Abstract
Contagious ovine digital dermatitis (CODD) is an important foot disease in sheep, with significant animal welfare and economic implications. It is thought that CODD emerged from bovine digital dermatitis (BDD) via treponemal bacteria. With wildlife species such as elk now suffering a CODD-like disease, it is imperative to clarify these disease etiologies. A large investigation into treponemal association with CODD is warranted. CODD lesions (n = 58) and healthy sheep foot tissues (n = 56) were analyzed by PCR for the three BDD-associated Treponema phylogroups and two other lameness-associated bacteria, Dichelobacter nodosus and Fusobacterium necrophorum. Spirochete culture was also attempted on CODD lesions. “Treponema medium/Treponema vincentii-like,” “Treponema phagedenis-like,” and Treponema pedis spirochetes were identified in 39/58 (67%), 49/58 (85%), and 41/58 (71%) of CODD lesions, respectively. One or more BDD-associated Treponema phylogroups were detected in 100% of CODD lesions. Healthy foot tissues did not amplify BDD-associated Treponema phylogroup DNA. D. nodosus and F. necrophorum were present in 34/58 (59%) and 41/58 (71%) of CODD lesions and 22/56 (39%) and 5/56 (9%) of healthy foot tissues, respectively. Thirty-two spirochetes were isolated from CODD lesions, with representatives clustering with, and indistinguishable from, each of the three BDD-associated Treponema phylogroups based on 16S rRNA gene comparisons. This study for the first time demonstrates a high-level association for BDD treponeme phylogroups in CODD and their absence from healthy tissues, supporting the hypothesis that BDD treponemes play a primary causative role in CODD and confirming that the specific PCR assays are an effective differential diagnostic tool for CODD.
INTRODUCTION
Digital dermatitis (DD) is an infectious lameness commonly found in dairy cattle worldwide, and it is known as bovine digital dermatitis (BDD) or papillomatous digital dermatitis (PDD). The disease was first reported in 1980 in the United States (1) and in the late 1980s in the United Kingdom (2). BDD has also been confirmed in beef cattle (3, 4), and over the last 30 years, the disease has been recognized as an important cause of bovine lameness (5).
Lameness in cattle and sheep has serious animal welfare and economic implications (6–9). The effects of lameness in cattle include a decrease in milk yield (9, 10) and fertility (8, 11–13) and an increase in rate of culling (12, 14). This has been found to be particularly true for cattle suffering from BDD (8, 15, 16) with a recent study of the cost of lameness in the United States estimating that on average, BDD costs $133 per case (17).
BDD is now a worldwide problem, and controlling BDD on dairy operations has proven difficult. Moreover, in the last 20 years, sheep in the United Kingdom have been identified with a form of DD termed contagious ovine digital dermatitis (CODD), which is rapidly emerging as a severe infectious foot disease since it was first reported in the United Kingdom in 1997 (18–20). Now, CODD has spread into the Republic of Ireland (20), and it was recently reported in dairy goats in the United Kingdom (21), indicating further cross-species transmission. The contagious nature of DD is also evident by the reports of a manifestation of the disease in a wildlife host, North American elk (Cervus elaphus) from Washington State (22). The reports of DD in previously unaffected species, including U.S. wildlife, suggests a much greater global threat of the disease than previously considered.
BDD in cattle manifests in several forms, but most frequently as an ulcerative lesion of the digital skin located immediately above the coronary band between the heel bulbs which results in severe lameness (23). The clinical features of CODD in sheep are slightly different, mainly because the initial lesion site on the sheep foot is different. CODD lesions commence at the coronary band and then run under the hoof horn capsule dorsally and abaxially (24). CODD frequently presents a particularly severe outcome where the whole horn capsule can be lost (18, 25–27). As a result of the severity of the lesions, sheep can be extremely lame, impacting the welfare of the affected sheep (28). This is concurrent with the lesion pathology identified in elk, described as erosive lesions on the coronary band which lead to underrunning of the hoof wall (22, 29).
The primary causative agents of BDD are considered to be spirochetal bacteria with a polytreponemal etiology suggested (30–32). Cloning bacterial 16S rRNA genes indicated five phylotypes of treponemes in BDD lesions from Germany (33). Three of these Treponema phylogroups have been isolated consistently from dairy cattle lesions in the United Kingdom and United States (34, 35). The phylogroups are described as “Treponema medium/Treponema vincentii-like,” “Treponema phagedenis-like” and “Treponema denticola/T. putidum-like” BDD spirochetes (34) with the latter now recognized as a new species, Treponema pedis (36). These three phylogroups of Treponema have also been reported in DD lesions in beef cattle (4) and also in DD lesions in goats (21) and elk (22). Due to the promiscuous nature of these treponemes and their growing host range, it is necessary to gain more information on their etiological role in currently infected species.
Previous studies have investigated the association of Treponema bacteria with CODD lesions (20, 37) and hypothesized that CODD was derived from BDD lesions in dairy cows (38). To date, little substantiating molecular evidence has been produced, and the possible involvement of other organisms such as Dichelobacter nodosus and Fusobacterium necrophorum has been proposed (37). In a previous study, D. nodosus, F. necrophorum, and also treponemal bacteria were detected in a considerable proportion of CODD lesions (74%, 86%, and 70%, respectively) (37), but these bacteria were also detected in a substantial proportion of healthy foot tissue (31%, 46%, and 38%, respectively). However, this study did not discriminate between Treponema species as their respective PCR assay detects both pathogenic and commensal treponemes, the latter of which are found in the ruminant gastrointestinal (GI) tract and feces (39). Additionally, healthy bovine foot tissue samples often amplify treponemal DNA using the genus-specific PCR assay but fail to produce products using DD treponeme phylogroup PCR assays (32, 39). Indeed, in a large survey of the dairy farm environment (39), nearly all samples were positive using the general Treponema PCR, suggesting it has little diagnostic value for clarifying relevant associations with clinical disease. There is a clear association with certain phylogroups of treponemes for BDD, and these treponemes are present only in BDD lesions and are completely absent from healthy bovine tissues and most environmental and fecal samples (32, 39). Therefore, the previous CODD study (37) provides no specific data on the presence/association of BDD-associated Treponema species with CODD lesions. Similarly, the remaining bacteriological data on CODD lesions comprised of a study based entirely on culture-dependent methods, using only 10 samples; only 7 treponemal cultures were produced from the 10 samples, and only 2 pure treponeme isolates were obtained (20).
Thus, while the role of treponemes as primary causative agents in BDD appears convincing, a comprehensive bacterial molecular survey of CODD lesions is justified to determine whether there is a shared spirochetal etiopathogenesis between BDD and CODD. The aim of this study was to further our understanding of CODD etiology by surveying a large number of lesions and healthy foot tissues for the presence of the three BDD treponeme phylogroups, as well as D. nodosus and F. necrophorum. Furthermore, given the small number of isolations thus far, in the present study, we attempted to determine the range of spirochetes present in CODD lesions from a number of farms across the United Kingdom and successfully isolate and characterize a large number of spirochete strains for comparison with other relevant treponemes.
MATERIALS AND METHODS
Sample collection.
Surgical biopsy samples were collected from 44 CODD lesions from six different farms between March 2013 and July 2014. These farms were in the following United Kingdom areas: Anglesey, Cheshire, Denbighshire, Shropshire, and Conwy. Genomic DNA from a further 14 CODD lesion biopsy samples collected during 2009 to 2010 from two farms in Cheshire and Gloucestershire in the United Kingdom were also included in this investigation. All farms had between 300 and 1,000 breeding ewes and were lowland farms except for a Conwy farm located on hill land. The sheep breeds on farms included Welsh Mountain, Scottish Blackface, Suffolk/Suffolk crosses, Lleyn/Lleyn crosses, Charolais crosses, and Easy Care. In addition to the lesional material, 54 healthy foot skin biopsy samples were collected. Of these samples, 16 were obtained from a farm in Meirionydd, United Kingdom, from each foot of four Balwen sheep, and 8 were obtained from non-CODD feet of three cross bred sheep with CODD lesions on their other feet sampled in this study (from a farm located in Cheshire, United Kingdom) (all euthanized for other reasons). The remaining 32 healthy foot tissue samples were obtained from sheep sent to slaughter that did not have any evidence of CODD or any other foot lesions. These unidentified sheep were sampled at an abattoir that received sheep from farms in Lancashire, Cheshire, and South Cumbria in the United Kingdom.
