Abstract
We developed an in-house assay for the direct identification, by matrix-assisted laser desorption ionization–time of flight (MALDI-TOF) mass spectrometry, of yeasts in blood culture. Sixty-one representative strains from 12 species were analyzed in spiked blood cultures. Our assay accurately identified 95 of 107 (88.8%) positive blood cultures and outperformed the commercial Sepsityper kit (81.7% identification).
TEXT
Prompt, appropriate, first-line antifungal treatment has been shown to improve outcomes in patients with fungemia, but identification is often delayed by the need for subculture from positive blood cultures (1, 2). Mass spectrometry (MS) techniques based on matrix-assisted laser desorption ionization–time of flight (MALDI-TOF) technology have made it possible to identify species directly from positive blood cultures (3). The sample contains human cells, and identification protocols must lyse these cells efficiently (without disrupting the microorganism) and eliminate the unwanted human proteins. In-house protocols based on the lytic agent saponin have been described for bacterial identification (4, 5), but only a few assays have been developed for yeast-positive blood cultures (6–9). Bruker produces an easy-to-use kit, Sepsityper, which has yielded good results for the characterization of both bacteria and yeasts (10, 11). However, this commercial test has never been compared, for yeast identification, with any in-house protocol. We developed an in-house protocol for the direct identification, by MALDI-TOF MS, of the yeast species commonly isolated from blood cultures. We compared the results obtained by our assay to those obtained by Sepsityper.
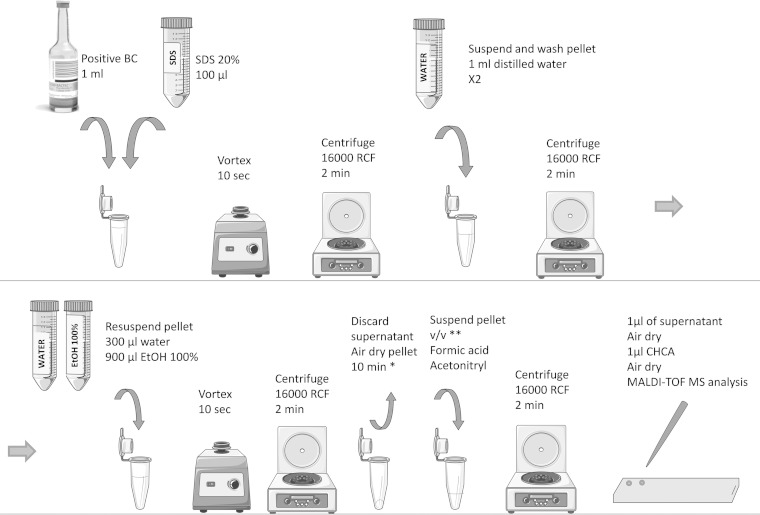
To facilitate the testing of large numbers of strains and species (12), we used blood collected from healthy volunteers and spiked the blood culture flasks with cultured yeasts. We used 61 strains formerly isolated in the context of invasive fungal infection (stored at – 80°C) and previously identified by MALDI-TOF MS. These strains were representative of 12 species commonly implicated in fungemia: Candida albicans (15), Candida glabrata (9), Candida parapsilosis (7), Candida kefyr (6), Candida krusei (6), Candida tropicalis (7), Cryptococcus neoformans (4), Candida dubliniensis (3), Candida utilis (1), Candida guilliermondii (1), Candida inconspicua (1), and Saccharomyces cerevisiae (1). Six were isolated directly from blood cultures from patients. Mycosis IC/F, a selective medium for the recovery of yeasts from blood, and Bactec Plus Aerobic/F or Bactec Plus Anaerobic/F (C. glabrata only) flasks, previously filled with 7 to 10 ml of healthy volunteers' blood, were inoculated (10 yeast cells/flask). Yeast suspensions, adjusted to one McFarland standard (approximately 10e7 CFU/ml), were serially diluted in sterile saline to obtain a final concentration of 100 CFU/ml. Then, 100 μl of this last dilution (average of 10 CFU) was inoculated in the blood culture and on a culture plate to control the inoculum. Flasks were incubated in Bactec FX (Becton Dickinson) until they tested positive. The average time to positivity depended on the species and on the type of culture flasks (from 21 to 66 h) (data not shown). Each positive flask was subjected to MALDI-TOF MS identification with a microflex MALDI-TOF