Summary
Inflammasome biology is one of the most exciting and rapidly growing areas in immunology. Over the past 10 years, inflammasomes have been recognized for their roles in the host defense against invading pathogens and in the development of cancer, autoinflammatory, metabolic, and neurodegenerative diseases. Assembly of an inflammasome complex requires cytosolic sensing of pathogen-associated molecular patterns or danger-associated molecular patterns by a nucleotide-binding domain and leucine-rich repeat receptor (NLR) or absent in melanoma 2-like receptor (ALR). NLRs and ALRs engage caspase-1, in most cases requiring the adapter protein apoptosis-associated speck-like protein containing a CARD (ASC), to catalyze proteolytic cleavage of pro-interleukin-1β (pro-IL-1β) and pro-IL-18 and drive pyroptosis. Recent studies indicate that caspase-8, caspase-11, IL-1R–associated kinases (IRAK), and receptor-interacting protein (RIP) kinases contribute to inflammasome functions. In addition, post-translational modifications, including ubiquitination, deubiquitination, phosphorylation, and degradation, control almost every aspect of inflammasome activities. Genetic studies indicate that mutations in NLRP1, NLRP3, NLRC4, and AIM2 are linked to the development of autoinflammatory diseases, enterocolitis, and cancer. Overall, these findings transform our understanding of the basic biology and clinical relevance of inflammasomes. In this review, we provide an overview of the latest development of inflammasome research and discuss how inflammasome activities govern health and disease.
Keywords: inflammasome, NLR, caspase-1, caspase-8, caspase-11, IL-1
Introduction
Inflammasomes are multimeric protein complexes that are formed in a cell to orchestrate host defense mechanisms against infectious agents and physiological aberration. Assembly of the inflammasome complex is initiated by nucleotide-binding domain and leucine-rich repeat receptors (NLRs) or absent in melanoma 2 (AIM2)-like receptors (ALRs). NLRs and ALRs mediate host recognition of pathogen-associated molecular patterns (PAMPs) released during bacterial, viral, fungal, and protozoan infections, or danger-associated molecular patterns (DAMPs) released during cellular damage (1). Since the discovery of the founding member of the NLR family, NOD1 (2, 3), 22 human NLRs and 34 mouse NLRs have been identified (4). In the ALR or the PYHIN protein family, 4 members in human and 14 in mouse have been found (5). In 2002, the ability of NLRP1 to form an inflammasome complex was described (6). A decade later, it is now clear that other members of the NLR and ALR family, including NLRP3, NLRC4, and AIM2, can also assemble the inflammasome (7). Emerging evidence now indicate that human NLRP2, NLRP7, IFI16, and Pyrin and mouse NLRP6, NLRP12, and Pyrin also activate caspase-1 (7, 8).
Activated NLRs and ALRs, in most cases, recruit a bipartite protein known as apoptosis-associated speck-like protein containing a caspase activation and recruitment domain (ASC) to engage caspase-1 activation. In macrophages or dendritic cells, inflammasome-forming NLRs and ALRs induce reorganization of cytoplasmic ASC into a single ‘speck’ of 0.8 to 1μM, which is considered a hallmark of inflammasome assembly (9–14) (Fig. 1). ASC is crucial for the recruitment of caspase-1 into the speck, where caspase-1 induces proteolytic processing of pro-IL-1β and pro-IL-18 (11–13).
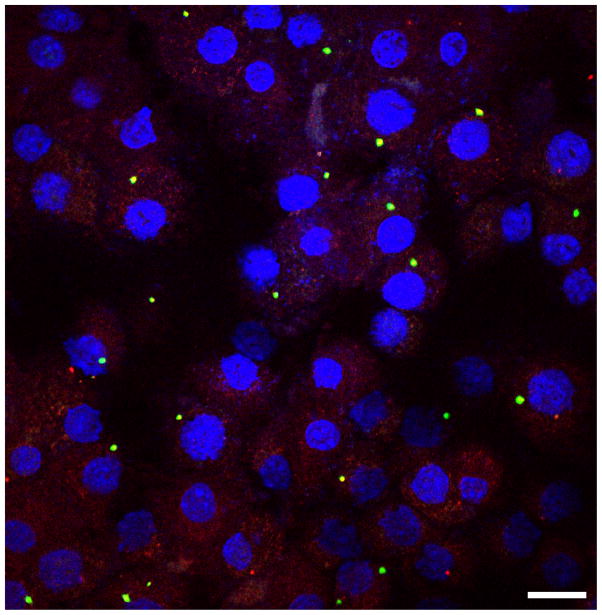
Fig. 1. An inflammasome complex can be visualized as a single distinct speck or focus in the cytoplasm of a cell.
Primary mouse bone-marrow derived macrophages transfected with the dsDNA ligand poly(dA:dT) and stained for ASC (red), caspase-1 (green) and DNA (blue). Bar, 20 μM.
Recent studies identified that previously generated caspase-1-deficent mouse lines were deficient in another member of the pro-inflammatory caspase family, caspase-11 (15). This finding led to further characterization of the distinct roles of caspase-1 and caspase-11 in inflammasome activation. Both of these pro-inflammatory caspases initiate pyroptosis, but the molecular and physiological consequences underlying caspase-1- and caspase-11-dependent pyroptosis are still unknown (7). Caspase-1 and caspase-11 activation drives the release of IL-1β and IL-18, but only caspase-1 directly cleaves IL-1β and IL-18.
The biological activities of IL-1β and IL-18 and pyroptosis are largely beneficial to the host during an infection. However, IL-1β and IL-18 induced by endogenous danger signals trigger sterile inflammation, a risk factor for the development of autoinflammatory and metabolic diseases. Therefore, activation of the inflammasome must be finely controlled to avoid overt tissue damage. These regulatory activities are governed by scaffolding proteins and post-translational modifications, which together, tightly control and modulate inflammasome activation. Here, we provide an overview of the recent advances in the field of inflammasome biology, with a particular emphasis on the regulation of inflammasome signaling in health and disease.
NLRP1 inflammasome
NLRP1 was the first member of the NLR family to be identified to form an inflammasome complex (6). The human NLRP1 protein contains an N-terminal pyrin domain (PYD), a nucleotide-binding domain (NBD), a leucine-rich repeat (LRR) domain, a ‘function to find’ (FIIND) domain, and a C-terminal caspase activation and recruitment domain (CARD) (6). Unlike the case in human, the mouse genome encodes three Nlrp1 paralogs, Nlrp1a, Nlrp1b, and Nlrp1c, all of which appear to lack a PYD (16). Mouse NLRP1b is highly polymorphic and is represented by five unique protein sequences across 18 inbred mouse strains. Two of these are associated with a susceptible phenotype and the other three are associated with a resistant phenotype to macrophage pyroptosis and caspase-1 activation induced by anthrax lethal toxin (16).
The anthrax lethal toxin produced by Bacillus anthracis is composed of a protective antigen and a lethal factor. The protective antigen generates pores on the host cell membrane, through which the lethal factor enters the cell. Further mechanistic studies found that the lethal factor cleaves mouse NLRP1b and rat NLRP1 to induce activation of the inflammasome (17, 18) (Fig. 2). A cleavage site within the N-terminal domain of mouse NLRP1b and rat NLRP1 was identified (17, 18). A subsequent study demonstrated that cleavage of mouse NLRP1b is sufficient to induce caspase-1 activation even in the absence of the lethal factor (19), suggesting NLRP1b may have the capacity to activate the inflammasome in response to any protein that is capable of inducing NLRP1b cleavage.
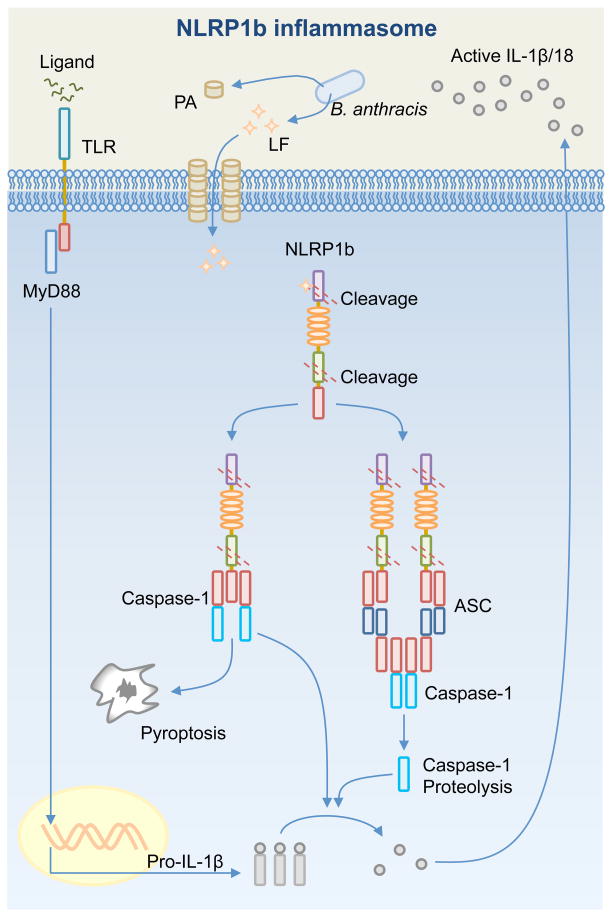
Fig. 2. Activation of the NLRP1b inflammasome.
Bacillus anthracis releases the anthrax lethal toxin, a bipartite toxin composed of a protective antigen (PA) and a lethal factor (LF). PA generates a pore on the host cell membrane, which is used by LF to enter the cell. Inflammasome responds to the presence of LF in the cytosol following LF-induced cleavage of NLRP1b at the N-terminal domain. Autoproteolytic cleavage at the FIIND domain of NLRP1b has also been observed. Cleavage of NLRP1b is sufficient to activate the inflammasome. In response to a high dose of LF, the CARD of NLRP1b binds the CARD of pro-caspase-1. This complex is sufficient to drive pro-IL-1β and pro-IL-18 processing and pyroptosis independently of ASC or caspases-1 self-proteolysis. In response to a low dose of LF, the CARD of NLRP1b recruits ASC to form a macromolecular cytoplasmic speck, where caspase-1 undergoes proteolysis and contributes to pro-IL-1β and pro-IL-18 processing.
