Abstract
Chlamydia pecorum is an important global pathogen of livestock, and it is also a significant threat to the long-term survival of Australia's koala populations. This study employed a culture-independent DNA capture approach to sequence C. pecorum genomes directly from clinical swab samples collected from koalas with chlamydial disease as well as from sheep with arthritis and conjunctivitis. Investigations into single-nucleotide polymorphisms within each of the swab samples revealed that a portion of the reads in each sample belonged to separate C. pecorum strains, suggesting that all of the clinical samples analyzed contained mixed populations of genetically distinct C. pecorum isolates. This observation was independent of the anatomical site sampled and the host species. Using the genomes of strains identified in each of these samples, whole-genome phylogenetic analysis revealed that a clade containing a bovine and a koala isolate is distinct from other clades comprised of livestock or koala C. pecorum strains. Providing additional evidence to support exposure of koalas to Australian livestock strains, two minor strains assembled from the koala swab samples clustered with livestock strains rather than koala strains. Culture-independent probe-based genome capture and sequencing of clinical samples provides the strongest evidence yet to suggest that naturally occurring chlamydial infections are comprised of multiple genetically distinct strains.
INTRODUCTION
Chlamydia pecorum is a widespread pathogen of economically important livestock species, such as cattle and sheep. In cattle, C. pecorum is associated with sporadic bovine encephalomyelitis (SBE), which presents as a fever followed by limb stiffness and staggering (1). In sheep, C. pecorum infections commonly are linked to polyarthritis and conjunctivitis, which can spread rapidly in a flock (2, 3). While these infections are economically relevant to producers, most C. pecorum infections in ruminants appear to be asymptomatic or subclinical, characterized by a consistent presence in the gastrointestinal tract (4, 5). While questions remain over the impact of these infections in livestock globally, the best example of the pathogenic potential of this obligate intracellular bacterium actually is found in koalas, a native Australian marsupial that continues to experience localized extinctions. C. pecorum infections in koalas can cause debilitating ocular and urogenital tract diseases (6, 7). Epidemiological questions have been raised about the relationships between C. pecorum strains infecting domesticated animals and the koala, with a recent C. pecorum multilocus sequence typing (MLST) study revealing the presence of identical sequence types in samples collected from each host (8). As a follow-up to these studies, we recently sequenced the genomes of several cultured koala C. pecorum isolates, revealing a high degree of synteny and sequence identity (98.5 to 98.8%) with C. pecorum genomes from European and U.S. cattle and sheep (9).
High-throughput comparative genome sequencing of cultured isolates has fundamentally changed our understanding of the biology and genetic relationships of chlamydial species infecting a wide range of human and animal hosts (10). To date, however, a major limitation of this approach has been the requirement for extensive passaging in tissue culture to generate a sufficient concentration of chlamydial DNA for the purpose of genome sequencing (11–14). Culturing also has the potential to produce bias, with a recent study revealing that extended in vitro culturing can introduce mutations in the genome sequence of chlamydial isolates, presumably due to a lack of host immune pressures (15). In a major breakthrough in the Chlamydia research field, culture-independent sequencing methods have been developed, with one of the first approaches involving immunomagnetic separation (IMS) of chlamydial cells using antibodies specific for the chlamydial lipopolysaccharide (16). Using IMS in conjunction with multiple displacement amplification (MDA) produced whole-genome sequences for C. trachomatis strains from low-volume archival samples and swab samples collected from patients (17, 18).
Sequence capture by hybridization also can be used to sequence chlamydial DNA without the need for cell culturing (19). The sequence capture method involves designing customized biotin-labeled RNA probes that can hybridize to a complete target genome sequence so that magnetic beads coated with the biotin-binding protein streptavidin can be used to extract the captured sequences (20). This sequence capture method originally was designed to enrich specific regions of large eukaryotic genomes for deep sequencing of a selected subset of genes (21, 22) but has since been used for other applications, including deep sequencing of viral genomes (23). In this study, we adapted this sequence capture technique to extract C. pecorum DNA sequence from clinical swabs and assess its feasibility as a novel culture-independent technique for high-throughput sequencing of C. pecorum strains. In doing so, we not only demonstrate that this technique is feasible for this chlamydial species but also reveal that it can be used to detect an unprecedented diversity of mixed infections in clinical samples, an observation that has ramifications for our understanding of the genetic diversity of naturally occurring infections for all chlamydial species.
MATERIALS AND METHODS
Clinical samples analyzed in this study.
C. pecorum PCR-positive samples (n = 6) and a cell-cultured C. pecorum isolate (n = 1) were selected. The strain names are abbreviations referring to geographical location, the animal name, and site of infection/disease (e.g., NSW/Bov/SBE is New South Wales/bovine/sporadic bovine encephalomyelitis). The collection of previously tested C. pecorum PCR-positive swab samples included two urogenital tract (UGT) swabs (SA/k2/UGT and Gun/koa1/Urethra [Ure]) collected from two different koalas, one from South Australia (SA) and one from Gunnedah (Gun) (8). The remaining C. pecorum PCR-positive swab samples (n = 4) were collected from previously tested Australian sheep from the NSW regions of Narromine (Nar) and Merriwa (Mer) (8, 24), including swab samples collected from (i) the eye (C. pecorum Nar/S22/RE) and the rectum (C. pecorum Nar/S22/Rec) (8) of a single sheep (S22) presenting with conjunctivitis; (ii) the left eye of a second sheep (S42) from the same flock (C. pecorum Nar/S42/LE), also presenting with conjunctivitis (24); and (iii) the joint of a third sheep (Ovi1) suffering from polyarthritis (C. pecorum Mer/Ovi1/Jnt) (8). All collected dry swabs were kept at 4°C until processing by vortexing and centrifugation, as previously described (8). The final sample analyzed (C. pecorum NSW/Bov/SBE) was a cell-cultured and purified strain isolated from the brain of an NSW calf with SBE (8). Briefly, the C. pecorum NSW/Bov/SBE isolate was propagated in Hep-2 cells and ultrapurified using density gradient centrifugation as previously described (9).
Quantification of C. pecorum in swab samples.
DNA was extracted from swab samples and from the C. pecorum NSW/Bov/SBE cell culture as previously described (8). DNA concentration for each sample was measured in duplicates on a NanoDrop instrument, and the presence of C. pecorum was requantified using a C. pecorum species-specific quantitative PCR (qPCR) assay (25). Samples with <10 copies of the C. pecorum 16S rRNA gene were considered negative. The total amounts of DNA and the genome copies used as the template for targeted enrichment are outlined in Table 1.