mass spectrometer (Bruker Daltonics, Germany). We first compared 0.8% saponin (as described in reference 4), 1% Triton X-100 and 1.8% SDS (SDS1.8) (two other commonly used lytic agents), and the Sepsityper kit on the first 17 strains (randomly chosen). Two washing steps were included after lysis, because the inclusion of an additional washing step had been shown to improve identification scores (11) (data not shown). Figure 1 resumes the workflow for the SDS1.8 protocol. Briefly, the lytic agent was added to 1 ml of medium from each positive flask, which was then vortexed and pelleted (16,000 relative centrifugal force [RCF], 2 min). The supernatant was discarded and the pellet was washed twice in sterile distilled water (saponin, Triton X-100, and SDS1.8) or Sepsityper washing buffer. The pellet was suspended in 300 μl of distilled water and 900 μl of 100% ethanol. After centrifugation (16,000 RCF, 2 min), the pellet was air dried for 5 to 10 min. It was then homogenized with an appropriate quantity of formic acid (from 2 μl to 15 μl, depending on pellet volume; mean of 5 μl), and an equal volume of acetonitrile was added. Following a final centrifugation (16,000 RCF, 2 min), 1 μl of supernatant was dispensed, in duplicate, on a polished steel target and covered with 1 μl α-cyano-4-hydroxycinnamic acid (CHCA) for MALDI-TOF MS analysis. The spectra obtained were analyzed with the Biotyper software (Bruker database version 3.1.2.0, containing 3,995 entries). In light of our results and as previously proposed by other authors in the context of blood culture identification, the thresholds for interpretation were adapted (8, 13, 14). Identification scores were classified into four categories: A, >2; B, 1.7 to 2; C, 1.4 to 1.7, if the same species was called 4 times in a row; D, <1.4 or not identified. The first three categories yielded acceptable results for identification to the species level. In total, 305 tests were carried out (Fig. 2). According to our adapted scores, 95/107 (88.8%), 94/115 (81.7%), 16/42 (38%), and 12/41 (29.2%) tests led to correct identification for the SDS1.8, Sepsityper, saponin, and Triton X-100 protocols, respectively. Protocols based on saponin and Triton X-100 were the least efficient for yeast identification in blood cultures, possibly because they lysed human cells less efficiently, leading to a mixed (yeast and human) and thus less specific spectrum profile. The spectrum obtained with the SDS1.8 protocol was very similar to that obtained with the Sepsityper kit (Fig. 3). Based on these results, we limited subsequent comparisons to the Sepsityper and SDS1.8 protocols.
FIG 1.
Workflow of the SDS1.8 protocol. BC, blood culture; SDS, sodium dodecyl sulfate; EtOH, ethanol; HCCA, α-cyano-4-hydroxycinnamic acid; *, the pellet should be as dry as possible with no residual ethanol; **, volume of formic acid depending on the size of the pellet (formic acid should entirely recover the pellet). If no pellet is clearly visible, suspend the residual biological material, which should be present on the side of the tube, with 2 μl of formic acid and proceed.
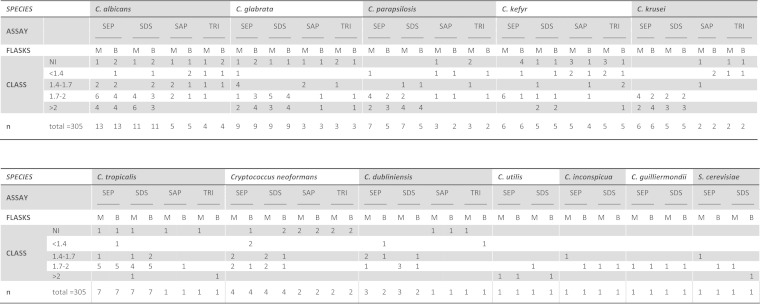
FIG 2.
Detailed results for each species and each assay. SEP, Sepsityper; SDS, 1.8% SDS; SAP, 0.8% saponin; TRI, 1% Triton X-100; M, mycosis medium; B, bacterial medium; NI, not identified.
