Abstract
Shiga toxin-producing Escherichia coli (STEC) is an enteropathogen of public health concern because of its ability to cause serious illness and outbreaks. In this prospective study, a diagnostic screening algorithm to categorize STEC infections into risk groups was evaluated. The algorithm consists of prescreening stool specimens with real-time PCR (qPCR) for the presence of stx genes. The qPCR-positive stool samples were cultured in enrichment broth and again screened for stx genes and additional virulence factors (escV, aggR, aat, bfpA) and O serogroups (O26, O103, O104, O111, O121, O145, O157). Also, PCR-guided culture was performed with sorbitol MacConkey agar (SMAC) and CHROMagar STEC medium. The presence of virulence factors and O serogroups was used for presumptive pathotype (PT) categorization in four PT groups. The potential risk for severe disease was categorized from high risk for PT group I to low risk for PT group III, whereas PT group IV consists of unconfirmed stx qPCR-positive samples. In total, 5,022 stool samples of patients with gastrointestinal symptoms were included. The qPCR detected stx genes in 1.8% of samples. Extensive screening for virulence factors and O serogroups was performed on 73 samples. After enrichment, the presence of stx genes was confirmed in 65 samples (89%). By culture on selective media, STEC was isolated in 36% (26/73 samples). Threshold cycle (CT) values for stx genes were significantly lower after enrichment compared to direct qPCR (P < 0.001). In total, 11 (15%), 19 (26%), 35 (48%), and 8 (11%) samples were categorized into PT groups I, II, III, and IV, respectively. Several virulence factors (stx2, stx2a, stx2f, toxB, eae, efa1, cif, espA, tccP, espP, nleA and/or nleB, tir cluster) were associated with PT groups I and II, while others (stx1, eaaA, mch cluster, ireA) were associated with PT group III. Furthermore, the number of virulence factors differed between PT groups (analysis of variance, P < 0.0001). In conclusion, a diagnostic algorithm enables fast discrimination of STEC infections associated with a high to moderate risk for severe disease (PT groups I and II) from less-virulent STEC (PT group III).
INTRODUCTION
Shiga toxin-producing Escherichia coli (STEC) is a zoonotic pathogen frequently identified as causative agent of acute diarrheal disease in humans. The outcomes of STEC infections may range from asymptomatic carriage and mild diarrhea to severe disease, such as hemorrhagic colitis (HC) and hemolytic-uremic syndrome (HUS) (1–3).
Based on pathogenic properties, a subgroup of STEC is also designated enterohemorrhagic E. coli (EHEC); this subgroup of stx-positive strains also contains the locus of enterocyte effacement (LEE) pathogenicity island (4). EHEC belongs to certain serotypes that are frequently associated with outbreaks and life-threatening illnesses (5). Worldwide, the most common EHEC serotype both in outbreaks and in sporadic cases of severe disease is E. coli O157:H7 (4, 6, 7). Consequently, public health and regulatory responses have been focused mainly on this serotype. However, due to increased surveillance with tests able to target all serotypes of STEC, evidence is accumulating that 30% to 60% of EHEC infections are caused by non-O157 strains (8, 9).
To aid in assessing the public health risks associated with STEC, an empirical seropathotype (SPT) classification of strains was proposed by Karmali et al. (5), based upon the reported frequency of STEC serotypes in human illness, their known association with outbreaks, and the severity of the outcome. Serotypes classified as SPT A (O157:H7 and O157:nonmotile [NM]) or SPT B (O26:H11/NM, O103:H2, O111:NM, O121:H19, and O145:NM) have been associated with outbreaks and severe disease; however, SPT A is more frequently reported. SPT C comprises serotypes (e.g., O91:H21, O113:H21, O5:NM, O104:H21, O121:NM, and O165:H25) that have been associated with sporadic cases of severe disease but not with outbreaks. SPT D includes STEC serotypes reported to cause sporadic disease that are associated with diarrhea but not severe disease. Serotypes included in SPT E have not been associated with human illness.
The identification of non-O157 EHEC serotypes remains challenging because of a lack of phenotypical characteristics that can distinguish these strains from less-virulent STEC serotypes and other E. coli that share the same environment. Furthermore, of all confirmed STEC infections in the European Union during 2007 to 2010, more than 85% of the isolates were not fully serotyped (9). As SPT classification requires fully serotyped isolates, the identification of non-O157 EHEC serotypes proves to be a major obstacle. Also, the 2011 O104:H4 EHEC outbreak has demonstrated that the emergence of new virulent strains is another limitation of the SPT classification proposed by Karmali et al., as these strains cannot be assigned to a specific SPT group (10, 11).
While it remains unclear which virulence factors (VF) precisely define STEC pathogenicity, the STEC serotypes that carry VF genes in addition to stx genes are more likely to be associated with HC and HUS (9, 12). These strains usually carry the LEE, a pathogenicity island (PAI) containing genes responsible for the characteristic attaching and effacing (A/E) lesions (4, 13). In addition, they can be characterized by non-LEE-encoded effector (nle) genes, which are harbored on other PAIs in the bacterial chromosome (5, 14, 15), and virulence plasmids encoding EHEC-hemolysin (EHEC-hlyA) that are widely distributed among EHEC of different serotypes (16–18). Enteroaggregative E. coli (EAEC)-STEC hybrid strains of serotypes other than O104:H4, such as O111:H2, O86:NM, O59:NM, and Orough:NM, have also been associated with sporadic cases and outbreaks of HUS and (bloody) diarrhea, advocating the incorporation of EAEC virulence markers for the categorization of STEC (19–21).
However, no single VF or combination of virulence factors precisely defines the potential of a STEC strain to cause more severe disease. While the stx subtypes stx2a and stx2c and the LEE-positive strains are associated with a high risk of more serious illness (9, 22–24), other virulence gene combinations (even in E. coli strains that lack the stx genes) may also be associated with severe disease, including HC and HUS (12, 25–27). Furthermore, patient characteristics and infectious doses also determine the outcome of disease (28).
Although the current approaches for detecting STEC in clinical microbiology laboratories still mainly rely on a conventional culture (e.g., sorbitol MacConkey agar [SMAC] or cefixime tellurite [CT-SMAC]) and to a lesser extent on Stx toxin-based assays, a trend toward PCR-based methods for the rapid detection of STEC (stx1 and stx2 genes) has been observed in recent years, resulting in improved detection rates (29, 30). Enhanced detection and reporting of STEC infections have as a drawback an increased workload for community health services, and the clinical and public health relevance of PCR findings solely based on the detection of the stx genes is unclear (31). Therefore, diagnostic approaches that can categorize STEC while avoiding the limitations of the SPT classification of Karmali et al. are needed.
In this study, we describe a rapid screening algorithm, including both molecular and conventional methods, to determine the pathogenic potential of STEC. The aim is to discriminate infections with less-virulent STEC from those with clinical relevance and risk for public health.
MATERIALS AND METHODS
Patient specimens.
Our laboratory serves a population of about 1 million inhabitants, including both community-based and hospitalized patients. From September 2012 through December 2012 a total of 5,022 stool samples were prospectively screened for the presence of enteric bacterial, protozoan, and viral pathogens. The samples originated from patients (n = 4,714) with infectious gastroenteritis (IG) included in their differential diagnosis. The mean age was 39 years (range, 0 to 101 years), and 1,985 (42.1%) patients were male. Clinical information addressing symptoms, use of antibiotics, and traveling history were obtained from the request form filled out by physicians. On receipt, all stool specimens were routinely examined by molecular methods (real-time PCR [qPCR]) for the presence of Campylobacter jejuni, Salmonella enterica, Shiga toxin-producing Escherichia coli (STEC), Shigella spp., enteroinvasive E. coli (EIEC), Cryptosporidium parvum, C. hominis, Dientamoeba fragilis, Giardia lamblia, and Entamoeba histolytica. Upon specific request of physicians, an examination for the presence of adenovirus (by enzyme immunoassay [EIA]), rotavirus (by EIA), norovirus (by qPCR), and Clostridium difficile toxins A and B (by EIA) was also performed.