On all farms from which CODD biopsy samples were obtained from live sheep, the sheep were forced to run, and lame animals were inspected for CODD lesions. A sheep was defined as having CODD if one or more feet had a clear lesion consistent with the clinical signs of CODD (40). These clinical signs can be varied, but they include an ulcerative or granulomatous lesion at the coronary band which may extend dorsally and abaxially under the hoof wall and in severe cases lead to avulsion of the hoof capsule. The affected digit may be swollen and shortened. Sheep identified with classic CODD lesions were examined, and the lesions were biopsied using a 3-mm biopsy punch and local anesthesia (34, 41). All CODD lesions biopsied were active lesions shown by tissue appearing hemorrhagic, granulomatous, and/or necrotic. Tissue biopsy samples were divided in two with half transferred into transport medium and placed on ice for subsequent Treponema culture. The transport medium consisted of oral treponeme enrichment broth (OTEB) (Anaerobe Systems, Morgan Hill, CA, USA) and contained the antibiotics rifampin (5 μg/ml) and enrofloxacin (5 μg/ml). The remaining tissues from lesions (for PCR analysis) were transported on ice and stored at −20°C. All samples from CODD lesions, including those from 2009 to 2010 and those collected in 2013, were used for DNA extraction and Treponema culture, whereas healthy foot tissue samples were used only for DNA extraction and subsequent PCR analysis.
Culture of spirochetes.
Bacterial isolation, specifically for treponemes, was attempted on all CODD lesion samples (n = 58). A piece of tissue (1 to 1.5 mm) was transferred from the transport medium into an anaerobic cabinet (85% N2, 10% H2, and 5% CO2; 36°C) and inoculated into OTEB with 10% fetal calf serum (FCS) and antibiotics rifampin (5 μg/ml) and enrofloxacin (5 μg/ml). After 2 to 5 days, bacteria were subcultured on fastidious anaerobe agar (FAA) plates (LabM, Bury, United Kingdom) with 5% defibrinated sheep blood, 10% FCS, and antibiotics as described above. After 1 to 2 weeks, single colonies were inoculated into growth medium and the culture was checked by phase-contrast microscopy. DNA was extracted from treponeme cultures, and the isolated organisms were identified using a 16S rRNA gene PCR as previously described (34). Bacterial culture for treponemes was not attempted for sheep healthy foot tissue.
DNA extraction.
For PCR analysis, tissues from the lesions and healthy tissues were thawed and DNA was extracted using a DNeasy kit (Qiagen, United Kingdom) according to the manufacturer's instructions, and genomic DNA was stored at −20°C.
Genus- and phylogroup-specific treponeme PCR assays.
Samples were subjected to nested PCR assays specific for the three DD-associated treponeme groups, “T. medium/T. vincentii-like,” “T. phagedenis-like,” and T. pedis (“T. denticola/T. putidum-like”), as described previously (32, 34), with the resulting PCR products encompassing 300 to 500 bp of the 16S rRNA gene. All foot samples were also subjected to the Treponema genus PCR assay which detects all Treponema species, both pathogenic and commensal (37). To ensure validity in each assay, water was used as a negative control, and positive controls included genomic DNA from each of the three treponeme phylogroups. Primer sequences are shown in Table 1.
TABLE 1.
Primers used to detect DD-specific treponeme phylogroups, all treponeme species, D. nodosus, and F. necrophorum
| Primer target | Primer namea (sequence) | Predicted band size (bp) | Gene targeted | Region of gene targeted (positions)b | Reference |
|---|---|---|---|---|---|
| Universal | 16S F (5′-AGAGTTTGATCCTGG-3′) | 7–26 | 42 | ||
| 16S R (5′-TACCTTGTTACGACTT-3′) | 1,526 | 16S | 1491–1506 | ||
| Group 1 (“T. medium/T. vincentii-like”) | TmF (5′-GAATGCTCATCTGATGACGGTAATCGACG-3′) | 472–500 | 32 | ||
| TmR (5′-CCGGCCTTATCTAAGACCTTCTACTAG-3′) | 475 | 16S | 1001–1029 | ||
| Group 2 (“T. phagedenis-like”) | TbF (5′-GAAATACTCAAGCTTAACTTGAGAATTGC-3′) | 612–640 | 32 | ||
| TbR (5′-CTACGCTACCATATCTCTATAATATTGC-3′) | 400 | 16S | 1006–1029 | ||
| Group 3 (T. pedis) | TpF (5′-GGAGATGAGGGAATGCGTCTTCGATG-3′) | 459–484 | 32 | ||
| TpR (5′-CAAGAGTCGTATTGCTACGCTGATATATC-3′) | 475 | 16S | 1017–1045 | ||
| Treponema sp. | TPF (5′-AARCATGCAAGTCGARCGGCAAG-3′) | 49–71 | 37 | ||
| TPR1 (5′-TCCATTGCGGAATATTCTTA-3′) | 335 | 16S | 365–384 | ||
| D. nodosus | DnF (5′-TGAAGAATGAAAGCGGGGGC −3′) | 179–198 | This study | ||
| DnR (5′-CTAATCCTGTTTGCTACCCACG-3′) | 583 | 16S | 762–783 | ||
| F. necrophorum | lktA-F (5′-ACAATCGGAGTAGTAGGTTC-3′) | 6332–6350 | 49 | ||
| lktA-R (5′-ATTTGGTAACTGCCACTGC-3′) | 402 | Lkta | 6715–6732 |
Dichelobacter nodosus-specific PCR assay.
All lesion and healthy tissue biopsy samples were subjected to a species-specific D. nodosus PCR assay developed in this study. Initial attempts to use a previously developed PCR (44) failed to produce control PCR products, and by using recent primer design programs, these primers were identified as having poorly matching characteristics. Instead, species-specific D. nodosus primers (Table 1) were designed based on available 16S rRNA gene GenBank sequences. Representatives of D. nodosus, along with their nearest relatives (as identified using the Basic Local Alignment Search Tool [BLAST] [45]) were aligned to identify unique primer regions using ClustalW (46) in Molecular Evolutionary Genetics Analysis 2 (MEGA2) (47). The PCR assay primers were designed to amplify a 586-bp region of the D. nodosus 16S rRNA gene with primer pairs matched for annealing temperatures and guanine-cytosine content using the oligonucleotide properties calculator OligoCalc (48).
PCR mixtures used Taq polymerase (Qiagen, Crawley, United Kingdom) according to the manufacturer's instructions, with 1 μl of the DNA template and 1.5 mM MgCl2 (Qiagen, Crawley, United Kingdom), per 25-μl reaction mixture volume. To ensure validity, water and genomic DNA from the two closest relatives to D. nodosus (based on 16S rRNA gene sequence similarity) were used as negative controls. The two closest relatives were Suttonella indologenes (DSM8309) (GenBank accession no. AJ247267) and Cardiobacterium hominis (DSM8339) (GenBank accession no. AY360343). The genomic DNA of D. nodosus (DSM23057) was used as a positive control. The genomic DNA of Suttonella indologenes, Cardiobacterium hominis, and D. nodosus were purchased from DSMZ, Germany.
PCR conditions were as follows: (i) incubation at 95°C for 3 min; (ii) 35 cycles, with 1 cycle consisting of 94°C for 1 min, 59°C for 1 min, and 72°C for 2 min; (iii) a final extension step at 72°C for 10 min. These PCR conditions were previously optimized using a Mastercycler gradient thermocycler (Eppendorf, Germany). To further ensure validity of the PCR assay, a subset of PCR products were sequenced to ensure that positive PCR bands were produced by the presence of D. nodosus.
Fusobacterium necrophorum-specific PCR assay.
All lesion and healthy tissue biopsy samples were subjected to a species-specific F. necrophorum PCR assay as described originally (49). The primers used in this assay (Table 1) target the lktA gene which appears to be unique to F. necrophorum as it is not present in other Fusobacterium species (50).
To ensure validity, water and genomic DNA of Fusobacterium varium, a closely related species of Fusobacterium isolated by our laboratory and subsequently gene sequenced, were used as negative controls. The genomic DNA of F. necrophorum subsp. necrophorum (DSM21784) (DSMZ, Germany) was used as a positive control.