Both the susceptible and resistant forms of mouse NLRP1b are cleaved by lethal factor, yet only macrophages harboring a susceptible form of NLRP1b undergo caspase-1 activation and pyroptosis (18). The failure of the resistant form of NLRP1b to engage inflammasome formation post-cleavage indicates that additional events may be necessary to fulfill the requirement for inflammasome assembly. Additional studies revealed that autoproteolytic cleavage at the FIIND domain of human NLRP1 or a lethal-toxin-susceptible form of mouse NLRP1b also leads to inflammasome activation (20–22). In contrast, the FIIND domain of the lethal-toxin-resistant form of mouse NLRP1b is not cleavable. Conversion of this form to a cleavable form by mutagenesis was unable to render it capable of activating caspase-1 (21), suggesting that differential susceptibility of NLRP1b to undergo proteolytic cleavage alone cannot explain the differences in susceptibility of macrophages to pyroptosis in response to anthrax lethal toxin.
In mouse macrophages, NLRP1b-mediated production of IL-1β and pyroptosis in response to anthrax lethal toxin occurs independently of ASC and ASC-dependent caspase-1 proteolysis (23). This activity is possible because the CARD and part of the FIIND domain of NLRP1, at least in the human protein, can directly interact with the CARD of pro-caspase-1 (21, 24). Reconstitution of caspase-1-deficient cells with a non-cleavable form of pro-caspase-1 confirmed that proteolysis of caspase-1 itself is not required for IL-1β processing and pyroptosis upon lethal toxin stimulation (25). However, ASC is still required for the assembly of the inflammasome speck and for caspase-1 proteolysis in response to lethal toxin stimulation. In this context, ASC partially contributes to IL-1β release in mouse macrophages stimulated with a low dose of lethal toxin (23), indicating that ASC provides NLRP1b an enhanced capacity to detect lethal toxin.
Mice harboring a susceptible NLRP1b variant that responds to lethal toxin are more protected against B. anthracis infection compared to mice harboring a resistant NLRP1b variant that fails to response to lethal toxin (26, 27), confirming physiological relevance of the NLRP1b inflammasome in the host defense against B. anthracis. In a mutagenesis screen, a mutation that causes hyperactivation of mouse NLRP1a, owing to a glutamine-to-proline substitution in the linker region between the NBD and LRR domain of NLRP1a, results in lethal systemic inflammation (28). Mice deficient in caspase-1 or IL-1β, but not ASC or caspase-11, in the presence of the hyperactive NLRP1a mutation were protected against disease, demonstrating a role for NLRP1a in driving inflammasome-associated pathology (28). In humans, genetic studies revealed that mutations in NLRP1 are linked to autoinflammatory diseases, including vitiligo and Addison’s disease (29). How these mutations lead to manifestation of autoinflammatory diseases remains to be defined. Adherent blood monocytes from individuals harboring a homozygous NLRP1 haplotype associated with susceptibility to vitiligo and other autoimmune diseases produced elevated levels of IL-1β (30), indicating that excessive IL-1β production as a result of NLRP1 inflammasome activity could be a contributing factor for the development of these diseases. Elucidation of the molecular pathway that governs the activation of the NLRP1 inflammasome is required to further understand its role in infectious and autoinflammatory diseases.
Canonical NLRP3 inflammasome
NLRP3 (also known as CIAS1, Cryopyrin, NALP3, and Pypaf1) responds to a wide range of PAMPs and DAMPs, including bacterial messenger RNA, bacterial DNA:RNA hybrids, muramyl dipeptide (MDP), DNA and RNA viruses, fungi, protozoa, ATP, uric acid crystals, silica, aluminium hydroxide, asbestos, and bee venom (31–46). Activation of the NLRP3 inflammasome in macrophages requires two signals (Fig. 3). The first signal, known as priming, is provided by Toll-like receptors (TLRs), NOD2, TNFR1, or TNFR2, which engages NF-κB-mediated expression of NLRP3 (47). The second signal is provided by a PAMP or DAMP that activates NLRP3 to trigger inflammasome assembly, IL-1β and IL-18 release, and pyroptosis. Only the first signal is required for IL-1β secretion in human monocytes, possibly owing to the expression of constitutively active caspase-1 in these cells (48).
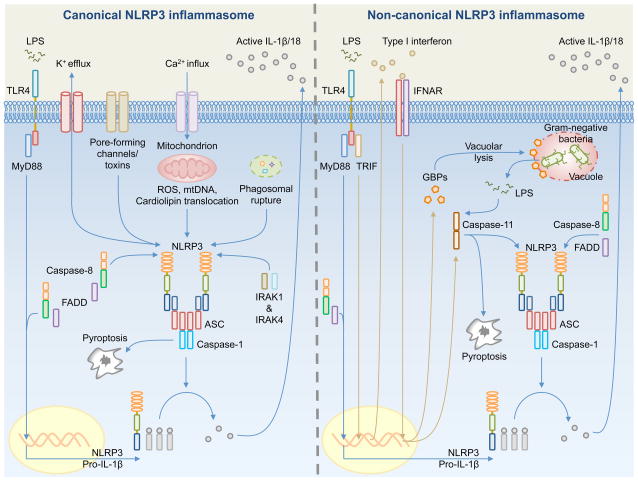
Fig. 3. Mechanisms of activation for the canonical and non-canonical NLRP3 inflammasomes.
(Left). Canonical NLRP3 inflammasome activation requires priming by a Toll-like receptor (TLR) ligand (e.g. LPS) – mediated by MyD88, caspase-8, FADD, and NF-κB – to induce the expression of pro-IL-1β and NLRP3. Pro-IL-18 is expressed constitutively in the cell. A number of mechanisms have been proposed to activate the canonical NLRP3 inflammasome, including K+ efflux, pore-forming channels or toxins, Ca2+ influx, mitochondrial reactive oxygen species (ROS), mitochondrial DNA (mtDNA), translocation of cardiolipin from the inner mitochondrial membrane to the outer mitochondrial membrane, and phagosomal destabilization. NLRP3, ASC and caspase-1 assemble the inflammasome, which leads to proteolytic cleavage of pro-IL-1β and pro-IL-18 for release and the induction of pyroptosis. A priming-independent pathway for canonical NLRP3 inflammasome activation has been described. This pathway requires IRAK1 and IRK4. (Right). Non-canonical NLRP3 inflammasome activation is activated by Gram-negative bacteria. Extracellular LPS induces the expression of pro-IL-1β and NLRP3 via the TLR4-MyD88-dependent pathway and type I interferon via the TLR4-TRIF-dependent pathway. Type I interferon provides a feedback loop and activates type I interferon receptor (IFNAR) to induce caspase-11 expression. Cytosolic Gram-negative bacteria deliver LPS into the cytosol when they escape the vacuole. Vacuolar Gram-negative bacteria release their LPS into the cytosol through a mechanism that requires vacuolar rupture mediated by interferon-inducible guanylate-binding proteins (GBPs). Caspase-11 is proposed to activate following its binding to cytosolic LPS. Caspase-11 then drives pyroptosis and activation of the non-canonical NLRP3 inflammasome.
The diverse PAMPs and DAMPs sensed by NLRP3 clearly indicate that NLRP3 is unlikely to interact directly with all these activators. Instead, a more likely scenario is that a common cellular event triggered by these stimuli induces a conformational change in NLRP3 that converts it from an inactive to an active form. Despite considerable efforts, there is no consensus for a unified mechanism for the activation of the NLRP3 inflammasome. A number of mechanisms have been proposed, including mitochondrial-associated dysfunction [mitochondrial reactive oxygen species (ROS) (49), oxidized mitochondrial DNA (50), translocation of cardiolipin from the inner to the outer mitochondrial membrane (51)], potassium efflux (52–54), an increase in intracellular calcium and a decrease in cellular cyclic AMP (55, 56), phagosomal destabilization (33), pore-forming actions driven by the host or bacteria (57–59), and changes in cell volume (60). A number of studies argued that mitochondrial perturbation, ROS, formation of large membrane pores and cell volume fluctuation are not required for the activation of the canonical NLRP3 inflammasome (53, 61, 62). Instead, a common event induced by classical canonical NLRP3 activators is a reduction in intracellular potassium concentration, which is responsible for activating the NLRP3 inflammasome (53). If this is the case, it would suggest that the mode of activation of the canonical NLRP3 inflammasome activation is similar to the apoptosome, as formation of this complex is also inhibited by an increase in intracellular potassium concentration (63).
A number of innate immune proteins have now been identified to provide scaffolding function for the canonical NLRP3 inflammasome. Work from our laboratory and others (61, 64, 65) has shown that caspase-8 and its adapter Fas-associated death domain (FADD) protein are required for both priming and activation of the NLRP3 inflammasome in macrophages. Because caspase-8 is essential for the priming signal, it was difficult to determine whether caspase-8 affects activation of the NLRP3 inflammasome. However, pharmacological inhibition of caspase-8 after priming inhibited canonical NLRP3 inflammasome activation (64). Further, recruitment of caspase-8 to the inflammasome speck and its ability to interact with NLRP3 provide evidence to support that caspase-8 is a component of the canonical NLRP3 inflammasome (11, 64). A requirement for caspase-8 in the activation of the NLRP3 inflammasome is also observed for the fungal cell wall component β-glucans and heat-killed Candida albicans (66). One study, however, suggests that caspase-8 suppresses NLRP3 activities in dendritic cells (67). Caspase-8-deficient dendritic cells release IL-1β following LPS stimulation independently of an NLRP3 activator (Signal 2), but instead, requires RIP1, RIP3, MLKL, and PGAM5 (68). The observation that caspase-8 could negatively regulate the canonical NLRP3 inflammasome may be specific to dendritic cells. Unlike in the case with dendritic cells, caspase-8 contributes to robust NF-κB activation in response to TLR stimulation in macrophages, T cells, B cells, and NK cells (11, 61, 64, 69, 70).
