Abstract
The existing data support Portugal as the western European country with the highest HIV-1 subtype diversity. However, detailed phylogenetic studies of Portuguese HIV-1 epidemics are still scarce. Thus, our main goal was to analyze the phylodynamics of a local HIV-1 infection in the Portuguese region of Minho. Molecular epidemiological analysis was applied to data from 289 HIV-1-infected individuals followed at the reference hospital of the province of Minho, Portugal, at which isolated viruses had been sequenced between 2000 and 2012. Viruses of the G (29.1%) and B (27.0%) subtypes were the most frequent, followed by recombinant forms (17.6%) and the C (14.5%), F1 (7.3%), and A1 (4.2%) subtypes. Multinomial logistic regression revealed that the odds of being infected with the A1 and F1 subtypes increased over the years compared with those with B, G, or C subtypes or recombinant viruses. As expected, polyphyletic patterns suggesting multiple and old introductions of the B and G subtypes were found. However, transmission clusters of non-B and non-G viruses among native individuals were also found, with the dates of the most recent common ancestor estimated to be in the early 2000s. Our study supports that the HIV-1 subtype diversity in the Portuguese region of Minho is high and has been increasing in a manner that is apparently driven by factors other than immigration and international travel. Infections with A1 and F1 viruses in the region of Minho are becoming established and are mainly found in sexually transmitted clusters, reinforcing the need for more efficacious control measures targeting this infection route.
INTRODUCTION
Globally, at the end of 2012, 35.3 million people were estimated to be infected with HIV-1, and AIDS remains one of the world's most serious health challenges (1). Phylogenetically, HIV-1 is divided into four groups: M, O, N, and P. Most HIV-1 infections are globally caused by M group viruses that can be further divided into at least nine subtypes, A to D, F to H, J, and K, as well as different circulating and unique recombinant forms (2). Although clinical evidence is still limited, and current antiretroviral regimens appear to have comparable efficacies with all subtypes, there is presently evidence showing that particular HIV-1 subtypes may have a transmission advantage (3–6), higher replicative efficiency, or altered drug susceptibilities (7–11). The geographic patterns of the M group subtypes are continuously changing in response to human population migrations and active transmission networks, thus inciting constant vigilance. Although several reports suggest that the prevalences of non-B subtypes are increasing in western Europe (12–17), the B subtype remains the most prevalent. Portugal contrasts with the rest of western Europe in its distribution of HIV-1 subtypes. In addition to the B subtype, Portugal also has a high prevalence of the G subtype (18–23). The high prevalences of the B and G subtypes are thought to have promoted the appearance among intravenous drug users (IDU) of different types of B/G recombinant strains, namely, CRF14_BG, which is considered to have emerged in Portugal in the early 1990s and then spread to Spain and other European countries (24–26). The association between HIV-1 subtype and risk-behavior patterns has been complex to define, mainly due to difficulties in obtaining large numbers of each viral subtype and transmission route within a homogeneous study population (27).
Portugal has one of the highest HIV-1 prevalence rates in western Europe and, following a decrease in the last decade of HIV-1 infection in IDU, heterosexual contact is now estimated to be the most relevant transmission route in Portugal (28). The reconstruction of viral transmission networks is a relevant tool to monitor disease transmission and the effectiveness of adopted preventive measures, as well as to suggest more adequate control strategies for specific populations. Several studies have shown that in addition to a patient interview, phylogenetic analysis of the genetic sequences from the isolated viruses can provide valuable insights to help identify events of onward transmission and evaluate the spread of the virus (29–31). Nonetheless, limited information is available to understand HIV-1 transmission clusters in Portugal.
The aim of this study was to perform a molecular epidemiologic characterization of a cohort of 289 patients followed in the reference hospital in Minho province, Portugal. Specifically, we aimed to identify local transmission networks and possible relationships with previously described transmission clusters. In line with previous studies in Portugal, we found a large diversity of subtypes. In addition, our analysis supports that the transmission of nonpredominant subtypes among the local population began more than a decade ago, providing valuable insights into the dynamics of infection in this geographic area.
MATERIALS AND METHODS
Study population.
From 2000 to 2012, a total of 792 HIV-1 patients were followed at Hospital de Braga (HB), representing 60.3% of the HIV-1-infected individuals in the Portuguese region of Minho (28). Two hundred eighty-nine individuals from this cohort were selected for this study, according to two criteria: (i) availability of a plasma sample or plasma-derived viral sequence sampled from 2000 to 2012 and (ii) the absence of previous antiretroviral treatment at the time of sampling. The following information was collected anonymously from the clinical files of each individual: presumed transmission route, gender, age, nationality, presumed country of infection, and date of diagnosis. HB is a university-affiliated hospital serving as the reference hospital for the 1,093,021 habitants of the northwest Portuguese province of Minho. The prevalence of HIV-1 infection in the Minho region (0.12%) is lower than the overall prevalence in Portugal (0.31%) (28). According to transmission mode, the HB population presented significant differences from the data for the Portuguese HIV-1-infected individuals, with more IDU and fewer men who have sex with men (MSM) being infected. Also, in HB, there were more men and fewer patients >40 years old who were infected (see Table S1 in the supplemental material). The frequencies of individuals reporting heterosexual transmission are similar in a comparison of HB HIV-1 patients with the overall country data (see Table S1).
Sequencing of viral samples.
Viral RNA was extracted using MagNA Pure total nucleic acid isolation kits (Roche Applied Science). Reverse transcriptase PCR (RT-PCR) and DNA sequencing were performed with the Trugene HIV-1 genotyping system (Siemens Healthcare Diagnostics). The sequenced regions include part of the coding sequences of Gag (492 to 501), p6 (44 to 53), Pol (60 to 402), p2p7p1p6 (129 to 138), protease (4 to 99), and reverse transcriptase (RT) (1 to 127) (the reported positions are amino acid positions relative to the protein start in the HXB2 reference genome, GenBank accession no. K03455.1). The subtyping of the 289 sequences was made using REGA 3.0 (32). The subtyping results were confirmed using SimPlot and RDP (33, 34). The sequences were uploaded to GenBank and assigned the accession numbers KM205831 to KM206119.
Phylogenetic analysis.