Design of the diagnostic algorithm and STEC risk assessment.
The algorithm consists of qPCR for the detection of the stx genes (stx1 and stx2) on stool samples, as described previously (30). The qPCR stx-negative stool samples were regarded as STEC negative. In case of a stx-positive result on qPCR, the stool sample was enriched in brilliant green bile (BGB) broth, followed by DNA extraction and multiplex qPCR for the detection of VF (stx1, stx2, stx2f, escV, aggR, and aat genes), and O-serogroup determination (wzxO26, wzxO103, wzxO104, wbdO111, wzxO121, ihpO145, and rfbO157). In order to obtain an isolate, the qPCR positive samples were cultured directly and after enrichment on STEC-selective media. Virulence determinants and O serogroups were confirmed by qPCR on suspicious colonies (or streaks) grown on STEC-selective media and by seroagglutination. Attempts to obtain an isolate were made up to a maximum of 5 colonies per agar plate. A schematic overview of the diagnostic algorithm is presented in Fig. 1.
FIG 1.
The STEC diagnostic algorithm consists of both molecular and conventional methods; when stx genes are detected with direct qPCR, the stool sample is enriched. DNA is isolated from the enriched broth and screened for the presence of stx genes and additional VF and O serogroups. VF and O serogroups are confirmed by qPCR on suspicious colonies grown on selective media and by seroagglutination. STEC isolates were fully serotyped (O:H typing) at the RIVM.
The risk assessment of STEC infection was performed using a molecular approach, as described previously (9). It delivers a scheme that describes the presumptive categorization of STEC according to the potential risk, using the presence of genes encoding VF in addition to the presence of the stx genes. Categorization of stx-positive PCR samples is based on the presence of the VF escV (LEE-positive) and/or aggR and/or aat (pAA-positive) and on the detection of O serogroups that are most frequently associated with severe human disease and outbreaks, e.g., O26, O103, O104, O111, O121, O145, and O157. The potential risk for diarrhea and severe disease has been categorized into pathotype (PT) group I (high risk for diarrhea and severe disease) to PT group III (moderate risk for diarrhea/low risk for severe disease), while PT group IV consists of stx-positive PCR samples that are not confirmed after enrichment (Table 1). In case an isolate was obtained and fully serotyped, a classification of the STEC isolate into a seropathotype (SPT) was made, as described previously (5). The stx subtyping and genetic characterization of cultured isolates were performed in order to confirm the validity of the proposed molecular-based PT approach for the risk assessment of STEC infections.
TABLE 1.
Proposed molecular approach for the presumptive categorization of STEC based on enriched BGB PCR results
| PT group | Direct PCR stx genes present | Enriched BGB PCR stx genes present | Additional genesa | Serogroup(s) | Potential risk |
|
|---|---|---|---|---|---|---|
| Diarrhea | HUS/HC | |||||
| I | Yes | Yes | escV positive or aggR and/or aat positive | O26, O103, O104, O111, O121, O145, O157 | High | High |
| II | Yes | Yes | escV positive or aggR and/or aat positive | Any other serogroup | High | Moderate |
| III | Yes | Yes | escV negative and aggR and/or aat negative | Any serogroup | Moderate | Low |
| IV | Yes | No | NA | NA | NA | NA |
escV gene, marker for presence of the LEE PAI; aggR and/or aat gene, markers for the presence of the pAA plasmid carried by EAEC; NA, not applicable.
PCR-guided culture.
For culture of STEC, the selective media SMAC and CHROMagar STEC (CHROMagar Microbiology, Paris, France) were used directly (24 h at 35°C) and after enrichment in BGB broth for approximately 16 h at 35°C. Identification of STEC- and/or EHEC O157-suspicious colonies (non-sorbitol-fermenting colonies on SMAC and mauve nonfluorescent colonies on CHROMagar STEC) and STEC and/or EHEC non-O157 (mauve fluorescent colonies on CHROMagar STEC) was carried out by the detection of virulence genes and serogenotyping with qPCR, performing an indole reaction and serological typing (serogroup O157 only). All genotypically/biochemically identified E. coli strains were confirmed using the Vitek 2 system (bioMérieux, Boxtel, The Netherlands). All culture and identification media were produced by Mediaproducts BV (Groningen, The Netherlands), whereas the E. coli O157 agglutination serum was from Oxoid (Basingstoke, Hampshire, England). Resistance profiling was performed with the Vitek 2 system. Furthermore, of qPCR positive samples with a threshold cycle (CT) value of <35, five E. coli colonies cultured on SMAC agar were subcultured and sent to the National Institute for Public Health and the Environment (RIVM, Bilthoven, The Netherlands) for genotying (stx1, stx2, stx2f, eae, EHEC-hlyA, and O157) and O:H-serotyping of the isolates as part of STEC national surveillance.
Molecular assays. (i) Specimen preparation and DNA extraction.
Specimen preparation followed by DNA extraction using the automated NucliSens easyMag (bioMérieux, Boxtel, The Netherlands) according to the manufacturer's instructions was performed as previously described (30). Briefly, for the DNA extraction from stool, 100 μl fecal suspension and 50 μl of enriched selenite broth were used as input. For DNA extraction from enriched BGB broth, 100 μl was used as input. In addition, approximately 6,000 copies of the Phocine herpesvirus 1 (PhHV), which served as the internal control (IC), were copurified. DNA was eluted in 110 μl of elution buffer.
For confirmation of suspicious colonies by qPCR, the DNA from isolates was extracted by heat lysis for 10 min at 95°C in the NucliSens easyMag elution buffer. For genetic characterization by microarray, the DNA extraction was performed using a DNeasy blood and tissue kit (Qiagen, GmbH, Germany) from the overnight culture of the pure isolates, according to the manufacturer's instructions.
(ii) Real-time PCR.
Real-time amplification was carried out on an AB 7500 sequence detection system (Applied Biosystems, Nieuwerkerk a/d IJssel, The Netherlands), as described previously (30). Each 25-μl reaction consisted of 5 μl template DNA, 1× TaqMan Universal PCR master mix, and 2.5 μg bovine serum albumin (Roche Diagnostics Netherlands B.V., Almere, the Netherlands). The primers and probes used for the detection of virulence determinants and O-serogroup-specific gene targets are listed in Table S1 in the supplemental material. Reactions were run under the following conditions: 50°C for 2 min, 95°C for 10 min, followed by 40 cycles of 95°C for 15 s, and 60°C for 1 min. In every PCR run, a negative extraction control (NEC) and positive extraction control (PEC) or PCR mix control (PMC) were included. A real-time PCR was considered inhibited when the CT value for the PhHV exceeded the mean CT value for uninhibited specimens + 2 standard deviations.
(iii) stx subtyping.
DNA isolates of the BGB broth and/or confirmed STEC isolates were sent to the University Medical Center Groningen (UMCG; Groningen, The Netherlands) for stx subtyping. Subtyping of the stx1 and stx2 genes was performed as described previously (32). Briefly, for stx1 subtyping, a triplex PCR was performed, and each 25-μl reaction mixture consisted of 2.5 μl PCR buffer 10× (Qiagen), 1 μl MgCl2 25 mM (Qiagen), 0.5 μl dNTP mix10 mM (Applied Biosystems), 0.25 μl HotStar polymerase 5 U/μl (Qiagen), 2 μl each of the two primers for stx1a, 1 μl each of the four primers for stx1c and stx1d (stock solution of all primers was 5 μM), and 5 μl template DNA.