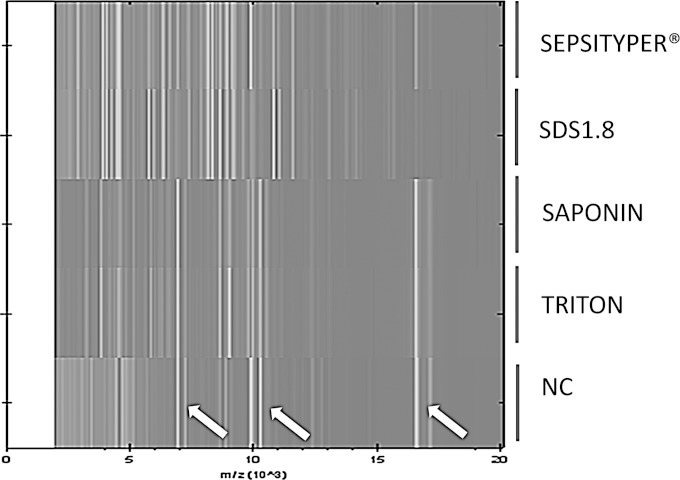
FIG 3.
Spectral profile obtained for Candida krusei isolated from mycosis blood culture and subjected to various lysis protocols. SDS1.8, 1.8% SDS; NC, negative control (mycosis blood culture negative on day 6). Arrows indicate the three principal nonspecific spectral signals present in negative controls but also identified in the saponin and Triton X-100 protocols.
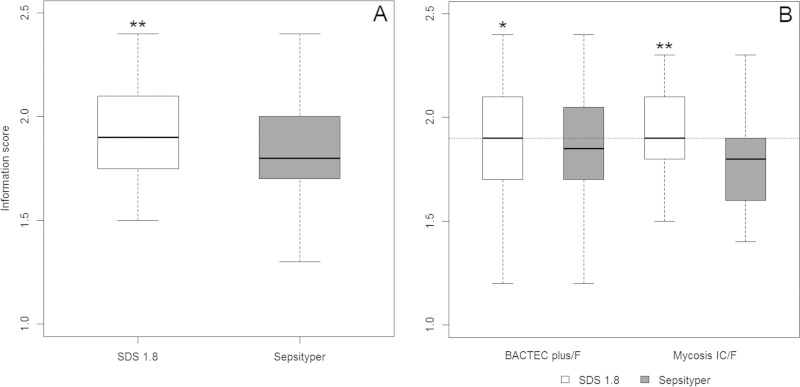
Among the 61 isolates, we analyzed 44 strains grown in both Mycosis IC/F and Bactec Plus/F flasks with the SDS1.8 and Sepsityper protocols (176 tests). Scores are reported as median values (interquartile range [IQR]). Scores were transformed into qualitative values, with four classes corresponding to the identification categories (see above). The Wilcoxon signed-rank sum test and Spearman's rank correlation analysis were used for quantitative scores, and Fisher's exact test was used for qualitative scores. The results obtained with the two protocols were well correlated (r = 0.70, P < 0.001). The median score was significantly higher with the SDS protocol (1.9 [IQR, 1.7 to 2.1]) than with Sepsityper (1.8 [IQR, 1.55 to 2.0]) (P = 0.003). This difference remained significant if separate analyses were carried out by vial type (Mycosis IC/F, P < 0.001; Bactec Plus, P = 0.04) (Fig. 4) and for the non-Candida albicans yeasts (C. albicans, P = 0.16; non-C. albicans yeasts, P < 0.001) (data not shown). A subgroup analysis by vial type showed that score class results were also better for the SDS assay than for the Sepsityper assay for Mycosis IC/F (P = 0.04) (Fig. 5).
FIG 4.
(A) Boxplots of scores by protocol: **, median score is significantly higher with the SDS protocol than with Sepsityper (P = 0.003). (B) Boxplots of scores by protocols and type of blood culture flasks: *, significant difference between the two protocols for Bactec Plus (P = 0.04); **, significant difference between the two protocols for Mycosis IC/F (P < 0.001).
FIG 5.