The PCR thermal profile consisted of an initial denaturation step at 94°C for 5 min, followed by 35 cycles, with 1 cycle consisting of 94°C for 30 s, 59°C for 30 s, and 72°C for 30 s. A final extension of 5 min at 72°C was performed.
To ensure validity of the PCR assay, a subset of PCR products were sequenced to ensure that positive PCR bands were produced by the presence of F. necrophorum.
Phylogenetic analysis of spirochete isolates.
To understand the relationship of the isolated spirochetes with other treponemes, a phylogenetic tree was produced from the aligned and trimmed nearly full-length 16S rRNA gene sequences of the isolates produced in this study together with relevant microorganisms available in GenBank and identified using BLAST (45). Sequences were assembled into a double-stranded consensus sequence using Chromas Pro 1.41 (Technelysium Pty Ltd.). Consensus sequences were aligned by ClustalW (46) in MEGA5.2 (51). For tree analysis, the most appropriate evolution model was predicted using “model test” as implemented in the Topali program (52). The final model for nucleotide substitutions chosen was the TrN model (53), used to infer a bootstrapped maximum likelihood tree (bootstrapping was performed 10,000 times).
Nucleotide sequence accession numbers.
The GenBank accession numbers for the 16S rRNA gene sequences of spirochetes isolated in this study are KP063152 to KP063183.
RESULTS
Genus- and phylogroup-specific treponeme PCR survey of CODD lesions and healthy foot tissues.
The results of the specific DD Treponema phylogroup PCR and Treponema genus-specific PCR assays of CODD lesions and healthy foot tissues are shown in Tables 2 and 3, respectively.
TABLE 2.
PCR detection of treponemes, D. nodosus, and F. necrophorum in CODD lesion biopsy samples
| Sample | Biopsy date (mo/yr) | Farm location | Sheep no.a | Treponeme(s) isolatedb | PCR result |
|||||
|---|---|---|---|---|---|---|---|---|---|---|
| PCR specific for groupc: |
Treponema PCR | F. necrophorum | D. nodosus | |||||||
| 1 | 2 | 3 | ||||||||
| 1 | 02/09 | Cheshire | 51 | G2S1F | + | + | + | + | + | − |
| 2 | 02/09 | Cheshire | 52 | G2S2R | + | + | + | + | + | − |
| 3 | 02/09 | Cheshire | 53 | G2S3R1 | + | + | + | + | + | − |
| 4 | 02/09 | Cheshire | 54 | G2S4F | + | + | + | + | + | − |
| 5 | 02/09 | Cheshire | 55 | IF | + | − | + | + | + | − |
| 6 | 08/09 | Gloucestershire | 11 | G1OV11 | + | + | − | + | − | − |
| 7 | 08/09 | Gloucestershire | 14 | IF | + | − | + | + | + | − |
| 8 | 08/09 | Gloucestershire | 17 | IF | − | + | + | + | + | − |
| 9 | 08/09 | Gloucestershire | 18 | IF | + | − | + | + | + | + |
| 10 | 08/09 | Gloucestershire | 20 | IF | − | + | + | + | + | + |
| 11 | 08/09 | Gloucestershire | 21 | IF | + | + | + | + | − | + |
| 12 | 08/09 | Gloucestershire | 22 | IF | + | − | − | + | − | + |
| 13 | 01/10 | Cheshire | 28 | IF | − | − | + | + | + | + |
| 14 | 01/10 | Cheshire | 29 | IF | + | + | − | + | + | + |
| 15 | 05/13 | Anglesey | 1 | G2SL1 | + | + | + | + | − | + |
| 16 | 06/13 | Anglesey | 97 | IF | − | + | − | + | − | + |
| 17 | 06/13 | Anglesey | 73 | IF | − | + | − | + | − | + |
| 18 | 06/13 | Anglesey | 30 | G2SL5 | + | + | + | + | + | + |
| 19 | 06/13 | Anglesey | 63 | G12F2 | + | + | + | + | + | + |
| 20 | 06/13 | Anglesey | 229 | G12F2, G23F1 | + | + | + | + | + | + |
| 21 | 06/13 | Anglesey | 36 back left | IF | + | + | + | + | + | + |
| 22 | 06/13 | Anglesey | 36 back right | IF | + | + | + | + | − | − |
| 23 | 06/13 | Anglesey | 2 | IF | + | + | + | + | + | + |
| 24 | 06/13 | Denbighshire | 3 | G16F2, G26F1 | + | + | − | + | + | − |
| 25 | 07/13 | Conwy farm 1 | 218 | IF | − | + | + | + | − | − |
| 26 | 07/13 | Conwy farm 1 | 10 | G2F2C10, G2ST24 | − | + | − | + | + | + |
| 27 | 07/13 | Conwyfarm 1 | 49 | G2F3C12, G2F3 | + | + | + | + | + | − |
| 28 | 07/13 | Conwy farm 1 | 4 | G2F4C4 | + | + | + | + | + | − |
| 29 | 07/13 | Conwy farm 1 | 53 | IF | + | + | + | + | + | − |
| 30 | 07/13 | Conwy farm 1 | 12 | G2F6C6 | + | + | + | + | + | − |
| 31 | 07/13 | Conwy farm 1 | 33 | G1F7C5 | + | + | + | + | + | − |
| 32 | 07/13 | Conwy farm 1 | 8 | IF | + | + | + | + | + | − |
| 33 | 07/13 | Conwy farm 1 | 86 | G1F9C27, G2F9 | + | + | + | + | − | − |
| 34 | 07/13 | Conwy farm 1 | 85 | G2F10C10 | + | + | + | + | + | − |
| 35 | 07/13 | Conwy farm 1 | 62 | G2F11C11 | + | + | + | + | − | − |
| 36 | 07/13 | Conwy farm 1 | 96 | IF | + | + | + | + | + | − |
| 37 | 08/13 | Conwy farm 2 | 5 | G2138C | + | + | + | + | − | + |
| 38 | 08/13 | Conwy farm 2 | 6 | G2148C | − | + | + | + | + | + |
| 39 | 08/13 | Conwy farm 2 | 900 | G2158C | − | + | + | + | + | + |
| 40 | 08/13 | Conwy farm 2 | 930 | IF | + | + | + | + | + | + |
| 41 | 08/13 | Anglesey | 38 | IF | − | + | − | + | − | + |
| 42 | 08/13 | Anglesey | 653 | IF | + | − | − | + | + | + |
| 43 | 08/13 | Anglesey | 58 | IF | + | + | − | + | + | + |
| 44 | 08/13 | Anglesey | 40 | IF | − | + | − | + | + | + |
| 45 | 08/13 | Anglesey | 74 | IF | + | + | + | + | + | + |
| 46 | 08/13 | Anglesey | 60 | G21C11 | − | + | − | + | + | − |
| 47 | 08/13 | Anglesey | 59 | G22C4 | + | + | + | + | − | − |
| 48 | 08/13 | Anglesey | 41 | IF | − | + | − | + | − | + |
| 49 | 08/13 | Anglesey | 39 | IF | + | + | − | + | + | + |
| 50 | 08/13 | Anglesey | 651 | IF | + | − | + | + | + | + |
| 51 | 08/13 | Anglesey | 652 | IF | − | + | − | + | + | − |
| 52 | 08/13 | Anglesey | 33 | IF | − | + | + | + | − | + |
| 53 | 12/13 | Cheshire | 101* | G21LJ | − | + | − | + | + | + |
| 54 | 12/13 | Cheshire | 102* | IF | − | + | − | + | − | − |
| 55 | 12/13 | Cheshire | 103 front left* | G23LJ | + | + | + | + | + | − |
| 56 | 12/13 | Cheshire | 103 back right* | IF | + | − | + | + | + | − |
| 57 | 07/14 | Shropshire | 1 | G3ST1 | − | − | + | + | − | + |
| 58 | 07/14 | Shropshire | 4 | G3S4S | − | + | + | + | + | + |
Sheep number given with additional foot information if animal had multiple feet sampled. Sheep from which healthy foot tissue was also obtained and investigated in this study are indicated by an asterisk. The corresponding results are shown in Table 3.
All isolations are shown for comparison to PCR results. Abbreviation: IF, isolation failed. If isolation was successful, the strain isolated is listed.