For stx2 subtyping PCR, each 20-μl reaction consisted of 2.5 μl PCR buffer 10× (Qiagen), 0.8 μl MgCl2 25 mM (Qiagen), 0.4 μl dNTP mix 10 mM (Applied Biosystem), 0.2 μl HotStar polymerase 5 U/μl (Qiagen), 1.25 μl each of the primers, and 5 μl template DNA. The stx2c and stx2e subtyping PCR was performed as a duplex PCR as well as with stx2f and stx2g. Reactions were run under the following conditions: 95°C for 15 min followed by 35 cycles of 94°C for 50 s, 64°C (hybridization was at 66°C for stx2d) for 40 s, and 72°C for 60 s, with a final extension at 72°C for 3 min.
(iv) Genetic characterization by DNA microarray.
Confirmed STEC isolates were sent to the University Medical Centre Groningen for genetic characterization using an E. coli genotyping combined assay kit according to the manufacturer's protocol (Clondiag; Alere Technologies, GmbH, Jena, Germany). The E. coli oligonucleotide array strips contain gene targets for the identification of virulence genes, antimicrobial resistance genes, and DNA-based serotyping genes. Briefly, multiplex linear DNA amplification and labeling were performed in a total volume of 10 μl containing 3.9 μl of 2× labeling buffer, 1 μl E. coli labeling primer mix, 0.1 μl DNA polymerase, and 5 μl genomic DNA (100 to 200 ng/μl). Reactions were run under the following conditions: 96°C for 5 min followed by 45 cycles of 50°C for 20 s, 72°C for 30 s, and 96°C for 20 s.
The hybridization and washing steps were performed using the Hybridization Plus kit according to the manufacturer's protocol. Visualization of hybridization was achieved using the ArrayMate instrument (CLONDIAG GmbH, Jena, Germany), and signals of the array spots were analyzed automatically. Ambiguous-called signals were rechecked visually in order to obtain a definite interpretation when possible. In case any signal remained inconclusive, they were regarded as negative.
Statistical analysis.
We used the Fisher exact method to test if the presence or absence of VF was associated with certain PT groups and whether growth of suspicious colonies on CHROMagar STEC was associated with the presence of the escV gene (LEE-positive) (JavaStat). The median CT values of subgroups were compared using the Wilcoxon rank sum test with NCSS version 2007 (NCSS statistical software, Kaysville, UT, USA). A one-way analysis of variance (ANOVA) was used to compare the total number of VF present in isolated strains that were assigned to PT groups. For all tests, statistical significance was indicated by a two-tailed P value of <0.05.
Furthermore, cluster analysis of VF with construction of dendrograms was performed with Bionumerics version 4.6 (Applied Maths NV, Sint-Martens-Latem, Belgium) using the dice correlation and the unweighted-pair group method using average linkages (UPGMA).
RESULTS
Detection frequency of stx genes in patient specimens.
A total of 5,022 stool specimens from 4,714 patients were examined using direct qPCR for the detection of the stx genes. In total, 90 samples (84 patients) were positive for the stx genes (1.8%). The diagnostic algorithm was applied on all samples, but all screening data were available for only 73 samples (70 patients); therefore, the remaining 17 samples were excluded from the analysis. Direct qPCR for the stx genes was confirmed by qPCR on enriched BGB in 65 samples (89%). In the remaining 8 samples (11%), no stx genes could be detected after broth enrichment, although the virulence factors aggR, aat, and/or escV and O104 serogroup were detected in one sample. These 8 samples initially had a relatively high CT value (CT, ≥34) in the direct qPCR.
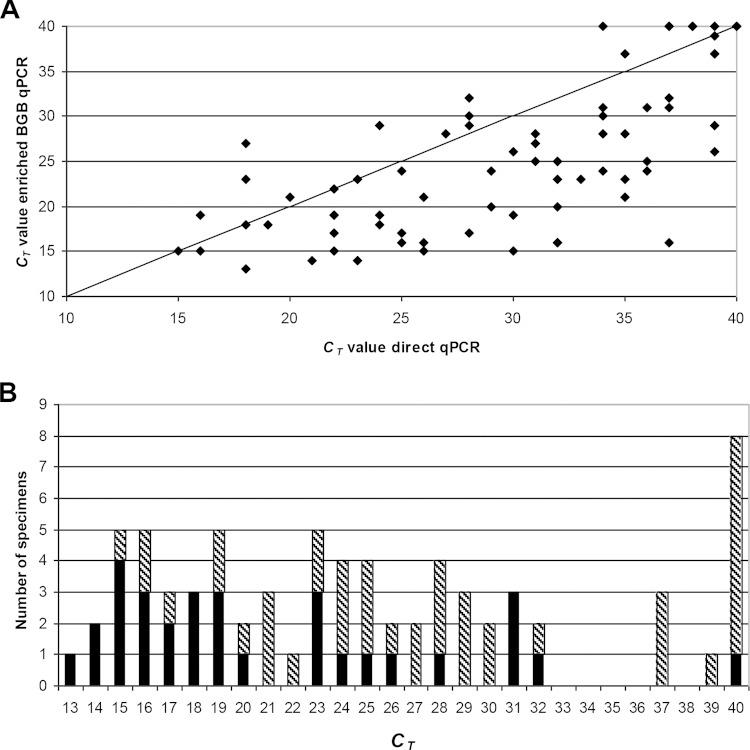
The stx ΔCT values for enriched BGB PCR and direct qPCR (ΔCT = CT BGB − CT direct) ranged from 9 to −21. In 55/65 (85%) of the enriched BGB samples, the ΔCT was ≤0, which was indicative for the presence of viable STEC; the CT values of the enriched BGB PCR (mean CT, 23.1) were significantly lower than the CT values of the direct qPCR (mean CT, 29.6) (Wilcoxon rank sum, P < 0.001) (Fig. 2A).
FIG 2.
Direct comparison of stx CT values for direct qPCR versus stx CT values for enriched BGB PCR (A). The solid line represents the hypothetical identical performance between both methods. The stx CT values for enriched BGB PCR were significantly lower (Wilcoxon rank sum, P < 0.001). Distribution of CT values for STEC isolates that were positive according to enriched BGB qPCR (B). The black bars represent the number of stool specimens positive in the PCR-guided culture. The dashed bars represent the additional qPCR-positive stool specimens.
The additional virulence genes escV, aggR and/or aat, and bfpA were detected in 49% (n = 36), 6% (n = 4), and 6% (n = 4) of qPCR-positive samples, respectively. The O145, O26, O157, O104, O121, and O111 serogroups were detected in 11.0% (n = 8), 8% (n = 6), 4% (n = 3), 3% (n = 2), 3% (n = 2), and 1% (n = 1) of qPCR-positive samples, respectively (Table 2).
TABLE 2.