The 289 HIV-1 sequences obtained in this study and 88 sequences from the databases, including the M group consensus and a previously defined set (32) of comprehensive subtype reference sequences with at least two reference sequences from each M group subtype (A1, A2, B, C, D, F1, F2, G, H, J, and K) and from 26 different circulating recombinant forms (CRF) (CRF01_AE, CRF02_AG, CRF03_AB, CRF04_CPX, CRF05_DF, CRF06_CPX, CRF10_CD, CRF11_CPX, CRF12_BF, CRF13_CPX, CRF14_BG, CRF18_CPX, CRF19_CPX, CRF20_BG, CRF24_BG, CRF25_CPX, CRF27_CPX, CRF29_BF, CRF31_BC, CRF35_AD, CRF37_CPX, CRF39_BF, CRF40_BF, CRF42_BF, and CRF47_BF), were aligned using MUSCLE (35). The phylogenetic analysis of the 377 sequences was conducted using RAxML 8.0.9 to produce a maximum likelihood tree using 1,000 bootstrapping replicates (36). The analysis was repeated with PhyML (37), computing the approximate likelihood ratio test (aLRT) support of all tree branches, and by Bayesian analysis using BEAST (38). The best-fitting nucleotide substitution model was estimated using jModeltest version 2.1.2 (39) to be the general time reversible (GTR) model, with a proportion of invariant site (I) and gamma distribution of rates (G), selected among 88 different models according to the Akaike information criterion (AIC), the Bayesian information criterion (BIC), and the decision theoretic framework (DT). An eventual bias introduced by convergent evolution due to the presence of drug-resistant mutations was discarded by repeating the analysis after the removal of codons associated with drug resistance in the standardized list of mutations for the surveillance of transmitted drug resistance established by the World Health Organization (40). The general topology of the trees and identification of the clustering remained unchanged. The identification of transmission clusters was based on the maximum likelihood tree, selecting the clusters from at least three individuals, with a bootstrap support of ≥95% and an average genetic distance of ≤0.03 substitutions per site (29, 41). We also used Bayesian inference, and all identified clusters showed a posterior probability equal to 1 (42).
Estimation of evolutionary dates.
Estimates of the time of the most recent common ancestor (MRCA) were performed by simultaneously inferring population parameters, substitution parameters, and tree topology using Bayesian Markov chain Monte Carlo (MCMC) inference, as implemented in BEAST version 1.8.0 (38). Three independent runs of 260 million replicates were performed under a Bayesian skyline relaxed molecular clock model, using a general time reversible nucleotide substitution model, with heterogeneity among the sites modeled with a gamma distribution. An examination of the MCMC samples with Tracer version 1.6 indicated convergence and adequate mixing of the Markov chains. After inspection with Tracer, we discarded an appropriate number of steps from each run as burn-in and combined the resulting MCMC tree samples for a subsequent estimation of posteriors using TreeAnnotator version 1.8.0. We summarized the MCMC samples using the maximum clade credibility tree, with the branch length depicted in years.
Statistical analysis.
To identify the main predictors of HIV-1 subtype group distribution, a multinomial logistic regression model was performed. Using this procedure, we assessed the association between the date of diagnosis and HIV-1 subtype group, controlling for other relevant variables. The HIV-1 subtypes with low numbers of cases were pooled, resulting in 5 groups that were used for statistical analysis: G (n = 84), B (n = 75), C (n = 42), other subtypes (A1, n = 12; F1, n = 21; total, n = 33), and recombinants (n = 42). The least represented subtypes, J (n = 1) and D (n = 3), had no influence on the results and were excluded from the analysis. The independent variables analyzed were date of diagnosis, age, gender, and transmission route. The IBM SPSS package (version 19) was used to conduct all statistical analysis, and the results were considered to be significant at a P value of <0.05.
Ethics.
The project was approved by the ethics committee of the HB. Written consent was obtained from all patients enrolled in the study. The clinical data were codified to ensure the confidentiality of the patients.
RESULTS
High HIV-1 subtype diversity.
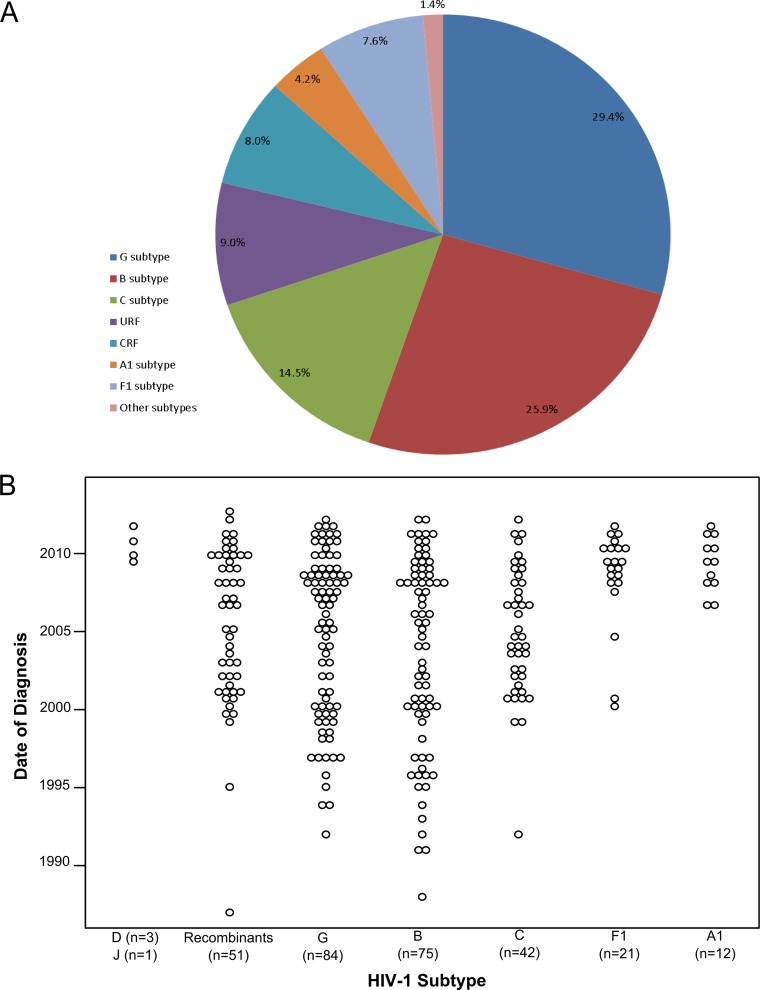
Among the 289 individuals who met the inclusion criteria, 76.8% were male, and the average age at diagnosis in the study population was 44.5 years (range, 18 to 87 years) (Table 1). The most frequently reported route of infection was heterosexual contact (n = 161 [55.7%]), followed by IDU (n = 99 [34.2%]) and MSM (n = 26 [9.0%]) (Table 1). In accordance with the population of Minho, the study population was highly homogeneous, with >90% of the individuals being Portuguese, of white ethnicity, and presumed to have been infected in Portugal. The most frequent subtypes found were G (n = 85 [29.4%]), B (n = 75 [26%]), and C (n = 42 [14.5%]), followed by the F1 (n = 22 [7.6%]) and A1 (n = 12 [4.2%]) subtypes. Only 1.4% of the studied individuals were infected with other “pure” subtypes (D, n = 3; J, n = 1). The most frequently found CRF was CRF14_BG (n = 15 [5.2%]), followed by CRF02_AG (n = 4 [1.4%]). Individuals infected with unique recombinant forms (URF) constituted 9.0% of the population (Fig. 1A).