TABLE 1.
C. pecorum clinical samples and sequencing output
| Strain | Host | Diseasea | Amt of DNA used as templateb (ng) | Total no. of input C. pecorum genome copiesc | Total no. of reads | Mapped readsd |
Mean read depth | Read coverage (SD) | Accession no. | |
|---|---|---|---|---|---|---|---|---|---|---|
| No. | % | |||||||||
| C. pecorum NSW/Bov/SBE | Cattle | SBE | 1,011.35 | 4.6 ×1012 | 37,110,550 | 36,996,629 | 99.69 | 2,632 | 250.81 | SRR1693788 |
| C. pecorum Gun/koa1/Ure | Koala | UTI | 209.90 | 1.6 ×105 | 13,281,012 | 13,023,096 | 98.06 | 184 | 33.70 | SRR1693763 |
| C. pecorum SA/k2/UGT | Koala | UTI | 453.92 | 2,253 | 4,623,524 | 3,874,162 | 83.79 | 44 | 18.70 | SRR1693792 |
| C. pecorum Mer/Ovi1/Jnt | Ovine | Polyarthritis | 157.63 | 2,628 | 1,105,062 | 1,063,335 | 96.22 | 12 | 29.90 | SRR1693791 |
| C. pecorum Nar/S22/RE | Ovine | Conjunctivitis | 173.21 | 10,299 | 4,890,048 | 3,576,983 | 73.15 | 11 | 139.47 | SRR1693793 |
| C. pecorum Nar/S22/Rec | Ovine | Conjunctivitis | 84.45 | 2,674 | 27,257,774 | 21,705,968 | 79.63 | 21 | 116.61 | SRR1693794 |
| C. pecorum Nar/S42/LE | Ovine | Conjunctivitis | 129.94 | 8,662 | 26,729,184 | 185,341 | 0.69 | 6 | 1.64 | SRR1693795 |
SBE, sporadic bovine encephalomyelitis; UTI, urinary tract infection.
DNA concentration includes host and other resident microbiota DNA.
The number of C. pecorum genome copies determined by qPCR that were used as input in the library preparation.
Reads were mapped to the complete C. pecorum E58 genome.
SureSelect target enrichment.
The 120-mer RNA probes were designed in-house at the Institute for Genome Sciences (IGS) to span across the forward strand of the C. pecorum E58 genome (accession number CP002608). The custom-designed C. pecorum probe library was uploaded to SureDesign (https://earray.chem.agilent.com/suredesign/) and synthesized by Agilent Technologies. DNA libraries were constructed using the TrueSeq library kit, and the hybridization was done using the C. pecorum-specific probes and SureSelectXT reagents.
Genome sequencing.
C. pecorum genomes were sequenced from the seven samples using Illumina HiSeq to produce paired-end 101-bp reads. Read quality was checked with FASTQC, and filtering and trimming were performed on the reads with Trimmomatic version 0.32 (26). Genomes were assembled using Spades 3.0.0 with k-mer values of 15, 21, 33, 51, and 71 (27).
Read mapping and SNP analysis.
Duplicate reads were removed using Picard (https://github.com/broadinstitute/picard), and read mapping was done using the very sensitive parameters for Bowtie-2 (28) with C. pecorum E58 as the reference. The read mapping was visualized using the BLAST ring image generator (BRIG) program (29). Single-nucleotide polymorphisms (SNPs) were detected using Freebayes with min-alternate-fraction (minimum allele frequency) set to 0.05 and ploidy set to 10 (30).
Detection of mixed infection in samples.
A custom Python script called groupFlows.py was used to assess the distribution of variants in the reads for the identification of separate strains in each swab sample (https://github.com/mjsull/HaploFlow). Briefly, for each site with two genomic variants (An and Bn) identified by Freebayes, groupFlows.py iterated through the reads that aligned to that position using the Python module, pysam, and sorted reads based on which variant is present in the reads. Once reads had been assigned to either variant, groupFlows.py determined how many of the assigned reads also were assigned to each variant at the site immediately upstream (An-1 and Bn-1). If either upstream variant shared two or more reads with the downstream variant, a link was created, with up to four links possibly being created (An-1-An, An-1-Bn, Bn-1-Bn, and Bn-1-An). A cutoff of two reads allowed us to reduce the amount of links being created incorrectly due to read error. In most cases, this resulted in variant A at site n linked to a single variant at site n-1 and variant B at site n linked to the alternate variant (i.e., if variant An-1 linked to variant Bn, then Bn-1 linked to An). If the distance between adjacent variant sites was too long to be spanned by paired-end reads, then it was not possible to form links. In regions where there is a large amount of sequencing or mapping errors, low-complexity regions where non-C. pecorum DNA is captured, and regions where additional subpopulations at low levels are observable, three or four links are found. When this occurs, the variants are marked as unlinkable. This process then was repeated for all variants in the genome, resulting in chains of linked variants separating into two distinct groups (e.g., An-4-Bn-3-An-2-An-1-Bn and Bn-4-An-3-Bn-2-Bn-1-An) separated by unlinkable regions or gaps that cannot be spanned by reads. These chains then were assigned to strains by looking at the prevalence of variants in the chain that have not undergone a coverage drop significant enough to make the coverage of two strains indiscrete (greater than half the median coverage of the genome). If there is contradictory information between reporting sites, groups are not assigned to strains (as coverage in the strains was completely distinct, all groups were assigned). If the distance from the first variant in the chain to the last variant in the chain was less than 100 bp and unlinkable regions were found on either side of the chain, the area was marked as “noisy” and the variants weren't assigned to strains. Once variants had been assigned to either the dominant or minor strain, all reads associated with those variants also were assigned and a BAM file of both the dominant and minor strain was created. SAMtools (31) then was used to call the consensus with bases with a PHRED quality score of less than 30 and identified noisy regions being masked. Finally, we iterated through each base in the original BAM file, and if there was no significant coverage drop (coverage of more than half the median coverage) and a 98% or greater consensus between reads mapping to that site, the base was called for the dominant strain. This allowed us to call bases for the dominant strain in regions with no interstrain variation.
Phylogenetic analyses.
