Abstract
Objective
To describe clinical and laboratory findings from the 2012 southeastern Minnesota pertussis outbreak.
Patients and Methods
Patients were selected for 2 parts of the study. In the first part, nasopharyngeal swabs from a convenience sample of 265 unique patients were used for both the clinician-requested polymerase chain reaction (PCR) test and culture. B pertussis isolates were tested for macrolide susceptibility and typed using whole genome sequencing and pulsed-field gel electrophoresis. Pertactin gene sequences were analyzed to identify pertactin-deficient B pertussis. In the second part, all patients seen at Mayo Clinic in Rochester, Minnesota, who had PCR results positive for Bordetella pertussis or Bordetella parapertussis between January 1, 2012, and December 31, 2012, were analyzed for patient demographic features and vaccination records.
Results
One hundred sixty patients had results positive for B pertussis, and 21 patients had results positive for B parapertussis. Among the 265 swabs cultured, B pertussis was detected by both culture and PCR in 11. One swab was positive for B pertussis by culture alone, and 13 were positive by PCR alone. Polymerase chain reaction detected B pertussis more frequently than did culture (P=.001). No macrolide resistance was detected. All 12 isolates tested had an altered pertactin gene, including 9 with a signal sequence deletion, 2 with insertion sequence disruptions, and 1 with a premature stop codon. Nine and 3 isolates were pertactin types prn1 and prn2, respectively. Whole genome sequencing and pulsed-field gel electrophoresis detected the presence of multiple B pertussis strains. The mean age of patients with pertussis was younger than that of those without pertussis (15.6 and 25.5 years, respectively; P=.002). Compared with those whose test results were negative for B pertussis, fewer patients with positive results had received whole-cell pertussis vaccine (P=.02). In the subgroup who had received acellular vaccine exclusively, the time since the most recent pertussis vaccination in those with results positive for B pertussis was longer than that in those with negative results (1363 vs 1010 days; P=.004).
Conclusion
The 2012 pertussis outbreak in southeastern Minnesota included multiple strains of B pertussis, all putatively lacking pertactin. Our findings may indicate decreased efficacy of (and waning immunity from) acellular vaccines as contributors to the outbreak.
Pertussis is a respiratory illness that begins with upper respiratory tract symptoms, progresses to severe paroxysms of cough, and evolves into a convalescent stage. Although infection can be associated with mild symptoms in adults and older children, neonatal infections may be deadly. The etiologic agent is Bordetella pertussis. Other less common Bordetella species (Bordetella parapertussis, Bordetella bronchiseptica, and Bordetella holmesii) have been implicated in similar illness. B pertussis is found exclusively in humans, with adolescents and adults likely serving as a source of infection of younger children and infants. Since the introduction of vaccines against B pertussis, the incidence of pertussis has declined, reaching a nadir in 1976.1,2 The yearly incidence of pertussis has steadily increased since the 1980s, and communities throughout the United States have experienced a resurgence of pertussis in recent years.2-5
Whole-cell pertussis vaccines (thermally or chemically inactivated B pertussis cells) were introduced in the 1940s and later combined with diphtheria and tetanus toxoids to form the “DTP” (diphtheria and tetanus toxoids and pertussis) vaccine.1 Although the vaccines were efficacious and immunogenic, tolerability was limited by vaccine reactions, including local reactions, fever, and febrile seizures. Acellular vaccines, composed of proteins purified from B pertussis cell lysates, were introduced in the 1990s. Compared with whole-cell vaccines, acellular vaccines have fewer adverse events.1,6 Several acellular pertussis vaccines have been used, all of which have contained pertussis toxin, with or without pertactin, filamentous hemagglutinin, and/or fimbrial proteins.1
Our medical center is a large, tertiary/quaternary referral center in Rochester, Minnesota, where polymerase chain reaction (PCR) has been used to diagnose pertussis since 1995. Rochester is a city of approximately 109,000 residents located in Olmsted County. In 2012, southeastern Minnesota experienced its largest pertussis outbreak in recent history. That year, Olmsted County reported 237 cases of pertussis (compared with 19 and 28 in 2011 and 2010, respectively). The outbreak occurred in a region with a relatively high vaccination rate compared with that reported in other studies7; children in Olmsted County have an 88% rate of acellular pertussis vaccination, higher than the state average of 77%.8 Herein, we report the epidemiology and clinical and microbiological characteristics of the 2012 pertussis outbreak in southeastern Minnesota and examine possible contributing factors.
PATIENTS AND METHODS
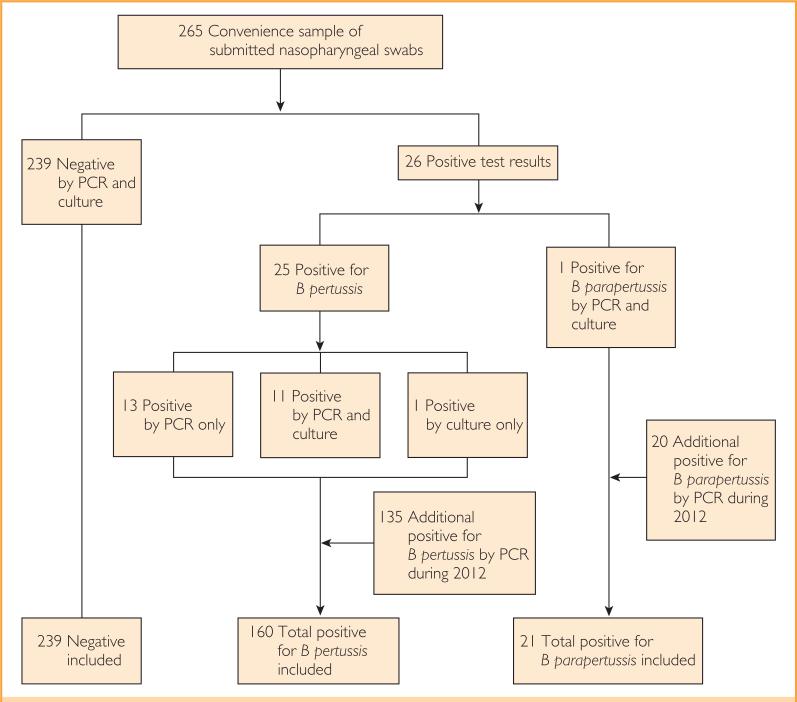
This study was approved by the Mayo Clinic Institutional Review Board. Patients were selected for 2 parts of the study (Figure 1). In the first part, detailed subsequently, a convenience sample of submitted nasopharyngeal swabs was used for both a clinician requested PCR test and an additional culture as part of the study. In the second part, the electronic charts of all patients seen at the Mayo Clinic in Rochester who had PCR results positive for B pertussis or B parapertussis between January 1, 2012, and December 31, 2012, were analyzed for patient demographic features and vaccination records. We defined a pertussis or parapertussis case as any patient with PCR or culture positive for B pertussis or B parapertussis. All patients were cross-referenced with the Minnesota research authorization status database and excluded if records indicated a request to be excluded from research studies.