Other studies have shown that caspase-8 and FADD are essential for caspase-1 processing and cell death induced by Yersinia infection (71, 72). During Yersinia infection, RIP1, but not RIP3, contributes to caspase-1 activation (71, 72). In response to vesicular stomatitis virus, an RNA virus, RIP1 and RIP3 form a complex to drive mitochondrial damage and ROS production that leads to activation of the NLRP3 inflammasome (73). RIP2, however, enhances autophagy of mitochondria or mitophagy to prevent accumulation of ROS and dampens activation of the canonical NLRP3 inflammasome during infection by influenza A virus (74). More recent studies identified a ‘priming-independent’ mode of canonical NLRP3 inflammasome activation that requires IRAK1 and IRAK4 and their respective kinase activities (75, 76). Unprimed macrophages co-stimulated with TLRs and NLRP3 activators (LPS+ATP/nigericin or Listeria infection) induced activation of the canonical NLRP3 inflammasome as early as 15 min (75, 76). In contrast, transcription-dependent activation of the canonical NLRP3 inflammasome, in LPS-primed cells followed by ATP treatment for example, occurs independently of IRAK1 and IRAK4 (75). These findings show that priming (Signal 1) and synthesis of new proteins are not always necessary for engaging the canonical NLRP3 inflammasome.
Post-translational modification of NLRP3 can either activate or suppress inflammasome activation. For example, deubiquitination of the LRR domain of NLRP3 by BRCC3, a JAMM domain-containing Zn2+ metalloprotease deubiquitinating enzyme, leads to activation of the inflammasome (77, 78). Conversely, nitric oxide induces S-nitrosylation of NLRP3 and caspase-1 and prevents assembly of the inflammasome (79–81). These studies collectively provide evidence to show that caspase-8, FADD, IRAK, RIP kinases, and post-translational modification tightly control the activation and suppression of the canonical NLRP3 inflammasome. How these regulators can be targeted and used for the development of immunotherapy remains to be explored.
Genetic studies in the human populations revealed that mutations in the NLRP3 gene are associated with the development of a group of autoinflammatory conditions known as Cryopyrin–associated periodic syndromes (CAPS) (82–84). These conditions include familial cold autoinflammatory syndrome, Muckle–Wells syndrome, and neonatal-onset multisystem inflammatory disease [(NOMID) or chronic infantile neurological cutaneous and articular syndrome (CINCA)]. Patients with CAPS produce increased amount of IL-1β compared to healthy individuals (85, 86). Studies using knockin mouse strains showed that CAPS-associated NLRP3 mutations, caspase-1, IL-1β and IL-18 collectively contribute to disease (87, 88). Other elegant mouse models have provided insights into the role of the NLRP3 inflammasome in gout, type 2 diabetes, obesity, atherosclerosis, rheumatoid arthritis, non-alcoholic fatty liver disease, bone disease, inflammatory bowel diseases, colorectal cancer, Alzheimer’s disease and aging (35, 89–101). For example, genetic deletion of NLRP3, caspase-1 and IL-1R protected mice against disease in a mouse model of spontaneous erosive polyarthritis caused by a specific deletion of the rheumatoid arthritis susceptibility gene A20/Tnfaip3 (93). This finding suggests a critical role for the NLRP3 inflammasome in driving arthritic inflammation. Recent studies from our lab identified a dual role for caspase-1 and caspase-8 in modulating IL-1β-mediated bone disease (94). Pstpip2cmo mice express a missense mutation in proline-serine-threonine phosphatase interacting protein 2 (PSTPIP2) spontaneously develop osteomyelitis, a condition which could be prevented by deletion of IL-1β alone or caspase-1, caspase-8 and RIP3 in combination (94, 102, 103). Since deletion of NLRP3 or caspase-1 alone still results in PSTPIP2-mediated osteomyelitis (102, 103), we suspect that NLRP3 in addition to other inflammasome receptors or proteases could function synergistically to produce caspase-1- and caspase-8-mediated IL-1β release that drives disease manifestation. Overall, the canonical NLRP3 inflammasome is now a recognized entity in the pathogenesis of autoinflammation, cancer and neurodegenerative diseases.
Caspase-11 and the non-canonical NLRP3 inflammasome
Most Gram-negative bacteria, including Citrobacter rodentium, Escherichia coli, and Vibrio cholerae, activate an alternative or ‘non-canonical’ NLRP3 inflammasome pathway, defined by its requirement for caspase-11 (15). The upstream mechanism governing activation of this pathway involves TLR4-TRIF-mediated recognition of extracellular LPS, which induces type I interferon signaling, at least in part, to engage caspase-11 expression (34, 104–106). Whether type I interferon is unequivocally required to induce caspase-11 expression is still under debate (107). During Citrobacter infection, NOD2 and RIP2 negatively regulate caspase-11 expression via a ROS- and c-Jun N-terminal kinase (JNK)-dependent pathway (108). The ability of one NLR to negatively regulate another highlights the dynamic and cooperative nature of innate immune sensors.
Caspase-11 is a crucial activator of the non-canonical NLRP3 inflammasome and induces pyroptosis and IL-1α and high-mobility group protein 1 (HMGB1) production (15, 109) (Fig. 3). Unlike caspase-1, caspase-11 does not directly cleave IL-1β and IL-18. Caspase-11 and the non-canonical NLRP3 inflammasome can be activated directly in mouse macrophages when LPS is transfected or electroporated into the cell (110, 111). Of particular importance is that LPS-induced caspase-11 activation occurs independently of TLR4 (110, 111). This seminal finding demonstrates that a cytosolic receptor, together with TLR4, provides dual recognition of LPS within and outside the cell. The partnership between TLR4 and the cytosolic LPS receptor is analogous to the recognition of flagellin by the TLR5 and Naip5-Naip6-NLRC4 receptors. Since caspase-11 contains a CARD, it was hypothesized that a CARD-bearing protein would interact with caspase-11 and act as an upstream sensor of LPS. Unexpectedly, a recent study showed that caspase-11 and human caspase-4 and caspase-5 bind LPS, implying that these pro-inflammatory caspases are direct sensors of LPS (112). Human caspase-4 and caspase-5 can activate murine caspase-1 in the absence of murine caspase-11 in vitro (112, 113), indicating that these pro-inflammatory caspases have interchangeable and cross-species functional activities. Caspase-11, but not caspase-1, confers LPS-induced septic shock and mortality (15, 114). In addition, induction of pro-caspase-11 expression by injecting poly(I:C) into mice renders them hypersusceptible to LPS-induced mortality, which is largely mediated by caspase-11, but not TLR4 (110).
Caspase-11 is important in conferring host protection against intestinal inflammation and bacterial infection. Three studies have shown that mice deficient in caspase-11 are highly susceptible to dextran sodium sulfate (DSS)-induced intestinal inflammation (115–117). One study observed similar levels of IL-1β and IL-18 in homogenized colon tissues from DSS-treated wildtype and caspase-11–deficient mice (115), whereas two studies found role for caspase-11 in mediating IL-1β and IL-18 production. Ex vivo organ culture of intestinal tissues harvested from DSS-treated mice revealed a caspase-11-dependency for the production of IL-1β and IL-18 (116). It is possible that an ex vivo culture system promoted the growth of specific intestinal cell populations in which caspase-11 plays a major role in driving IL-1β and IL-18 production. Exogenous IL-18 administration decreases the severity of DSS-induced colitis in caspase-11–deficient mice (117). Interestingly, type II interferon, rather than type I interferon and TRIF, mediates caspase-11 expression during DSS-induced colitis (117).
Caspase-11 detects LPS carried into the cytoplasm by cytosolic Gram-negative pathogens, including Burkholderia thailandensis and Burkholderia pseudomallei (118). A recent study shed light on how LPS from vacuolar-restricted pathogens, such as Salmonella, might enter the cytoplasm to activate caspase-11. This study showed that the guanylate-binding protein (GBP) member GBP2 contributes to activation of caspase-11 and the non-canonical NLRP3 inflammasome in response to Gram-negative bacteria (119). During Salmonella infection of a macrophage, GBP2 is recruited to the Salmonella-containing vacuole (SCV), where it mediates the rupture of the SCV to allow the release of LPS into the cytoplasm for detection by caspase-11. The nature of how GBP2 induces lysis of the SCV is unknown. Interestingly, lysis of the SCV enhances autophagy of cytosolic salmonellae, a mechanism which is proposed to restrict further caspase-11 activation (119). Caspase-11 (or caspase-4) also induces IL-18 secretion and pyroptosis in Salmonella-infected epithelial cells, the latter functions to extrude and remove infected cells from the polarized cell layer (120). Another vacuolar pathogen Legionella pneumophila also triggers caspase-11-dependent pyroptosis, but through a network of GBPs encoded on chromosome 3 (GBP1, 2, 3, 5 and 7) rather than a single GBP (121). Furthermore, caspase-11 promotes the fusion of the vacuole with lysosomes by modulating actin polymerization to restrict Legionella replication (122).
Another GBP, GBP5, was shown to promote the assembly of the NLRP3 inflammasome in response to canonical (ATP and nigericin) and non-canonical (stationary phase grown S. Typhimurium) triggers (123). In contrast, another study examined two other independently generated Gbp5-deficient mouse lines and showed that GBP5 is not required for canonical or non-canonical NLRP3 inflammasome activation in vitro (119). Further studies are required to unravel the role of GBPs in the activation of the NLRP3 inflammasome. Overall, major advances have been made to unravel the functions of the non-canonical NLRP3 inflammasome and the pathways that regulate its activation. These studies will enable development of therapies to enhance host resistance against sepsis or bacterial infection.
NAIP-NLRC4 inflammasome
NLRC4 (also known as IPAF, CARD12, or CLAN) was reported in 2001 and was shown to interact with pro-caspase-1 and induce caspase-1 proteolysis and caspase-1–dependent cell death in 293T cells (124). The mouse NLRC4 inflammasome is activated by flagellin (125, 126) and the inner rod proteins of the Type III secretion system of S. Typhimurium (PrgJ), B. pseudomallei (BsaK), E. coli (EprJ and EscI), Shigella flexneri (MxiI), and Pseudomonas aeruginosa (PscI) (127). Given no direct interaction between NLRC4 and the proposed ligands has been observed, it was speculated that one or more sensors upstream of NLRC4 were responsible for mediating ligand recognition (Fig. 4).