A whole-genome alignment of the six dominant C. pecorum draft genomes (excluding C. pecorum Nar/S42/LE), two minor C. pecorum strains, and the eight publicly available C. pecorum genomes (E58, PV3056, W73, P787, VR629, IPTaLE, DBDeUG, and MC/Marsbar) (32, 33) was performed using Mugsy with default settings (34). Syntenic alignments across all 16 genomes were concatenated using a custom Perl script to construct a core alignment where poorly aligned regions were filtered from the alignment using GBLOCKS version 0.91b with the minimum length of a block set to 5, and no gap positions were allowed. A phylogenetic tree was constructed using the core genome alignment of the 16 C. pecorum genomes with PhyML 3.1 using the generalized time reversible (GTR) model, and bootstrap values were calculated using 1,000 replicates.
Short read sequence accession numbers.
The sequence data were submitted to the National Center for Biotechnology Information (NCBI) short read archive (SRA) database, and the accession numbers are listed in Table 1.
RESULTS
Probe-based sequence capture enables sequencing of C. pecorum strains directly from clinical samples.
SureSelectXT sequence capture was performed using C. pecorum-specific probes on extracted DNA from six clinical swab samples and a single cell-cultured sample. To determine the efficacy of the SureSelectXT sequence capture method to enrich for C. pecorum DNA in samples without prior genome purification, we compared the total number of C. pecorum genomes copies to the number of reads that mapped to the complete C. pecorum E58 genome and the mean read depth (summarized in Table 1). Cultured C. pecorum NSW/Bov/SBE resulted in the highest percentage of mapped reads (99.69%) as well as the highest mean read depth (2,632×). The most likely factor contributing to the high yield of the C. pecorum reads in this sample was the high genome copy number of 14.6 × 1012 as a result of extensive passaging through cell culture.
For the swab samples, C. pecorum Gun/koa1/Ure had the highest genome copy number (1.6 × 105 copies), and 98.06% of the reads mapped to C. pecorum E58 with a mean read depth of 184×. The C. pecorum SA/k2/UGT, C. pecorum Mer/Ovi1/Jnt, C. pecorum Nar/S22/RE, and C. pecorum Nar/S22/Rec samples each had a comparatively lower number of genome copies (2,253 to 10,299 copies), but the sequencing still yielded >71% of reads that mapped to C. pecorum E58; however, the mean read depth varied greatly among these samples. C. pecorum SA/k2/UGT had a mean read depth of 44×, while C. pecorum strains Mer/Ovi1/Jnt, Nar/S22/RE, and Nar/S22/Rec had a mean read depth of 12×, 11×, and 21×, respectively. However, the standard deviations of read depth for Mer/Ovi1/Jnt, Nar/S22/RE, and Nar/S22/Rec are higher than the mean read depth, suggesting there is atypical distribution of read depth in these samples. Read quality checking revealed that these three samples have a large number of duplicated reads (>90%) occurring at a specific location, which can be seen in Fig. 1 as coverage spikes. The coverage spikes can be observed in the other samples but the reads are more evenly distributed, resulting in better read depth across the rest of the genome. The coverage spikes correlated with the presence of genes containing tandem repeats (data not shown). Lastly, while the C. pecorum Nar/S42/LE sample had a genome copy number of 8,662 copies, only 0.69% of the reads mapped to C. pecorum E58; however, a mean read depth of 6× was achieved.
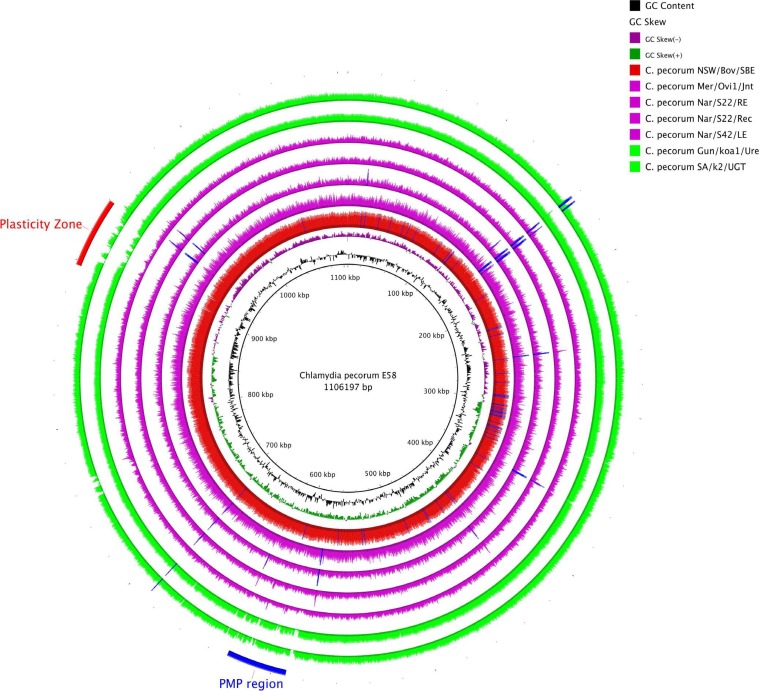
FIG 1.
Coverage of C. pecorum E58 reference genome. BRIG image shows C. pecorum E58 as the reference and the rings (from inner to outer) for GC content, GC skew, and filtered reads mapped against the reference with Bowtie2. The bovine strain C. pecorum NSW/Bov/SBE is shown as the red ring. The ovine strains C. pecorum Mer/Ovi1/Jnt, C. pecorum Nar/S22/RE, C. pecorum Nar/S22/Rec, and C. pecorum Nar/S42/LE are shown as purple rings. The green rings represent the koala strains C. pecorum SA/k2/UGT and C. pecorum Gun/Koa1/Ure. Blue spikes represent read-enriched regions that cause large coverage spikes. Only coverage across the reference genome is shown, and read depth is not to scale.
The read mapping revealed that 100% coverage of the reference genome was achieved for the four C. pecorum samples collected from sheep (C. pecorum Mer/Ovi1/Jnt, C. pecorum Nar/S22/RE, C. pecorum Nar/S22/Rec, and C. pecorum Nar/s42/LE) and the cattle sample, C. pecorum NSW/Bov/SBE (Fig. 1). For both koala C. pecorum strains, SA/k2/UGT and Gun/koa1/Ure, approximately 95% of the reference genome was covered. Gaps in the coverage were found in a hypervariable region called the plasticity zone (PZ) and in the polymorphic membrane protein (PMP) gene cluster. The cultured C. pecorum NSW/Bov/SBE genome was assembled into two contigs, while C. pecorum Gun/koa1/Ure, Nar/S22/Rec, SA/K2/UGT, and Mer/Ovi1/Jnt were assembled into 8 contigs, 15 contigs, 17 contigs, and 27 contigs, respectively. Lastly, the assemblies for C. pecorum Nar/S22/RE and Nar/S42/LE were poorer, and both assembled into >554 contigs.