TABLE 1.
Demographics of the study population
| Variablea | No. | % |
|---|---|---|
| Gender | ||
| Male | 222 | 76.8 |
| Female | 67 | 23.2 |
| Age at diagnosis (yr) | ||
| ≤20 | 17 | 5.9 |
| 21–40 | 190 | 65.7 |
| 41–50 | 41 | 14.2 |
| >50 | 41 | 14.2 |
| Patient nationality | ||
| Portuguese | 260 | 90.0 |
| Other | 29 | 10.0 |
| Ethnicity | ||
| White | 274 | 94.8 |
| Black | 15 | 5.2 |
| Presumed country of infection | ||
| Portugal | 280 | 96.9 |
| Other | 9 | 3.1 |
| Route of transmission | ||
| Heterosexual contact | 161 | 55.7 |
| MSM | 26 | 9.0 |
| IDU | 99 | 34.2 |
| Other | 3 | 1.1 |
| Total | 289 | 100 |
IDU, intravenous drug users; MSM, men who have sex with men.
FIG 1.
HIV-1 subtype diversity and temporal distribution of the date of diagnosis in the cohort of 289 HIV-1-infected individuals from the province of Minho, Portugal.
Increasing incidence of infection with A1 and F1 HIV-1 subtypes.
The dates of HIV-1 diagnosis in the study population spanned the period 1987 through 2012. We investigated the proportion of infections diagnosed each year with different subtypes. The less frequent subtypes were pooled to allow for statistical analysis (Fig. 1B), and a significant multinomial logistic regression model was obtained (χ2(df = 20) = 77.3; P < 0.001; pseudo-R2Nagelkerke, 0.245). The model (Table 2) shows that the chance of being infected with the A1 or F1 subtype increased during the period of time studied compared to that with the other subtypes (odds ratio [OR], 0.852 and P < 0.05 for recombinant viruses; OR, 0.824 and P < 0.01 for G subtypes; OR, 0.781 and P < 0.001 for B subtypes; and OR, 0.862 and P < 0.05 for C subtypes). Overall, these data support the hypothesis that infection with A1 and F1 subtypes began to appear recently in Minho province but are becoming established.
TABLE 2.
Multinomial logistic regression model relating HIV-1 subtypes with date of diagnosis, age, gender, and presumed transmission mode
| Subtype comparison | Variable | Be | SE | Walda | ORb | 95% CIc |
|
|---|---|---|---|---|---|---|---|
| LB | UB | ||||||
| Recombinants vs A1/F1 | Days (or yr) since date of diagnosis | −0.0004 | 0.000 | 6.278* | 1.000 (0.852) | 0.999 | 1.000 |
| Age | −0.006 | 0.019 | 0.091 | 0.994 | 0.959 | 1.031 | |
| Male | 0.298 | 0.638 | 0.218 | 1.347 | 0.386 | 4.699 | |
| Heterosexual | −0.808 | 0.735 | 1.207 | 0.446 | 0.106 | 1.884 | |
| IDUd | −0.346 | 0.820 | 0.178 | 0.708 | 0.142 | 3.533 | |
| G vs A1/F1 | Days (or yr) since date of diagnosis | −0.001 | 0.000 | 10.774** | 0.999 (0.824) | 0.999 | 1.000 |
| Age | 0.016 | 0.016 | 1.100 | 1.016 | 0.986 | 1.048 | |
| Male | −0.847 | 0.476 | 3.167 | 0.429 | 0.169 | 1.090 | |
| Heterosexual | 1.450 | 1.161 | 1.559 | 4.264 | 0.438 | 41.537 | |
| IDU | 2.012 | 1.214 | 2.747 | 7.475 | 0.693 | 80.672 | |
| B vs A1/F1 | Days (or yr) since date of diagnosis | −0.001 | 0.000 | 17.319*** | 0.999 (0.781) | 0.999 | 1.000 |
| Age | −0.016 | 0.017 | 0.932 | 0.984 | 0.952 | 1.017 | |
| Male | −0.473 | 0.512 | 0.856 | 0.623 | 0.229 | 1.698 | |
| Heterosexual | −1.234 | 0.663 | 3.459 | 0.291 | 0.079 | 1.069 | |
| IDU | −1.593 | 0.771 | 4.269* | 0.203 | 0.045 | 0.921 | |
| C vs A1/F1 | Days (or yr) since date of diagnosis | −0.000 | 0.000 | 5.154* | 1.000 (0.862) | 0.999 | 1.000 |
| Age | 0.017 | 0.019 | 0.815 | 1.017 | 0.980 | 1.056 | |
| Male | −0.646 | 0.579 | 1.242 | 0.524 | 0.168 | 1.632 | |
| Heterosexual | −0.267 | 0.949 | 0.079 | 0.766 | 0.119 | 4.919 | |
| IDU | 1.216 | 0.996 | 1.491 | 3.374 | 0.479 | 23.754 | |
*, P < 0.05; **, P < 0.01; ***, P < 0.001.
OR, odds ratio.
95% CI, 95% confidence interval; LB, lower bound; UB, upper bound.
IDU, intravenous drug users.
B, regression coefficient.
Evidence for local transmission clusters.