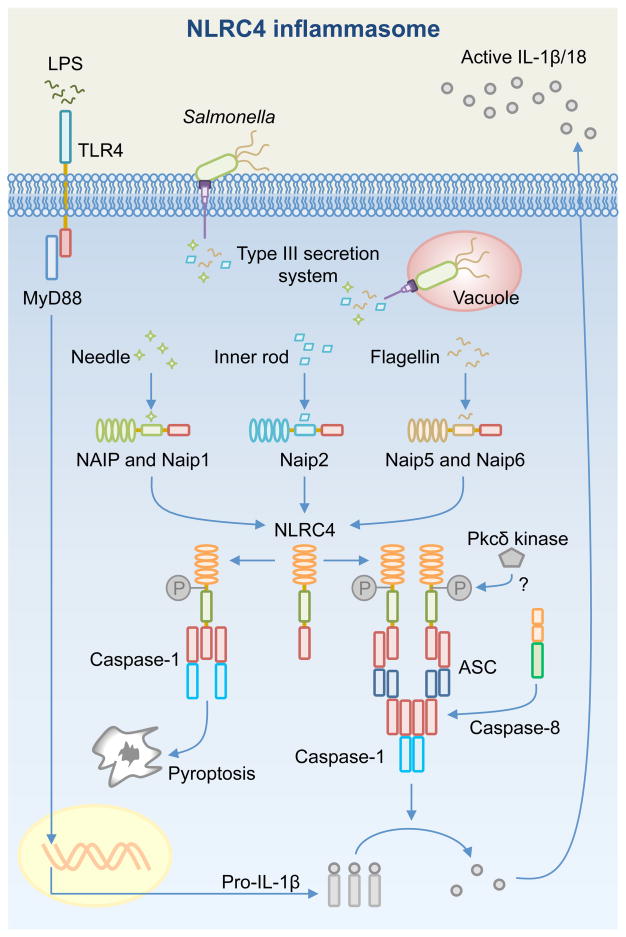
Fig. 4. Activation of the NAIP-NLRC4 inflammasome.
Toll-like receptors (e.g. TLR4) mediate the production of pro-IL-1β via MyD88 and NF-κB. Certain pathogenic bacteria use the Type III secretion system (T3SS) to deliver effector proteins to subvert host cell functions (e.g. Salmonella enterica serovar Typhimurium uses the Salmonella Pathogenicity Island-1 or SPI-1– Type III secretion system). In doing so, bacterial proteins such as the T3SS needle protein, inner rod protein, and flagellin are injected into the cytoplasm. These proteins are detected by NAIPs to activate the NLRC4 inflammasome, which results in pro-IL-1β and pro-IL-18 processing via an ASC-dependent mechanism. Caspase-1 and caspase-8 are recruited to the ASC inflammasome independently of each other during early infection. Caspase-8 is speculated to enhance delayed processing of pro-IL-1β and pro-IL-18 and induces delayed cell death. NLRC4 induces caspase-1-dependent pyroptosis via an ASC-independent mechanism. Phosphorylation of NLRC4 by the Pkcδ kinase is proposed to contribute to the activation of the NLRC4 inflammasome.
Members of the NAIP (NLR family, apoptosis inhibitory protein) family were later identified to be the upstream sensors of the NLRC4 inflammasome. The mouse genome encodes seven Naip paralogs (Naip1-7) (128). Mouse Naip5 and Naip6 recognize cytosolic flagellin, whereas mouse Naip1 and Naip2 recognize bacterial needle and inner rod proteins of the Type III secretion system, respectively (129–132). The functional roles of mouse Naip3, Naip4, and Naip7 remain to be elucidated. Interestingly, analysis of mouse Naip chimeras revealed that the LRR domain, typically considered to be important for ligand recognition, is not required for ligand binding (133). Instead, α-helical domains within the NBD are required to confer ligand specificity (133). Unlike the case in mouse, only one functional NAIP is encoded in the human genome. NAIP recognizes the needle protein of the Type III secretion system from S. Typhimurium (PrgI) and the homologous needle subunits found in Chromobacterium violaceum (CprI), S. flexneri (MxiH), P. aeruginosa (PscF), B. thailandensis (BsaL), and enterohaemorrhagic E. coli (EprI) (130). The ability to recognize flagellin and components of the bacterial secretion systems enables the NLRC4 inflammasome to provide host defense to a range of pathogens, including S. Typhimurium (10, 12, 134–136), P. aeruginosa (136–138), S. flexneri (127, 139, 140), C. rodentium (141, 142), L. pneumopila (143, 144), Listeria monocytogenes (145), Klebsiella pneumoniae (146), Yersinia (136, 147), and C. albicans (148). In some cases, excessive inflammation driven by the NLRC4 inflammasome in response to bacterial infection is detrimental to the host (149, 150).
NLRC4 contains a CARD that can directly interact with the CARD of caspase-1 in the absence of ASC (124). This interaction may explain why NLRC4 is able to induce pyroptosis independently of ASC (13, 151). However, ASC serves to enhance NLRC4-mediated IL-1β and IL-18 release in the cell by aggregating into a large speck that recruits caspase-1 for proteolytic cleavage of IL-1β (11, 12). In macrophages infected with S. Typhimurium, endogenous ASC forms an outer core surrounding an inner ring of NLRC4, with caspase-1 in the center of the speck (10). The NLRC4 inflammasome recruits multiple effector proteins into the complex, including caspase-8, upon infection of macrophages with S. Typhimurium (11). Caspase-8 is speculated to induce cell death functions and promote NLRP3 and pro-IL-1β expression (11). In addition, caspase-7 is activated downstream of the NLRC4 inflammasome to facilitate lysosome fusion with vacuoles containing L. pneumophila (152). Induction of NLRC4 inflammasome assembly in response to S. Typhimurium infection requires Pkcδ kinase-induced phosphorylation of a single evolutionarily conserved residue, Ser533, found between the NBD and the C-terminal LLR domain of NLRC4 (153). Another report, however, argues that the Pkcδ kinase is dispensable for NLRC4 inflammasome activation in response to Salmonella or Shigella infection (139). The reason for the discrepancy between these two studies is unknown and additional experiments are required to resolve whether Pkcδ kinase is involved in licensing NLRC4 inflammasome activation.
NLRC4 has important cell type-specific functions in the host. Activation of the NLRC4 inflammasome by Salmonella infection results in cellular stiffness of macrophages, which has important and diverse cellular consequences (154). These include a reduced capacity of the macrophage to take up more bacteria, enhanced ROS production to kill resident bacteria, and reduced macrophage movement to prevent bacterial dissemination (154). In addition, NLRC4 inflammasome-driven pyroptosis of macrophages releases intracellular bacteria for uptake by neutrophils, which mediates bacterial killing independently of inflammasome-associated IL-1β and IL-18 production (134). In mouse neutrophils, activation of the NLRC4 inflammasome by S. Typhimurium leads to robust IL-1β secretion without pyroptosis (155), indicating that pyroptosis is a cell type-specific feature that is not always induced by caspase-1 activation. The mechanism preventing caspase-1-dependent pyroptosis in neutrophils remains to be determined. Bacteria can inhibit the expression of NLRC4 inflammasome components, an observation which has been reported in mouse B cells infected with S. Typhimurium (156). The NLRC4 inflammasome also mediates sensing of S. Typhimurium in CD8α+ dendritic cells, resulting in the release of IL-18 to induce IFN-γ production by memory CD8+ T cells to control the infection (136).
Additional studies have investigated cell type-specific roles of NLRC4 inflammasome in the gastrointestinal tract. Unlike mouse bone marrow–derived macrophages, intestinal mononuclear phagocytes induce activation of the NLRC4 inflammasome, but do not release TNF-α, in response to S. Typhimurium and P. aeruginosa infection (135). Furthermore, Salmonella-infected epithelial cells are extruded from the intestinal epithelium in a process that requires the Naip1-6 locus and NLRC4 (157). However, deficiency in IL-1α and IL-1β or IL-18 does not lead to a failure to control intraepithelial bacterial burden (157). Further studies on the role of NLRC4 in the gastrointestinal tract revealed that NLRC4 confers protection against colorectal cancer induced by azoxymethane (AOM) and DSS (158). In contrast, NLRC4 inflammasome activities increase susceptibility to DSS-induced sepsis (159). In this context, intestinal dysbiosis driven by broad-spectrum antibiotic treatment in mice leads to expansion and extraintestinal colonization of a multidrug-resistant E. coli pathobiont. Recognition of this pathobiont by the Naip5-NLRC4 inflammasome renders the host highly susceptible to sepsis (159). In addition to its role in driving disseminating infection, systemic delivery of cytosolic flagellin activates the Naip5-NLRC4 inflammasome and causes rapid death of the mice in 30 min (160). This striking observation is unrelated to IL-1β or IL-18 production. Instead, inflammasome-driven production of eicosanoids, inflammatory lipid mediators including prostaglandins and leukotrienes, promptly initiates inflammation and loss of fluid into the intestine and peritoneal cavity that ultimately result in the rapid demise of the host (160).
The availability of a crystal structure of mouse NLRC4 has provided vital clues to unraveling the mechanism governing the autoinhibitory nature of this NLR. The mouse NLRC4 is composed of a CARD, a NBD, a winged-helix domain (WHD), a helicase domain 1 (HD1), a HD2, and a LRR (161). In the absence of the CARD, the mouse NLRC4 crystal forms a solenoid shape (161). The NBD and the winged-helix domain (WHD) of the protein interact to maintain autoinhibition, while the C-terminal LRR domain sequesters NLRC4 in a monomeric state (161).