Culture-independent sequencing of C. pecorum revealed mixed infections.
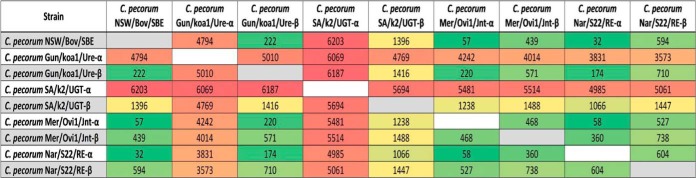
An SNP analysis revealed that four of the seven samples contained heterogeneous sites in the read depth data, indicating the presence of multiple C. pecorum strains. This was determined by the identification of a set of variants that occur only in a subset of reads: 8.0% of reads in C. pecorum SA/k2/UGT, 13.4% of reads in C. pecorum Gun/koa1/Ure, and 23.4% of reads in C. pecorum Mer/Ovi1/Jnt (Table 2 and Fig. 2). Although a high level of heterogeneity was detected in C. pecorum Nar/S22/Rec, suggesting the potential presence of >2 separate C. pecorum strains, the read depth of 21× was too low to accurately predict the prevalence of different strains in this sample. The low read coverage in C. pecorum Nar/S22/RE and C. pecorum Nar/S42/LE swab samples meant that we also were unable to reliably identify variant reads. We did not detect any variants in the C. pecorum NSW/Bov/SBE cultured isolate, suggesting it is a clonal isolate. Table 2 summarizes the number of SNPs identified in the dominant and minor strains of all samples compared to the reference C. pecorum E58 genome. The dominant strain is identified by the Greek letter alpha at the end of the strain name (e.g., C. pecorum Mer/Ovi1/Jnt-α), while the minor strain is designated by the letter beta at the end of the strain name (e.g., C. pecorum Mer/Ovi1/Jnt-β). The cultured C. pecorum NSW/Bov/SBE and C. pecorum E58 genomes from the brains of cattle with SBE in Australia and the United States, respectively, differ by 61 SNPs. Likewise, both dominant ovine strains (C. pecorum Mer/Ovi1/Jnt-α and Nar/S22/RE-α) differed from C. pecorum E58 by 59 SNPs. Higher numbers of SNPs were detected in the ovine minor strains (C. pecorum Mer/Ovi1/Jnt-β and Nar/S22/RE-β), ranging from 449 to 589 SNPs. The dominant koala strains, C. pecorum SA/k2/UGT-α and C. pecorum Gun/koa1/Ure-α, differ from C. pecorum E58 by 4,775 and 6,191 SNPs, respectively. However, there are fewer predicted SNPs (267 SNPs for C. pecorum Gun/koa1/Ure-β and 1,391 SNPs for C. pecorum SA/k2/UGT-β) in the minor koala C. pecorum strains. Figure 3 shows all of the heatmap comparisons of the number of variant differences between each of the C. pecorum strains identified from the swab samples, including dominant and minor strains. The number of different variant sites between each of the minor strains is 571 to 1,488 SNPs, suggesting that it is unlikely that the presence of these minor strains was caused by cross-contamination of the samples. The high number of different variant sites between the dominant strains and the minor strains from the same swab samples is further evidence for the presence of multiple genetically distinct C. pecorum strains coinfecting the same anatomical sites in both koalas and sheep.
TABLE 2.
Prevalence of the dominant strain and the total number of SNPs in dominant and minor strains using C. pecorum E58 as a reference
| Strain | No. of SNPs in: |
Prevalence rate (%) of dominant strain (based on assigned reads) | |
|---|---|---|---|
| Dominant strain | Minor strain | ||
| C. pecorum NSW/Bov/SBE | 61 | NAa | 100.0 |
| C. pecorum Gun/koa1/Ure | 4775 | 267 | 86.6 |
| C. pecorum SA/k2/UGT | 6191 | 1391 | 92.0 |
| C. pecorum Mer/Ovi1/Jnt | 59 | 449 | 76.6 |
| C. pecorum Nar/S22/RE | 59 | 589 | Unknownb |
NA, not applicable.
Read coverage was too low to accurately determine the prevalence of the dominant and minor strains.
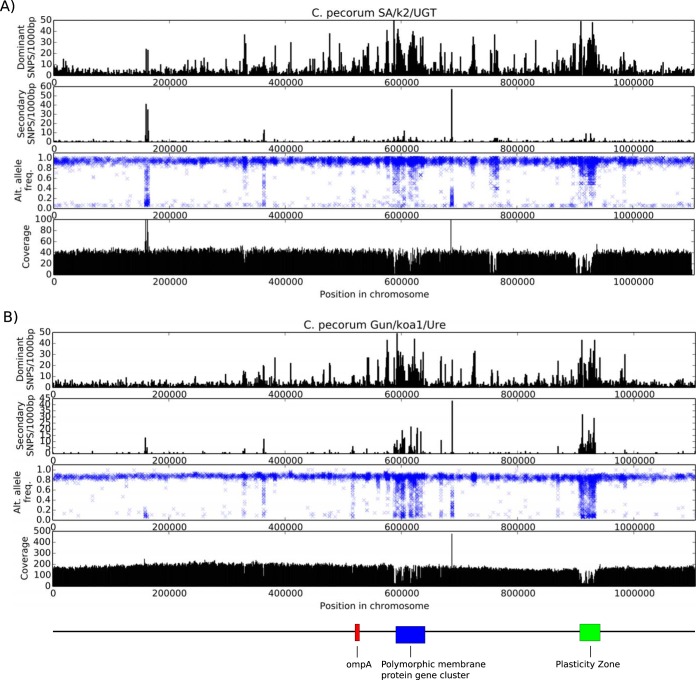
FIG 2.
SNP frequency and coverage for C. pecorum SA/k2/UTG (A) and C. pecorum Gun/koa1/Ure (B). The first graph shows frequency per 1,000 bp of SNPs with an allele frequency of ≥0.5. The second graph shows frequency per 1,000 bp of SNPs with an allele frequency of <0.5. The third graph plots a cross for each SNP variant in the genome. The x axis represents the position in the genome, and the y axis indicates the frequency with which the SNP appears in the raw reads. The bottom graph shows average read coverage along the genome.
FIG 3.
Heatmap of the number of nucleotide variant differences between each C. pecorum strain identified in the swab samples.
Reassembly of the dominant and minor C. pecorum strain reads.