Groups 1, 2, and 3 are “T. medium/T. vincentii-like,” “T. phagedenis-like,” and T. pedis spirochetes, respectively, which are routinely found in bovine DD lesions.
TABLE 3.
PCR detection of treponemes, D. nodosus and F. necrophorum in healthy sheep foot tissue biopsies
| Sample | Biopsy date | Farm locationa | Sheep no.b | PCR result |
|||||
|---|---|---|---|---|---|---|---|---|---|
| Specific PCR for groupc: |
Treponema PCR | F. necrophorum | D. nodosus | ||||||
| 1 | 2 | 3 | |||||||
| 1 | 09/13 | Meirionydd | 1 front left | − | − | − | + | − | + |
| 2 | 09/13 | Meirionydd | 1 front right | − | − | − | + | − | + |
| 3 | 09/13 | Meirionydd | 1 back left | − | − | − | + | − | + |
| 4 | 09/13 | Meirionydd | 1 back right | − | − | − | + | − | + |
| 5 | 09/13 | Meirionydd | 2 front left | − | − | − | + | − | − |
| 6 | 09/13 | Meirionydd | 2 front right | − | − | − | + | − | − |
| 7 | 09/13 | Meirionydd | 2 back left | − | − | − | + | − | − |
| 8 | 09/13 | Meirionydd | 2 back right | − | − | − | + | − | − |
| 9 | 09/13 | Meirionydd | 3 front left | − | − | − | + | − | + |
| 10 | 09/13 | Meirionydd | 3 front right | − | − | − | + | − | + |
| 11 | 09/13 | Meirionydd | 3 back left | − | − | − | + | − | − |
| 12 | 09/13 | Meirionydd | 3 back right | − | − | − | + | − | + |
| 13 | 09/13 | Meirionydd | 4 front left | − | − | − | + | + | − |
| 14 | 09/13 | Meirionydd | 4 front right | − | − | − | + | − | − |
| 15 | 09/13 | Meirionydd | 4 back left | − | − | − | + | − | − |
| 16 | 09/13 | Meirionydd | 4 back right | − | − | − | + | − | − |
| 17 | 12/13 | Cheshire | 101 front left* | − | − | − | + | − | + |
| 18 | 12/13 | Cheshire | 101 back left* | − | − | − | + | − | + |
| 19 | 12/13 | Cheshire | 101 back right* | − | − | − | + | − | + |
| 20 | 12/13 | Cheshire | 102 front left* | − | − | − | + | − | + |
| 21 | 12/13 | Cheshire | 102 front right* | − | − | − | + | − | + |
| 22 | 12/13 | Cheshire | 102 back right* | − | − | − | − | + | − |
| 23 | 12/13 | Cheshire | 103 front right* | − | − | − | − | + | + |
| 24 | 12/13 | Cheshire | 103 back left* | − | − | − | + | − | − |
| 25 | 03/14 | 26 front left | − | − | − | + | − | − | |
| 26 | 03/14 | 26 front right | − | − | − | + | − | − | |
| 27 | 03/14 | 26 back left | − | − | − | − | − | − | |
| 28 | 03/14 | 26 back right | − | − | − | − | − | − | |
| 29 | 03/14 | 83 front left | − | − | − | + | + | − | |
| 30 | 03/14 | 83 front right | − | − | − | + | − | − | |
| 31 | 03/14 | 31 front left | − | − | − | + | − | − | |
| 32 | 03/14 | 31 back left | − | − | − | + | − | − | |
| 33 | 03/14 | 89 front left | − | − | − | − | − | − | |
| 34 | 03/14 | 89 back left | − | − | − | − | − | − | |
| 35 | 04/14 | 8 | − | − | − | − | − | − | |
| 36 | 04/14 | 5 | − | − | − | − | − | − | |
| 37 | 04/14 | 6 | − | − | − | − | − | − | |
| 38 | 04/14 | 79 front left | − | − | − | − | − | − | |
| 39 | 04/14 | 79 back right | − | − | − | − | − | − | |
| 40 | 04/14 | 80 front left | − | − | − | − | − | − | |
| 41 | 04/14 | 80 front right | − | − | − | − | − | − | |
| 42 | 04/14 | 80 back left | − | − | − | − | − | − | |
| 43 | 04/14 | 80 back right | − | − | − | + | − | − | |
| 44 | 04/14 | 7 front left | − | − | − | − | − | − | |
| 45 | 04/14 | 7 front right | − | − | − | + | − | + | |
| 46 | 04/14 | 7 back left | − | − | − | + | − | + | |
| 47 | 04/14 | 7 back right | − | − | − | + | − | + | |
| 48 | 04/14 | 87 front left | − | − | − | + | + | + | |
| 49 | 04/14 | 87 front right | − | − | − | + | − | + | |
| 50 | 04/14 | 87 back left | − | − | − | − | − | − | |
| 51 | 04/14 | 87 back right | − | − | − | + | − | − | |
| 52 | 04/14 | 987 front left | − | − | − | + | − | + | |
| 53 | 04/14 | 987 front right | − | − | − | − | − | + | |
| 54 | 04/14 | 987 back left | − | − | − | + | − | + | |
| 55 | 04/14 | 987 back right | − | − | − | + | − | + | |
| 56 | 04/14 | 9 | − | − | − | − | − | − | |
Farm location listed for sheep foot samples not obtained from sheep at the abattoir.
Sheep number given with additional foot information if animal had multiple feet sampled. Sheep which also had a CODD lesion present on a different foot were also investigated in this study (indicated by an asterisk) with corresponding results shown in Table 2.
Groups 1, 2, and 3 are “T. medium/T. vincentii-like,” “T. phagedenis-like,” and T. pedis spirochetes, respectively, which are routinely found in bovine DD lesions.
All CODD lesions (n = 58) were found to be positive for general Treponema DNA. The phylogroup-specific PCR for “T. medium/T. vincentii-like,” “T. phagedenis-like,” and T. pedis DD spirochetes showed that they were present in 39/58 (67%), 49/58 (85%), and 41/58 (71%) of CODD lesions, respectively. All CODD lesions (100%) were positive for at least one or more of the DD-associated Treponema phylogroups, with 27/58 (47%) of CODD lesions positive for all three DD-associated Treponema phylogroups. Of the healthy foot tissues sampled (n = 56), 38/56 (68%) were positive for the presence of general treponemes (Treponema genus-specific PCR). However, all healthy foot tissues were negative for the three DD-associated Treponema phylogroups.
D. nodosus- and F. necrophorum-specific PCR survey of CODD lesions and healthy foot tissue.
The D. nodosus- and F. necrophorum-specific PCR results for CODD lesions and healthy foot tissues are shown in Tables 2 and 3, respectively.
D. nodosus was present in 34/58 (59%) of CODD lesions. In healthy tissues surveyed, D. nodosus was present in 22/56 (39%) of samples.
F. necrophorum was present in 41/58 (71%) of CODD lesions and present in only 5/56 (9%) of healthy foot tissues.
Isolation of spirochetes and subsequent phylogenetic analysis.
As part of this study, spirochetes were successfully isolated from a high proportion of CODD lesions (Table 2). In several cases, multiple isolates were obtained from a single CODD lesion biopsy specimen. In total, 32 spirochetes were successfully isolated from 27/58 CODD lesions (47%). Many of these isolates (n = 24; 75%), were identified as belonging to the “T. phagedenis-like” spirochete group, with 23/24 sharing 100% 16S rRNA gene sequence identity with the “T. phagedenis-like” DD spirochete strain T320A (GenBank accession no. EF061261), previously isolated from a dairy cow DD lesion in the United Kingdom (34). The remaining “T. phagedenis-like” DD spirochete isolate shared a higher sequence identity (100%) with the human T. phagedenis strain CIP62.29 (GenBank accession no. EF645248) which both differ from the dairy cow DD isolate, “T. phagedenis-like” DD spirochete strain T320A, by a single-nucleotide substitution.
Six isolates (19%) belonged to the “T. medium/T. vincentii-like” spirochetes and showed 100% 16S rRNA gene sequence identity with “T. medium-like” DD spirochete strain T19 (GenBank accession no. EF061249) previously isolated from a dairy cow DD lesion in the United Kingdom (34).