FIGURE 1.
Study flowchart. B pertussis = Bordetella pertussis; B parapertussis Bordetella parapertussis; PCR = polymerase chain reaction.
Culture and B pertussis Identification
Nasopharyngeal swabs received for Bordetella PCR testing were cultured on Regan-Lowe charcoal media with cephalexin (Hardy Diagnostics). Colonies suspected to represent B pertussis were subjected to matrix-assisted laser desorption ionization time-of-flight mass spectrometry,9 with identification confirmed phenotypically. B pertussis isolates were frozen in Brucella broth on freezer beads (Hardy Diagnostics).
Real-time PCR for B pertussis and B parapertussis
Nasopharyngeal swab samples were placed into LightCycler Advanced lysis tubes (Roche Molecular Diagnostics) and subjected to heat lysis on a Thermomixer R (Eppendorf AG) for 6 minutes at 99°C and 1400 rpm, followed by centrifugation for 20 seconds at 20,800g. Then 5 μL of the supernatant was combined with 15 μL of PCR master mix and tested using a previously described duplex PCR assay targeting IS481 and IS1001 of B pertussis and B parapertussis, respectively.10
Macrolide Resistance Detection in B pertussis
Isolates of B pertussis were tested for phenotypic and genotypic macrolide resistance. 0.5 McFarland suspensions of each isolate were prepared in normal saline. Using the prepared suspensions, 2 Regan-Lowe agar plates without cephalexin (Hardy Diagnostics) were inoculated for a lawn of growth and allowed to acclimate. A 15-μg erythromycin disk (Becton Dickinson and Company) was placed on one plate and an erythromycin Etest strip (bioMérieux, Inc) on the other. Plates were incubated for 5 days at 35°C in room air. Disk inhibition zone diameters were measured with a micrometer, and Etest minimum inhibitory concentration values were determined following the manufacturer's recommendations. Polymerase chain reaction targeted to the 23S ribosomal RNA gene followed by bidirectional sequencing of the ampli-fied product was performed to detect the A-to-G sequence variation at position 2058 (Escherichia coli numbering) associated with macrolide resistance in B pertussis.11 For resistance studies, American Type Culture Collection strains BAA-1335 and 9797 were included as positive and negative controls, respectively.
Pulsed-Field Gel Electrophoresis
Pulsed-field gel electrophoresis (PFGE) of B pertussis isolates, including the control isolate Salmonella Braenderup H9812 (kindly provided by the Minnesota Department of Health), was performed using XbaI restriction enzyme (Roche Applied Science) as previously described.12,13 The gel image was captured on a Gel Doc XR system (Bio-Rad Laboratories, Inc). Analysis of PFGE patterns was performed with GelCompar II software (Applied Maths). Similarity coefficients were calculated using the Dice algorithm.
Whole Genome Sequencing
B pertussis isolates were treated with 5 mg/mL lysozyme and genomic DNA extracted using a Maxwell 16 tissue DNA purification kit (Promega Corporation) and prepared using a genomic DNA Clean & Concentrator-10 kit (Zymo Research Corp). Sequencing of paired-end and Nextera mate-pair libraries was performed using a MiSeq platform (Illumina, Inc) with a 300-cycle kit, resulting in an average coverage of 133X ± 10X genomes per isolate. Reads were processed for adaptor removal and quality filtering using Trimmomatic 0.32 with parameters “ILLUMINACLIP: adapters.fasta LEADING:3 TRAILING:3MAX-INFO:220:0.1 MINLEN:70.”14 Assemblies were generated using Velvet 1.2.10.15 Average nucleotide identity was generated using JSpecies.16 A neighbor-joining tree was calculated and visualized using the APE (Analysis of Phylogenetics and Evolution) package version 3.1-4 in R language.17 Pertactin gene sequences were identi-fied from open reading frames using Prodigal 2.6018 alongside manual curation for insertion sequence elements and premature stops. Alignment was performed using MAFFT.19 Allele typing based on pertactin sequences was done by examination of region 1 repeats.20
Medical Record Review and Analysis
A single reviewer (A.G.T.) surveyed the electronic medical record of included patients and collected the following data: age at the time of nasopharyngeal swab collection, date of birth, sex, duration of symptoms before presentation to a health care professional, reported contacts with pertussis, documented symptoms including cough (and whether the cough was productive), nasal symptoms (rhinorrhea or congestion), sore throat, fever, and posttussive emesis. In addition, history of chronic respiratory conditions (asthma or chronic obstructive pulmonary disease) and exposure to tobacco smoke were recorded. Dates of pertussis vaccine administration, age at the time of vaccination, type of vaccine administered (whole-cell killed vs acellular), and influenza vaccination for the 2011-2012 influenza seasons were also recorded for each patient.