In order to understand the phylogenetic relationships between the dominant and minor strains, the raw reads needed to be separated and the consensus sequences called individually. For the most part, an analysis of genetic variants in the sequence reads for each swab sample revealed the presence of two discrete populations of variants that could be separated easily into dominant and minor strains (Fig. 2). There were two exceptions to this observation, however, including regions that are (i) low-complexity regions where non-C. pecorum DNA potentially has been captured by probes, (ii) reads belonging to additional putative subpopulations, and (iii) regions with low read coverage. Strain-specific coverage drops occurred when one strain in the sample varied too much from the sequence capture probes at the nucleotide level, resulting in less sequence from that area of the strain being captured. As the minor strain may have high similarity in these regions, we predicted that the dominant strain often would be less prevalent at these locations.
Minor C. pecorum strains were successfully separated from the dominant C. pecorum Gun/koa1/Ure-α and SA/k2/UGT-α strains in the koala UGT swab samples. The minor strain, C. pecorum Gun/koa1/Ure-β, was assembled into 648 contigs with a total combined sequence of 915,868 bp. The other minor strain, C. pecorum SA/k2/UGT-β, was assembled into 2,070 contigs, with a combined length of 519,725 bp. The assemblies of Gun/koa1/Ure-β and SA/k2/UGT-β are less than the 1.1-Mbp chromosome size of C. pecorum E58, suggesting that only partial genomes were acquired for these minor strains. The successful identification of the minor strains for both koala swab samples was possible because of the high starting DNA concentration and the high number of sequence reads generated. In contrast, the low coverage of chlamydial reads in both of the Nar/S22 swab samples made it impossible to separate a minor strain. There also was difficulty with identifying a minor strain in the C. pecorum Mer/Ovi1/Jnt sample because of the relatively small amount of sequence reads produced for this sample.
Phylogenetic analysis revealed genetically distinct C. pecorum strains coinfecting the same anatomical site.
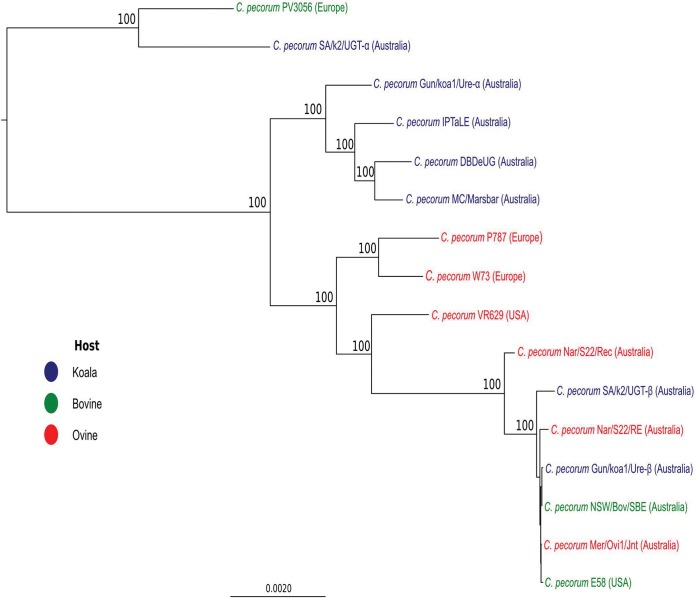
A core genome alignment of 279,714 bp was built using all sequenced C. pecorum genomes, including the two minor strains, C. pecorum Gun/koa1/Ure-β and SA/k2/UGT-β. Since only partial genomes could be acquired for the minor strains, the resulting core alignment only accounts for 25% of the C. pecorum chromosome. A maximum likelihood tree showed that all of the koala and livestock C. pecorum strains form separate, well-supported clades, with the exception of the koala C. pecorum SA/k2/UGT and cattle C. pecorum PV3056 genomes, which cluster together in a distant clade (Fig. 4). This suggests that the population structure of koala C. pecorum strains is more complex than what was initially established in other phylogenetic analyses, which showed all koala C. pecorum strains forming a single lineage (8, 9). Remarkably, both koala minor strains (C. pecorum SA/k2/UGT-β and C. pecorum Gun/koa1/Ure-β) belong to the clade comprised of livestock C. pecorum E58 and other Australian C. pecorum livestock strains, which are different from that of the associated dominant strains, indicating an unexpected degree of diversity in C. pecorum strains infecting the same anatomical sites of animals. In order to evaluate the topology of this tree, a maximum likelihood tree constructed using the partial gene fragments of seven C. pecorum housekeeping genes targeted in our previous C. pecorum MLST study (8) were extracted from the genome sequences analyzed in this study and previously (9, 33), revealing topology similar to that observed from our whole-genome phylogeny (see Fig. S1 in the supplemental material).
FIG 4.
Maximum-likelihood tree based on 279,714-bp genome alignment. The phylogenetic tree was constructed using PhyML with the GTR substitution model. Bootstrap values are shown as percentages of 1,000 replicates. Bootstrap values lower than 85% are not shown.
DISCUSSION
High-throughput sequencing of C. pecorum genomes is limited by cell culturing, which is time- and labor-intensive and not always successful. In addition, it has been shown that extended in vitro culturing of other chlamydial species may alter the genotype of these isolates (15). Due to these issues, culture-independent methods have been devised to sequence Chlamydia genomes directly from clinical swabs (17, 18). In this study, a probe hybridization sequence capture method was used to extract C. pecorum DNA from swab samples collected from koalas and Australian sheep and a cell-cultured Australian bovine isolate (20). The sequence capture resulted in sequence data for the five livestock C. pecorum genomes that enabled complete coverage across the reference genome of C. pecorum E58. Complete coverage was almost achieved for the two koala C. pecorum genomes (SA/k2/UGT and Gun/koa1/Ure); however, the coverage decreased at the PZ and PMP region. The PZ encodes several virulence factors that have been linked to host-pathogen interactions and appear to undergo rapid evolution (35). The PMPs are a family of autotransporter surface proteins that are thought to be important for adhesion to host cells and, like the PZ, are a major source of diversity in Chlamydia genomes (36). Between koala and livestock C. pecorum strains, there is a significant degree of nucleotide variability in both PZ (678 SNPs over 42 kb) and the PMP gene cluster (894 SNPs over 36 kb) (9). Since the probes were designed using the sequence of the cattle C. pecorum E58 strain, the decrease in coverage at the PZ and PMP cluster likely is caused by reduced binding affinity of the probes. Interestingly, this was not an issue for another traditionally hypervariable C. pecorum gene, ompA (37), encoding the chlamydial major outer membrane (data not shown), as full-length sequences could be retrieved for each of the completely assembled genomes. One possible explanation for the success in sequencing this variable gene, however, was that the ompA sequence from all of the livestock strains sequenced was indeed identical to the E58 ompA sequence, while the other two ompA genotypes, detected in each of the dominant koala C. pecorum genome sequences, contained 84 to 133 different SNPs compared to the E58 sequence.