Two isolates (6%) belonged to the T. pedis spirochetes. Spirochete isolate G3ST1 (GenBank accession no. KP063171) showed 100% 16S rRNA gene sequence identity with T. pedis T3552B (GenBank accession no. EF061268) previously isolated from a dairy cow DD lesion in the United Kingdom (34). The other T. pedis spirochete isolate from this study, G3S4S (GenBank accession no. KP063170), was found to show 100% 16S rRNA gene sequence identity with Treponema sp. strain G179 (GenBank accession no. AF363634), which was similarly isolated from a sheep CODD lesion in the United Kingdom (41). These two T. pedis spirochete groups are separated by just three nucleotide substitutions.
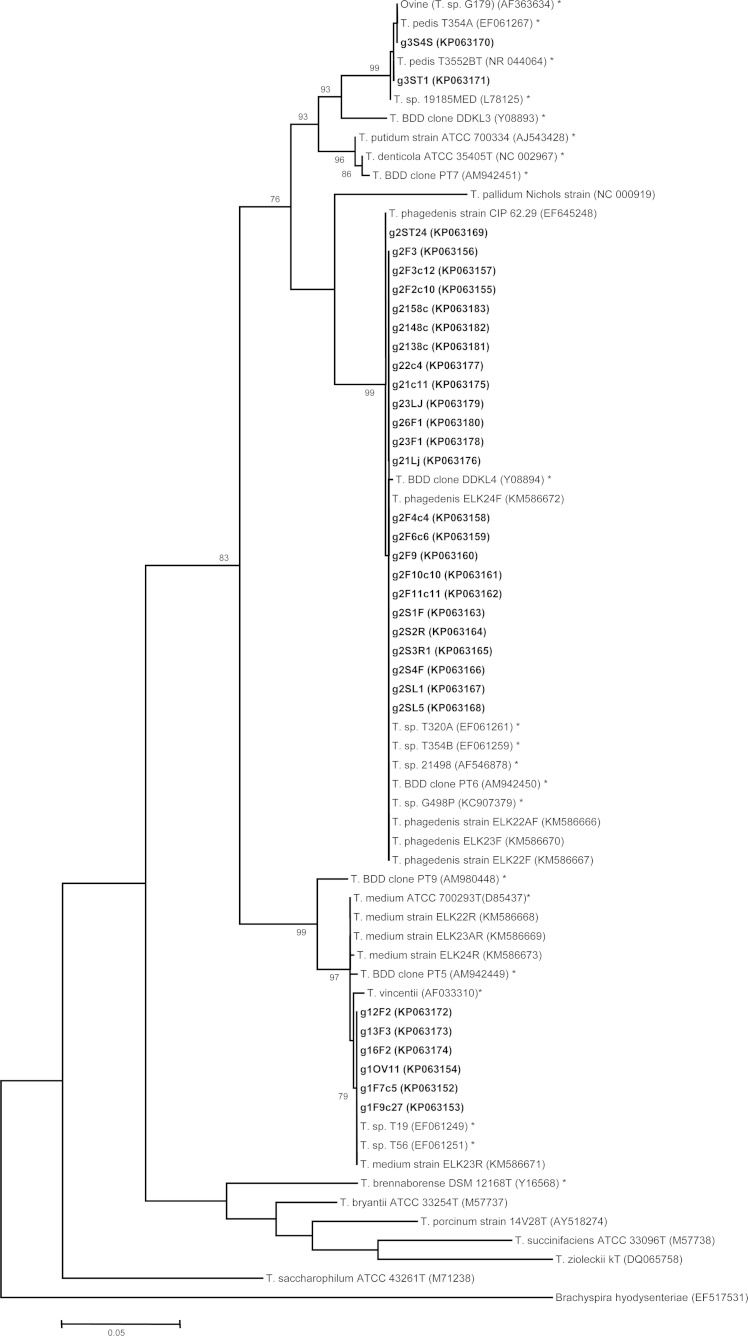
On phylogenetic tree analysis, the 32 CODD treponeme isolates separated into three distinct phylogroups corresponding exactly to the three Treponema phylogroups commonly isolated from BDD lesions (Fig. 1).
FIG 1.
A maximum likelihood tree based on 16S rRNA gene sequence comparisons of ∼1,200 aligned bases showing the relationship between the Treponema strains isolated here (shown in bold type) from sheep foot CODD lesions and other DD-associated and commensal treponeme 16S rRNA gene sequences. Accession numbers are shown in parentheses. Bootstrapping was performed 10,000 times, and for clarity, only bootstrap values above 70% are shown. Previously reported 16S rRNA gene sequences from BDD lesions are indicated by an asterisk.
DISCUSSION
With the emergence of CODD in dairy goats in the United Kingdom (21) and foot disease in elk in the United States recently reported as both clinically and etiologically similar (22), CODD is clearly increasing in importance and geographical spread. It is very clear that CODD leads to severe animal welfare issues for the animal involved and leads to significant financial implications for farmers (54) and hence is a food security issue. Thus, understanding the etiopathogenesis of CODD is key to developing the means of managing and preventing the spread of this debilitating disease. Since the first CODD report in 1997 (18), it has become apparent that there is an infective component and that the specific treponemes closely associated with DD in dairy cattle (30, 32, 34, 35) and beef cattle (4) are clearly involved in CODD and may be a primary initiating agent (38, 55). However, the available microbiological data produced thus far have been limited and not sufficient for proving or disproving a causative association. The current study is a comprehensive attempt to consider the link between BDD treponemes and CODD and to address the role of other bacteria frequently detected in infectious lameness issues in sheep.
What is clear from the study is that the BDD treponemes (individually and frequently collectively) are present in all CODD lesions, whereas in contrast, they are totally absent in samples from healthy sheep foot tissue. This is very strong data supportive of a primary infective etiology for CODD by these organisms. Interestingly, our data also show that other bacteria frequently associated with other sheep foot infections may also be commonly detected in CODD lesions, though with a far less striking frequency than BDD treponemes when comparing sheep feet with CODD lesions and healthy sheep feet.
Thus, a key question is whether BDD treponemes are the primary or secondary infection leading to the development of CODD lesions. What is clear is that they are present in all CODD lesions, which adds weight to the hypothesis that they are primary etiological agents. However, they may also be secondary infections to other, possibly noninfective, lesions of sheep feet. It has become apparent that the BDD treponemes must be considered promiscuous and opportunistic infective agents, as it has been clearly demonstrated that they can invade other (noninfective) lesions in cattle feet, such as white line disease and sole ulcers and clinically manifest as new serious infectious diseases which are very difficult to treat (56). Previously, only a 70% association of treponemes with CODD lesions was found (37), much lower than the 100% reported in this study (albeit by different methodologies) which is in accordance with the 100% association shown in dairy cattle DD lesions (32). Additionally, the previous study (37) also detected Treponema DNA in 38% of healthy foot tissue samples. It should be noted that the previous study (37) used a general treponeme PCR assay, which is not specific for the DD-associated Treponema phylogroups but targets all Treponema species. Therefore, this previous study gave no indication as to whether the Treponema species detected were pathogenic DD-associated Treponema species found commonly in dairy cattle BDD lesions or simply commensal treponemes found in the environment or a ruminant's GI tract (39). As seen in our present study, using the general treponeme PCR assay, treponemes were detected in 68% of healthy foot tissue samples; however, all of these healthy tissue samples were negative for the three DD-associated Treponema phylogroups tested for.
In cattle BDD samples, it was found that “T. medium/T. vincentii-like,” “T. phagedenis-like,” and T. pedis spirochetes were present in 96.1%, 98%, and 76.5% of DD lesions, respectively (32). These results are similar to the findings of this CODD study in terms of the detection of the three Treponema phylogroups, with 67%, 85%, and 71% of CODD lesions positive for “T. medium/T. vincentii-like,” “T. phagedenis-like,” and T. pedis spirochetes, respectively. In both cattle and sheep DD lesions, “T. phagedenis-like” spirochetes seem to be the most commonly detected spirochetes. However, “T. medium/T. vincentii-like” spirochetes appear to be less prevalent in CODD lesions than in BDD lesions. Importantly, one or more of the BDD treponemes are found in all cattle (32) and sheep DD lesions using the group-specific nested PCR assays.