With regard to vaccination status, patients were considered “up-to-date” if they had received the correct number of vaccinations at Centers for Disease Control and Prevention (CDC)–defined intervals by 2 weeks (14 days) before the reported date on which symptoms developed and had not passed the last date suggested for a given vaccine without having received it. For example, 12-year-old children would be counted as up-to-date if they had received the 2-month, 4-month, 6-month, 15- to 18-month, and 4- to 6-year acellular vaccines even if the 11- to 12-year-old vaccine had not been administered (as they are not yet 13 years of age). If complete pertussis vaccination records were unavailable in the electronic medical record, available data were collected from the Minnesota Immunization Information Connection. Vaccines were counted as valid if given at an age different from the recommended schedule as long as the CDC-recommended intervals for “catch up” schedules were followed (eg, 1 month between the first and second doses and 6 months between the fourth and fifth doses of acellular vaccine). Vaccinations documented during an era when they were unavailable were corrected to the vaccine type available at the time documented (eg, a patient documented as having received an acellular vaccine in 1988 was considered to have received a whole-cell vaccine). All patients were included in the final analysis regardless of vaccine record availability.
Statistical Analyses
Descriptive summaries are reported as frequencies and percentages for categorical variables and as mean (SD) and (minimum, maximum) for continuous variables. Comparisons between the groups (eg, results positive for B pertussis vs positive for B parapertussis or positive for B pertussis vs negative for B pertussis or B parapertussis) were performed using the χ2 test, Fisher exact test, or Wilcoxon rank sum test, as appropriate. All tests were 2-sided, and P<.05 was considered statistically significant.
RESULTS
Study Population
In total, 159 patients (24 from the convenience sample) with positive and 239 patients (all from the convenience sample) with negative PCR test results for B pertussis were studied (Table 1). One patient with results positive for B pertussis by culture but negative by PCR was also included, as were 21 (1 from the convenience sample) with positive B parapertussis PCR test results.
TABLE 1.
Baseline Characteristics, Symptoms, and Vaccination Status of Patients Who Underwent Testing for Bordetella pertussis and Bordetella parapertussha,b
| Variable | Negative for B pertussis and B parapertussis (N=239) | B pertussis cases (N=160) | B parapertussis cases (N=21) |
|---|---|---|---|
| Demographic characteristics | |||
| Female | 142 (59.4) | 75 (46.9) | 12 (57.1) |
| Age at presentation (y), mean (SD; range) | 25.5 (23.5; 8 d-90 y) | 15.6 (17.7; 35 d-99 y) | 3.8 (2.1; 6 mo-10y) |
| Time to testing (d), mean (SD; range) | 16.4 (40.6; 1-560) | 13.1 (9.1; 2-56) | 15.4 (14.3; 1-56) |
| Presumed B pertussis or B parapertussis contact recorded | 44 (18.4) | 62 (38.8) | 11 (52.4) |
| Physician-recorded symptoms | |||
| Cough | 232 (97.1) | 159 (99.4) | 21 (100) |
| Cough productive of sputum | 63 (26.4) | 42 (26.2) | 5 (23.8) |
| Nasal symptoms | 118 (49.4) | 57 (35.6) | 11 (52.4) |
| Sore throat | 73 (30.5) | 28 (17.5) | 5 (23.8) |
| Fever | 60 (25.1) | 17 (10.6) | 5 (23.8) |
| Posttussive emesis | 33 (13.8) | 29 (18.1) | 5 (23.8) |
| Documented history of asthma or COPD | 37 (15.5) | 27 (16.9) | 1 (4.8) |
| Tobacco smoke exposure | 31 (13.0) | 13 (8.1) | 1 (4.8) |
| Azithromycin treatment | 88 (36.8) | 158 (98.8) | 19 (90.5) |
| Hospitalization | 11 (4.6) | 6 (3.8) | 0(0) |
| Vaccination history | |||
| Childhood pertussis vaccination records available | 143 (59.8) | 135 (84.4) | 21 (100) |
| Pertussis vaccine up-to-date | 124 (51.9) | 122 (76.2) | 21 (100) |
| Received whole-cell pertussis vaccine | 40 (16.7) | 13 (8.1) | 0 (0) |
| Received 2011-2012 season influenza vaccine | 141 (59.0) | 87 (54.4) | 15 (71.4) |
COPD = chronic obstmctive pulmonary disease.
Data are presented as No. (percentage) unless indicated othewise.
Real-time PCR and Culture
We studied 265 nasopharyngeal swab samples from unique patients in the convenience sample. Culture and PCR detected 11 B pertussis– and 1 B parapertussis–positive swabs. Culture recovered B pertussis in a single sample that was negative by PCR, and PCR detected IS481 in 13 swabs from which B pertussis failed to grow in culture. Polymerase chain reaction detected B pertussis more frequently (N=24) than did culture (N=12) (P=.001). Among the PCR- and culture-positive samples, the average real-time PCR crossing point was 29.03 cycles, whereas among the culture-negative samples, the average crossing point was 34.63 cycles (P=.01), indicating the presence of more B pertussis DNA in the culture-positive than the culture-negative samples.
Macrolide Resistance Detection in B pertussis
Neither phenotypic nor genotypic macrolide resistance was detected in any of the 12 B pertussis isolates tested. Erythromycin disk diameters were 52 to 64 mm, and minimum inhibitory concentrations were 0.016 to 0.064 μg of erythromycin per milliliter. No mutation at base position 2058 (A-to-G) of the 23S ribosomal RNA gene was detected in any isolate. Together, these data indicate the absence of macrolide resistance.
B pertussis Pertactin Gene Sequencing
Nine and 3 isolates were pertactin types prn1 and prn2, respectively. Based on in silico analysis, all 12 isolates tested had an altered or absent pertactin membrane protein (Table 2). A previously described 84–base pair signal sequence deletion (prn1ΔSS) was present in 9 isolates.21 The pertactin gene was disrupted by IS481 at nucleotide 1613 in 2 isolates (prn2::IS481), the sequences of which closely match those reported by Queenan et al.22 Finally, a previously described premature stop codon was present at nucleotide 1273 in one isolate.22,23
TABLE 2.