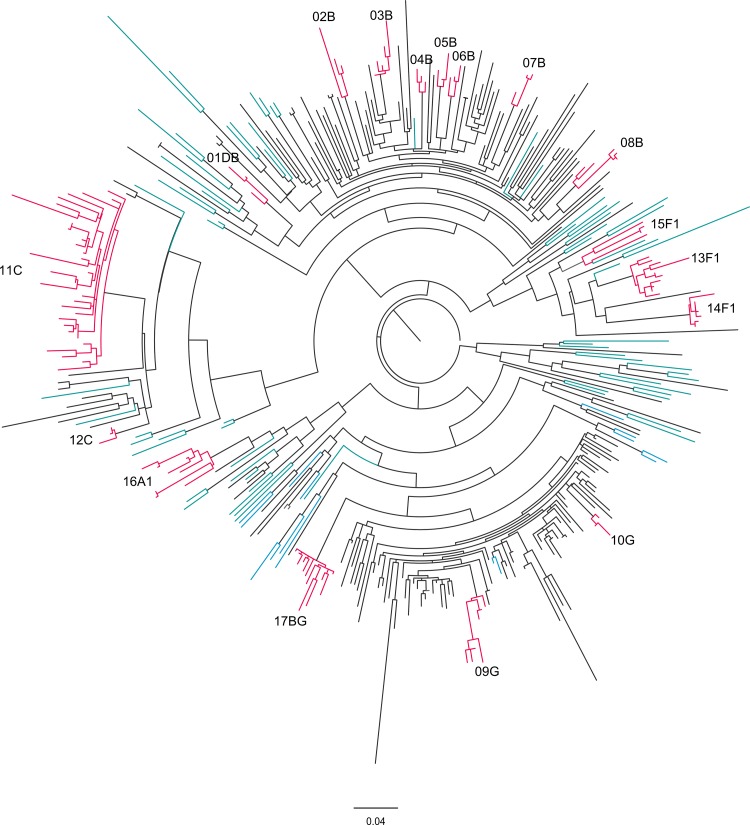
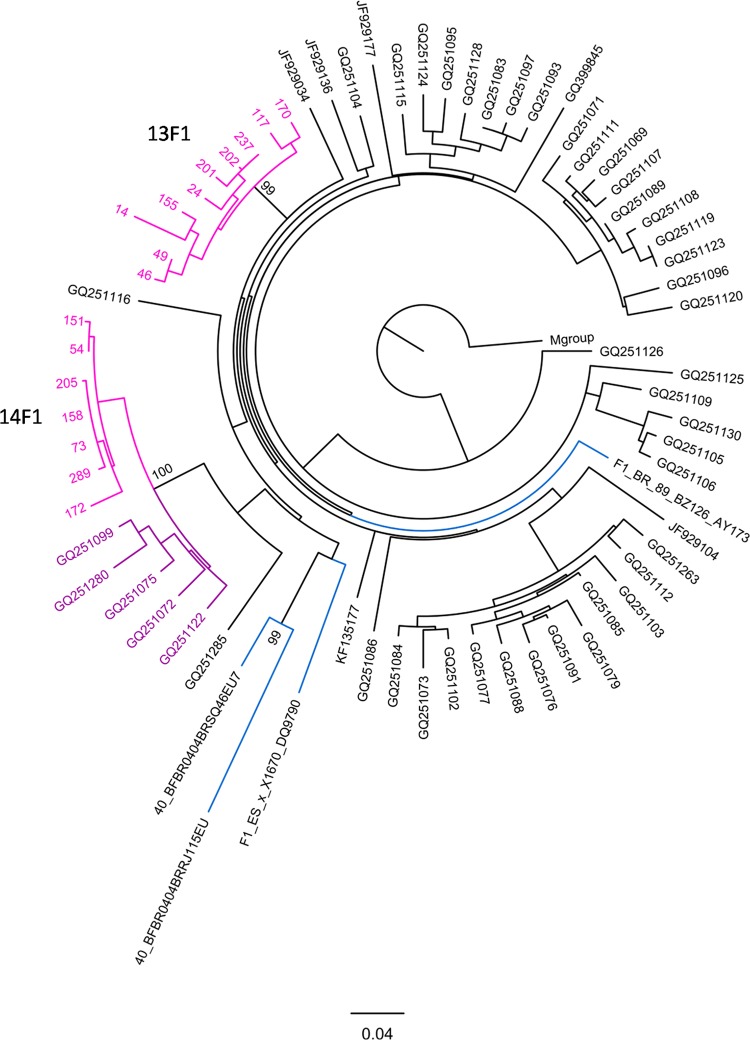
The phylogenetic analysis of the viral sequences allowed the identification of 17 transmission clusters (Fig. 2). This analysis was performed using a maximum likelihood tree and confirmed by Bayesian inference (see Table S2 in the supplemental material). Of all the sequences, 39.4% grouped in the 17 transmission clusters. In terms of HIV-1 subtype distribution, 7 clusters included subtype B viruses, three included subtype F1, two included subtype G, two included subtype C, one included subtype A1, one included CRF14_BG, and one included DB URF (Table 3). The distribution of the clustered sequences per subtype showed that 95.5% of all F1 sequences (21 out of 22) were incorporated in clusters. Respectively, 66.7%, 80.9%, and 80% of all A1, C, and CRF14_BG sequences were found in clusters. In contrast, the B and G subtype clusters include only 35.9% and 11.8% of the B and G subtype sequences, respectively. The mean number of individuals per cluster was 6.9. Among the clusters with a number of individuals that was above the mean, the largest cluster was 11C. It was composed almost entirely of IDU (27 out of 31 individuals), and 4 of these individuals reported sharing injection material. The second largest cluster was 17BG, and it was also composed mainly of IDU (11 out of 12 individuals), with the non-IDU individual in the cluster reporting sexual contact with one of the other members of the cluster. For all F1 clusters (13F1, 14F1, and 15F1), the transmission mode was predominantly sexual (Table 3). In the 13F1 cluster, 1 patient out of 10 was of Brazilian nationality. Furthermore, the phylogenetic analysis shows that the viral sequences from the 13F1 and 14F1 clusters have common ancestors with reference sequences collected in Brazil (GenBank accession no. AY173957.1, EU735538.1, and EU735540.1). Additionally, BLAST analysis identified 5 sequences highly related to the 14F1 cluster (GenBank accession no. GQ251099, GQ251280, GQ251075, GQ251072, and GQ251122; Fig. 3) viruses that were isolated in Italy and phylogenetically linked to those in Brazil (43). Cluster 16A1 was a sexually transmitted cluster (5 heterosexual and 3 MSM).
FIG 2.
Phylogenetic relationships among the HIV-1 sequences isolated from 289 infected individuals from Minho province, Portugal. Maximum likelihood (ML) phylogenetic analysis was performed using 289 partial HIV-1 sequences obtained in this study and 89 subtype reference sequences (colored in blue) and rooted using the M-group consensus sequence. The branch lengths are expressed as the number of nucleotide substitutions per site. The transmission clusters (colored in pink) were supported by an ML bootstrap support of ≥95% based on 1,000 replicates, an average genetic distance of ≤0.03 substitutions per site, and a Bayesian posterior probability equal to 1.
TABLE 3.