Overall results of risk categorization of STEC-positive stool samples
| No. of samples by PT group | qPCR |
stx subtyping | Serotyping of cultured isolatesa | Total no. of virulence factors by DNA array | |
|---|---|---|---|---|---|
| Serogenotype | Additional virulence factor(s) | ||||
| I (n = 11) | |||||
| 1 | O157 | escV | stx1a + stx2c | O157:H7 | 32 |
| 1 | O157 | escV, aggR and/or aat | stx2c | Not cultured | |
| 1 | O26 | escV | stx1a + stx2b + stx2c | O26:H11 | 35 |
| 1 | O26 | escV | stx2a | O26:H11 | 31 |
| 2 | O26 | escV (n = 2) | stx1a | O26:H11 | 29/29 |
| 1 | O26/O121/O145 | escV | stx2b + stx2c | O26 (streak)b | |
| 1 | O145 | escV | stx1a + stx2a | O145:NM | 28 |
| 1 | O145 | escV | stx2 not typeable | O145:NMc | 34 |
| 1 | O157/O26/O145 | escV | stx2a + stx2d + stx2e | Not cultured | |
| 1 | O121/O111/O145 | escV | stx1a | Not cultured | |
| II (n = 19) | |||||
| 2 | escV (n = 2) | stx1a + stx2a | O165:NM | 33 | |
| O182:H25 | 25 | ||||
| 4 | escV (n = 4), aggR and/or aat (n = 1) | stx1a | Not cultured | ||
| 1 | escV | stx2a | O182:H25 | 27 | |
| 1 | escV | stx2a + stx2c | Not typedd | 33 | |
| 1 | escV | stx2c | Not cultured | ||
| 1 | escV | stx2f | O63:H6 (n = 1) | 18 | |
| 1 | escV | stx2f | O125:H6 (n = 1) | 16 | |
| 1 | escV, bfpA | stx2f | O88:H25 (EPEC)e | 15 | |
| 6 | escV (n = 6) | stx2f | Not cultured | ||
| 1 | escV | stx1c | Not cultured | ||
| III (n = 35) | |||||
| 3 | escV (n = 1), bfpA (n = 1) | stx1a | O91:NM (n = 3)f | 9/9/10 | |
| 2 | stx1a | O91:H14 (n = 2) | 9/11 | ||
| 1 | stx1a | ONT:NM | 8 | ||
| 1 | escV | stx1a | O117:H7f | 4 | |
| 1 | stx1a | Culture positive (streak) | |||
| 11 | O145 (n = 1) | stx1a | Not cultured | ||
| 1 | escV, bfpA | stx1c + stx2b | O128:H2f | 17 | |
| 1 | stx1c + stx2b | O76:H19 | 20 | ||
| 1 | aggR and/or aat | stx1c + stx2b | O146:H21f | 19 | |
| 1 | stx1c + stx2b | Not cultured | |||
| 1 | stx2b | O7:H6 | 2 | ||
| 1 | stx2b | ONT:H31 (aEPEC)e | 12 | ||
| 1 | stx2b | Not cultured | |||
| 2 | stx2c | ONT:H28 (n = 1) | 11 | ||
| 1 | stx2d | Not cultured | |||
| 1 | stx2e | Not cultured | |||
| 1 | stx1c | Not cultured | |||
| 1 | stx1 not typeable | O16:H5 | 13 | ||
| 3 | O145 (n = 1) | escV, bfpA (n = 1) | Not typeable | Not cultured | |
| IV (n = 8) | |||||
| 1 | O104 | escV, aggR and/or aat | Not typed | O104:H4 (EAEC)e | 10 |
| 7 | O145/O104 (n = 1) | Not typed | Not cultured | ||
NM, nonmotile; ONT, O-serogroup O1 to O187 negative.
PCR-positive culture by screening DNA isolated from a loopful of bacterial growth of the first streaking area of culture plates.
Coinfection with an ONT:H45 (stx2f positive) isolate. This isolate was not genetically characterized.
The isolate could not be recultured after transportation to the RIVM for genotyping/serotyping.
Isolates did not contain stx genes and were designated EPEC (O88:H25; escV, bfpA), atypical EPEC (aEPEC [ONT:H31; escV]), and EAEC (O104:H4; aggR and/or aat).
The isolates did not contain the additional virulence factors escV or aggR and/or aat.
PCR-guided culture.
The PCR-guided culture yielded a positive result in 42.5% (31/73) of direct qPCR-positive samples. A STEC isolate was obtained in 35.6% (26/73) of the samples; from one sample, two STEC isolates were obtained. Two additional samples (3%) were streak-positive for stx genes on PCR. The serotypes that were identified are listed in Table 2. Using the seropathotype concept of Karmali et al., one STEC isolate (O157:H7) could be assigned to SPT group A; six STEC isolates (O26:H11, n = 4; O145:NM, n = 2), to SPT group B; and two STEC isolates (O117:H7 and O146:H21), to SPT group D. The other 17 STEC isolates (65.4%) could not be assigned to a SPT group. One isolate and one streak-positive PCR sample could not be serotyped.
From the remaining three culture positive samples, an enteropathogenic E. coli (EPEC) (n = 2; O88:H25 and ONT:H31) or an EAEC (n = 1; O104:H4) was isolated. The isolation yield of the SMAC medium was higher (21/73) than that of the CHROMagar STEC medium (15/73), although five isolates and one streak-positive PCR sample were only identified with the CHROMagar STEC. Furthermore, the growth of suspicious colonies on the CHROMagar STEC was highly associated with the presence of the escV gene (LEE-positive) detected by the enriched BGB PCR (19/23 versus 17/50) (Fisher exact test, P < 0.0001).
The CT values (stx1 and/or stx2) of samples in which the PCR-guided culture remained negative were significantly higher than in samples with positive guided-culture results (Wilcoxon rank sum, P < 0.0003). This difference in the CT value between the PCR-guided culture negative and positive groups remained significant when comparing the CT values of the enriched BGB PCR (Wilcoxon rank sum, P < 0.001). The distribution of the CT values of the enriched BGB PCR on which the guided culture was performed is shown in Fig. 2B.
stx subtyping of clinical samples.
Subtyping of stx genes was performed on DNA isolates of enriched BGB broths that were PCR positive for stx genes. Of these 65 positive samples, 30 (46%) were stx1 positive, 26 (40%) were stx2 positive, and 9 (14%) were stx1 and stx2 positive. Two stx1 subtypes (stx1a and stx1c) and six stx2 subtypes (stx2a, stx2b, stx2c, stx2d, stx2e, and stx2f) were detected, with a total of 15 different stx1 and stx2 subtype combinations. The most frequently detected subtype variants were stx1a (40%), stx2f (14%), stx1c + stx2b (6%), stx2c (6%), stx2b (5%), and stx1a + stx2a (5%), accounting for 49 samples (75%). For three samples, subtyping results remained negative, although the DNA load seemed to be sufficient. For two of these samples, subtyping remained negative after DNA isolation from the obtained STEC isolate. For an additional three samples, no stx subtype could be obtained due to a low DNA load (all CT values were ≥32 in an enriched BGB PCR).
Risk categorization of STEC and distribution of virulence factors between PT groups.
Samples were presumptively categorized in four pathotype (PT) groups based on the enriched BGB PCR results. A total of 11 samples (15%), 24 samples (33%), 30 samples (41%), and 8 samples (11%) were categorized in PT groups I, II, III, and IV, respectively. However, based on the presence of the additional virulence factor bfpA and on screening of VF in the cultured isolates, a total of 5 samples (7%) were recategorized from PT group II to PT group III; 4 STEC isolates (O91:NM, O117:H7, O128:H2, and ONT:H31) did not contain the escV gene, and there was no correlation in the CT value of one sample for stx (39) and the other VF escV (19) and bfpA (18). The final risk categorizations were 11 samples (15%), 19 samples (26%), 35 samples (48%), and 8 samples (11%) for PT groups I, II, III, and IV, respectively (Table 2). The presence of stx genes for samples categorized in PT group IV could not be confirmed after enrichment, thereby excluding them from further analysis.
The studied virulence factors (VF) differed with respect to their distribution among the different pathotype groups (Table 3). Compared to the PT group that is associated with a moderate risk for diarrhea and low risk for severe disease (III), PT groups that are associated with a high risk for diarrhea and higher risk for severe disease (I + II, combined) exhibited a significantly higher prevalence of various analyzed VF (specifically, stx2, stx2a, stx2f, toxB, eae, efa1, cif, espA, tccP, espP, nleA and/or nleB, and the tir cluster). Although not significant, stx2c was more prevalent in PT groups I + II (odds ratio [OR], 4.1; 95% confidence interval [CI] = 0.7 to 32.7). Inversely, stx1, the mch cluster, ireA, and eaaA were significantly more prevalent in PT group III (Table 3). Interestingly, the adhesion-encoding gene iha was present in all PT group I isolates and in almost all isolates in PT group III (92%); however, there was no significant association between the presence of iha and the PT groups. Furthermore, all LEE-positive STEC isolates contained the EHEC-hlyA gene, with the exception of the two stx2f STEC isolates. Of note, certain VF were also highly associated with PT group I (specifically, stx2c, toxB, eae, efa1, cif, tccP, nleA and/or nleB, katP, and the tir cluster).
TABLE 3.