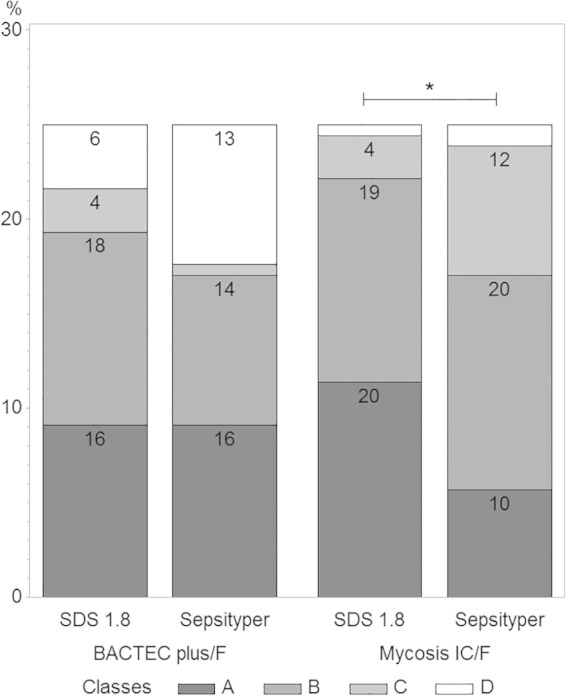
Diagram showing the distribution of identification score classes of 44 strains grown in both Mycosis IC/F and Bactec Plus/F flasks, with the SDS1.8 and Sepsityper protocols. The classes correspond to the following scores: A, >2; B, 1.7 to 2; C, >1.4; D, <1.4. *, Subgroup analysis: class-to-class comparison was significantly different between SDS1.8 and Sepsityper in the mycosis medium (P = 0.04).
This is the first comparison of an in-house protocol with the Sepsityper kit for the MALDI-TOF MS identification of yeasts in positive blood cultures. We found that lysis with 1.8% SDS was globally superior to the Sepsityper kit, particularly for yeasts isolated from Mycosis IC/F medium and for non-albicans species of Candida. The two protocols had similar durations (30 min), but the SDS1.8 protocol was cheaper than the Sepsityper protocol (by a factor of 220 [0.025 compared to 5.5 euros per test]). As the 0.8% saponin procedure gave poorer results than previously reported for bacteria, rapid identification procedures for positive blood cultures will probably need to be adapted according to the results of direct examination. The SDS1.8 protocol appears to be an effective, cheap alternative for yeast identification in positive blood cultures.
ACKNOWLEDGMENT
We have no conflicts of interest to declare.
REFERENCES
- 1.Puig-Asensio M, Pemán J, Zaragoza R, Garnacho-Montero J, Martín-Mazuelos E, Cuenca-Estrella M, Almirante B, Prospective Population Study on Candidemia in Spain (CANDIPOP) Project Hospital Infection Study Group (GEIH), Medical Mycology Study Group (GEMICOMED) of the Spanish Society of Infectious Diseases and Clinical Microbiology (SEIMC), Spanish Network for Research in Infectious Diseases. 2014. Impact of therapeutic strategies on the prognosis of candidemia in the ICU. Crit Care Med 42:1423–1432. doi: 10.1097/CCM.0000000000000221. [DOI] [PubMed] [Google Scholar]
- 2.Puig-Asensio M, Padilla B, Garnacho-Montero J, Zaragoza O, Aguado JM, Zaragoza R, Montejo M, Muñoz P, Ruiz-Camps I, Cuenca-Estrella M, Almirante B, CANDIPOP Project, GEIH-GEMICOMED (SEIMC), REIPI . 2014. Epidemiology and predictive factors for early and late mortality in Candida bloodstream infections: a population-based surveillance in Spain. Clin Microbiol Infect 20:O245–O254. doi: 10.1111/1469-0691.12380. [DOI] [PubMed] [Google Scholar]
- 3.Posteraro B, De Carolis E, Vella A, Sanguinetti M. 2013. MALDI-TOF mass spectrometry in the clinical mycology laboratory: identification of fungi and beyond. Expert Rev Proteomics 10:151–164. doi: 10.1586/epr.13.8. [DOI] [PubMed] [Google Scholar]
- 4.Martiny D, Dediste A, Vandenberg O. 2012. Comparison of an in-house method and the commercial SepsityperTM kit for bacterial identification directly from positive blood culture broths by matrix-assisted laser desorption-ionisation time-of-flight mass spectrometry. Eur J Clin Microbiol Infect Dis 31:2269–2281. doi: 10.1007/s10096-012-1566-1. [DOI] [PubMed] [Google Scholar]
- 5.Meex C, Neuville F, Descy J, Huynen P, Hayette M-P, De Mol P, Melin P. 2012. Direct identification of bacteria from BacT/ALERT anaerobic positive blood cultures by MALDI-TOF MS: MALDI Sepsityper kit versus an in-house saponin method for bacterial extraction. J Med Microbiol 61:1511–1516. doi: 10.1099/jmm.0.044750-0. [DOI] [PubMed] [Google Scholar]