Treponemes are anaerobic, highly fastidious bacteria and notoriously difficult to grow in culture (57). Despite this, treponemes were isolated from a significant number of CODD lesions in this study, giving evidence to the aforementioned molecular data and further highlighting their abundance in these lesions. In agreement with previous BDD studies, the “T. phagedenis-like” spirochetes were the most commonly isolated treponeme group from BDD and CODD lesions (32). This is also consistent with our PCR detection findings of an increased prevalence of this phylogroup in CODD lesions.
The phylogenetic analysis of Treponema isolates from CODD lesions here highlights the close clustering of these isolates with the previously isolated BDD and elk lesion treponemes. This strong phylogenetic relationship further strengthens the case for CODD and BDD having a shared treponemal etiopathogenesis. Interestingly, epidemiological data demonstrate an association between the presence of CODD in sheep with cattle on farms (55), and the study areas that allowed for identification of a CODD-like manifestation in U.S. elk were cograzed by cattle and sheep (22). Taken together, these observations suggest this disease is managing to transmit between host species effectively on shared farmland.
The phylogeny data clearly show that treponemes isolated from BDD, CODD, and also from the manifestation of DD in elk, fall into one of three well-defined groups and are completely different from those identified in GI tract samples (39). Unsurprisingly, one of the CODD isolates (G3S4S) belonging to the T. pedis spirochete group showed 100% 16S rRNA gene sequence identity to the previous T. pedis spirochete isolated from a CODD lesion (41). More interestingly, T. pedis spirochete isolate G3ST1 shared 100% 16S rRNA gene sequence identity with T. pedis T3552B (GenBank accession no. NR044064), which was isolated from a dairy cow DD lesion (34), suggesting that these treponemes cannot be distinguished upon between host species. These results provide further evidence toward a shared etiopathogenesis and for treponeme transmission between different host species. Further studies are needed to delineate whether there are specific changes to the three DD treponeme phylogroups associated with adaptation to different hosts and to underpin specific transmission cycles.
The CODD lesions were often associated with F. necrophorum, which was present in 71% of CODD lesions, versus a 9% prevalence in healthy foot tissue samples. This is consistent with previous data (37) where F. necrophorum was detected in a very low number of healthy sheep foot samples compared with CODD lesions. In that study, F. necrophorum subsp. necrophorum was present in 86% of CODD lesions compared with 46% of healthy feet, and F. necrophorum subsp. funduliforme was present in 28% of CODD lesions compared with 0% in healthy foot samples (37).
In the current study, D. nodosus, interestingly, was present in a similar number (59%) of CODD lesions as F. necrophorum. However, D. nodosus had a much higher detection rate in healthy tissues (39%) than F. necrophorum (9%). D. nodosus has previously been detected in CODD lesions with a prevalence of 74% compared to 31% in healthy foot tissues (37). These findings support our data and suggest that D. nodosus does not have a primary infective role in CODD lesions but may have a secondary role.
It is generally thought that D. nodosus plays a primary role and that F. necrophorum plays a secondary role in lesion development in sheep footrot (58–60). The current study shows that F. necrophorum has a strong association with CODD (9% prevalence in healthy tissues versus a 71% prevalence in CODD lesions), though not as strong as the BDD treponemes. However, it is possible that in CODD, like in footrot, F. necrophorum may act as a secondary invader to the DD-associated treponeme infection, as it apparently does to D. nodosus in footrot. This role of F. necrophorum is supported by recent research which highlights F. necrophorum as a key but secondary invader in footrot (60). This finding is consistent with the existing understanding of the role of Fusobacterium spp. in other diseases. F. necrophorum and other Fusobacteria are found in polymicrobial infections causing lesions and abscesses (61, 62) and are thought to progress disease severity through relationships with other pathogens (63, 64).
This study included a large number of farms in order to investigate CODD lesions from a large geographical area. However, due to the obvious ethical limitations in obtaining healthy foot tissue samples, a study design including healthy animal foot tissue from animals on the same farms from which CODD foot samples were obtained was not possible in all cases. This could introduce a weakness in the sampling strategy used, as it could be the case that sheep sampled for CODD lesions, and therefore from a farm with CODD present, could have bacteria present on their feet due to environmental contamination. However, the inclusion of healthy foot tissue from a small number of animals which also had a CODD lesion on another one of their feet (therefore healthy tissue obtained from a CODD-positive farm) limits this bias in sampling strategy.
Interestingly, the CODD lesions obtained from the sheep that also had their remaining healthy foot tissues sampled, were all positive for at least one of the DD-associated Treponema phylogroups; however, all the remaining healthy feet from each animal appeared negative for DD-associated treponemes. When looking at the presence of D. nodosus and F. necrophorum in these animals, no consistent finding was observed. For example, D. nodosus and F. necrophorum were found to be present in the CODD lesion obtained from sheep 101; however, its healthy feet were all negative for D. nodosus and all positive for F. necrophorum, whereas the CODD lesion from sheep 102 appeared negative for both D. nodosus and F. necrophorum, but 1/3 and 2/3 of its remaining healthy feet were positive for D. nodosus and F. necrophorum, respectively. This provides more evidence toward these two bacterial species playing a lesser role in CODD lesions than treponemal bacteria, which are present only in the lesions and not healthy foot tissue.
The fastidious nature of treponemes means it is extremely hard to successfully isolate them from tissue samples. However, DD-associated treponemes were isolated from a large number of CODD lesions and included all three DD-associated Treponema phylogroups. This, together with PCR data showing that all CODD lesions contained at least one or more DD-associated treponemes, supports the hypothesis that treponeme species, specifically the ones associated with BDD, are likely to be important etiological agents in CODD lesions. Our data also indicate that F. necrophorum may contribute to lesion pathogenesis in many CODD lesions.
From this study, it would appear that DD treponemes are likely to be the primary infective agent in CODD lesions, although the promiscuous nature of the DD treponemes and their wide range of tissue tropisms must be considered when making such conclusions. Additionally, the data presented here further confirm that the three DD treponeme-specific PCR assays can be used as a differential diagnostic tool for CODD and other DD manifestations from different geographical regions and hosts. Further molecular studies on Treponema isolates obtained from DD lesions of different animal species are necessary to compare their genetic relatedness and therefore help determine routes of transmission of this disease.
ACKNOWLEDGMENTS
This work was funded by QMS, HCC, and EBLEX, a division of the Agriculture and Horticulture Development Board, United Kingdom.