Pathotype distribution of virulence factors and stx subtypes
| Virulence genotype | Total no. (%) | No. (%)a of isolates or enriched BGB broths for PT: |
Statistical comparison of PT I + PT II vs PT IIIb |
|||
|---|---|---|---|---|---|---|
| I | II | III | Pc | OR (95% CI) | ||
| Enriched BGB broths (n = 65) | ||||||
| stx1 (all) | 39 (60) | 6 (55) | 8 (42) | 25 (71) | 0.021 | 0.3 (0.08–0.9) |
| stx1a | 31 (48) | 6 (55) | 7 (37) | 18 (51) | ||
| stx1c | 6 (9) | 0 | 1 (5) | 5 (14) | ||
| stx2 (all) | 35 (54) | 8 (73) | 14 (74) | 13 (37) | 0.006 | 4.7 (1.4–15.6) |
| stx2a | 7 (11) | 3 (27) | 4 (21) | 0 | 0.003 | ∞ (1.7–∞) |
| stx2b | 9 (14) | 2 (18) | 0 | 7 (20) | ||
| stx2c | 8 (12) | 4 (36) | 2 (11) | 2 (6) | 4.1 (0.7–32.7) | |
| stx2d | 2 (3) | 1 (9) | 0 | 1 (3) | ||
| stx2e | 2 (3) | 1 (9) | 0 | 1 (3) | ||
| stx2f | 9 (14) | 0 | 9 (47) | 0 | <0.0001 | ∞ (2.5–∞) |
| stx2a or stx2c | 14 (22) | 7 (64) | 5 (26) | 2 (6) | 0.002 | 11.0 (2.0–80.5) |
| Isolates (n = 26) | ||||||
| astA | 7 (27) | 3 (43) | 3 (50) | 1 (8) | ||
| EHEC-hlyA | 20 (77) | 7 (100) | 4 (67) | 9 (69) | ||
| toxB | 7 (27) | 6 (86) | 1 (17) | 0 | 0.005 | ∞ (1.9–∞) |
| mch cluster | 9 (35) | 0 | 0 | 9 (69) | <0.0001 | <0.0001 (0–0.3) |
| ireA | 8 (31) | 0 | 0 | 8 (62) | 0.002 | <0.0001 (0–0.4) |
| eae | 13 (50) | 7 (100) | 6 (100) | 0 | <0.0001 | ∞ (18.4–∞) |
| efa | 7 (27) | 6 (86) | 1 (17) | 0 | 0.005 | ∞ (1.9–∞) |
| iha | 20 (77) | 7 (100) | 1 (17) | 12 (92) | ||
| lpfA | 16 (62) | 3 (43) | 3 (50) | 8 (62) | ||
| iss | 15 (58) | 5 (71) | 0 | 10 (77) | ||
| cif | 9 (35) | 6 (86) | 3 (50) | 0 | <0.0001 | ∞ (3.5–∞) |
| espA | 17 (65) | 6 (86) | 6 (100) | 5 (38) | 0.011 | 19.2 (1.5–537.4) |
| tccP | 12 (46) | 7 (100) | 5 (83) | 0 | <0.0001 | ∞ (10.4–∞) |
| eaaA | 10 (39) | 0 | 0 | 10 (77) | <0.0001 | <0.0001 (0–0.2) |
| espP | 11 (42) | 5 (71) | 4 (67) | 2 (15) | 0.015 | 12.4 (1.4–142.1) |
| nleA and/or nleB | 12 (46) | 7 (100) | 5 (83) | 0 | <0.0001 | ∞ (10.4–∞) |
| etpD | 4 (15) | 2 (29) | 2 (33) | 0 | ||
| katP | 7 (27) | 5 (71) | 1 (17) | 1 (8) | ||
| tir cluster | 10 (39) | 6 (86) | 4 (67) | 0 | <0.0001 | ∞ (4.8–∞) |
Total no. of enriched BGB broths: PT I, 11; PT II, 19; PT III, 35. Total no. of isolates: PT I, 7; PT II, 6; PT III, 13.
PT I and II combined (associated with high risk for diarrhea and high/moderate risk for severe disease) are compared to PT III (lower risk for diarrhea and severe disease). ∞, infinite.
P values (from the Fisher exact test) are shown only if the P value was <0.05.
The total number of VF present in the STEC isolates also showed a significant, nonrandom distribution between PT groups (Table 2); the number of VF differed significantly between PT group I (VFmean = 31; 95% CI = 27 to 35), PT group II (VFmean = 25; 95% CI = 21 to 30), and PT group III (VFmean = 11; 95% CI = 8 to 13) (ANOVA, P < 0.0001, F = 38.5). Interestingly, the total number of VF present in the two cultured stx2f STEC isolates (O63:H6 and O125:H6) that were categorized in PT group II were considerably lower than the other 4 STEC isolates categorized in this PT group (Table 2). By cluster analysis of potential VF, escV-positive and escV-negative isolates were separated into two main clusters (Fig. 3). The escV-negative cluster included all PT group III isolates and a stx-negative isolate, EAEC O104:H4 (PT group IV). The escV-positive cluster included all PT groups I + II isolates and two stx-negative EPEC isolates (O88:H25 and ONT:H31) that clustered in a distinct branch with the two stx2f-positive STEC isolates.
FIG 3.
Cluster analysis of potential virulence genes in cultured strains. The escV-negative cluster included all PT group III isolates and an stx-negative isolate, EAEC O104:H4 (PT group IV). The escV-positive cluster included all PT groups I + II isolates and two stx-negative EPEC isolates (O88:H25 and ONT:H31) that clustered in a distinct branch with the two stx2f-positive STEC isolates. Spots depicted in gray represent ambiguous results that were regarded as negative.
Clinical symptoms of patients.
Diarrhea was reported by 80%, 44%, 57%, and 75% of patients in PT groups I, II, III, and IV, respectively. Bloody diarrhea was reported by 20%, 6%, 3%, and 0% of patients in PT groups I, II, III, and IV, respectively. Patients in PT group I presented significantly more often with (bloody) diarrhea compared to PT groups II + III (Fisher exact test, P = 0.006). One patient categorized in PT group I developed HC (serotype O26:H11), and family members of another patient categorized in PT group I (serotype O26:H11) also had gastrointestinal complaints. Interestingly, symptoms reported by patients that are not associated with acute disease, such as persistent diarrhea and/or abdominal complaints without loose stools, were absent in patients categorized in PT group I (0%) but present in patients in PT groups II, III, and IV (33%, 29%, and 25%, respectively). The age distribution of patients did not differ between PT groups (PT group I: mean age = 27 years; 95% CI = 10 to 44 years; PT group II: mean age = 36 years; 95% CI = 23 to 48 years; PT group III: mean age = 41 years; 95% CI = 32 to 50 years) (ANOVA, P = 0.33; F = 1.1), although the median age of patients in PT group I was considerably lower (15 years) compared to PT group II (33 years) and PT group III (39 years).
DISCUSSION
We here present the first prospective study that uses a diagnostic algorithm directly applied on stool samples of patients presenting with gastrointestinal complaints to asses the public health risk of STEC. Although the disease severity and incidence of STEC are not based solely on the pathogenic potential of the organism but also on host-associated and environmental factors, enough information has accumulated that the presence of virulence factors (VF) carried in addition to the stx genes varies considerably between STEC strains and, therefore, could be used to categorize the potential risk of STEC (5, 17, 33–36).
The detection frequency of the stx genes observed in this study (1.8%) was comparable with previous studies performed in The Netherlands (30, 37). The diagnostic algorithm enabled the categorization of STEC infections into 4 pathotype (PT) groups. The majority of the initial stx-positive PCR samples (48%) were categorized in PT group III, while 15% and 26% of stx-positive PCR samples were categorized in PT group I and PT group II, respectively; both PT groups I and II had high risks for diarrhea and moderate to high risks for severe disease. The presence of stx genes could not be confirmed after enrichment in 11% of samples, and these were categorized in PT group IV. The stx subtyping and genetic characterization were performed in order to confirm the validity of the proposed categorization of STEC infections.