Nevertheless, we anticipate that this limitation can be overcome in future iterations of the capture probe systems by designing additional probes that capture the sequence diversity present in previously sequenced koala C. pecorum genomes (9). The percentage of mapped reads is proportional to the total number of genome copies, which also has been observed when this culture-independent sequencing method was applied to Chlamydia trachomatis (19). In this study, it was observed that a genome copy number greater than 100,000 results in >98% of reads mapping to C. pecorum E58 and a mean read depth of >100×. However, samples with a genome copy number as low as 2,253 still can result in >71% of the reads mapping to the reference and a mean read depth of at least 10× even if high numbers of reads are the result of read duplication.
Using this culture-independent technique, we demonstrated compelling evidence to suggest that, at least in the case of C. pecorum, naturally occurring chlamydial infections are comprised of multiple infections by genetically distinct strains. Mixed infections previously have been detected in other bacterial species, such as Clostridium difficile, where a dominant strain and at least one minor strain were identified in 15 clinical samples (38). Limited evidence of mixed infections also exists for C. trachomatis, where DNA was extracted using IMS-MDA (17). One of the C. trachomatis samples that were extracted with IMS-MDA showed a high level of heterogeneity in the read sequences, with a major variant appearing in ∼85% of the reads and a minor variant appearing in ∼15%, which is indicative of a mixed infection by two C. trachomatis strains (17). However, there is no data on the clinical ramifications of these mixed infections.
In the case of C. pecorum specifically, a previous MLST study on livestock C. pecorum strains showed that a single animal could be infected by multiple strains at different anatomical sites, with speculation only on shedding multiple strains from the same site (8, 39). However, this is the first study to reveal the presence of multiple C. pecorum strains at the same anatomical site in both koalas and sheep. C. pecorum MLST was performed with bidirectional dideoxy Sanger sequencing of target amplicons resulting in a pair of single chromatograms, making it unlikely to detect the signals from minor genotypes. In this study, using culture-independent and deep whole-genome sequencing, we were able to capture and confirm the presence of minor strains. The DNA for at least two separate and genetically distinct C. pecorum strains were captured via probe hybridization for each of the clinical swab samples; however, no additional strains were detected in the cell-cultured C. pecorum NSW/Bov/SBE sample. The most likely explanation for this is that the C. pecorum NSW/Bov/SBE isolate cultured was the dominant strain found in the clinical sample at the time of culturing, and its growth was favored to the exclusion of others through subsequent passaging. Alternatively, since this was a brain isolate, it also might reflect the success of this strain in invading the central nervous system over other strains infecting the animal at the same time, a possibility given our recent observation of genetically distinct strains present in the gastrointestinal tract versus central nervous system tissues of calves with SBE (39).
The observation of mixed infections in the C. pecorum-positive swab samples analyzed provides further significant insight into the complex genetic relationships that exist between C. pecorum strains detected in koalas and Australian sheep. While this analysis revealed one more example (Gun/koa1/Ure-α) of a C. pecorum strain that is phylogenetically related to other koala strains (Fig. 4), phylogenetic analysis of SA/k2/UGT-α, SA/k2/UGT-β, and Gun/koa1/Ure-β provided examples of koala strains more similar to Australian and European livestock C. pecorum strains than to other koala strains (Fig. 4). Our group previously provided evidence to suggest that genetically similar, if not identical, strains of C. pecorum can be found in Australian sheep, cattle, and koalas (6, 8, 39). The whole-genome phylogenies constructed for the strains sequenced from northern New South Wales (Gun/koa1/Ure) and South Australian koalas (SA/k2/UTG) appear to confirm this finding. Although the small pool of samples analyzed obviously limits firm conclusions, it is tantalizing to speculate on what this means for our understanding of the potential origin of koala C. pecorum infections. In terms of the C. pecorum strains present in the major koala clade, it is possible that these strains are the result of millions of years of evolution or by cross-host transmission from Australian cattle and sheep in the past following European colonization. Furthermore, the presence of minor koala C. pecorum strains in clades dominated by Australian livestock supports ongoing exposure and potential cross-transmission of livestock strains into koalas from sheep and cattle. Development of a molecular clock for C. pecorum and extensive sampling of Australian sheep and cattle and koalas from sympatric regions is warranted to confirm these hypotheses and to gain insight into the origin of this pathogen in Australian native animals.
This study also led to the development of a new approach for detecting mixed infections from bacterial whole-genome sequencing data based on detecting variants within the raw reads (groupFlows.py). This was possible due to the presence of only two observable strains with distinct levels of coverage in the sample and a significant amount of variation between strains across the entire genome. If these two conditions are met, this application could be applied to nonrepetitive regions in other mixed-strain samples.
In conclusion, this study highlights the efficiency of the probe-based sequence capture method for rapidly acquiring genome sequences of C. pecorum strains directly from clinical swabs. In doing so, we have provided compelling evidence to show that we can reliably detect the genomes of at least two genetically distinct C. pecorum strains infecting an animal at the same anatomical site. The presence of multiple strains in a single host suggests that the chlamydial infection process is more complicated than initially expected and has ramifications for our broader understanding of chlamydial infections in a variety of hosts. The mixed population of C. pecorum strains will need to be investigated further to determine if they affect pathogenesis. In order to unravel these potentially complex mixtures of strains, we also developed a method for separating reads belonging to different C. pecorum strains, which will be critical for future studies relying on this sequence capture method. While these experiments were largely successful and provided powerful new data on the genetic diversity of C. pecorum infections, we were able to extract only minor strains from two samples, indicating that a significant amount of starting DNA is required for the minor strain to be assembled with acceptable coverage. It is more than likely that additional strains also are present, but higher sequence coverage would be required to identify them. Further optimization of the sequence capture method and increasing the depth of the sequencing runs will confirm if the diversity of C. pecorum strains in a single swab sample extends beyond two strains.