Genotypic and Phenotypic Antimicrobial Susceptibility Characteristics of Bordetella pertussis Isolatesa
| Isolate | Pertactin allele | Pertactin sequence variation | NCBI GenBank database closest match | 23S sequence variation | Erythromycin disk diffusion diameter (mm) | MIC (Etest) (μg/mL) |
|---|---|---|---|---|---|---|
| BP010 | prn1 | Signal sequence deletion | AB670735.1 | Not present | 57 | 0.016 |
| BP022 | prn1 | Signal sequence deletion | AB670735.1 | Not present | 59 | 0.023 |
| BP034 | prn1 | Signal sequence deletion | AB670735.1 | Not present | 52 | 0.023 |
| BP296 | prn1 | Signal sequence deletion | AB670735.1 | Not present | 57 | 0.064 |
| BP120 | prn1 | Signal sequence deletion | AB670735.1 | Not present | 57 | 0.023 |
| BP172 | prn1 | Signal sequence deletion | AB670735.1 | Not present | 58 | 0.023 |
| BP179 | prn1 | Signal sequence deletion | AB670735.1 | Not present | 56 | 0.016 |
| BP186 | prn1 | Signal sequence deletion | AB670735.1 | Not present | 60 | 0.023 |
| BP207 | prn1 | Signal sequence deletion | AB670735.1 | Not present | 64 | 0.016 |
| BP191 | prn2 | S481 disruption | KC445197.1 | Not present | 60 | 0.016 |
| BP279 | prn2 | S481 disruption plus nonsynonymous sequence variationb | KC445197.1 | Not present | 58 | 0.064 |
| BP101 | prn2 | Stop codon | KC445199.1 | Not present | 59 | 0.016 |
MIC = minimum inhibitory concentration; NCBI = National Center for Biotechnology Information.
BP279 has a nonsynonymous sequence variation (C1577G) compared with KC445197.1.
Typing of B pertussis Isolates
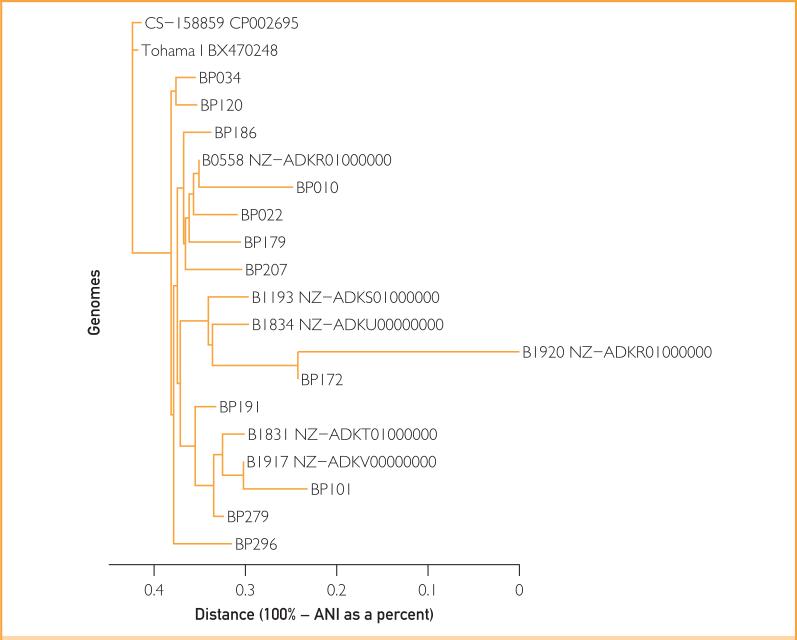
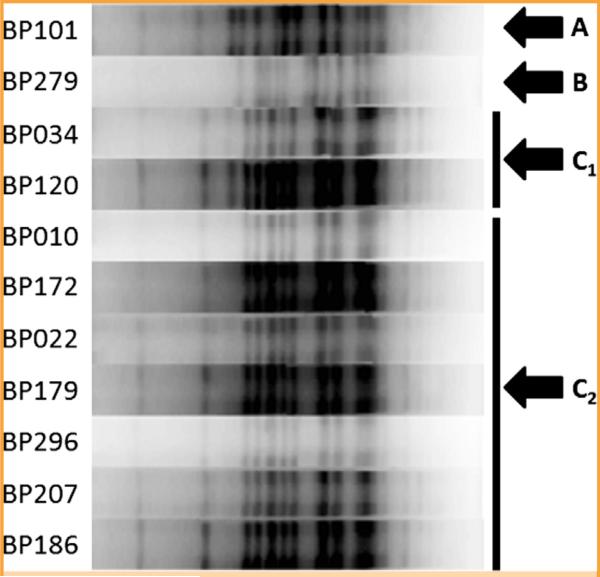
Whole genome sequencing was performed on all 12 isolates (Figure 2). BP279, BP101, and BP191 clustered together and away from the remainder of the isolates, consistent with their pertactin gene sequences. All but BP191 produced readable PFGE profiles (Figure 3). Pulsed-field gel electrophoresis similarity coefficient values ranged from 55% to 100%; using a similarity cutoff of 90%, there were 4 PFGE patterns, with that of BP279 being most different from the rest, consistent with results of whole genome and pertactin sequence analysis. Together, these results indicate the presence of multiple B pertussis strains.
FIGURE 2.
Phylogenetic tree of genomic sequences of the 12 study Bordetella pertussis isolates compared with strains from GenBank (analysis based on average nucleotide identity [ANI]).
FIGURE 3.
Pulsed-field gel electrophoresis of 11 Bordetella pertussis isolates. Four patterns, A, B, C1, and C2, were observed, with B (associated with BP279) being the most different from the rest.
Vaccination Status
As shown in Table 1, 84.5% of patients with and 59.8% of patients without B pertussis had some documentation of childhood pertussis immunization, with 76.2% and 51.9% considered upto-date at presentation. Patients with B pertussis were less likely to have received whole-cell pertussis vaccine than those without B pertussis (8.1% vs 16.7%, respectively; P=.02), despite similar proportions having received the 2011-2012 influenza vaccine (54.4% vs 59.0%, respectively; P=.36).