REFERENCES
- 1.Rebhun WC, Payne RM, King JM, Wolfe M, Begg SN. 1980. Interdigital papillomatosis in dairy cattle. J Am Vet Med Assoc 177:437–440. [PubMed] [Google Scholar]
- 2.Blowey RW, Sharp MW. 1988. Digital dermatitis in dairy cattle. Vet Rec 122:505–508. doi: 10.1136/vr.122.21.505. [DOI] [PubMed] [Google Scholar]
- 3.Brown CC, Kilgo PD, Jacobsen KL. 2000. Prevalence of papillomatous digital dermatitis among culled adult cattle in the southeastern United States. Am J Vet Res 61:928–930. doi: 10.2460/ajvr.2000.61.928. [DOI] [PubMed] [Google Scholar]
- 4.Sullivan LE, Carter SD, Blowey R, Duncan JS, Grove-White D, Evans NJ. 2013. Digital dermatitis in beef cattle. Vet Rec 173:582. doi: 10.1136/vr.101802. [DOI] [PubMed] [Google Scholar]
- 5.Bruijnis MR, Hogeveen H, Stassen EN. 2010. Assessing economic consequences of foot disorders in dairy cattle using a dynamic stochastic simulation model. J Dairy Sci 93:2419–2432. doi: 10.3168/jds.2009-2721. [DOI] [PubMed] [Google Scholar]
- 6.Marshall DJ, Walker RI, Cullis BR, Luff MF. 1991. The effect of footrot on body weight and wool growth of sheep. Aust Vet J 68:45–49. doi: 10.1111/j.1751-0813.1991.tb03126.x. [DOI] [PubMed] [Google Scholar]
- 7.Enting H, Kooija D, Dijkhuizen AA, Huirnea RBM, Noordhuizen-Stassenb EN. 1997. Economic losses due to clinical lameness in dairy cattle. Livest Prod Sci 49:259–267. doi: 10.1016/S0301-6226(97)00051-1. [DOI] [Google Scholar]
- 8.Hernandez J, Shearer JK, Webb DW. 2001. Effect of lameness on the calving-to-conception interval in dairy cows. J Am Vet Med Assoc 218:1611–1614. doi: 10.2460/javma.2001.218.1611. [DOI] [PubMed] [Google Scholar]
- 9.Warnick LD, Janssen D, Guard CL, Grohn YT. 2001. The effect of lameness on milk production in dairy cows. J Dairy Sci 84:1988–1997. doi: 10.3168/jds.S0022-0302(01)74642-5. [DOI] [PubMed] [Google Scholar]
- 10.Rajala-Schultz PJ, Gröhn YT, McCulloch CE. 1999. Effects of milk fever, ketosis, and lameness on milk yield in dairy cows. J Dairy Sci 82:288–294. doi: 10.3168/jds.S0022-0302(99)75235-5. [DOI] [PubMed] [Google Scholar]
- 11.Lucey S, Rowlands GJ, Russell AM. 1986. The association between lameness and fertility in dairy cows. Vet Rec 118:628–631. doi: 10.1136/vr.118.23.628. [DOI] [PubMed] [Google Scholar]
- 12.Collick DW, Ward WR, Dobson H. 1989. Associations between types of lameness and fertility. Vet Rec 125:103–106. doi: 10.1136/vr.125.5.103. [DOI] [PubMed] [Google Scholar]
- 13.Lee LA, Ferguson JD, Galligan DT. 1989. Effect of disease on days open assessed by survival analysis. J Dairy Sci 72:1020–1026. doi: 10.3168/jds.S0022-0302(89)79197-9. [DOI] [PubMed] [Google Scholar]
- 14.Sogstad ÅM, Østerås O, Fjeldaas O, Nafstad T. 2007. Bovine claw and limb disorders related to culling and carcass characteristics. Livest Sci 106:87–95. doi: 10.1016/j.livsci.2006.07.003. [DOI] [Google Scholar]
- 15.Argaez-Rodriguez FJ, Hird DW, Hernandez J, Read DH, Rodriguez-Lainz A. 1997. Papillomatous digital dermatitis on a commercial dairy farm in Mexicali, Mexico: incidence and effect on reproduction and milk production. Prev Vet Med 32:275–286. doi: 10.1016/S0167-5877(97)00031-7. [DOI] [PubMed] [Google Scholar]
- 16.Relun A, Lehebel A, Chesnin A, Guatteo R, Bareille N. 2013. Association between digital dermatitis lesions and test-day milk yield of Holstein cows from 41 French dairy farms. J Dairy Sci 96:2190–2200. doi: 10.3168/jds.2012-5934. [DOI] [PubMed] [Google Scholar]
- 17.Cha E, Hertl JA, Bar D, Gröhn YT. 2010. The cost of different types of lameness in dairy cows calculated by dynamic programming. Prev Vet Med 97:1–8. doi: 10.1016/j.prevetmed.2010.07.011. [DOI] [PubMed] [Google Scholar]
- 18.Harwood DG, Cattell JH, Lewis CJ, Naylor R. 1997. Virulent foot rot in sheep. Vet Rec 140:687. [PubMed] [Google Scholar]
- 19.Davies IH, Naylor RD, Martin PK. 1999. Severe foot lesions in sheep. Vet Rec 145:646. [PubMed] [Google Scholar]
- 20.Sayers G, Marques PX, Evans NJ, O'Grady L, Doherty ML, Carter SD, Nally JE. 2009. Identification of spirochetes associated with contagious ovine digital dermatitis. J Clin Microbiol 47:1199–1201. doi: 10.1128/JCM.01934-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sullivan LE, Evans NJ, Clegg SR, Carter SD, Horsfield JE, Grove-White D, Duncan JS. 26 November 2014. Digital dermatitis treponemes associated with a severe foot disease in dairy goats. Vet Rec doi: 10.1136/vr.102858. [DOI] [PubMed] [Google Scholar]
- 22.Clegg SR, Mansfield KG, Newbrook K, Sullivan LE, Blowey RW, Carter SD, Evans NJ. 2015. Isolation of digital dermatitis treponemes from hoof lesions in wild North American elk (Cervus elaphus) in Washington State, USA. J Clin Microbiol 53:88–94. doi: 10.1128/JCM.02276-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cheli R, Mortellaro C. 1974. Digital dermatitis in cattle, p 208–213. In Gallarati P. (ed), Proceedings of the 8th International Conference on Diseases of Cattle, Milan, Italy. [Google Scholar]
- 24.Duncan JS, Angell JW, Carter SD, Evans NJ, Sullivan LE, Grove-White DH. 2014. Contagious ovine digital dermatitis: an emerging disease. Vet J 201:265–268. doi: 10.1016/j.tvjl.2014.06.007. [DOI] [PubMed] [Google Scholar]
- 25.Naylor RD, Martin PK, Jones JR, Burnell MC. 1998. Isolation of spirochetes from an incident of severe virulent ovine footrot. Vet Rec 143:690–691. [PubMed] [Google Scholar]
- 26.Wassink GJ, Moore LJ, Grogono-Thomas R, Green LE. 2003. Exploratory findings on the prevalence of contagious ovine digital dermatitis in sheep in England and Wales during 1999 to 2000. Vet Rec 152:504–506. doi: 10.1136/vr.152.16.504. [DOI] [PubMed] [Google Scholar]
- 27.Winter AC. 2008. Lameness in sheep. Small Ruminant Res 76:149–153. doi: 10.1016/j.smallrumres.2007.12.008. [DOI] [Google Scholar]
- 28.Duncan JS, Grove-White D, Oultram JW, Phythian CJ, Dijk JV, Carter SD, Cripps PJ, Williams HJ. 2011. Effects of parenteral amoxicillin on recovery rates and new infection rates for contagious ovine digital dermatitis in sheep. Vet Rec 169:606. doi: 10.1136/vr.d4394. [DOI] [PubMed] [Google Scholar]
- 29.Han S, Mansfield KG. 2014. Severe hoof disease in free-ranging Roosevelt elk (Cervus elaphus roosevelti) in southwestern Washington, USA. J Wildl Dis 50:259–270. doi: 10.7589/2013-07-163. [DOI] [PubMed] [Google Scholar]
- 30.Klitgaard K, Boye M, Capion N, Jensen TK. 2008. Evidence of multiple Treponema phylogroups involved in bovine digital dermatitis as shown by 16S rRNA gene analysis and fluorescence in situ hybridization. J Clin Microbiol 46:3012–3020. doi: 10.1128/JCM.00670-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nordhoff M, Moter A, Schrank K, Wieler LH. 2008. High prevalence of treponemes in bovine digital dermatitis – a molecular epidemiology. Vet Microbiol 131:293–300. doi: 10.1016/j.vetmic.2008.04.019. [DOI] [PubMed] [Google Scholar]
- 32.Evans NJ, Brown JM, Demirkan I, Singh P, Getty B, Timofte D, Vink WD, Murray RD, Blowey RW, Birtles RJ, Hart CA, Carter SD. 2009. Association of unique, isolated treponemes with bovine digital dermatitis lesions. J Clin Microbiol 47:689–696. doi: 10.1128/JCM.01914-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Choi BK, Nattermann H, Grund S, Haider W, Gobel UB. 1997. Spirochetes from digital dermatitis lesions in cattle are closely related to treponemes associated with human periodontitis. Int J Syst Bacteriol 47:175–181. doi: 10.1099/00207713-47-1-175. [DOI] [PubMed] [Google Scholar]