- 6.Marinach-Patrice C, Fekkar A, Atanasova R, Gomes J, Djamdjian L, Brossas J-Y, Meyer I, Buffet P, Snounou G, Datry A, Hennequin C, Golmard J-L, Mazier D. 2010. Rapid species diagnosis for invasive candidiasis using mass spectrometry. PLoS One 5:e8862. doi: 10.1371/journal.pone.0008862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ferroni A, Suarez S, Beretti J-L, Dauphin B, Bille E, Meyer J, Bougnoux M-E, Alanio A, Berche P, Nassif X. 2010. Real-time identification of bacteria and Candida species in positive blood culture broths by matrix-assisted laser desorption ionization-time of flight mass spectrometry. J Clin Microbiol 48:1542–1548. doi: 10.1128/JCM.02485-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ferreira L, Sánchez-Juanes F, Porras-Guerra I, García-García MI, García-Sánchez JE, González-Buitrago JM, Muñoz-Bellido JL. 2011. Microorganisms direct identification from blood culture by matrix-assisted laser desorption/ionization time-of-flight mass spectrometry: blood culture pathogens direct identification by MALDI-TOF. Clin Microbiol Infect 17:546–551. doi: 10.1111/j.1469-0691.2010.03257.x. [DOI] [PubMed] [Google Scholar]
- 9.Spanu T, Posteraro B, Fiori B, D'Inzeo T, Campoli S, Ruggeri A, Tumbarello M, Canu G, Trecarichi EM, Parisi G, Tronci M, Sanguinetti M, Fadda G. 2012. Direct MALDI-TOF mass spectrometry assay of blood culture broths for rapid identification of Candida species causing bloodstream infections: an observational study in two large microbiology laboratories. J Clin Microbiol 50:176–179. doi: 10.1128/JCM.05742-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Buchan BW, Riebe KM, Ledeboer NA. 2012. Comparison of the MALDI Biotyper system using Sepsityper specimen processing to routine microbiological methods for identification of bacteria from positive blood culture bottles. J Clin Microbiol 50:346–352. doi: 10.1128/JCM.05021-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yan Y, He Y, Maier T, Quinn C, Shi G, Li H, Stratton CW, Kostrzewa M, Tang Y-W. 2011. Improved identification of yeast species directly from positive blood culture media by combining Sepsityper specimen processing and microflex analysis with the matrix-assisted laser desorption ionization Biotyper system. J Clin Microbiol 49:2528–2532. doi: 10.1128/JCM.00339-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fricker-Hidalgo H, Lebeau B, Pelloux H, Grillot R. 2004. Use of the BACTEC 9240 system with Mycosis-IC/F blood culture bottles for detection of fungemia. J Clin Microbiol 42:1855–1856. doi: 10.1128/JCM.42.4.1855-1856.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.La Scola B, Raoult D. 2009. Direct identification of bacteria in positive blood culture bottles by matrix-assisted laser desorption ionisation time-of-flight mass spectrometry. PLoS One 4:e8041. doi: 10.1371/journal.pone.0008041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Idelevich EA, Grunewald CM, Wüllenweber J, Becker K. 2014. Rapid identification and susceptibility testing of Candida spp. from positive blood cultures by combination of direct MALDI-TOF mass spectrometry and direct inoculation of Vitek 2. PLoS One 9:e114834. doi: 10.1371/journal.pone.0114834. [DOI] [PMC free article] [PubMed] [Google Scholar]