Characterization of the 17 HIV-1 transmission clusters identified in the study population of the Portuguese region of Minho
| Cluster name | No. of individuals | HIV-1 subtype | Yr of diagnosis (mean, range) | Route of transmission (n)a | Time of MRCA |
|
|---|---|---|---|---|---|---|
| Yr | 95% HPD | |||||
| 01DB | 3 | DB URF | 2009, 2006–2012 | IDU (4), heterosexual (1) | 2002 | 1998.2–2006.1 |
| 02B | 3 | B | 2006, 2003–2008 | IDU (2), heterosexual (1) | 1999 | 1994.2–2003.3 |
| 03B | 5 | B | 2003, 2000–2009 | Heterosexual (2), MSM (1) | 2004 | 2001.9–2006.5 |
| 04B | 3 | B | 2007, 2006–2009 | Heterosexual (4) | 2004 | 2000.7–2006.9 |
| 05B | 3 | B | 2006, 2003–2008 | MSM (3), heterosexual (2) | 2003 | 2000.0–2005.6 |
| 06B | 3 | B | 2008, 2005–2010 | Heterosexual (3) | 2002 | 1998.4–2004.8 |
| 07B | 3 | B | 2009, 2008–2010 | Heterosexual (3) | 2004 | 2000.0–2006.9 |
| 08B | 5 | B | 2007, 2000–2010 | Heterosexual (2), MSM (1) | 2002 | 1998.1–2004.4 |
| 09G | 7 | G | 2006, 1999–2009 | Heterosexual (5), IDU (2) | 2000 | 1997.1–2003.2 |
| 10G | 3 | G | 2010, 2008–2011 | Heterosexual (2), IDU (1) | 2008 | 2004.8–2010.4 |
| 11C | 31 | C | 2004, 1999–2011 | IDU (27), heterosexual (4) | 1994 | 1990.4–1998.1 |
| 12C | 3 | C | 2006, 2001–2009 | Heterosexual (3) | 2005 | 2001.6–2007.6 |
| 13F1 | 10 | F1 | 2006, 2000–2011 | Heterosexual (7), IDU (3) | 2000 | 1993.2–2003.5 |
| 14F1 | 7 | F1 | 2009, 2007–2011 | Heterosexual (6), IDU (1) | 2005 | 2002.7–2007.2 |
| 15F1 | 4 | F1 | 2010, 2009–2010 | Heterosexual (4) | 1994 | 1987.5–2001.6 |
| 16A1 | 8 | A1 | 2009, 2006–2011 | Heterosexual (5), MSM (3) | 1999 | 1994.0–2003.3 |
| 17BG | 12 | CRF14_BG | 2002, 2000–2006 | IDU (10), heterosexual (2) | 1999 | 1994.5–2002.8 |
IDU, intravenous drug users; MSM, men who have sex with men.
MRCA, most recent common ancestor; HPD, high posterior density.
FIG 3.
Phylogenetic relationships between the 13F1 and 14F1 transmission cluster sequences and public sequences available in the databases. Maximum likelihood (ML) phylogenetic analysis was performed using 13F1 and 14F1 sequences (pink), 4 subtype references (blue), and the 50 most similar sequences obtained in the BLAST query of all publically available sequences. Highlighted in purple are the public sequences that are included in cluster 14F1. The tree was rooted using the M-group consensus.
Established local transmission of non-B/G HIV-1 subtypes.
In order to estimate the evolutionary dates of the reported transmission clusters, we performed a Bayesian MCMC analysis. All independent runs converged to almost identical values for all parameters (data not shown). The mean substitution rate was 2.16 × 10−3 (95% highest posterior density [HPD] interval, 1.85 × 10−3 to 2.49 × 10−3) substitutions per site per year. The date of the most recent common ancestor (MRCA) was determined for all the clusters and ranged from 1994 to 2008 (Table 3). There were no marked differences in the MRCA dates among the different subtypes. Among the clusters of nonprevalent HIV-1, the 11C, 15F1, 16A1, and 17BG subtypes were the ones with older MRCA dates, ranging from 1994 to 1999. These clusters are composed of viruses from Portuguese individuals presumed to be infected in Portugal (Table 3). These results support the existence of clusters of nonprevalent subtypes that started transmitting among the local population more than a decade ago. To gain insights into the activities of the clusters over the years, the date of HIV-1 diagnosis for all cluster-included individuals was analyzed. For the majority of the clusters, the results are suggestive of continuous onward transmission (Table 3). A possible exception is cluster 17BG, since it showed an earlier mean date of diagnosis (2002) and since in the 6 years from 2006 to 2012, no viruses belonging to this cluster were found in the study population, thus suggesting a decrease in transmission.
DISCUSSION
Since its origin in Africa approximately 100 years ago, HIV-1 has continuously been undergoing genetic diversification that is enhanced by the massive globalization of the human population (21). Despite the perception that current antiretroviral regimens have comparable efficacies across existing diverse HIV-1 subtypes, there is evidence showing that some HIV-1 subtypes may have a transmission advantage, higher replicative efficiency, or even altered drug susceptibility (7–11), raising awareness on the relevance of investigating HIV-1 diversity. Furthermore, for the effective targeting of preventive measures, it is very relevant to perform persistent monitoring of the HIV-1 pandemic using phylogenetic and epidemiological data analyses as tools in the reconstruction of viral transmission networks (27–29).
Existing data show that Portugal contrasts with the rest of western Europe in its distribution of HIV-1 subtypes (18, 21). It is important to gain a further understanding on the causes underlying this difference, also in light of the evidence supporting a recent increase in infections with non-B HIV-1 subtypes in several western European countries (15–17, 44). In this study, we analyzed 289 HIV-1-infected individuals from Minho province, Portugal. Collectively, the results obtained are consistent with previous studies in Portugal showing a high prevalence of non-B subtypes (73.0%), mainly viruses of the G subtype (29.4%), followed by those of the C (14.5%) subtype. In our study population, the heterogeneity of HIV-1 subtypes is attributable to Portuguese-born individuals presumed to be infected in the region, with only 3.8% of the cases occurring in immigrants or individuals from Portugal presumed to be infected elsewhere. Contrarily, the rising prevalence of non-B HIV-1 subtypes in western Europe has been attributed to the growing number of immigrants from sub-Saharan Africa and South America, where these variants are prevalent. As an example, 27% of the HIV-1 cases diagnosed in 2007 in Spain were of non-B subtypes, and 90% of these cases were African and South American immigrants (45). A phylogenetic analysis of our study population indicates high interindividual genetic distances for the most prevalent subtypes, B and G, suggesting old and multiple introductions of viruses of these subtypes in the region. This observation is in accordance with the predominance of the B subtype in western Europe, having probably been introduced on several occasions in the late 1970s and early 1980s (46). As for the G subtypes, it is possible that the intense human migrations between Portugal and its former African colonies in the 1970s and 1980s, due mostly to the independence wars (47), contributed to its early introduction in Portugal. These intense human migrations may have also contributed to the early introduction of other HIV-1 subtypes, namely in the case of the migration connection with Angola, where there is a large HIV-1 genetic diversity (48). The distribution of clustered sequences per subtype showed that the vast majority of the F1, C, A1, and CRF14_BG sequences were incorporated in clusters. Contrarily, the B and G subtype clusters include only 35.9% and 11.8% of the respective subtype sequences, likely due to the long-time circulation of B and G subtypes among the studied population. Interestingly, clusters of non-B and non-G subtypes were also found, namely those of the C, F1, A1, and CRF14_BG viruses, with MRCA dates in the late 1990s, even when considering only Portuguese-born individuals presumed to be infected in the region. This supports the hypothesis that these HIV-1 subtypes were introduced in Minho more than one decade ago. The two largest transmission clusters, 11C and 17BG, are in a large majority (>87%) composed of IDU, reflecting the compartmentalization and closed character of the transmission among individuals from this risk group. The analysis of the date of diagnosis of the individuals in these clusters suggests that its transmission might be decreasing, namely in the case of 17BG, since in the 6 years ranging from 2006 to 2012, no HIV-1 infection with viruses from this cluster were found in the study population. In the last decade, other local epidemics with CRF14_BG have been described among IDU in Spain and Portugal (20, 22). Our results are in line with the data showing a decrease in the prevalence of HIV-1 among IDU in Portugal (28), consequently reducing the transmission of BG recombinant viruses. Importantly, our data support an increased incidence of infection with the F1 and A1 subtypes in the study population. The occurrence of onward transmission events of F1 and A1 HIV-1 subtypes in our study population was strongly linked to sexual transmission. We identified three F1 clusters, mainly formed by individuals who report heterosexual contact as the presumed viral transmission route. The phylogenetic analysis shows that the viral sequences from clusters 13F1 and 14F1 have common ancestors with reference sequences collected in Brazil. The analysis of the public databases allowed the identification of 5 sequences isolated in Italy that belong to the 14F1 cluster, thus suggesting a large geographic range of this transmission network, contrary to what was found in all the other clusters. In common with the 13F1 and 14F1 clusters, the previously described Italian cluster was also phylogenetically linked to Brazil (43), suggesting this country as the origin of subtype F1 viruses in southern Europe. We also identified one sexually transmitted A1 subtype cluster (16A1) that was epidemiologically linked to Mozambique, since a viral sequence isolated from one Mozambican presumed to be infected prior to immigrating to Portugal had a common ancestor with the cluster.