- 34.Evans NJ, Brown JM, Demirkan I, Murray RD, Vink WD, Blowey RW, Hart CA, Carter SD. 2008. Three unique groups of spirochetes isolated from digital dermatitis lesions in UK cattle. Vet Microbiol 130:141–150. doi: 10.1016/j.vetmic.2007.12.019. [DOI] [PubMed] [Google Scholar]
- 35.Stamm LV, Bergen HL, Walker RL. 2002. Molecular typing of papillomatous digital dermatitis-associated Treponema isolates based on analysis of 16S-23S ribosomal DNA intergenic spacer regions. J Clin Microbiol 40:3463–3469. doi: 10.1128/JCM.40.9.3463-3469.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Evans NJ, Brown JM, Demirkan I, Murray RD, Birtles RJ, Hart CA, Carter SD. 2009. Treponema pedis sp. nov., a spirochete isolated from bovine digital dermatitis lesions. Int J Syst Evol Microbiol 59:987–991. doi: 10.1099/ijs.0.002287-0. [DOI] [PubMed] [Google Scholar]
- 37.Moore LJ, Woodward MJ, Grogono-Thomas R. 2005. The occurrence of treponemes in contagious ovine digital dermatitis and the characterisation of associated Dichelobacter nodosus. Vet Microbiol 111:199–209. doi: 10.1016/j.vetmic.2005.10.016. [DOI] [PubMed] [Google Scholar]
- 38.Dhawi A, Hart CA, Demirkan I, Davies IH, Carter SD. 2005. Bovine digital dermatitis and severe virulent ovine foot rot: a common spirochaetal pathogenesis. Vet J 169:232–241. doi: 10.1016/j.tvjl.2004.01.029. [DOI] [PubMed] [Google Scholar]
- 39.Evans NJ, Timofte D, Isherwood DR, Brown JM, Williams JM, Sherlock K, Lehane MJ, Murray RD, Birtles RJ, Hart CA, Carter SD. 2012. Host and environmental reservoirs of infection for bovine digital dermatitis treponemes. Vet Microbiol 156:102–109. doi: 10.1016/j.vetmic.2011.09.029. [DOI] [PubMed] [Google Scholar]
- 40.Winter AC. 2004. Lameness in sheep 2. Treatment and control. In Practice 26:130–139. doi: 10.1136/inpract.26.3.130. [DOI] [Google Scholar]
- 41.Demirkan I, Carter SD, Winstanley C, Bruce KD, McNair NM, Woodside M, Hart CA. 2001. Isolation and characterisation of a novel spirochete from severe virulent ovine foot rot. J Med Microbiol 50:1061–1068. [DOI] [PubMed] [Google Scholar]
- 42.Rurangirwa FR, Dilbeck PM, Crawford TB, McGuire TC, McElwain TF. 1999. Analysis of the 16S rRNA gene of microorganism WSU 86-1044 from an aborted bovine foetus reveals that it is a member of the order Chlamydiales: proposal of Waddliaceae fam. nov., Waddlia chondrophila gen. nov., sp. nov. Int J Syst Bacteriol 49:577–581. doi: 10.1099/00207713-49-2-577. [DOI] [PubMed] [Google Scholar]
- 43.Ehresmann C, Stiegler P, Mackie GA, Zimmermann RA, Ebel JP, Fellner P. 1975. Primary sequence of the 16S ribosomal RNA of Escherichia coli. Nucleic Acids Res 2:265–278. doi: 10.1093/nar/2.2.265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.La Fontaine S, Egerton JR, Rood JI. 1993. Detection of Dichelobacter nodosus using species-specific oligonucleotides as PCR primers. Vet Microbiol 35:101–117. doi: 10.1016/0378-1135(93)90119-R. [DOI] [PubMed] [Google Scholar]
- 45.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. 1990. Basic local alignment search tool. J Mol Biol 215:403–410. [DOI] [PubMed] [Google Scholar]
- 46.Thompson JD, Higgins DG, Gibson TJ. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res 22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kumar S, Tamura K, Jakobsen IB, Nei M. 2001. MEGA2: Molecular Evolutionary Genetics Analysis software. Bioinformatics 17:1244–1245. doi: 10.1093/bioinformatics/17.12.1244. [DOI] [PubMed] [Google Scholar]
- 48.Kibbe WA. 2007. OligoCalc: an online oligonucleotide properties calculator. Nucleic Acids Res 35(Suppl 2):W43–W46. doi: 10.1093/nar/gkm234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bennett G, Hickford J, Sedcole R, Zhou H. 2009. Dichelobacter nodosus, Fusobacterium necrophorum and the epidemiology of footrot. Anaerobe 15:173–176. doi: 10.1016/j.anaerobe.2009.02.002. [DOI] [PubMed] [Google Scholar]
- 50.Oelke AM, Nagaraja TG, Wilkerson MJ, Stewart GC. 2005. The leukotoxin operon of Fusobacterium necrophorum is not present in other species of Fusobacterium. Anaerobe 11:123–129. doi: 10.1016/j.anaerobe.2004.10.003. [DOI] [PubMed] [Google Scholar]
- 51.Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. 2011. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 28:2731–2739. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Milne I, Lindner D, Bayer M, Husmeier D, McGuire G, Marshall DF, Wright F. 2009. TOPALi v2: a rich graphical interface for evolutionary analyses of multiple alignments on HPC clusters and multi-core desktops. Bioinformatics 25:126–127. doi: 10.1093/bioinformatics/btn575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tamura K, Nei M. 1993. Estimation of the number of nucleotide substitutions in the control region of mitochondrial DNA in humans and chimpanzees. Mol Biol Evol 10:512–526. [DOI] [PubMed] [Google Scholar]
- 54.Phythian CJ, Michalopoulou E, Jones PH, Winter AC, Clarkson MJ, Stubbings LA, Grove-White D, Cripps PJ, Duncan JS. 2011. Validating indicators of sheep welfare through a consensus of expert opinion. Animal 5:943–952. doi: 10.1017/S1751731110002594. [DOI] [PubMed] [Google Scholar]
- 55.Angell JW, Duncan JS, Carter SD, Grove-White DH. 2014. Farmer reported prevalence and factors associated with contagious ovine digital dermatitis in Wales: a questionnaire of 511 sheep farmers. Prev Vet Med 113:132–138. doi: 10.1016/j.prevetmed.2013.09.014. [DOI] [PubMed] [Google Scholar]
- 56.Evans NJ, Blowey RW, Timofte D, Isherwood DR, Brown JM, Murray R, Paton RJ, Carter SD. 2011. Association between bovine digital dermatitis treponemes and a range of ‘non-healing’ bovine hoof disorders. Vet Rec 168:214. doi: 10.1136/vr.c5487. [DOI] [PubMed] [Google Scholar]
- 57.Norris SJ, Paster BJ, Smibert RM. 2010. Genus IV. Treponema Schaudinn 1905, 1728AL (Spironema Vuillemin 1905, 1568; Microspironema Stiles and Pfender 1905, 936), p 501–531. In Krieg NR, Ludwig W, Whitman W, Hedlund BP, Paster BJ, Staley JT, Ward N, Brown D, Parte A (ed), Bergey's manual of systematic bacteriology, 2nd ed, vol 4 Springer, New York, NY. [Google Scholar]
- 58.Beveridge WIB. 1941. Footrot in sheep: a transmissible disease due to infection with Fusiformis nodosus: studies on its cause, epidemiology and control. CSIRO Aust Bull 140:1–56. [Google Scholar]
- 59.Kennan RM, Dhungyel OP, Whittington RJ, Egerton JR, Rood JI. 2001. The type IV fimbrial subunit gene (fimA) of Dichelobacter nodosus is essential for virulence, protease secretion, and natural competence. J Bacteriol 183:4451–4458. doi: 10.1128/JB.183.15.4451-4458.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Witcomb LA, Green LE, Kaler J, Ul-Hassan A, Calvo-Bado LA, Medley GF, Grogono-Thomas R, Wellington EM. 2014. A longitudinal study of the role of Dichelobacter nodosus and Fusobacterium necrophorum load in initiation and severity of footrot in sheep. Prev Vet Med 115:48–55. doi: 10.1016/j.prevetmed.2014.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Brook I. 2002. Secondary bacterial infections complicating skin lesions. J Med Microbiol 51:808–812. [DOI] [PubMed] [Google Scholar]
- 62.Hofstad T. 2006. The genus Fusobacterium, p 1016–1027. In Dworkin M, Falkow S (ed), The prokaryotes. Springer, Berlin, Germany. doi: 10.1007/0-387-30747-8_51. [DOI] [Google Scholar]
- 63.Brook I, Walker RI. 1986. The relationship between Fusobacterium species and other flora in mixed infection. J Med Microbiol 21:93–100. doi: 10.1099/00222615-21-2-93. [DOI] [PubMed] [Google Scholar]
- 64.Tan ZL, Nagaraja TG, Chengappa MM. 1996. Fusobacterium necrophorum infections: virulence factors, pathogenic mechanism and control measures. Vet Res Commun 20:113–140. doi: 10.1007/BF00385634. [DOI] [PubMed] [Google Scholar]