen AL, Legler TC, Kitano TK, Hodel MR, Petersen JF, Wyatt PW, Steenblock ER, Shah PH, Bousse LJ, Troup CB, Mellen JC, Wittmann DK, Erndt NG, Cauley TH, Koehler RT, So AP, Dube S, Rose KA, Montesclaros L, Wang S, Stumbo DP, Hodges SP, Romine S, Milanovich FP, White HE, Regan JF, Karlin-Neumann GA, Hindson CM, Saxonov S, Colston BW. 2011. High-throughput droplet digital PCR system for absolute quantitation of DNA copy number. Anal Chem 83:8604–8610. doi: 10.1021/ac202028g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sedlak RH, Jerome KR. 2013. Viral diagnostics in the era of digital polymerase chain reaction. Diagn Microbiol Infect Dis 75:1–4. doi: 10.1016/j.diagmicrobio.2012.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sykes PJ, Neoh SH, Brisco MJ, Hughes E, Condon J, Morley AA. 1992. Quantitation of targets for PCR by use of limiting dilution. Biotechniques 13:444–449. [PubMed] [Google Scholar]
- 18.Vogelstein B, Kinzler KW. 1999. Digital PCR. Proc Natl Acad Sci U S A 96:9236–9241. doi: 10.1073/pnas.96.16.9236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Haynes RJ, Kline MC, Toman B, Scott C, Wallace P, Butler JM, Holden MJ. 2013. Standard reference material 2366 for measurement of human cytomegalovirus DNA. J Mol Diagn 15:177–185. doi: 10.1016/j.jmoldx.2012.09.007. [DOI] [PubMed] [Google Scholar]
- 20.Caliendo AM, Shahbazian MD, Schaper C, Ingersoll J, Abdul-Ali D, Boonyaratanakornkit J, Pang XL, Fox J, Preiksaitis J, Schonbrunner ER. 2009. A commutable cytomegalovirus calibrator is required to improve the agreement of viral load values between laboratories. Clin Chem 55:1701–1710. doi: 10.1373/clinchem.2009.124743. [DOI] [PubMed] [Google Scholar]
- 21.Cattozzo G, Franzini C. 2013. Commutability: a peculiar property of calibration and control materials. definition and evaluation. Clin Chem Lab Med 51:e167–e168. [DOI] [PubMed] [Google Scholar]
- 22.Hayden RT, Shahbazian MD, Valsamakis A, Boonyaratanakornkit J, Cook L, Pang XL, Preiksaitis JK, Schonbrunner ER, Caliendo AM. 2013. Multicenter evaluation of a commercial cytomegalovirus quantitative standard: effects of commutability on interlaboratory concordance. J Clin Microbiol 51:3811–3817. doi: 10.1128/JCM.02036-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.