Demographic Characteristics and Clinical Data
The age distribution of those with pertussis is shown in Supplemental Figure 1 (available online at http://www.mayoclinicproceedings.org). Patients who had pertussis were younger than those without pertussis (15.6 vs 25.5 years; P=.002). The mean time from symptom onset to Bordetella PCR testing was 13.1 and 16.4 days, respectively, in the 2 groups (P=.08). Patients with and without B pertussis had similar rates of hospitalization (3.8% vs 4.6%; P=.68) and previous respiratory illness (asthma or chronic obstructive pulmonary disease; 16.9% vs 15.5%; P=.71). Rates of post-tussive emesis were similar between those with positive and negative PCR findings (18.1% vs 13.8%; P=.24). No deaths were attributable to pertussis.
Time Since Last Vaccination in Patients Who Received Acellular Pertussis Vaccine Only
The time from the date of the most recent acellular pertussis vaccination to the date a patient underwent testing for B pertussis was examined among those who had received 5 or more doses of acellular pertussis vaccine (and no whole-cell vaccine). The mean (SD) time since the most recent pertussis vaccination in those with B pertussis (n=93) was 1363 days (818 days), compared with 1010 days (879 days) among those without B pertussis (n=51; P=.004).
B parapertussis Cases
Twenty-one patients had B parapertussis, all of whom had been age-appropriately vaccinated against B pertussis. Patients with B parapertussis were younger than those with B pertussis (mean ages, 3.8 and 15.6 years, respectively; P<.0001). No hospitalizations or deaths were attributed to B parapertussis.
DISCUSSION
In 2012, southeastern Minnesota experienced its largest recent epidemic of pertussis. Our data support several possible explanations for the resurgence of pertussis our region, including genetic changes in the etiologic agent (B pertussis) associated with vaccine escape, waning of immunity conferred by acellular vaccines, and decreased efficacy of acellular compared with the whole-cell vaccines previously in use. The outbreak, which occurred in a geographically restricted area over a short time span, consisted of multiple circulating strains, consistent with findings in other pertussis outbreaks.24
All B pertussis isolates recovered from the outbreak (which represent a subset of patients studied) putatively lack expression of pertactin, an important component of acellular pertussis vaccines currently in use. Pertactin-negative B pertussis has been increasingly reported,22,25-28 but to our knowledge, this is the first time that only pertactin-negative strains have been recovered during an outbreak. The finding of 9 isolates with the prn1 allele with the 5’ signal sequence deletion is unusual among B pertussis isolates from the United States.23 The recovery of pertactin-negative strains in the setting of widespread vaccine utilization implies selective pressure and microbial adaptation. Because pertactin is a component of all currently used acellular vaccines, the presence of pertactin-negative B pertussis has potential implications for future vaccine design. Interestingly, pertactin-deficient B pertussis can outcompete pertactin-producing B pertussis, suggesting that pertactin deficiency may not negatively affect microbial fitness.21
The immunity conferred by any vaccine, including pertussis vaccine, wanes over time.5,29 We found that among patients who had received at least 5 doses of acellular pertussis vaccine, the time since the last acellular vaccination was more than 350 days longer (on average) in those who had test results positive for B pertussis than in those who had negative results. These data suggest waning immunity from the currently used vaccines as a contributor to the outbreak.
Growing evidence supports the decreased efficacy of acellular compared with whole-cell pertussis vaccines, and there is evidence that receipt of whole-cell pertussis vaccines is associated with a lower risk of pertussis.30,31 This evidence is supported by our finding that proportionally fewer patients with B pertussis had received whole-cell vaccine.
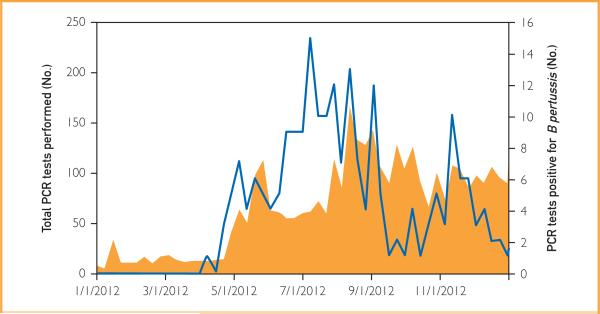
Beyond these explanations, other theories have been offered for the increase in pertussis in recent years, including increased clinical awareness and availability of more sensitive diagnostic tests.5,32 Polymerase chain reaction testing is more rapid and has increased sensitivity compared with culture. These advantages have clinical relevance because, given the clinical course of pertussis, the microbial load may be low by the time a diagnosis of pertussis is entertained.10 The increased sensitivity of PCR over culture is supported by our data. In our convenience sample, PCR detected more cases (N=24) than did culture (N=12) with only one case detected by culture and not PCR. The lower real-time PCR crossing point of the culture-positive vs the culture-negative specimens suggests that the increased detection by PCR vs culture relates to the ability of the former to detect specimens containing low numbers of organisms. We do not believe that the use of this more sensitive test accounted for the outbreak, although it most certainly enabled its recognition. We have been using PCR routinely to diagnose pertussis since 1995. As shown in Figure 4, the increase in the number of PCR test results positive for B pertussis preceded the increase in clinical ordering of PCR testing, suggesting that increased utilization of PCR was not exclusively responsible for the increased number of cases diagnosed.
FIGURE 4.
Total number of polymerase chain reaction (PCR) tests for Bordetella pertussis and Bordetella parapertussis performed at Mayo Clinic in Rochester, Minnesota, in 2012 (orange area, left y-axis) and the number of PCR tests positive for B pertussis by month (blue line, right y-axis).