Previous studies have indicated that the subtype of Shiga toxin produced may influence the clinical outcome of STEC infections (23, 24). STEC harboring stx2a or stx2c are associated with HUS and bloody diarrhea, while strains carrying stx1c or stx2b have often been isolated from patients with milder infections (38). Although STEC strains carrying stx2d usually predict a milder disease, strains that produce elastase-activatable stx2d may predict a severe clinical outcome of the infection (39). Other variants, such as stx2e and stx2f, have been associated with animals and are rarely isolated from humans (24, 40).
In our study, there was a strong association between the presence of stx2, in particular stx2a or stx2c, and samples categorized in PT groups I + II (LEE-positive), while the presence of the stx1 gene was associated with samples categorized in PT group III. Similar to a previous study performed in Belgium, stx1a was the most detected subtype (41). Furthermore, the detection frequency of the stx2f gene in our study (12.3%) was comparable with previous studies (41, 42). stx2f was, together with stx2b, the first-most detected stx2 subtype among samples that were serogroup O157 negative on PCR in this study. Similar to previous studies, all stx2f-positive PCR samples also contained the escV gene (LEE-positive) (41–43). With the exception of stx2b, the detection frequencies of subtypes stx1c (9%) and stx2e (3%) that are associated with milder disease or asymptomatic carriage were similar to the incidence detected in Belgium (41).
Furthermore, cluster analysis of VF clearly showed a separation into an escV-negative cluster (PT group III) and escV-positive cluster (PT groups I + II), with a significant difference in the number of accessory virulence factors (VF) present between these PT groups. Furthermore, VF that play an important role in toxin production and attachment to host cells were highly associated with PT groups I + II or PT group I alone, while other VF were associated with PT group III. Previous studies also reported that the number of VF present in STEC isolates increases the pathogenic potential of STEC and the strong association of certain accessory VF with severe illness and outbreaks (12, 17, 22, 27, 33, 34, 36, 44). Interestingly, the accessory virulence gene content of the stx2f-positive STEC isolates that clustered in a distinct branch with two stx-negative EPEC isolates was lower than the other STEC isolates categorized in PT group II. Others have also reported that stx2f STEC isolates form a distinct group within STEC with regard to virulence genes and their association with a relatively mild disease (41, 42).
Our findings with respect to the main clinical features of STEC infection are consistent with those of others (9, 40). Patients with STEC infections categorized in PT group I presented significantly more often with (bloody) diarrhea, suggesting that the pathogenic potential of STEC in this group is higher than that of STEC categorized in PT groups II + III, as was confirmed by stx subtyping and genetic characterization. Although there was no clear association between patient age and PT groups in our study, the age distribution of patients in PT group I was considerably lower than that of patients categorized in PT group II and PT group III. Others also reported a close relation between a patient's young age and infection with more virulent (LEE-positive) STEC strains (40). Furthermore, this study revealed a high number (45%) of other enteric pathogens detected in individual stx-positive PCR samples (data not shown). However, the clinical relevance of these mixed infections was beyond the scope of this study.
Although stx subtyping and genetic characterization confirmed the validity of the PT classification, categorization with this molecular-based PT approach should be regarded as presumptive. Additional subtyping of stx genes, genetic characterization, and O:H serotyping of STEC isolates will provide a clearer assessment of the potential public health risk. Hence, a high culture yield remains important for facilitating these laboratory procedures.
An important step in the diagnostic algorithm is the use of an enrichment step, which was performed on the initial stx-positive PCR stool samples. Performing this step has several advantages. First, confirmation by performing PCR on the enrichment broth increases the positive predictive value for the detection of STEC; 89% of the initial stx-positive PCR results could be confirmed. In the majority of the samples (85%), the stx CT values were lower after enrichment, which suggests the presence of viable STEC. In a part of the samples (11%) no stx genes could be detected after enrichment (PT group IV). The stx CT values for direct qPCR were relatively high in all of these samples, which was suggestive for the presence of low loads of nonviable STEC or free stx DNA. Another possibility would be the detection of free stx phages in the stool of these patients, which has been described previously in stool of healthy individuals (45).
Second, stx subtyping was performed directly from the DNA isolate obtained from the enriched broth. To our knowledge, subtyping of stx genes is only being performed on STEC isolates, which will take additional time for obtaining final subtyping results. Third, although not statistically proven, culture yield will improve using enrichment; in the majority of the initial stx-positive PCR samples, the stx gene load increased after enrichment, which is suggestive for the presence of viable STEC. Larger amounts of STEC bacteria in the background of intestinal flora will increase the odds of isolation by culture. The culture yield in our study (38%) was lower than other studies (24, 41). However, the amount of colonies screened with PCR (maximum of 10) in our study was considerably lower than those studies. Hence, increasing the total amount of colonies screened, and routinely screening DNA isolated from a loopful of bacteria growth from the first streaking area of culture plates (as was performed for two samples in this study), would increase the probability of obtaining an isolate or at least would confirm the growth of STEC.
Furthermore, due to easier identification of suspicious colonies, the CHROMagar STEC medium proved to be an effective supplemental medium for isolation, especially of more virulent (LEE-positive) STEC serotypes, as described previously by others (46–48). The medium also supported the growth of EAEC (O104:H4) and EPEC (O88:H25 and ONT:H31), suggesting that it could also be a useful tool for the support of EAEC and EPEC isolation, as described previously (49).
Unfortunately, our diagnostic algorithm only includes direct molecular screening for the stx genes, rendering the detection of Shiga-toxin-lost (STL) EHEC impossible (25, 26, 50). Another limitation of this study was the concise amount of clinical information that was available on request forms that may have influenced clinical associations. Furthermore, the number of STEC isolates that were characterized was limited.
In conclusion, the proposed diagnostic algorithm for risk categorization of STEC infection offers a rapid testing format that could be easily implemented in laboratories that already perform qPCR-based detection of STEC. It enables stx-positive PCR stool samples to be categorized for the potential risk to public health. This risk assessment may provide valuable information to aid community health services in estimating the level of action required (with regard to source/contact tracing and intervention measures to minimize secondary transmission) to address the potential threat and may be a useful tool for public health surveillance. However, the proposed risk categorization should be regarded as presumptive and interpreted with care, as infections with STEC serotypes categorized in PT group III, such as O117:H7, can still pose a public health concern, as has been shown recently (51). Currently, a multicenter, prospective, cohort study is being conducted that will verify the performance of the proposed molecular-based pathotyping approach on a larger scale in order to justify its application in the case of STEC infections for determining if swift action by community health services is warranted or not.
Supplementary Material
ACKNOWLEDGMENTS
We thank Alex W. Friedrich (Department of Medical Microbiology, University of Groningen, University Medical Center Groningen, Groningen, the Netherlands) for critically reviewing the manuscript and for his helpful suggestions. We also thank Evert van Zanten (Certe Laboratory for Infectious Disease, Department of Research and Development, Groningen, the Netherlands) for help with the construction of dendrograms for the cluster analysis of VF.
This study formed part of a larger multidisciplinary research initiative called STEC-ID-net.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JCM.03590-14.