Overall, our study of the local HIV-1 epidemic in the Portuguese region of Minho supports that it contrasts with other regions of western Europe in its high HIV-1 diversity. This diversity is increasing due to transmission among native individuals and not in a manner driven by immigration or international travel. Our molecular and epidemiologic analyses highlight the onward transmission of the F1 and A1 subtype viruses via sexual transmission routes, supporting the need for continuous monitoring and strengthening of preventive strategies targeted at this transmission mode.
Supplementary Material
ACKNOWLEDGMENTS
N.S.O. is supported by the FCT Investigator Program (IF/00474/2014). M.S. is an Associate FCT-Investigator. This work is cofunded by the Programa Operacional Regional do Norte (ON.2 – O Novo Norte), Quadro de Referência Estratégico Nacional (QREN), through the Fundo Europeu de Desenvolvimento Regional (FEDER).
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JCM.03611-14.
REFERENCES
- 1.Joint United Nations Programme on HIV/AIDS (UNAIDS). 2013. Global report: UNAIDS report on the global AIDS epidemic 2013. Joint United Nations Programme on HIV/AIDS (UNAIDS), Geneva, Switzerland: http://www.unaids.org/sites/default/files/en/media/unaids/contentassets/documents/epidemiology/2013/gr2013/UNAIDS_Global_Report_2013_en.pdf. [Google Scholar]
- 2.Hemelaar J, Gouws E, Ghys PD, Osmanov S. 2006. Global and regional distribution of HIV-1 genetic subtypes and recombinants in 2004. AIDS 20:W13–W23. doi: 10.1097/01.aids.0000247564.73009.bc. [DOI] [PubMed] [Google Scholar]
- 3.Morison L, Buve A, Zekeng L, Heyndrickx L, Anagonou S, Musonda R, Kahindo M, Weiss HA, Hayes RJ, Laga M, Janssens W, van der Groen G, Study Group on Heterogeneity of HIV Epidemics in African Cities. 2001. HIV-1 subtypes and the HIV epidemics in four cities in sub-Saharan Africa. AIDS 15:S109–S116. doi: 10.1097/00002030-200108004-00012. [DOI] [PubMed] [Google Scholar]
- 4.Hudgens MG, Longini IM Jr, Vanichseni S, Hu DJ, Kitayaporn D, Mock PA, Halloran ME, Satten GA, Choopanya K, Mastro TD. 2002. Subtype-specific transmission probabilities for human immunodeficiency virus type 1 among injecting drug users in Bangkok, Thailand. Am J Epidemiol 155:159–168. doi: 10.1093/aje/155.2.159. [DOI] [PubMed] [Google Scholar]
- 5.Bobkov AF, Kazennova EV, Sukhanova AL, Bobkova MR, Pokrovsky VV, Zeman VV, Kovtunenko NG, Erasilova IB. 2004. An HIV type 1 subtype A outbreak among injecting drug users in Kazakhstan. AIDS Res Hum Retroviruses 20:1134–1136. doi: 10.1089/aid.2004.20.1134. [DOI] [PubMed] [Google Scholar]
- 6.Kiwanuka N, Laeyendecker O, Quinn TC, Wawer MJ, Shepherd J, Robb M, Kigozi G, Kagaayi J, Serwadda D, Makumbi FE, Reynolds SJ, Gray RH. 2009. HIV-1 subtypes and differences in heterosexual HIV transmission among HIV-discordant couples in Rakai, Uganda. AIDS 23:2479. doi: 10.1097/QAD.0b013e328330cc08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Atlas A, Granath F, Lindström A, Lidman K, Lindbäck S, Alaeus A. 2005. Impact of HIV type 1 genetic subtype on the outcome of antiretroviral therapy. AIDS Res Hum Retroviruses 21:221–227. doi: 10.1089/aid.2005.21.221. [DOI] [PubMed] [Google Scholar]
- 8.Keller M, Lu Y, Lalonde RG, Klein MB. 2009. Impact of HIV-1 viral subtype on CD4+ T-cell decline and clinical outcomes in antiretroviral naive patients receiving universal healthcare. AIDS 23:731–737. doi: 10.1097/QAD.0b013e328326f77f. [DOI] [PubMed] [Google Scholar]
- 9.Easterbrook PJ, Smith M, Mullen J, O'Shea S, Chrystie I, de Ruiter A, Tatt ID, Geretti AM, Zuckerman M. 2010. Impact of HIV-1 viral subtype on disease progression and response to antiretroviral therapy. J Int AIDS Soc 13:4. doi: 10.1186/1758-2652-13-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Santos AF, Soares MA. 2010. HIV genetic diversity and drug resistance. Viruses 2:503–531. doi: 10.3390/v2020503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Theys K, Vercauteren J, Snoeck J, Zazzi M, Camacho RJ, Torti C, Schülter E, Clotet B, Sönnerborg A, De Luca A, Grossman Z, Struck D, Vandamme AM, Abecasis AB. 2013. HIV-1 subtype is an independent predictor of reverse transcriptase mutation K65R in HIV-1 patients treated with combination antiretroviral therapy including tenofovir. Antimicrob Agents Chemother 57:1053–1056. doi: 10.1128/AAC.01668-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Descamps D, Chaix M-L, André P, Brodard V, Cottalorda J, Deveau C, Harzic M, Ingrand D, Izopet J, Kohli E, Masquelier B, Mouajjah S, Palmer P, Pellegrin I, Plantier JC, Poggi C, Rogez S, Ruffault A, Schneider V, Signori-Schmück A, Tamalet C, Wirden M, Rouzioux C, Brun-Vezinet F, Meyer L, Costagliola D. 2005. French national sentinel survey of antiretroviral drug resistance in patients with HIV-1 primary infection and in antiretroviral-naive chronically infected patients in 2001–2002. J Acquir Immune Defic Syndr 38:545–552. doi: 10.1097/01.qai.0000155201.51232.2e. [DOI] [PubMed] [Google Scholar]
- 13.Martinez-Picado J, Gutiérrez C, de Mendoza C, Erkicia I, Domingo P, Camino X. 2005. Surveillance of drug resistance and HIV subtypes in newly diagnosed patients in Spain during 2004. Antivir Ther 10:S137. [Google Scholar]