Human genetic studies demonstrate that gain-of-function mutations in NLRC4 are associated with autoinflammation and enterocolitis (162–164). One report showed that several members of the same family with a p.Val341Ala substitution mutation within the NLRC4 HD1 domain experience recurrent autoinflammation and enterocolitis (162). A newborn from this family with the same mutation suffered severe gastrointestinal complications, fever, and systemic inflammation and died 23 days after birth (162). A second report found a heterozygous de novo mutation resulting in a p.Thr337Ser substitution in the NBD of NLRC4 (163). This mutation is linked to macrophage activation syndrome (MAS) in a seven-year-old patient and has been suggested to play a role in destabilizing the interaction between the NBD and HD1 domains that critically confers autoinhibition of NLRC4. The patient suffered from recurrent fever, splenomegaly, gastrointestinal pathology, and systemic elevation of inflammatory markers. A third report describes a heterozygous missense mutation in five patients within the same family (164). This mutation causes an A>C transversion (1589A>C mutation) in the NBD of NLRC4. A transgenic mouse strain carrying the 1589A>C mutation developed dermatitis and arthritis, conditions which were accelerated by exposure to cold temperature (164). The NLRC4 mutations described in all three reports contribute to spontaneous activation of caspase-1, ASC speck formation, elevated levels of pyroptosis and enhanced production of IL-1β and IL-18 in monocytes or macrophages (162–164). The patient with MAS responded to anakinra (163), suggesting that recombinant IL-1 receptor antagonist therapy could be used to treat those with autoinflammatory diseases associated with excessive NLRC4 inflammasome activation. These recent discoveries of disease-associated mutations of NLRC4 indicate that additional research efforts to unravel the regulatory mechanism governing NLRC4 inflammasome activation are required.
The AIM2 inflammasome
AIM2 is an inflammasome receptor for double-stranded DNA (dsDNA) (165–168). AIM2 consists of a HIN-200 domain and a PYD. Structural analysis demonstrated that the positively charged HIN-200 domain embraces the dsDNA, whereas the PYD recruits ASC for caspase-1 activation (169). Cryo-electron microscopy analysis revealed that ASC and caspase-1 form filamentous structures following nucleation initiated by AIM2 or NLRP3 (170). These filamentous structures are likely to ultimately form a single inflammasome speck observed in primary macrophages or dendritic cells (171, 172).
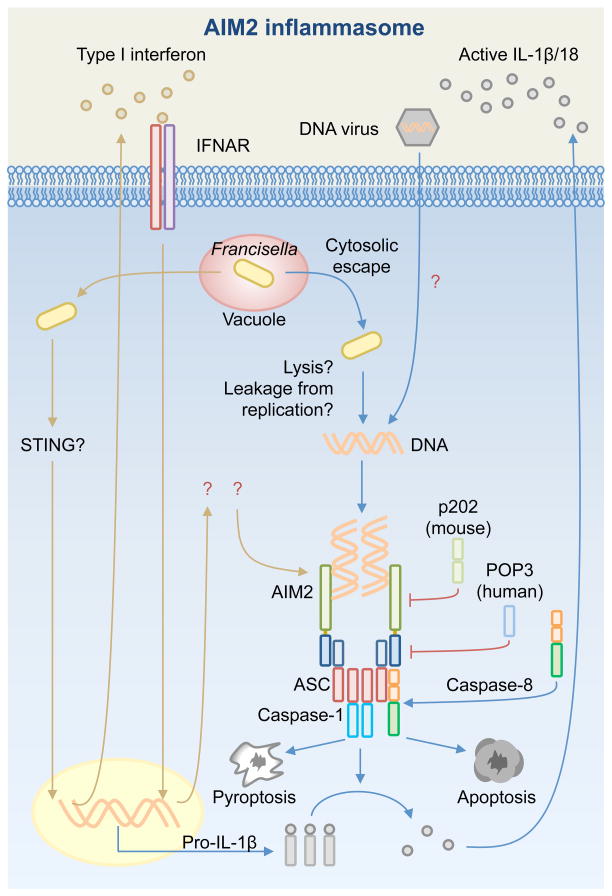
The AIM2 inflammasome contributes to the host defense against a subset of bacterial and viral pathogens (165–168, 173, 174). These includes Francisella tularensis (172–174), L. monocytogenes (145, 175–178), Streptococcus pneumoniae (179), Mycobacterium tuberculosis (180), cytomegalovirus (CMV), and vaccinia virus (174). The molecular mechanism governing DNA sensing by AIM2 during bacterial or viral infection is not clear. One difference between bacterial and viral engagement of the AIM2 inflammasome is that type I interferon signaling contributes to AIM2 inflammasome activation in response to bacterial pathogens, such as Francisella, but not in response to mouse CMV infection (173, 181). In addition, bacterial escape from the vacuole and bacteriolysis are required to engage the AIM2 inflammasome during Francisella infection (Fig. 5) (171, 176, 182). Since DNA is found in all bacterial pathogens and DNA viruses, another unresolved question is why only a small subset of DNA-containing pathogens activates the AIM2 inflammasome. Recent studies indicate that certain bacteria encode virulence factors to evade detection of the AIM2 inflammasome. F. tularensis subspecies novicida encodes a Clustered, regularly interspaced, short palindromic repeats-CRISPR associated (CRISPR-Cas) system, which is proposed to strengthen the integrity of the bacterial membrane to prevent excessive DNA release, thereby, minimizing the amount of ligands available for AIM2 in the cytoplasm (183). L. pneumophila encodes an effector protein SdhA, as part of the Dot/Icm type IVB secretion system, to prevent rupture of the Legionella-containing vacuole. This strategy prevents DNA release into the cytosol and circumvents detection by AIM2 (184).
Fig. 5. Activation of the AIM2 inflammasome.
AIM2 is activated by cytosolic bacteria, such as Francisella tularensis, and DNA viruses, such as cytomegalovirus. During Francisella infection, type I interferon provides a feedback loop and activates type I interferon receptor (IFNAR). The downstream signaling of IFNAR is unknown. Francisella escapes the vacuole and replicates in the cytosol. It has been proposed that DNA release by means of bacteriolysis or bacterial replication in the cytosol activates AIM2. The mechanism leading to viral DNA recognition by AIM2 is less clear. The HIN-200 domain of AIM2 directly binds dsDNA and the pyrin domain recruits ASC. Caspase-1 and caspase-8 are recruited to the ASC inflammasome, where caspase-1 mediates pro-IL-1β and pro-IL-18 processing and pyroptosis and caspase-8 induces apoptosis. The pyrin-containing human protein POP3 and the HIN-domains-containing mouse p202 protein interact with AIM2 to inhibit inflammasome activation.
A number of PYD-containing proteins have been found to inhibit the AIM2 inflammasome. In human, the PYD-containing protein, POP3, interacts with the PYD domain of AIM2 and competes with ASC to inhibit AIM2 inflammasome activation in response to poly(dA:dT), mouse CMV, and modified vaccinia virus Ankara (185). Similarly, mouse p202, a bipartite protein containing two HIN domains, inhibits the AIM2 inflammasome through a mechanism requiring homotypic interaction between one of the HIN domains of p202 and the HIN domain of AIM2 (168, 186). In addition to pyroptosis, AIM2 contributes to apoptotic cell death responses. During Francisella infection, caspase-8 is recruited to the AIM2 inflammasome, where it has been proposed to interact with the PYD domain of ASC (187, 188). In the absence of caspase-1, caspase-8 drives apoptotic cell death via an ASC-dependent mechanism in response to Francisella infection (188).
Dysregulated AIM2 expression in human cells is associated with a number of diseases. Increased AIM2 expression is associated with the development of psoriasis, abdominal aortic aneurysm, and systemic lupus erythematosus (189–191), whereas reduced AIM2 expression is linked to colorectal and prostate cancer (192, 193). Whether the development of these diseases is a result of aberrant inflammasome function is unclear. Nevertheless, it has been shown that DNA accumulated in keratinocytes activates the AIM2 inflammasome to drive the release of IL-1β in psoriatic lesions (189), providing evidence to suggest that AIM2 may respond to self-DNA released during damage of a cell to drive chronic inflammatory disease. Analysis of AIM2 expression in 414 colorectal tumors and matching control tissues revealed that 67% of the tumors displayed reduced AIM2 expression when compared to controls (194). Nearly 50% of patients whose tumor cells completely lacked AIM2 expression died from cancer within five years after diagnosis, whereas over 70% of patients whose tumor cells retained some AIM2 expression survived beyond five years after diagnosis (194). Why there is a link between AIM2 expression in tumor cells and patient survival is unknown. Nevertheless, therapeutic inhibition of dysregulated AIM2 expression or prevention of inappropriate DNA sensing by AIM2 (e.g. self DNA) could have a profound impact on disease progression.
Functional roles and regulation of ASC
NLRP3 and AIM2 contain a PYD and do not interact with caspase-1 directly. Instead, the PYD of these inflammasome receptors interacts with the PYD of ASC. The CARD domain of ASC then binds the CARD of caspase-1 via CARD-CARD interaction. Therefore, ASC is a central component for a number of inflammasomes. Two complementary reports provide elegant biochemical evidence to show that NLRP3 or AIM2 induces ASC polymerization in prion-like manner, culminating in the recruitment and activation of caspase-1 (170, 195). Visualization of overexpressed NLRP3 and AIM2 inflammasomes showed filamentous structures, but how these filaments assemble into an endogenous macromolecular speck visualized in primary cells is unknown. Interestingly, cells infected by pathogens that activate multiple NLRs (e.g. S. Typhimurium) still generate a single inflammasome speck per cell, with each speck arranged in a multi-layered ring-like structure (10). In a Salmonella-infected macrophage, the outer core of an endogenous speck is comprised of ASC, which surrounds an inner layer of NLRC4 and NLRP3. Active caspase-1 and caspase-8 are located in the core of the ASC inflammasome where pro-IL-1β molecules are found (10). The concentric arrangement of the endogenous ASC inflammasome suggests that it is a dynamic unit where different effectors such as caspase-1 and caspase-8 are recruited at different times depending on the stimuli countered by the cell during microbial infection.
Extracellular ASC specks have been detected in the serum of patients with CAPS and in the extracellular milieu of lungs from patients with chronic obstructive pulmonary disease and pneumonia (196, 197). It has been proposed pyroptosis mediates the release of ASC specks from macrophages (196, 197). Intriguingly, extracellular ASC specks recruit and activate pro-caspase-1 and cleave pro-IL-1β in the extracellular environment. In addition, they are phagocytosed by neighboring macrophages and serve as an endogenous danger signal to trigger a second wave of inflammasome activation (196, 197).