Interestingly, although there was an overall increase in the number of pertussis cases detected by PCR in 2012, infections in patients less than 1 year old declined while infections diagnosed by PCR increased among those aged 5 to 19 years when compared with national trends in the late 1990s. That there were no cases of pertussis in neonates may be due to recent changes in recommendations for the immunization of pregnant women. In 2011, the Advisory Committee on Immunization Practices of the CDC recommended that pregnant women who were not yet vaccinated with the tetanus toxoid, reduced diphtheria toxoid, and acellular pertussis (Tdap) vaccine receive it in the late part of the second trimester or the third trimester of their pregnancy or in the early postpartum period.33 Since the initial recommendation, repeated Tdap vaccinations with each subsequent pregnancy have been advocated by the Advisory Committee on Immunization Practices.34 The lower number of pertussis cases in infants in our study than reported in previous eras1 (Supplemental Figure 2, available online at http://www.mayoclinicproceedings.org) may be due at least in part to changes in vaccination protocols for pregnant women. This finding may be helpful because there is little available data supporting the efficacy of the new Advisory Committee on Immunization Practices recommendation.35
The PCR assay used in our laboratory detects both B pertussis and B parapertussis. Interestingly, 21 cases of B parapertussis infection were detected by PCR in 2012. Pertussis vaccines are unlikely to provide protection against B parapertussis, which can cause illness similar to that caused by B pertussis.36 Consistent with lack of protection from the vaccine, our patients infected with B parapertussis were younger than those infected with B pertussis (P<.001), and all patients with B parapertussis had been appropriately vaccinated for pertussis. The mean age of patients with B parapertussis was 3.8 years, which is similar to that recently reported by Spicer et al37 in Ohio (4.2 years). The reason(s) for the cocirculation of B pertussis and B parapertussis is unclear, but studies in a rodent model have suggested that acellular pertussis vaccination may impair clearance of B parapertussis.38
There are multiple limitations to our study. The vaccination history was incomplete for some patients, especially those with results negative for B pertussis and B parapertussis. These patients tended to be older and were therefore more likely to have been born during the whole-cell vaccine era. The lack of childhood immunization data for a larger proportion of those with negative than with positive results for B pertussis may have led to a falsely lower documented rate of vaccination with the whole-cell vaccine in those with negative results. Had complete records been available, it is likely we would have detected an even larger difference between the groups with respect to the proportion vaccinated with whole-cell vaccine. A second limitation is that culture of nasopharyngeal swabs was performed using Regan-Lowe medium and there was variable time between specimen collection and culture. Enhanced culture techniques (eg, cough plates, bedside plating, Bordet-Gengou medium) may have improved the sensitivity of culture. A final limitation is the use of a PCR assay for B pertussis targeting the IS481 insertion sequence, which is also present in B holmesii. B holmesii was first characterized in the 1990s as a cause of bacteremia and sepsis and has subsequently been found to cause a respiratory illness similar to B pertussis.1 IS481 is present in higher copy numbers in B pertussis (50-238 copies per genome) than in B holmesii (5-8 copies per genome).39 Several studies have attempted to characterize the role of B holmesii in reported pertussis outbreaks and have come to varying conclusions. Estimates in smaller studies vary from 0.6% (Chile, 2010-2011) to 20% (France, 2009-2010) to 29% (Ohio, 2010-2011) of pertussislike illnesses.37,39-41 However, a retrospective study of over 11,000 PCR tests of patients from Finland and the Netherlands with suspected pertussis (1992-2005) detected no B holmesii, despite finding 1856 PCR tests positive for pertussis using the IS481 target.42 We are unable to estimate what, if any, proportion of our cases were due to B holmesii; we were unable to isolate B holmesii because Regan-Lowe media contains cephalexin, which is inhibitory to B holmesii. However, given the above cited reports suggesting that B holmesii accounts for fewer than 30% of pertussis cases, the outbreak reported herein would still involve a greater number of cases than in previous years. Further, 12 of the B pertussis cases were confirmed by culture.
CONCLUSION
In 2012, our region experienced its largest local pertussis outbreak in recent history. The outbreak included multiple circulating strains, all of which putatively lack expression of pertactin, a target of the acellular pertussis vaccine. Fewer patients who had pertussis had received whole-cell pertussis vaccine, and the time since the most recent dose of acellular pertussis vaccine was longer in those patients with pertussis than in those without. Taken together, these data support decreased efficacy of acellular compared with whole-cell pertussis vaccine and waning immunity from acellular vaccines as possible contributors to this outbreak. We also found circumstantial evidence that recent increased vaccination efforts in pregnant women may have impacted neonatal pertussis.
Although the current vaccine may be suboptimal with respect to immunogenicity and durability of induced immunity, it is unlikely there will be a return to the use of the whole-cell vaccine given its association with adverse events and reactogenicity. In the absence of changes in vaccine strategy, we anticipate ongoing endemic pertussis activity with peaks in the late summer months and epidemics every 2 to 5 years, as occurred in the prevaccine era. Areas of the world where acellular vaccines were adopted later than in our area should anticipate the possibility of upcoming pertussis outbreaks. Acellular vaccines are safe; given possible success in the reduction in neonatal pertussis, decreases in pertussis incidence may be realized with increased frequency of acellular vaccine administration and/or deployment of improved pertussis vaccines.
Supplementary Material
ACKNOWLEDGMENTS
We acknowledge Ms Emily A. Vetter and Ms Stefanea L. Rucinski for assistance in retrieving data from the records of the Mayo Clinic Clinical Microbiology Laboratory and the outstanding technologists of the Mayo Clinic Bacteriology Laboratory for performing the PCR testing.
Grant Support: This work was supported by the Mayo Clinic Division of Clinical Microbiology, Department of Laboratory Medicine and Pathology; the Mayo Clinic Center for Individualized Medicine; and the Mayo-Illinois Strategic Alliance for Technology-Based Healthcare (P.R.J.).
Abbreviations and Acronyms
- CDC
Centers for Disease Control and Prevention
- PCR
polymerase chain reaction
- PFGE
pulsed-field gel electrophoresis
- Tdap
tetanus toxoid, reduced diphtheria toxoid, and acellular pertussis
Footnotes
Potential Competing Interests: Dr Patel has a patent on Bordetella pertussis/parapertussis polymerase chain reaction. The other authors have no related disclosures.
Data Previously Presented: These data were presented in part at the 53rd Interscience Conference on Antimicrobial Agents and Chemotherapy in Denver, Colorado, September 10-13, 2013.
SUPPLEMENTAL ONLINE MATERIAL
Supplemental material can be found online at http://www.mayoclinicproceedings.org.