REFERENCES
- 1.Paton JC, Paton AW. 1998. Pathogenesis and diagnosis of Shiga toxin-producing Escherichia coli infections. Clin Microbiol Rev 11:450–479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Griffin PM, Tauxe RV. 1991. The epidemiology of infections caused by Escherichia coli O157:H7, other enterohemorrhagic E. coli, and the associated hemolytic uremic syndrome. Epidemiol Rev 13:60–98. [DOI] [PubMed] [Google Scholar]
- 3.Gyles CL. 2007. Shiga toxin-producing Escherichia coli: an overview. J Anim Sci 85:E45-62. doi: 10.2527/jas.2006-508. [DOI] [PubMed] [Google Scholar]
- 4.Kaper JB, Nataro JP, Mobley HL. 2004. Pathogenic Escherichia coli. Nat Rev Microbiol 2:123–140. doi: 10.1038/nrmicro818. [DOI] [PubMed] [Google Scholar]
- 5.Karmali MA, Mascarenhas M, Shen S, Ziebell K, Johnson S, Reid-Smith R, Isaac-Renton J, Clark C, Rahn K, Kaper JB. 2003. Association of genomic O island 122 of Escherichia coli EDL 933 with verocytotoxin-producing Escherichia coli seropathotypes that are linked to epidemic and/or serious disease. J Clin Microbiol 41:4930–4940. doi: 10.1128/JCM.41.11.4930-4940.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Karmali MA. 1989. Infection by verocytotoxin-producing Escherichia coli. Clin Microbiol Rev 2:15–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rangel JM, Sparling PH, Crowe C, Griffin PM, Swerdlow DL. 2005. Epidemiology of Escherichia coli O157:H7 outbreaks, United States, 1982 to 2002. Emerg Infect Dis 11:603–609. doi: 10.3201/eid1104.040739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Scientific Opinion of the Panel on Biological Hazards. 2007. Monitoring of verotoxigenic Escherichia coli (VTEC) and identification of human pathogenic VTEC types. EFSA J 579:1–61. [Google Scholar]
- 9.EFSA Panel on Biological Hazards. 2013. Scientific opinion on VTEC-seropathotype and scientific criteria regarding pathogenicity assessment. EFSA J 11:3138. [Google Scholar]
- 10.Bielaszewska M, Mellmann A, Zhang W, Kock R, Fruth A, Bauwens A, Peters G, Karch H. 2011. Characterisation of the Escherichia coli strain associated with an outbreak of haemolytic uraemic syndrome in Germany, 2011: a microbiological study. Lancet Infect Dis 11:671–676. doi: 10.1016/S1473-3099(11)70165-7. [DOI] [PubMed] [Google Scholar]
- 11.Piérard D, De Greve H, Haesebrouck F, Mainil J. 2012. O157:H7 and O104:H4 Vero/Shiga toxin-producing Escherichia coli outbreaks: respective role of cattle and humans. Vet Res 43:13. doi: 10.1186/1297-9716-43-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Prager R, Annemuller S, Tschape H. 2005. Diversity of virulence patterns among Shiga toxin-producing Escherichia coli from human clinical cases-need for more detailed diagnostics. Int J Med Microbiol 295:29–38. doi: 10.1016/j.ijmm.2004.12.009. [DOI] [PubMed] [Google Scholar]
- 13.Schmidt MA. 2010. LEEways: tales of EPEC, ATEC and EHEC. Cell Microbiol 12:1544–1552. doi: 10.1111/j.1462-5822.2010.01518.x. [DOI] [PubMed] [Google Scholar]
- 14.Karch H, Schubert S, Zhang D, Zhang W, Schmidt H, Olschlager T, Hacker J. 1999. A genomic island, termed high-pathogenicity island, is present in certain non-O157 Shiga toxin-producing Escherichia coli clonal lineages. Infect Immun 67:5994–6001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Makino S, Tobe T, Asakura H, Watarai M, Ikeda T, Takeshi K, Sasakawa C. 2003. Distribution of the secondary type III secretion system locus found in enterohemorrhagic Escherichia coli O157:H7 isolates among Shiga toxin-producing E. coli strains. J Clin Microbiol 41:2341–2347. doi: 10.1128/JCM.41.6.2341-2347.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Brunder W, Schmidt H, Frosch M, Karch H. 1999. The large plasmids of Shiga-toxin-producing Escherichia coli (STEC) are highly variable genetic elements. Microbiology 145 (Part 5):1005–1014. [DOI] [PubMed] [Google Scholar]
- 17.Bugarel M, Martin A, Fach P, Beutin L. 2011. Virulence gene profiling of enterohemorrhagic (EHEC) and enteropathogenic (EPEC) Escherichia coli strains: a basis for molecular risk assessment of typical and atypical EPEC strains. BMC Microbiol 11:142. doi: 10.1186/1471-2180-11-142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Newton HJ, Sloan J, Bulach DM, Seemann T, Allison CC, Tauschek M, Robins-Browne RM, Paton JC, Whittam TS, Paton AW, Hartland EL. 2009. Shiga toxin-producing Escherichia coli strains negative for locus of enterocyte effacement. Emerg Infect Dis 15:372–380. doi: 10.3201/eid1502.080631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Iyoda S, Tamura K, Itoh K, Izumiya H, Ueno N, Nagata K, Togo M, Terajima J, Watanabe H. 2000. Inducible stx2 phages are lysogenized in the enteroaggregative and other phenotypic Escherichia coli O86:HNM isolated from patients. FEMS Microbiol Lett 191:7–10. doi: 10.1111/j.1574-6968.2000.tb09311.x. [DOI] [PubMed] [Google Scholar]
- 20.Morabito S, Karch H, Mariani-Kurkdjian P, Schmidt H, Minelli F, Bingen E, Caprioli A. 1998. Enteroaggregative, Shiga toxin-producing Escherichia coli O111:H2 associated with an outbreak of hemolytic-uremic syndrome. J Clin Microbiol 36:840–842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Prager R, Lang C, Aurass P, Fruth A, Tietze E, Flieger A. 2014. Two Novel EHEC/EAEC hybrid strains isolated from human infections. PLoS One 9:e95379. doi: 10.1371/journal.pone.0095379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Boerlin P, McEwen SA, Boerlin-Petzold F, Wilson JB, Johnson RP, Gyles CL. 1999. Associations between virulence factors of Shiga toxin-producing Escherichia coli and disease in humans. J Clin Microbiol 37:497–503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Persson S, Olsen KE, Ethelberg S, Scheutz F. 2007. Subtyping method for Escherichia coli Shiga toxin (verocytotoxin) 2 variants and correlations to clinical manifestations. J Clin Microbiol 45:2020–2024. doi: 10.1128/JCM.02591-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Friedrich AW, Bielaszewska M, Zhang WL, Pulz M, Kuczius T, Ammon A, Karch H. 2002. Escherichia coli harboring Shiga toxin 2 gene variants: frequency and association with clinical symptoms. J Infect Dis 185:74–84. doi: 10.1086/338115. [DOI] [PubMed] [Google Scholar]
- 25.Bielaszewska M, Kock R, Friedrich AW, von Eiff C, Zimmerhackl LB, Karch H, Mellmann A. 2007. Shiga toxin-mediated hemolytic uremic syndrome: time to change the diagnostic paradigm? PLoS One 2:e1024. doi: 10.1371/journal.pone.0001024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Friedrich AW, Zhang W, Bielaszewska M, Mellmann A, Kock R, Fruth A, Tschape H, Karch H. 2007. Prevalence, virulence profiles, and clinical significance of Shiga toxin-negative variants of enterohemorrhagic Escherichia coli O157 infection in humans. Clin Infect Dis 45:39–45. doi: 10.1086/518573. [DOI] [PubMed] [Google Scholar]
- 27.Haugum K, Brandal LT, Lindstedt BA, Wester AL, Bergh K, Afset JE. 2014. PCR-based detection and molecular characterization of Shiga toxin-producing Escherichia coli strains in a routine microbiology laboratory over 16 years. J Clin Microbiol 52:3156–3163. doi: 10.1128/JCM.00453-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Todd WT, Dundas S. 2001. The management of VTEC O157 infection. Int J Food Microbiol 66:103–110. doi: 10.1016/S0168-1605(00)00527-4. [DOI] [PubMed] [Google Scholar]