Supplementary Material
ACKNOWLEDGMENTS
We thank Oliver Funnell and Evelyn Walker for their assistance in swab collection from koalas and livestock and Tamieka Fraser for her initial screening of the koala samples. We also thank Helena Seth-Smith and Jacques Ravel for their critical reading of the manuscript during its preparation.
This work was supported financially by an Australian Research Council Discovery Grant (DP130102066) and a Queensland Department of Environment and Heritage Protection Koala Research Grant awarded to A.P. and P.T.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JCM.03534-14.
REFERENCES
- 1.Kessell AE, Finnie JW, Windsor PA. 2011. Neurological diseases of ruminant livestock in Australia. III Bacterial and protozoal infections. Aust Vet J 89:289–296. doi: 10.1111/j.1751-0813.2011.00807.x. [DOI] [PubMed] [Google Scholar]
- 2.Fukushi H, Hirai K. 1992. Proposal of Chlamydia pecorum sp. nov. for Chlamydia strains derived from ruminants. Int J Syst Bacteriol 42:306–308. doi: 10.1099/00207713-42-2-306. [DOI] [PubMed] [Google Scholar]
- 3.Polkinghorne A, Borel N, Becker A, Lu ZH, Zimmermann DR, Brugnera E, Pospischil A, Vaughan L. 2009. Molecular evidence for chlamydial infections in the eyes of sheep. Vet Microbiol 135:142–146. doi: 10.1016/j.vetmic.2008.09.034. [DOI] [PubMed] [Google Scholar]
- 4.Reinhold P, Sachse K, Kaltenboeck B. 2011. Chlamydiaceae in cattle: commensals, trigger organisms, or pathogens? Vet J 189:257–267. doi: 10.1016/j.tvjl.2010.09.003. [DOI] [PubMed] [Google Scholar]
- 5.Jee J, Degraves FJ, Kim T, Kaltenboeck B. 2004. High prevalence of natural Chlamydophila species infection in calves. J Clin Microbiol 42:5664–5672. doi: 10.1128/JCM.42.12.5664-5672.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jackson M, White N, Giffard P, Timms P. 1999. Epizootiology of Chlamydia infections in two free-range koala populations. Vet Microbiol 65:255–264. doi: 10.1016/S0378-1135(98)00302-2. [DOI] [PubMed] [Google Scholar]
- 7.Polkinghorne A, Hanger J, Timms P. 2013. Recent advances in understanding the biology, epidemiology and control of chlamydial infections in koalas. Vet Microbiol 165:214–223. doi: 10.1016/j.vetmic.2013.02.026. [DOI] [PubMed] [Google Scholar]
- 8.Jelocnik M, Frentiu FD, Timms P, Polkinghorne A. 2013. Multi-locus sequence analysis provides insights into the molecular epidemiology of Chlamydia pecorum infections in Australian sheep, cattle and koalas. J Clin Microbiol 51:2625–2632. doi: 10.1128/JCM.00992-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bachmann NL, Fraser TA, Bertelli C, Jelocnik M, Gillett A, Funnell O, Flanagan C, Myers GS, Timms P, Polkinghorne A. 2014. Comparative genomics of koala, cattle and sheep strains of Chlamydia pecorum. BMC Genomics 15:667. doi: 10.1186/1471-2164-15-667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bachmann NL, Polkinghorne A, Timms P. 2014. Chlamydia genomics: providing novel insights into chlamydial biology. Trends Microbiol 22:464–472. doi: 10.1016/j.tim.2014.04.013. [DOI] [PubMed] [Google Scholar]
- 11.Moulder JW. 1991. Interaction of chlamydiae and host cells in vitro. Microbiol Rev 55:143–190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Stephens RS, Kalman S, Lammel C, Fan J, Marathe R, Aravind L, Mitchell W, Olinger L, Tatusov RL, Zhao Q, Koonin EV, Davis RW. 1998. Genome sequence of an obligate intracellular pathogen of humans: Chlamydia trachomatis. Science 282:754–759. doi: 10.1126/science.282.5389.754. [DOI] [PubMed] [Google Scholar]
- 13.Carlson JH, Porcella SF, McClarty G, Caldwell HD. 2005. Comparative genomic analysis of Chlamydia trachomatis oculotropic and genitotropic strains. Infect Immun 73:6407–6418. doi: 10.1128/IAI.73.10.6407-6418.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Seth-Smith HM, Harris SR, Persson K, Marsh P, Barron A, Bignell A, Bjartling C, Clark L, Cutcliffe LT, Lambden PR, Lennard N, Lockey SJ, Quail MA, Salim O, Skilton RJ, Wang Y, Holland MJ, Parkhill J, Thomson NR, Clarke IN. 2009. Co-evolution of genomes and plasmids within Chlamydia trachomatis and the emergence in Sweden of a new variant strain. BMC Genomics 10:239. doi: 10.1186/1471-2164-10-239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Borges V, Ferreira R, Nunes A, Sousa-Uva M, Abreu M, Borrego MJ, Gomes JP. 2013. Effect of long-term laboratory propagation on Chlamydia trachomatis genome dynamics. Infect Genet Evol 17:23–32. doi: 10.1016/j.meegid.2013.03.035. [DOI] [PubMed] [Google Scholar]
- 16.Seth-Smith HM, Harris SR, Scott P, Parmar S, Marsh P, Unemo M, Clarke IN, Parkhill J, Thomson NR. 2013. Generating whole bacterial genome sequences of low-abundance species from complex samples with IMS-MDA. Nat Protoc 8:2404–2412. doi: 10.1038/nprot.2013.147. [DOI] [PubMed] [Google Scholar]
- 17.Seth-Smith HM, Harris SR, Skilton RJ, Radebe FM, Golparian D, Shipitsyna E, Duy PT, Scott P, Cutcliffe LT, O'Neill C, Parmar S, Pitt R, Baker S, Ison CA, Marsh P, Jalal H, Lewis DA, Unemo M, Clarke IN, Parkhill J, Thomson NR. 2013. Whole-genome sequences of Chlamydia trachomatis directly from clinical samples without culture. Genome Res 23:855–866. doi: 10.1101/gr.150037.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Putman TE, Suchland RJ, Ivanovitch JD, Rockey DD. 2013. Culture-independent sequence analysis of Chlamydia trachomatis in urogenital specimens identifies regions of recombination and in-patient sequence mutations. Microbiology 159:2109–2117. doi: 10.1099/mic.0.070029-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Christiansen MT, Brown AC, Kundu S, Tutill HJ, Williams R, Brown JR, Holdstock J, Holland MJ, Stevenson S, Dave J, Tong CY, Einer-Jensen K, Depledge DP, Breuer J. 2014. Whole-genome enrichment and sequencing of Chlamydia trachomatis directly from clinical samples. BMC Infect Dis 14:591. doi: 10.1186/s12879-014-0591-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gnirke A, Melnikov A, Maguire J, Rogov P, LeProust EM, Brockman W, Fennell T, Giannoukos G, Fisher S, Russ C, Gabriel S, Jaffe DB, Lander ES, Nusbaum C. 2009. Solution hybrid selection with ultra-long oligonucleotides for massively parallel targeted sequencing. Nat Biotechnol 27:182–189. doi: 10.1038/nbt.1523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bundock PC, Casu RE, Henry RJ. 2012. Enrichment of genomic DNA for polymorphism detection in a non-model highly polyploid crop plant. Plant Biotech J 10:657–667. doi: 10.1111/j.1467-7652.2012.00707.x. [DOI] [PubMed] [Google Scholar]