REFERENCES
- 1.Mattoo S, Cherry JD. Molecular pathogenesis, epidemiology, and clinical manifestations of respiratory infections due to Bordetella pertussis and other Bordetella subspecies. Clin Microbiol Rev. 2005;18(2):326–382. doi: 10.1128/CMR.18.2.326-382.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rohani P, Drake JM. The decline and resurgence of pertussis in the US. Epidemics. 2011;3(3-4):183–188. doi: 10.1016/j.epidem.2011.10.001. [DOI] [PubMed] [Google Scholar]
- 3.Eshofonie AO, Lin H, Valcin RP, Martin LR, Grunenwald PE. An outbreak of pertussis in rural Texas: an example of the resurgence of the disease in the United States. J Community Health. doi: 10.1007/s10900-014-9902-2. [published online ahead of print June 14, 2014] http://dx.doi.org/10.1007/s10900-014-9902-2. [DOI] [PubMed]
- 4.Centers for Disease Control and Prevention (CDC) Pertussis epidemic—Washington, 2012. MMWR. Morb Mortal Wkly Rep. 2012;61(28):517–522. [PubMed] [Google Scholar]
- 5.Tartof SY, Lewis M, Kenyon C, et al. Waning immunity to pertussis following 5 doses of DTaP. Pediatrics. 2013;131(4):e1047–e1052. doi: 10.1542/peds.2012-1928. [DOI] [PubMed] [Google Scholar]
- 6.Cherry JD. Historical review of pertussis and the classical vaccine. J Infect Dis. 1996;174(suppl 3):S259–S263. doi: 10.1093/infdis/174.supplement_3.s259. [DOI] [PubMed] [Google Scholar]
- 7.Liko J, Robison SG, Cieslak PR. Pertussis vaccine performance in an epidemic year—Oregon, 2012. Clin Infect Dis. 2014;59(2):261–263. doi: 10.1093/cid/ciu273. [DOI] [PubMed] [Google Scholar]
- 8.Childhood Immunizations [July 31, 2014];Minnesota Department of Health website. 2013 https://apps.health.state.mn.us/mndata/immunization_map.
- 9.Theel ES, Schmitt BH, Hall L, et al. Formic acid-based direct, on-plate testing of yeast and Corynebacterium species by Bruker Biotyper matrix-assisted laser desorption ionization-time of flight mass spectrometry. J Clin Microbiol. 2012;50(9):3093–3095. doi: 10.1128/JCM.01045-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sloan LM, Hopkins MK, Mitchell PS, et al. Multiplex LightCycler PCR assay for detection and differentiation of Bordetella pertussis and Bordetella parapertussis in nasopharyngeal specimens. J Clin Microbiol. 2002;40(1):96–100. doi: 10.1128/JCM.40.1.96-100.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bartkus JM, Juni BA, Ehresmann K, et al. Identification of a mutation associated with erythromycin resistance in Bordetella pertussis: implications for surveillance of antimicrobial resistance. J Clin Microbiol. 2003;41(3):1167–1172. doi: 10.1128/JCM.41.3.1167-1172.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mooi FR, Hallander H, Wirsing von König CH, Hoet B, Guiso N. Epidemiological typing of Bordetella pertussis isolates: recommendations for a standard methodology. Eur J Clin Micro-biol Infect Dis. 2000;19(3):174–181. doi: 10.1007/s100960050455. [DOI] [PubMed] [Google Scholar]
- 13.Centers for Disease Control and Prevention . Standard operating procedure for Pulsenet PFGE of Escherichia coli O157: H7, Escherichia coli non-O157 (STEC), Salmonella serotypes, Shigella sonnei and Shigella flexneri. Centers for Disease Control and Prevention website; [August 17, 2014]. http://www.cdc.gov/pulsenet/PDF/ecoli-shigella-salmonella-pfge-protocol-508c.pdf. Updated April 2013. [Google Scholar]
- 14.Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30(15):2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zerbino DR, Birney E. Velvet: algorithms for de novo short read assembly using de Bruijn graphs. Genome Res. 2008;18(5):821–829. doi: 10.1101/gr.074492.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Richter M, Rosselló-Móra R. Shifting the genomic gold standard for the prokaryotic species definition. Proc Natl Acad Sci U S A. 2009;106(45):19126–19131. doi: 10.1073/pnas.0906412106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Paradis E, Claude J, Strimmer K. APE: Analyses of phylogenetics and evolution in R language. Bioinformatics. 2004;20(2):289–290. doi: 10.1093/bioinformatics/btg412. [DOI] [PubMed] [Google Scholar]
- 18.Hyatt D, Chen GL, Locascio PF, Land ML, Larimer FW, Hauser LJ. Prodigal: prokaryotic gene recognition and translation initiation site identification. BMC Bioinformatics. 2010;11:119. doi: 10.1186/1471-2105-11-119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Katoh K, Toh H. Recent developments in the MAFFT multiple sequence alignment program. Brief Bioinform. 2008;9(4):286–298. doi: 10.1093/bib/bbn013. [DOI] [PubMed] [Google Scholar]
- 20.Muyldermans G, Piérard D, Hoebrekx N, et al. Simple algorithm for identification of Bordetella pertussis pertactin gene variants. J Clin Microbiol. 2004;42(4):1614–1619. doi: 10.1128/JCM.42.4.1614-1619.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Otsuka N, Han HJ, Toyoizumi-Ajisaka H, et al. Prevalence and genetic characterization of pertactin-deficient Bordetella pertussis in Japan. PloS one. 2012;7(2):e31985. doi: 10.1371/journal.pone.0031985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Queenan AM, Cassiday PK, Evangelista A. Pertactin-negative variants of Bordetella pertussis in the United States [letter]. N Engl J Med. 2013;368(6):583–584. doi: 10.1056/NEJMc1209369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pawloski LC, Queenan AM, Cassiday PK, et al. Prevalence and molecular characterization of pertactin-deficient Bordetella pertussis in the United States. Clin Vaccine Immunol. 2014;21(2):119–125. doi: 10.1128/CVI.00717-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bowden KE, Williams MM, Cassiday PK, et al. Molecular epidemiology of pertussis epidemic—Washington state. J Clin Microbiol. 2012 doi: 10.1128/JCM.01189-14. [published online ahead of print July 16, 2014]. http:// dx.doi.org/10.1128/JCM.01189-14. [DOI] [PMC free article] [PubMed]