Given the remarkably diverse role of ASC in amplifying inflammasome-dependent responses, a number of regulatory mechanisms have been identified that target ASC to modulate inflammasome activities. Syk or Jnk induces phosphorylation of ASC, directly or indirectly at residue Tyr144, to enhance activation of NLRP3 and AIM2 but not the NLRC4 inflammasomes in macrophages (198). Linear ubiquitination of ASC in macrophages is also required for NLRP3 and AIM2 inflammasome activation (199), suggesting that ASC must be modified by different cellular processes prior to assembly. Activation of the AIM2 or NLRP3 inflammasomes results in polyubiquitination of ASC, which induces the recruitment of the autophagic marker, p62, to target the inflammasome for autophagic degradation (200). This activity indicates that resolution of the inflammatory responses also requires modification of ASC. These post-translational modifications provide multiple ways to license the activation and degradation of many ASC-dependent inflammasomes.
The relationship between other pattern-recognition receptors and caspase-1 activation
In addition to the well-established inflammasome receptors NLRP1, NLRP3, NLRC4, and AIM2, there are growing numbers of studies demonstrating that other pattern-recognition receptors can assemble the inflammasome complex. Latest findings indicate that human and mouse Pyrin have the capacity to assemble an inflammasome complex (201–203). The Pyrin inflammasome is induced by the cytotoxin TcdB from Clostridium difficile and other Rho-inactivating toxins from Vibrio parahaemolyticus, Histophilus somni and Clostridium botulinum. Although by distinct modes of modification, the actions of these toxins culminate in modification of a switch-I residue of Rho GTPases, probably indicating that the Pyrin inflammasome indirectly senses modifications of Rho GTPases (203).
A number of other innate immune receptors have multifaceted functions. Both NLRP6 and NLRP12 have been shown to activate caspase-1 or negatively regulate inflammation in a context-specific manner. NLRP6 is required for host protection against DSS-induced colitis and Citrobacter infection (204, 205). Susceptibility to DSS-induced intestinal inflammation in mice lacking NLRP6 is owing to their impaired ability to produce IL-18, suggesting a role for NLRP6 in orchestrating inflammasome responses (204). During Citrobacter infection, NLRP6, ASC and caspase-1/11 were all required for host protection in mice (205). However, mice lacking NLRP6 produced normal levels of IL-1β and IL-18 over the course of the infection (205), suggesting that it may well be possible that NLRP6 exerts an inflammasome-independent role during Citrobacter infection. Indeed, NLRP6 suppresses mitogen-activated protein kinase (MAPK) and the canonical NF-κB pathway in macrophages infected with L. monocytogenes, E. coli and S. Typhimurium (206).
NLRP12 activates caspase-1 activation in response to Yersinia and Plasmodium infection (207, 208). However, NLRP12 does not play a role in activating the inflammasome in response to other pathogens, including Salmonella, Klebsiella, Escherichia, Mycobacterium, and Listeria species (178, 209, 210). Instead, NLRP12 negatively regulates NF-κB-dependent inflammatory responses during Salmonella infection and colorectal cancer (209, 211, 212). Whether there is a molecular switch that determines the ability of NLRP6 and NLRP12 to activate caspase-1 or induce anti-inflammatory functions in response to a specific stimulus is unknown.
Human IFI16 orchestrates inflammasome-dependent and inflammasome-independent functions in response to virus infections. IFI16 is a member of the ALR family which predominantly resides in the nucleus and binds to dsDNA (169, 213). IFI16 initiates inflammasome activities in response to Kaposi sarcoma-associated herpesvirus (214), Epstein-Barr virus (215), and herpes simplex virus-1 (HSV-1) (216). Intriguingly, IFI16 also activates the STING and TBK1 signaling axis to drive IFN-β induction in cells infected with HSV-1 and human CMV (213, 217). Identification of the upstream molecular events that activate NLRP6, NLRP12 and IFI16 would elucidate the specific factors that determine the functional outcome of these receptors.
Conclusions and future perspectives
A little more than a decade since the discovery of the inflammasome, the research field has undergone rapid expansion. The clinical relevance of inflammasomes and their connection with IL-1 cytokines are now deeply ingrained in infectious diseases, cancer, autoinflammatory, metabolic and neurodegenerative diseases. As we enter a new era in inflammasome biology, research efforts will focus on understanding the regulation of established inflammasomes and identification of the functional roles of new inflammasome receptors. Of the 22 NLRs in human and 34 NLRs in mouse (4, 5), the functions of most of these are unknown. A major goal in the field is to determine how many receptors are capable of forming inflammasomes, what their respective ligands are, and importantly, what their functional roles are in health and disease. Furthermore, how will we be able to use the knowledge we have now and in the future to design and formulate new vaccines and immunotherapies that target different components of the inflammasome for use in the fight against diseases? A combined effort from basic scientists, clinicians and academic and industry leaders is required to tackle these emerging challenges.
Acknowledgments
S.M.M. is a recipient of the Neoma Boadway Endowed Fellowship funded by the St. Jude Children’s Research Hospital and the Australian National Health and Medical Research Council RG Menzies Early Career fellowship. Work from the lab is supported by grants from the National Institutes of Health (grants AR056296, CA163507 and AI101935) and the American Lebanese Syrian Associated Charities (to T.-D.K).
Footnotes
The authors have no conflicts of interest to declare.
References
- 1.Schroder K, Tschopp J. The inflammasomes. Cell. 2010;140:821–832. doi: 10.1016/j.cell.2010.01.040. [DOI] [PubMed] [Google Scholar]
- 2.Bertin J, et al. Human CARD4 protein is a novel CED-4/Apaf-1 cell death family member that activates NF-kappaB. The Journal of biological chemistry. 1999;274:12955–12958. doi: 10.1074/jbc.274.19.12955. [DOI] [PubMed] [Google Scholar]
- 3.Inohara N, et al. Nod1, an Apaf-1-like activator of caspase-9 and nuclear factor-kappaB. The Journal of biological chemistry. 1999;274:14560–14567. doi: 10.1074/jbc.274.21.14560. [DOI] [PubMed] [Google Scholar]
- 4.Harton JA, Linhoff MW, Zhang J, Ting JP. Cutting edge: CATERPILLER: a large family of mammalian genes containing CARD, pyrin, nucleotide-binding, and leucine-rich repeat domains. Journal of immunology. 2002;169:4088–4093. doi: 10.4049/jimmunol.169.8.4088. [DOI] [PubMed] [Google Scholar]
- 5.Cridland JA, et al. The mammalian PYHIN gene family: phylogeny, evolution and expression. BMC evolutionary biology. 2012;12:140. doi: 10.1186/1471-2148-12-140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Martinon F, Burns K, Tschopp J. The inflammasome: a molecular platform triggering activation of inflammatory caspases and processing of proIL-beta. Molecular cell. 2002;10:417–426. doi: 10.1016/s1097-2765(02)00599-3. [DOI] [PubMed] [Google Scholar]
- 7.Lamkanfi M, Dixit VM. Mechanisms and functions of inflammasomes. Cell. 2014;157:1013–1022. doi: 10.1016/j.cell.2014.04.007. [DOI] [PubMed] [Google Scholar]
- 8.Lupfer C, Kanneganti TD. Unsolved Mysteries in NLR Biology. Frontiers in immunology. 2013;4:285. doi: 10.3389/fimmu.2013.00285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fernandes-Alnemri T, et al. The pyroptosome: a supramolecular assembly of ASC dimers mediating inflammatory cell death via caspase-1 activation. Cell death and differentiation. 2007;14:1590–1604. doi: 10.1038/sj.cdd.4402194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Man SM, et al. Inflammasome activation causes dual recruitment of NLRC4 and NLRP3 to the same macromolecular complex. Proceedings of the National Academy of Sciences of the United States of America. 2014;111:7403–7408. doi: 10.1073/pnas.1402911111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Man SM, Tourlomousis P, Hopkins L, Monie TP, Fitzgerald KA, Bryant CE. Salmonella infection induces recruitment of Caspase-8 to the inflammasome to modulate IL-1beta production. Journal of immunology. 2013;191:5239–5246. doi: 10.4049/jimmunol.1301581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Broz P, Newton K, Lamkanfi M, Mariathasan S, Dixit VM, Monack DM. Redundant roles for inflammasome receptors NLRP3 and NLRC4 in host defense against Salmonella. The Journal of experimental medicine. 2010;207:1745–1755. doi: 10.1084/jem.20100257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Broz P, von Moltke J, Jones JW, Vance RE, Monack DM. Differential requirement for Caspase-1 autoproteolysis in pathogen-induced cell death and cytokine processing. Cell host & microbe. 2010;8:471–483. doi: 10.1016/j.chom.2010.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Huang MT, et al. Critical role of apoptotic speck protein containing a caspase recruitment domain (ASC) and NLRP3 in causing necrosis and ASC speck formation induced by Porphyromonas gingivalis in human cells. Journal of immunology. 2009;182:2395–2404. doi: 10.4049/jimmunol.0800909. [DOI] [PubMed] [Google Scholar]
- 15.Kayagaki N, et al. Non-canonical inflammasome activation targets caspase-11. Nature. 2011;479:117–121. doi: 10.1038/nature10558. [DOI] [PubMed] [Google Scholar]
- 16.Boyden ED, Dietrich WF. Nalp1b controls mouse macrophage susceptibility to anthrax lethal toxin. Nature genetics. 2006;38:240–244. doi: 10.1038/ng1724. [DOI] [PubMed] [Google Scholar]