- 14.Monno L, Brindicci G, Lo Caputo S, Punzi G, Scarabaggio T, Riva C, Di Bari C, Pierotti P, Saracino A, Lagioia A, Mazzotta F, Balotta C, Angarano G. 2005. HIV-1 subtypes and circulating recombinant forms (CRFs) from HIV-infected patients residing in two regions of central and southern Italy. J Med Virol 75:483–490. doi: 10.1002/jmv.20300. [DOI] [PubMed] [Google Scholar]
- 15.Snoeck J, Van Laethem K, Hermans P, Van Wijngaerden E, Derdelinckx I, Schrooten Y, van de Vijver DA, De Wit S, Clumeck N, Vandamme A-M. 2004. Rising prevalence of HIV-1 non-B subtypes in Belgium: 1983–2001. J Acquir Immune Defic Syndr 35:279–285. doi: 10.1097/00126334-200403010-00009. [DOI] [PubMed] [Google Scholar]
- 16.Chaix M-L, Seng R, Frange P, Tran L, Avettand-Fenoël V, Ghosn J, Reynes J, Yazdanpanah Y, Raffi F, Goujard C, Rouzioux C, Meyer L, ANRS PRIMO Cohort Study Group. 2013. Increasing HIV-1 non-B subtype primary infections in patients in France and effect of HIV subtypes on virological and immunological responses to combined antiretroviral therapy. Clin Infect Dis 56:880–887. doi: 10.1093/cid/cis999. [DOI] [PubMed] [Google Scholar]
- 17.Ciccozzi M, Santoro MM, Giovanetti M, Andrissi L, Bertoli A, Ciotti M. 2012. HIV-1 non-B subtypes in Italy: a growing trend. New Microbiol 35:377–386. [PubMed] [Google Scholar]
- 18.Esteves A, Parreira R, Venenno T, Franco M, Piedade J, Germano de Sousa J, Canas-Ferreira WF. 2002. Molecular epidemiology of HIV type 1 infection in Portugal: high prevalence of non-B subtypes. AIDS Res Hum Retroviruses 18:313–325. doi: 10.1089/088922202753519089. [DOI] [PubMed] [Google Scholar]
- 19.Esteves A, Parreira R, Piedade J, Venenno T, Franco M, Germano de Sousa J, Patrício L, Brum P, Costa A, Canas-Ferreira WF. 2003. Spreading of HIV-1 subtype G and envB/gagG recombinant strains among injecting drug users in Lisbon, Portugal. AIDS Res Hum Retroviruses 19:511–517. doi: 10.1089/088922203766774568. [DOI] [PubMed] [Google Scholar]
- 20.Freitas FB, Esteves A, Piedade J, Parreira R. 2013. Novel multiregion hybridization assay for the identification of the most prevalent genetic forms of the human immunodeficiency virus type 1 circulating in Portugal. AIDS Res Hum Retroviruses 29:318–328. doi: 10.1089/AID.2012.0025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Abecasis AB, Martins A, Costa I, Carvalho AP, Diogo I, Gomes P, Camacho RJ, Portuguese HIV-1 Resistance Study Group. 2011. Molecular epidemiological analysis of paired pol/env sequences from Portuguese HIV type 1 patients. AIDS Res Hum Retroviruses 27:803–805. doi: 10.1089/aid.2010.0312. [DOI] [PubMed] [Google Scholar]
- 22.Parreira R, Pádua E, Piedade J, Venenno T, Paixão MT, Esteves A. 2005. Genetic analysis of human immunodeficiency virus type 1 nef in Portugal: subtyping, identification of mosaic genes, and amino acid sequence variability. J Med Virol 77:8–16. doi: 10.1002/jmv.20408. [DOI] [PubMed] [Google Scholar]
- 23.Palma AC, Araújo F, Duque V, Borges F, Paixão MT, Camacho R, Portuguese SPREAD Network. 2007. Molecular epidemiology and prevalence of drug resistance-associated mutations in newly diagnosed HIV-1 patients in Portugal. Infect Genet Evol 7:391–398. doi: 10.1016/j.meegid.2007.01.009. [DOI] [PubMed] [Google Scholar]
- 24.Delgado E, Thomson MM, Villahermosa ML, Sierra M, Ocampo A, Miralles C, Rodríguez-Pérez R, Diz-Aren J, Ojea-de Castro R, Losada E, Cuevas MT, Vazquez-de-Parga E, Carmona R, Perez-Alvarez L, Medrano L, Cuevas L, Taboada JA, Najera R. 2002. Identification of a newly characterized HIV-1 BG intersubtype circulating recombinant form in Galicia, Spain, which exhibits a pseudotype-like virion structure. J Acquir Immune Defic Syndr 29:536–543. doi: 10.1097/00126334-200204150-00016. [DOI] [PubMed] [Google Scholar]
- 25.Bártolo I, Abecasis AB, Borrego P, Barroso H, McCutchan F, Gomes P, Camacho R, Taveira N. 2011. Origin and epidemiological history of HIV-1 CRF14_BG. PLoS One 6:e24130. doi: 10.1371/journal.pone.0024130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Paraskevis D, Nikolopoulos G, Tsiara C, Paraskeva D, Antoniadou A, Lazanas M, Gargalianos P, Psychogiou M, Malliori M, Kremastinou J, Hatzakis A. 2011. HIV-1 outbreak among injecting drug users in Greece, 2011: a preliminary report. Euro Surveill 16:pii=19962 http://www.eurosurveillance.org/ViewArticle.aspx?ArticleId=19962. [DOI] [PubMed] [Google Scholar]
- 27.Taylor BS, Sobieszczyk ME, McCutchan FE, Hammer SM. 2008. The challenge of HIV-1 subtype diversity. N Engl J Med 358:1590–1602. doi: 10.1056/NEJMra0706737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Instituto Nacional de Saúde Doutor Ricardo Jorge (INSA). 2012. Infeção VIH/SIDA: a situação em Portugal a 31 de Dezembro de 2011. Instituto Nacional de Saúde Doutor Ricardo Jorge, Lisbon, Portugal: http://repositorio.insa.pt/bitstream/10400.18/984/3/relatorio%20VIH%20SIDA%202011%20versao%20web.pdf. [Google Scholar]
- 29.Hué S, Clewley JP, Cane PA, Pillay D. 2004. HIV-1 pol gene variation is sufficient for reconstruction of transmissions in the era of antiretroviral therapy. AIDS 18:719–728. doi: 10.1097/00002030-200403260-00002. [DOI] [PubMed] [Google Scholar]