- 25.Barkoff AM, Mertsola J, Guillot S, Guiso N, Berbers G, He Q. Appearance of Bordetella pertussis strains not expressing the vaccine antigen pertactin in Finland [letter]. Clin Vaccine Immunol. 2012;19(10):1703–1704. doi: 10.1128/CVI.00367-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hegerle N, Paris AS, Brun D, et al. Evolution of French Bordetella pertussis and Bordetella parapertussis isolates: increase of Bordetellae not expressing pertactin. Clin Microbiol Infect. 2012;18(9):E340–E346. doi: 10.1111/j.1469-0691.2012.03925.x. [DOI] [PubMed] [Google Scholar]
- 27.Quinlan T, Musser KA, Currenti SA, Zansky SM, Halse TA. Pertactin-negative variants of Bordetella pertussis in New York State: a retrospective analysis, 2004-2013. Mol Cell Probes. 2014;28(4):138–140. doi: 10.1016/j.mcp.2013.12.003. [DOI] [PubMed] [Google Scholar]
- 28.Lam C, Octavia S, Ricafort L, et al. Rapid increase in pertactindeficient Bordetella pertussis isolates, Australia. Emerg Infect Dis. 2014;20(4):626–633. doi: 10.3201/eid2004.131478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Misegades LK, Winter K, Harriman K, et al. Association of childhood pertussis with receipt of 5 doses of pertussis vaccine by time since last vaccine dose, California, 2010. JAMA. 2012;308(20):2126–2132. doi: 10.1001/jama.2012.14939. [DOI] [PubMed] [Google Scholar]
- 30.Liko J, Robison SG, Cieslak PR. Priming with whole-cell versus acellular pertussis vaccine [letter]. N Engl J Med. 2013;368(6):581–582. doi: 10.1056/NEJMc1212006. [DOI] [PubMed] [Google Scholar]
- 31.Klein NP, Bartlett J, Fireman B, Rowhani-Rahbar A, Baxter R. Comparative effectiveness of acellular versus whole-cell pertussis vaccines in teenagers. Pediatrics. 2013;131(6):e1716–e1722. doi: 10.1542/peds.2012-3836. [DOI] [PubMed] [Google Scholar]
- 32.Cherry JD. Epidemic pertussis in 2012—the resurgence of a vaccine-preventable disease. N Engl J Med. 2012;367(9):785–787. doi: 10.1056/NEJMp1209051. [DOI] [PubMed] [Google Scholar]
- 33.Centers for Disease Control and Prevention (CDC) Updated recommendations for use of tetanus toxoid, reduced diphtheria toxoid and acellular pertussis (Tdap) vaccine from the Advisory Committee on Immunization Practices, 2010. MMWR Morb Mortal Wkly Rep. 2011;60(1):13–15. [PubMed] [Google Scholar]
- 34.Centers for Disease Control and Prevention (CDC) Updated recommendations for use of tetanus toxoid, reduced diphtheria toxoid, and acellular pertussis vaccine (Tdap) in pregnant women—Advisory Committee on Immunization Practices (ACIP), 2012. MMWR Morb Mortal Wkly Rep. 2013;62(7):131–135. [PMC free article] [PubMed] [Google Scholar]
- 35.Jiménez-Truque N, Edwards KM. Maternal pertussis immunization: can it help infants [editorial]? JAMA. 2014;311(17):1736–1737. doi: 10.1001/jama.2014.3555. [DOI] [PubMed] [Google Scholar]
- 36.Khelef N, Danve B, Quentin-Millet MJ, Guiso N. Bordetella pertussis and Bordetella parapertussis: two immunologically distinct species. Infect Immun. 1993;61(2):486–490. doi: 10.1128/iai.61.2.486-490.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Spicer KB, Salamon D, Cummins C, Leber A, Rodgers LE, Marcon MJ. Occurrence of 3 Bordetella species during an outbreak of cough illness in Ohio: epidemiology, clinical features, laboratory findings and antimicrobial susceptibility. Pediatr Infect Dis J. 2014;33(7):e162–e167. doi: 10.1097/INF.0000000000000262. [DOI] [PubMed] [Google Scholar]
- 38.Long GH, Karanikas AT, Harvill ET, Read AF, Hudson PJ. Acellular pertussis vaccination facilitates Bordetella parapertussis infection in a rodent model of bordetellosis. Proc Biol Sci. 2010;277(1690):2017–2025. doi: 10.1098/rspb.2010.0010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rodgers L, Martin SW, Cohn A, et al. Epidemiologic and laboratory features of a large outbreak of pertussis-like illnesses associated with cocirculating Bordetella holmesii and Bordetella pertussis—Ohio, 2010-2011. Clin Infect Dis. 2013;56(3):322–331. doi: 10.1093/cid/cis888. [DOI] [PubMed] [Google Scholar]
- 40.Miranda C, Wozniak A, Castillo C, et al. Presence of Bordetella holmesii in an outbreak of pertussis in Chile [in Spanish]. Rev Chilena Infectol. 2013;30(3):237–243. doi: 10.4067/S0716-10182013000300001. [DOI] [PubMed] [Google Scholar]
- 41.Njamkepo E, Bonacorsi S, Debruyne M, Gibaud SA, Guillot S, Guiso N. Significant finding of Bordetella holmesii DNA in nasopharyngeal samples from French patients with suspected pertussis. J Clin Microbiol. 2011;49(12):4347–4348. doi: 10.1128/JCM.01272-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Antila M, He Q, de Jong C, et al. Bordetella holmesii DNA is not detected in nasopharyngeal swabs from Finnish and Dutch patients with suspected pertussis. J Med Microbiol. 2006;55(pt 8):1043–1051. doi: 10.1099/jmm.0.46331-0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.