- 17.Levinsohn JL, et al. Anthrax lethal factor cleavage of Nlrp1 is required for activation of the inflammasome. PLoS pathogens. 2012;8:e1002638. doi: 10.1371/journal.ppat.1002638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hellmich KA, et al. Anthrax lethal factor cleaves mouse nlrp1b in both toxin-sensitive and toxin-resistant macrophages. PloS one. 2012;7:e49741. doi: 10.1371/journal.pone.0049741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chavarria-Smith J, Vance RE. Direct proteolytic cleavage of NLRP1B is necessary and sufficient for inflammasome activation by anthrax lethal factor. PLoS pathogens. 2013;9:e1003452. doi: 10.1371/journal.ppat.1003452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.D’Osualdo A, Weichenberger CX, Wagner RN, Godzik A, Wooley J, Reed JC. CARD8 and NLRP1 undergo autoproteolytic processing through a ZU5-like domain. PloS one. 2011;6:e27396. doi: 10.1371/journal.pone.0027396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Frew BC, Joag VR, Mogridge J. Proteolytic processing of Nlrp1b is required for inflammasome activity. PLoS pathogens. 2012;8:e1002659. doi: 10.1371/journal.ppat.1002659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Finger JN, et al. Autolytic proteolysis within the function to find domain (FIIND) is required for NLRP1 inflammasome activity. The Journal of biological chemistry. 2012;287:25030–25037. doi: 10.1074/jbc.M112.378323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Van Opdenbosch N, Gurung P, Vande Walle L, Fossoul A, Kanneganti TD, Lamkanfi M. Activation of the NLRP1b inflammasome independently of ASC-mediated caspase-1 autoproteolysis and speck formation. Nature communications. 2014;5:3209. doi: 10.1038/ncomms4209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jin T, Curry J, Smith P, Jiang J, Xiao TS. Structure of the NLRP1 caspase recruitment domain suggests potential mechanisms for its association with procaspase-1. Proteins. 2013;81:1266–1270. doi: 10.1002/prot.24287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Guey B, Bodnar M, Manie SN, Tardivel A, Petrilli V. Caspase-1 autoproteolysis is differentially required for NLRP1b and NLRP3 inflammasome function. Proceedings of the National Academy of Sciences of the United States of America. 2014 doi: 10.1073/pnas.1415756111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Terra JK, et al. Cutting edge: resistance to Bacillus anthracis infection mediated by a lethal toxin sensitive allele of Nalp1b/Nlrp1b. Journal of immunology. 2010;184:17–20. doi: 10.4049/jimmunol.0903114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Moayeri M, et al. Inflammasome sensor Nlrp1b-dependent resistance to anthrax is mediated by caspase-1, IL-1 signaling and neutrophil recruitment. PLoS pathogens. 2010;6:e1001222. doi: 10.1371/journal.ppat.1001222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Masters SL, et al. NLRP1 inflammasome activation induces pyroptosis of hematopoietic progenitor cells. Immunity. 2012;37:1009–1023. doi: 10.1016/j.immuni.2012.08.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jin Y, et al. NALP1 in vitiligo-associated multiple autoimmune disease. The New England journal of medicine. 2007;356:1216–1225. doi: 10.1056/NEJMoa061592. [DOI] [PubMed] [Google Scholar]
- 30.Levandowski CB, et al. NLRP1 haplotypes associated with vitiligo and autoimmunity increase interleukin-1beta processing via the NLRP1 inflammasome. Proceedings of the National Academy of Sciences of the United States of America. 2013;110:2952–2956. doi: 10.1073/pnas.1222808110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mariathasan S, et al. Cryopyrin activates the inflammasome in response to toxins and ATP. Nature. 2006;440:228–232. doi: 10.1038/nature04515. [DOI] [PubMed] [Google Scholar]
- 32.Dostert C, Petrilli V, Van Bruggen R, Steele C, Mossman BT, Tschopp J. Innate immune activation through Nalp3 inflammasome sensing of asbestos and silica. Science. 2008;320:674–677. doi: 10.1126/science.1156995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hornung V, et al. Silica crystals and aluminum salts activate the NALP3 inflammasome through phagosomal destabilization. Nature immunology. 2008;9:847–856. doi: 10.1038/ni.1631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sander LE, et al. Detection of prokaryotic mRNA signifies microbial viability and promotes immunity. Nature. 2011;474:385–389. doi: 10.1038/nature10072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Martinon F, Petrilli V, Mayor A, Tardivel A, Tschopp J. Gout-associated uric acid crystals activate the NALP3 inflammasome. Nature. 2006;440:237–241. doi: 10.1038/nature04516. [DOI] [PubMed] [Google Scholar]
- 36.Kanneganti TD, et al. Bacterial RNA and small antiviral compounds activate caspase-1 through cryopyrin/Nalp3. Nature. 2006;440:233–236. doi: 10.1038/nature04517. [DOI] [PubMed] [Google Scholar]
- 37.Martinon F, Agostini L, Meylan E, Tschopp J. Identification of bacterial muramyl dipeptide as activator of the NALP3/cryopyrin inflammasome. Current biology : CB. 2004;14:1929–1934. doi: 10.1016/j.cub.2004.10.027. [DOI] [PubMed] [Google Scholar]
- 38.Marina-Garcia N, et al. Pannexin-1-mediated intracellular delivery of muramyl dipeptide induces caspase-1 activation via cryopyrin/NLRP3 independently of Nod2. Journal of immunology. 2008;180:4050–4057. doi: 10.4049/jimmunol.180.6.4050. [DOI] [PubMed] [Google Scholar]
- 39.Kailasan Vanaja S, et al. Bacterial RNA:DNA hybrids are activators of the NLRP3 inflammasome. Proceedings of the National Academy of Sciences of the United States of America. 2014;111:7765–7770. doi: 10.1073/pnas.1400075111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sha W, et al. Human NLRP3 inflammasome senses multiple types of bacterial RNAs. Proceedings of the National Academy of Sciences of the United States of America. 2014 doi: 10.1073/pnas.1412487111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Franchi L, et al. Cytosolic Double-Stranded RNA Activates the NLRP3 Inflammasome via MAVS-Induced Membrane Permeabilization and K+ Efflux. Journal of immunology. 2014;193:4214–4222. doi: 10.4049/jimmunol.1400582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Palm NW, Medzhitov R. Role of the inflammasome in defense against venoms. Proceedings of the National Academy of Sciences of the United States of America. 2013;110:1809–1814. doi: 10.1073/pnas.1221476110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Franchi L, Muñoz-Planillo R, Núñez G. Sensing and reacting to microbes through the inflammasomes. Nature immunology. 2012;13:325–332. doi: 10.1038/ni.2231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lupfer C, Kanneganti TD. The expanding role of NLRs in antiviral immunity. Immunological reviews. 2013;255:13–24. doi: 10.1111/imr.12089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kanneganti TD. Central roles of NLRs and inflammasomes in viral infection. Nature reviews Immunology. 2010;10:688–698. doi: 10.1038/nri2851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Thomas PG, et al. The intracellular sensor NLRP3 mediates key innate and healing responses to influenza A virus via the regulation of caspase-1. Immunity. 2009;30:566–575. doi: 10.1016/j.immuni.2009.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bauernfeind FG, et al. Cutting edge: NF-kappaB activating pattern recognition and cytokine receptors license NLRP3 inflammasome activation by regulating NLRP3 expression. Journal of immunology. 2009;183:787–791. doi: 10.4049/jimmunol.0901363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Netea MG, et al. Differential requirement for the activation of the inflammasome for processing and release of IL-1beta in monocytes and macrophages. Blood. 2009;113:2324–2335. doi: 10.1182/blood-2008-03-146720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhou R, Yazdi AS, Menu P, Tschopp J. A role for mitochondria in NLRP3 inflammasome activation. Nature. 2011;469:221–225. doi: 10.1038/nature09663. [DOI] [PubMed] [Google Scholar]
- 50.Shimada K, et al. Oxidized Mitochondrial DNA Activates the NLRP3 Inflammasome during Apoptosis. Immunity. 2012;36:401–414. doi: 10.1016/j.immuni.2012.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Iyer SS, et al. Mitochondrial cardiolipin is required for Nlrp3 inflammasome activation. Immunity. 2013;39:311–323. doi: 10.1016/j.immuni.2013.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Perregaux D, Gabel CA. Interleukin-1 beta maturation and release in response to ATP and nigericin. Evidence that potassium depletion mediated by these agents is a necessary and common feature of their activity. The Journal of biological chemistry. 1994;269:15195–15203. [PubMed] [Google Scholar]
- 53.Munoz-Planillo R, Kuffa P, Martinez-Colon G, Smith BL, Rajendiran TM, Nunez G. K(+) efflux is the common trigger of NLRP3 inflammasome activation by bacterial toxins and particulate matter. Immunity. 2013;38:1142–1153. doi: 10.1016/j.immuni.2013.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Petrilli V, Papin S, Dostert C, Mayor A, Martinon F, Tschopp J. Activation of the NALP3 inflammasome is triggered by low intracellular potassium concentration. Cell death and differentiation. 2007;14:1583–1589. doi: 10.1038/sj.cdd.4402195. [DOI] [PubMed] [Google Scholar]
- 55.Murakami T, et al. Critical role for calcium mobilization in activation of the NLRP3 inflammasome. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:11282–11287. doi: 10.1073/pnas.1117765109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lee GS, et al. The calcium-sensing receptor regulates the NLRP3 inflammasome through Ca2+ and cAMP. Nature. 2012;492:123–127. doi: 10.1038/nature11588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Qu Y, et al. Pannexin-1 is required for ATP release during apoptosis but not for inflammasome activation. Journal of immunology. 2011;186:6553–6561. doi: 10.4049/jimmunol.1100478. [DOI] [PubMed] [Google Scholar]
- 58.Gurcel L, Abrami L, Girardin S, Tschopp J, van der Goot FG. Caspase-1 activation of lipid metabolic pathways in response to bacterial pore-forming toxins promotes cell survival. Cell. 2006;126:1135–1145. doi: 10.1016/j.cell.2006.07.033. [DOI] [PubMed] [Google Scholar]
- 59.Harder J, Franchi L, Munoz-Planillo R, Park JH, Reimer T, Nunez G. Activation of the Nlrp3 inflammasome by Streptococcus pyogenes requires streptolysin O and NF-kappa B activation but proceeds independently of TLR signaling and P2X7 receptor. Journal of immunology. 2009;183:5823–5829. doi: 10.4049/jimmunol.0900444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Compan V, et al. Cell volume regulation modulates NLRP3 inflammasome activation. Immunity. 2012;37:487–500. doi: 10.1016/j.immuni.2012.06.013. [DOI] [PubMed] [Google Scholar]