- 22.Varela I, Tarpey P, Raine K, Huang D, Ong CK, Stephens P, Davies H, Jones D, Lin ML, Teague J, Bignell G, Butler A, Cho J, Dalgliesh GL, Galappaththige D, Greenman C, Hardy C, Jia M, Latimer C, Lau KW, Marshall J, McLaren S, Menzies A, Mudie L, Stebbings L, Largaespada DA, Wessels LF, Richard S, Kahnoski RJ, Anema J, Tuveson DA, Perez-Mancera PA, Mustonen V, Fischer A, Adams DJ, Rust A, Chan-On W, Subimerb C, Dykema K, Furge K, Campbell PJ, Teh BT, Stratton MR, Futreal PA. 2011. Exome sequencing identifies frequent mutation of the SWI/SNF complex gene PBRM1 in renal carcinoma. Nature 469:539–542. doi: 10.1038/nature09639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Depledge DP, Palser AL, Watson SJ, Lai IY, Gray ER, Grant P, Kanda RK, Leproust E, Kellam P, Breuer J. 2011. Specific capture and whole-genome sequencing of viruses from clinical samples. PLoS One 6:e27805. doi: 10.1371/journal.pone.0027805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jelocnik M, Walker E, Pannekoek Y, Ellem J, Timms P, Polkinghorne A. 2014. Evaluation of the relationship between Chlamydia pecorum sequence types and disease using a species-specific multi-locus sequence typing scheme (MLST). Vet Microbiol 174:214–222. doi: 10.1016/j.vetmic.2014.08.018. [DOI] [PubMed] [Google Scholar]
- 25.Marsh J, Kollipara A, Timms P, Polkinghorne A. 2011. Novel molecular markers of Chlamydia pecorum genetic diversity in the koala (Phascolarctos cinereus). BMC Microbiol 11:77. doi: 10.1186/1471-2180-11-77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bolger AM, Lohse M, Usadel B. 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, Pyshkin AV, Sirotkin AV, Vyahhi N, Tesler G, Alekseyev MA, Pevzner PA. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol 19:455–477. doi: 10.1089/cmb.2012.0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Langmead B, Salzberg SL. 2012. Fast gapped-read alignment with Bowtie 2. Nat Methods 9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Alikhan NF, Petty NK, Ben Zakour NL, Beatson SA. 2011. BLAST Ring Image Generator (BRIG): simple prokaryote genome comparisons. BMC Genomics 12:402. doi: 10.1186/1471-2164-12-402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Garrison E, Marth G. 2012. Haplotype-based variant detection from short-read sequencing. arXiv 2012:1207.3907. [Google Scholar]
- 31.Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R. 2009. The Sequence Alignment/Map format and SAMtools. Bioinformatics 25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mojica S, Huot Creasy H, Daugherty S, Read TD, Kim T, Kaltenboeck B, Bavoil P, Myers GS. 2011. Genome sequence of the obligate intracellular animal pathogen Chlamydia pecorum E58. J Bacteriol 193:3690. doi: 10.1128/JB.00454-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sait M, Livingstone M, Clark EM, Wheelhouse N, Spalding L, Markey B, Magnino S, Lainson FA, Myers GS, Longbottom D. 2014. Genome sequencing and comparative analysis of three Chlamydia pecorum strains associated with different pathogenic outcomes. BMC Genomics 15:23. doi: 10.1186/1471-2164-15-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Angiuoli SV, Salzberg SL. 2011. Mugsy: fast multiple alignment of closely related whole genomes. Bioinformatics 27:334–342. doi: 10.1093/bioinformatics/btq665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Read TD, Brunham RC, Shen C, Gill SR, Heidelberg JF, White O, Hickey EK, Peterson J, Utterback T, Berry K, Bass S, Linher K, Weidman J, Khouri H, Craven B, Bowman C, Dodson R, Gwinn M, Nelson W, DeBoy R, Kolonay J, McClarty G, Salzberg SL, Eisen J, Fraser CM. 2000. Genome sequences of Chlamydia trachomatis MoPn and Chlamydia pneumoniae AR39. Nucleic Acids Res 28:1397–1406. doi: 10.1093/nar/28.6.1397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Stephens RS, Lammel CJ. 2001. Chlamydia outer membrane protein discovery using genomics. Curr Opin Microbiol 4:16–20. doi: 10.1016/S1369-5274(00)00158-2. [DOI] [PubMed] [Google Scholar]
- 37.Kollipara A, Polkinghorne A, Wan C, Kanyoka P, Hanger J, Loader J, Callaghan J, Bell A, Ellis W, Fitzgibbon S, Melzer A, Beagley K, Timms P. 2013. Genetic diversity of Chlamydia pecorum strains in wild koala locations across Australia and the implications for a recombinant C. pecorum major outer membrane protein based vaccine. Vet Microbiol 167:513–522. doi: 10.1016/j.vetmic.2013.08.009. [DOI] [PubMed] [Google Scholar]
- 38.Eyre DW, Cule ML, Griffiths D, Crook DW, Peto TE, Walker AS, Wilson DJ. 2013. Detection of mixed infection from bacterial whole genome sequence data allows assessment of its role in Clostridium difficile transmission. PLoS Comput Biol 9:e1003059. doi: 10.1371/journal.pcbi.1003059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jelocnik M, Forshaw D, Cotter J, Roberts D, Timms P, Polkinghorne A. 2014. Molecular and pathological insights into Chlamydia pecorum-associated sporadic bovine encephalomyelitis (SBE) in Western Australia. BMC Vet Res 10:121. doi: 10.1186/1746-6148-10-121. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.