- 30.Brenner BG, Roger M, Routy J-P, Moisi D, Ntemgwa M, Matte C, Baril J-G, Thomas R, Rouleau D, Bruneau J, Leblanc R, Legault M, Tremblay C, Charest H, Wainberg MA, Quebec Primary HIV Infection Study Group. 2007. High rates of forward transmission events after acute/early HIV-1 infection. J Infect Dis 195:951–959. doi: 10.1086/512088. [DOI] [PubMed] [Google Scholar]
- 31.Lewis F, Hughes GJ, Rambaut A, Pozniak A, Brown AJL. 2008. Episodic sexual transmission of HIV revealed by molecular phylodynamics. PLoS Med 5:e50. doi: 10.1371/journal.pmed.0050050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pineda-Peña A-C, Faria NR, Imbrechts S, Libin P, Abecasis AB, Deforche K, Gómez-López A, Camacho RJ, de Oliveira T, Vandamme A-M. 2013. Automated subtyping of HIV-1 genetic sequences for clinical and surveillance purposes: performance evaluation of the new REGA version 3 and seven other tools. Infect Genet Evol 19:337–348. doi: 10.1016/j.meegid.2013.04.032. [DOI] [PubMed] [Google Scholar]
- 33.Lole KS, Bollinger RC, Paranjape RS, Gadkari D, Kulkarni SS, Novak NG, Ingersoll R, Sheppard HW, Ray SC. 1999. Full-length human immunodeficiency virus type 1 genomes from subtype C-infected seroconverters in India, with evidence of intersubtype recombination. J Virol 73:152–160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Martin D, Rybicki E. 2000. RDP: detection of recombination amongst aligned sequences. Bioinformatics 16:562–563. doi: 10.1093/bioinformatics/16.6.562. [DOI] [PubMed] [Google Scholar]
- 35.Edgar RC. 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30:1312–1313. doi: 10.1093/bioinformatics/btu033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Guindon S, Dufayard JF, Lefort V, Anisimova M, Hordijk W, Gascuel O. 2010. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst Biol 59:307–321. doi: 10.1093/sysbio/syq010. [DOI] [PubMed] [Google Scholar]
- 38.Drummond AJ, Rambaut A. 2007. BEAST: Bayesian Evolutionary Analysis by Sampling Trees. BMC Evol Biol 7:214. doi: 10.1186/1471-2148-7-214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Darriba D, Taboada GL, Doallo R, Posada D. 2012. jModelTest 2: more models, new heuristics and parallel computing. Nat Methods 9:772–772. doi: 10.1038/nmeth.2109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bennett DE, Camacho RJ, Otelea D, Kuritzkes DR, Fleury H, Kiuchi M, Heneine W, Kantor R, Jordan MR, Schapiro JM, Vandamme AM, Sandstrom P, Boucher CA, van de Vijver D, Rhee SY, Liu TF, Pillay D, Shafer RW. 2009. Drug resistance mutations for surveillance of transmitted HIV-1 drug-resistance: 2009 update. PLoS One 4:e4724. doi: 10.1371/journal.pone.0004724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kaye M, Chibo D, Birch C. 2008. Phylogenetic investigation of transmission pathways of drug-resistant HIV-1 utilizing pol sequences derived from resistance genotyping. J Acquir Immune Defic Syndr 49:9–16. doi: 10.1097/QAI.0b013e318180c8af. [DOI] [PubMed] [Google Scholar]
- 42.Chalmet K, Staelens D, Blot S, Dinakis S, Pelgrom J, Plum J, Vogelaers D, Vandekerckhove L, Verhofstede C. 2010. Epidemiological study of phylogenetic transmission clusters in a local HIV-1 epidemic reveals distinct differences between subtype B and non-B infections. BMC Infect Dis 10:262. doi: 10.1186/1471-2334-10-262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Torti C, Lapadula G, Izzo I, Brindicci G, Labbate G, Quiros-Roldan E, Diallo I, Gargiulo F, Castelnuovo F, Calabresi A, Carosi G, Manca N, Monno L. 2010. Heterogeneity and penetration of HIV-1 non-subtype B viruses in an Italian province: public health implications. Epidemiol Infect 138:1298–1307. doi: 10.1017/S0950268810000166. [DOI] [PubMed] [Google Scholar]
- 44.von Wyl V, Kouyos RD, Yerly S, Böni J, Shah C, Bürgisser P, Klimkait T, Weber R, Hirschel B, Cavassini M, Staehelin C, Battegay M, Vernazza PL, Bernasconi E, Ledergerber B, Bonhoeffer S, Gunthard HF, Swiss HIV Cohort Study. 2011. The role of migration and domestic transmission in the spread of HIV-1 non-B subtypes in Switzerland. J Infect Dis 204:1095–1103. doi: 10.1093/infdis/jir491. [DOI] [PubMed] [Google Scholar]
- 45.Holguin A, de Mulder M, Yebra G, Lopez M, Soriano V. 2008. Increase of non-B subtypes and recombinants among newly diagnosed HIV-1 native Spaniards and immigrants in Spain. Curr HIV Res 6:327–334. doi: 10.2174/157016208785132455. [DOI] [PubMed] [Google Scholar]
- 46.Vermund SH, Leigh-Brown AJ. 2012. The HIV epidemic: high-income countries. Cold Spring Harb Perspect Med 2:a007195. doi: 10.1101/cshperspect.a007195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bender GJ, Yoder PS. 1974. Whites in Angola on the eve of independence: the politics of numbers. Africa Today 23–37. [Google Scholar]
- 48.Bártolo I, Rocha C, Bartolomeu J, Gama A, Marcelino R, Fonseca M, Mendes A, Epalanga M, Silva PC, Taveira N. 2009. Highly divergent subtypes and new recombinant forms prevail in the HIV/AIDS epidemic in Angola: new insights into the origins of the AIDS pandemic. Infect Genet Evol 9:672–682. doi: 10.1016/j.meegid.2008.05.003. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.