- 29.Vallières E, Saint-Jean M, Rallu F. 2013. Comparison of three different methods for detection of Shiga toxin-producing Escherichia coli in a tertiary pediatric care center. J Clin Microbiol 51:481–486. doi: 10.1128/JCM.02219-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.de Boer RF, Ott A, Kesztyus B, Kooistra-Smid AM. 2010. Improved detection of five major gastrointestinal pathogens by use of a molecular screening approach. J Clin Microbiol 48:4140–4146. doi: 10.1128/JCM.01124-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lede IO, Kraaij-Dirkzwager MM, van den Kerkhof JHTC, Notermans DW. 2012. Lack of uniformity with notifications of Shiga-toxin producing Escherichia coli and Shigella towards and by community health services. Infectieziekten Bulletin 23:116–118. (In Dutch.) [Google Scholar]
- 32.Scheutz F, Teel LD, Beutin L, Pierard D, Buvens G, Karch H, Mellmann A, Caprioli A, Tozzoli R, Morabito S, Strockbine NA, Melton-Celsa AR, Sanchez M, Persson S, O'Brien AD. 2012. Multicenter evaluation of a sequence-based protocol for subtyping Shiga toxins and standardizing stx nomenclature. J Clin Microbiol 50:2951–2963. doi: 10.1128/JCM.00860-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Brandt SM, King N, Cornelius AJ, Premaratne A, Besser TE, On SL. 2011. Molecular risk assessment and epidemiological typing of Shiga toxin-producing Escherichia coli by using a novel PCR binary typing system. Appl Environ Microbiol 77:2458–2470. doi: 10.1128/AEM.02322-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Girardeau JP, Dalmasso A, Bertin Y, Ducrot C, Bord S, Livrelli V, Vernozy-Rozand C, Martin C. 2005. Association of virulence genotype with phylogenetic background in comparison to different seropathotypes of Shiga toxin-producing Escherichia coli isolates. J Clin Microbiol 43:6098–6107. doi: 10.1128/JCM.43.12.6098-6107.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Toma C, Martinez Espinosa E, Song T, Miliwebsky E, Chinen I, Iyoda S, Iwanaga M, Rivas M. 2004. Distribution of putative adhesins in different seropathotypes of Shiga toxin-producing Escherichia coli. J Clin Microbiol 42:4937–4946. doi: 10.1128/JCM.42.11.4937-4946.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wickham ME, Lupp C, Mascarenhas M, Vazquez A, Coombes BK, Brown NF, Coburn BA, Deng W, Puente JL, Karmali MA, Finlay BB. 2006. Bacterial genetic determinants of non-O157 STEC outbreaks and hemolytic-uremic syndrome after infection. J Infect Dis 194:819–827. doi: 10.1086/506620. [DOI] [PubMed] [Google Scholar]
- 37.van Duynhoven YT, Friesema IH, Schuurman T, Roovers A, van Zwet AA, Sabbe LJ, van der Zwaluw WK, Notermans DW, Mulder B, van Hannen EJ, Heilmann FG, Buiting A, Jansen R, Kooistra-Smid AM. 2008. Prevalence, characterisation and clinical profiles of Shiga toxin-producing Escherichia coli in The Netherlands. Clin Microbiol Infect 14:437–445. doi: 10.1111/j.1469-0691.2008.01963.x. [DOI] [PubMed] [Google Scholar]
- 38.Friedrich AW, Borell J, Bielaszewska M, Fruth A, Tschape H, Karch H. 2003. Shiga toxin 1c-producing Escherichia coli strains: phenotypic and genetic characterization and association with human disease. J Clin Microbiol 41:2448–2453. doi: 10.1128/JCM.41.6.2448-2453.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bielaszewska M, Friedrich AW, Aldick T, Schurk-Bulgrin R, Karch H. 2006. Shiga toxin activatable by intestinal mucus in Escherichia coli isolated from humans: predictor for a severe clinical outcome. Clin Infect Dis 43:1160–1167. doi: 10.1086/508195. [DOI] [PubMed] [Google Scholar]
- 40.Beutin L, Krause G, Zimmermann S, Kaulfuss S, Gleier K. 2004. Characterization of Shiga toxin-producing Escherichia coli strains isolated from human patients in Germany over a 3-year period. J Clin Microbiol 42:1099–1108. doi: 10.1128/JCM.42.3.1099-1108.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Buvens G, De Gheldre Y, Dediste A, de Moreau AI, Mascart G, Simon A, Allemeersch D, Scheutz F, Lauwers S, Pierard D. 2012. Incidence and virulence determinants of verocytotoxin-producing Escherichia coli infections in the Brussels Capital Region, Belgium, in 2008 to 2010. J Clin Microbiol 50:1336–1345. doi: 10.1128/JCM.05317-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Friesema I, van der Zwaluw K, Schuurman T, Kooistra-Smid M, Franz E, van Duynhoven Y, van Pelt W. 2014. Emergence of Escherichia coli encoding Shiga toxin 2f in human Shiga toxin-producing E. coli (STEC) infections in the Netherlands, January 2008 to December 2011. Euro Surveill 19:26–32. http://www.eurosurveillance.org/ViewArticle.aspx?ArticleId=20787. [PubMed] [Google Scholar]
- 43.Prager R, Fruth A, Siewert U, Strutz U, Tschape H. 2009. Escherichia coli encoding Shiga toxin 2f as an emerging human pathogen. Int J Med Microbiol 299:343–353. doi: 10.1016/j.ijmm.2008.10.008. [DOI] [PubMed] [Google Scholar]
- 44.Bosilevac JM, Koohmaraie M. 2011. Prevalence and characterization of non-O157 shiga toxin-producing Escherichia coli isolates from commercial ground beef in the United States. Appl Environ Microbiol 77:2103–2112. doi: 10.1128/AEM.02833-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Martinez-Castillo A, Quiros P, Navarro F, Miro E, Muniesa M. 2013. Shiga toxin 2-encoding bacteriophages in human fecal samples from healthy individuals. Appl Environ Microbiol 79:4862–4868. doi: 10.1128/AEM.01158-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hirvonen JJ, Siitonen A, Kaukoranta SS. 2012. Usability and performance of CHROMagar STEC medium in detection of Shiga toxin-producing Escherichia coli strains. J Clin Microbiol 50:3586–3590. doi: 10.1128/JCM.01754-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wylie JL, Van Caeseele P, Gilmour MW, Sitter D, Guttek C, Giercke S. 2013. Evaluation of a new chromogenic agar medium for detection of Shiga toxin-producing Escherichia coli (STEC) and relative prevalences of O157 and non-O157 STEC in Manitoba, Canada. J Clin Microbiol 51:466–471. doi: 10.1128/JCM.02329-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tzschoppe M, Martin A, Beutin L. 2012. A rapid procedure for the detection and isolation of enterohaemorrhagic Escherichia coli (EHEC) serogroup O26, O103, O111, O118, O121, O145, and O157 strains and the aggregative EHEC O104:H4 strain from ready-to-eat vegetables. Int J Food Microbiol 152:19–30. doi: 10.1016/j.ijfoodmicro.2011.10.009. [DOI] [PubMed] [Google Scholar]
- 49.Hauswaldt SI, Rodloff AC, Solbach W, Knobloch JKM. 2012. Improving diagnostics of diarrheagenic Escherichia coli by use of a new chromogenic medium, poster P1760. 22nd European Congress of Clinical Microbiology and Infectious Diseases, London, United Kingdom: http://www.chromagar.com/fichiers/1336122885ECCMID_Poster_STEC_Hauswaldt.pdf. [Google Scholar]
- 50.Schmidt H, Scheef J, Huppertz HI, Frosch M, Karch H. 1999. Escherichia coli O157:H7 and O157:H(-) strains that do not produce Shiga toxin: phenotypic and genetic characterization of isolates associated with diarrhea and hemolytic-uremic syndrome. J Clin Microbiol 37:3491–3496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Simms I, Gilbart VL, Byrne L, Jenkins C, Adak GK, Hughes G, Crook PD. 2014. Identification of verocytotoxin-producing Escherichia coli O117:H7 in men who have sex with men, England, November 2013 to August 2014. Euro Surveill 19:pii=20946 http://www.eurosurveillance.org/ViewArticle.aspx?ArticleId=20946. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.