- 61.Allam R, et al. Mitochondrial apoptosis is dispensable for NLRP3 inflammasome activation but non-apoptotic caspase-8 is required for inflammasome priming. EMBO reports. 2014;15:982–990. doi: 10.15252/embr.201438463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Bauernfeind F, Bartok E, Rieger A, Franchi L, Nunez G, Hornung V. Cutting edge: reactive oxygen species inhibitors block priming, but not activation, of the NLRP3 inflammasome. Journal of immunology. 2011;187:613–617. doi: 10.4049/jimmunol.1100613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Cain K, Langlais C, Sun XM, Brown DG, Cohen GM. Physiological concentrations of K+ inhibit cytochrome c-dependent formation of the apoptosome. The Journal of biological chemistry. 2001;276:41985–41990. doi: 10.1074/jbc.M107419200. [DOI] [PubMed] [Google Scholar]
- 64.Gurung P, et al. FADD and caspase-8 mediate priming and activation of the canonical and noncanonical Nlrp3 inflammasomes. Journal of immunology. 2014;192:1835–1846. doi: 10.4049/jimmunol.1302839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Vince JE, et al. Inhibitor of apoptosis proteins limit RIP3 kinase-dependent interleukin-1 activation. Immunity. 2012;36:215–227. doi: 10.1016/j.immuni.2012.01.012. [DOI] [PubMed] [Google Scholar]
- 66.Ganesan S, et al. Caspase-8 modulates dectin-1 and complement receptor 3-driven IL-1beta production in response to beta-glucans and the fungal pathogen, Candida albicans. Journal of immunology. 2014;193:2519–2530. doi: 10.4049/jimmunol.1400276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kang TB, Yang SH, Toth B, Kovalenko A, Wallach D. Caspase-8 Blocks Kinase RIPK3-Mediated Activation of the NLRP3 Inflammasome. Immunity. doi: 10.1016/j.immuni.2012.1009.1015. [DOI] [PubMed] [Google Scholar]
- 68.Kang TB, Yang SH, Toth B, Kovalenko A, Wallach D. Caspase-8 blocks kinase RIPK3-mediated activation of the NLRP3 inflammasome. Immunity. 2013;38:27–40. doi: 10.1016/j.immuni.2012.09.015. [DOI] [PubMed] [Google Scholar]
- 69.Su H, et al. Requirement for caspase-8 in NF-kappaB activation by antigen receptor. Science. 2005;307:1465–1468. doi: 10.1126/science.1104765. [DOI] [PubMed] [Google Scholar]
- 70.Lemmers B, et al. Essential role for caspase-8 in Toll-like receptors and NFkappaB signaling. The Journal of biological chemistry. 2007;282:7416–7423. doi: 10.1074/jbc.M606721200. [DOI] [PubMed] [Google Scholar]
- 71.Weng D, et al. Caspase-8 and RIP kinases regulate bacteria-induced innate immune responses and cell death. Proceedings of the National Academy of Sciences of the United States of America. 2014;111:7391–7396. doi: 10.1073/pnas.1403477111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Philip NH, et al. Caspase-8 mediates caspase-1 processing and innate immune defense in response to bacterial blockade of NF-kappaB and MAPK signaling. Proceedings of the National Academy of Sciences of the United States of America. 2014;111:7385–7390. doi: 10.1073/pnas.1403252111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Wang X, et al. RNA viruses promote activation of the NLRP3 inflammasome through a RIP1-RIP3-DRP1 signaling pathway. Nature immunology. 2014 doi: 10.1038/ni.3015. [DOI] [PubMed] [Google Scholar]
- 74.Lupfer C, et al. Receptor interacting protein kinase 2-mediated mitophagy regulates inflammasome activation during virus infection. Nature immunology. 2013;14:480–488. doi: 10.1038/ni.2563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Lin KM, et al. IRAK-1 bypasses priming and directly links TLRs to rapid NLRP3 inflammasome activation. Proceedings of the National Academy of Sciences of the United States of America. 2014;111:775–780. doi: 10.1073/pnas.1320294111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Fernandes-Alnemri T, Kang S, Anderson C, Sagara J, Fitzgerald KA, Alnemri ES. Cutting edge: TLR signaling licenses IRAK1 for rapid activation of the NLRP3 inflammasome. Journal of immunology. 2013;191:3995–3999. doi: 10.4049/jimmunol.1301681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Juliana C, Fernandes-Alnemri T, Kang S, Farias A, Qin F, Alnemri ES. Non-transcriptional priming and deubiquitination regulate NLRP3 inflammasome activation. The Journal of biological chemistry. 2012;287:36617–36622. doi: 10.1074/jbc.M112.407130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Py BF, Kim MS, Vakifahmetoglu-Norberg H, Yuan J. Deubiquitination of NLRP3 by BRCC3 critically regulates inflammasome activity. Molecular cell. 2013;49:331–338. doi: 10.1016/j.molcel.2012.11.009. [DOI] [PubMed] [Google Scholar]
- 79.Kim YM, Talanian RV, Li J, Billiar TR. Nitric oxide prevents IL-1beta and IFN-gamma-inducing factor (IL-18) release from macrophages by inhibiting caspase-1 (IL-1beta-converting enzyme) Journal of immunology. 1998;161:4122–4128. [PubMed] [Google Scholar]
- 80.Mishra BB, et al. Nitric oxide controls the immunopathology of tuberculosis by inhibiting NLRP3 inflammasome-dependent processing of IL-1beta. Nature immunology. 2013;14:52–60. doi: 10.1038/ni.2474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Hernandez-Cuellar E, et al. Cutting edge: nitric oxide inhibits the NLRP3 inflammasome. Journal of immunology. 2012;189:5113–5117. doi: 10.4049/jimmunol.1202479. [DOI] [PubMed] [Google Scholar]
- 82.Hoffman HM, Mueller JL, Broide DH, Wanderer AA, Kolodner RD. Mutation of a new gene encoding a putative pyrin-like protein causes familial cold autoinflammatory syndrome and Muckle-Wells syndrome. Nature genetics. 2001;29:301–305. doi: 10.1038/ng756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Feldmann J, et al. Chronic infantile neurological cutaneous and articular syndrome is caused by mutations in CIAS1, a gene highly expressed in polymorphonuclear cells and chondrocytes. Am J Hum Genet. 2002;71:198–203. doi: 10.1086/341357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Neven B, et al. Molecular basis of the spectral expression of CIAS1 mutations associated with phagocytic cell-mediated autoinflammatory disorders CINCA/NOMID, MWS, and FCU. Blood. 2004;103:2809–2815. doi: 10.1182/blood-2003-07-2531. [DOI] [PubMed] [Google Scholar]
- 85.Agostini L, Martinon F, Burns K, McDermott MF, Hawkins PN, Tschopp J. NALP3 forms an IL-1beta-processing inflammasome with increased activity in Muckle-Wells autoinflammatory disorder. Immunity. 2004;20:319–325. doi: 10.1016/s1074-7613(04)00046-9. [DOI] [PubMed] [Google Scholar]
- 86.Aksentijevich I, et al. De novo CIAS1 mutations, cytokine activation, and evidence for genetic heterogeneity in patients with neonatal-onset multisystem inflammatory disease (NOMID): a new member of the expanding family of pyrin-associated autoinflammatory diseases. Arthritis Rheum. 2002;46:3340–3348. doi: 10.1002/art.10688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Brydges SD, et al. Inflammasome-mediated disease animal models reveal roles for innate but not adaptive immunity. Immunity. 2009;30:875–887. doi: 10.1016/j.immuni.2009.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Brydges SD, Broderick L, McGeough MD, Pena CA, Mueller JL, Hoffman HM. Divergence of IL-1, IL-18, and cell death in NLRP3 inflammasomopathies. The Journal of clinical investigation. 2013;123:4695–4705. doi: 10.1172/JCI71543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Wen H, Ting JP, O’Neill LA. A role for the NLRP3 inflammasome in metabolic diseases--did Warburg miss inflammation? Nature immunology. 2012;13:352–357. doi: 10.1038/ni.2228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Man SM, Kaakoush NO, Mitchell HM. The role of bacteria and pattern-recognition receptors in Crohn’s disease. Nat Rev Gastroenterol Hepatol. 2011;8:152–168. doi: 10.1038/nrgastro.2011.3. [DOI] [PubMed] [Google Scholar]
- 91.Wen H, et al. Fatty acid-induced NLRP3-ASC inflammasome activation interferes with insulin signaling. Nature immunology. 2011;12:408–415. doi: 10.1038/ni.2022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Masters SL, et al. Activation of the NLRP3 inflammasome by islet amyloid polypeptide provides a mechanism for enhanced IL-1beta in type 2 diabetes. Nature immunology. 2010;11:897–904. doi: 10.1038/ni.1935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Vande Walle L, et al. Negative regulation of the NLRP3 inflammasome by A20 protects against arthritis. Nature. 2014;512:69–73. doi: 10.1038/nature13322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Lukens JR, et al. Dietary modulation of the microbiome affects autoinflammatory disease. Nature. 2014 doi: 10.1038/nature13788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Heneka MT, et al. NLRP3 is activated in Alzheimer’s disease and contributes to pathology in APP/PS1 mice. Nature. 2012;493:674–678. doi: 10.1038/nature11729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Duewell P, et al. NLRP3 inflammasomes are required for atherogenesis and activated by cholesterol crystals. Nature. 2010;464:1357–1361. doi: 10.1038/nature08938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Henao-Mejia J, et al. Inflammasome-mediated dysbiosis regulates progression of NAFLD and obesity. Nature. 2012;482:179–185. doi: 10.1038/nature10809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Huber S, et al. IL-22BP is regulated by the inflammasome and modulates tumorigenesis in the intestine. Nature. 2012;491:259–263. doi: 10.1038/nature11535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Zaki MH, Vogel P, Body-Malapel M, Lamkanfi M, Kanneganti TD. IL-18 production downstream of the Nlrp3 inflammasome confers protection against colorectal tumor formation. Journal of immunology. 2010;185:4912–4920. doi: 10.4049/jimmunol.1002046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Allen IC, et al. The NLRP3 inflammasome functions as a negative regulator of tumorigenesis during colitis-associated cancer. The Journal of experimental medicine. 2010;207:1045–1056. doi: 10.1084/jem.20100050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Youm YH, et al. Canonical Nlrp3 inflammasome links systemic low-grade inflammation to functional decline in aging. Cell metabolism. 2013;18:519–532. doi: 10.1016/j.cmet.2013.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Lukens JR, et al. Critical role for inflammasome-independent IL-1beta production in osteomyelitis. Proceedings of the National Academy of Sciences of the United States of America. 2014;111:1066–1071. doi: 10.1073/pnas.1318688111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Cassel SL, et al. Inflammasome-independent IL-1beta mediates autoinflammatory disease in Pstpip2-deficient mice. Proceedings of the National Academy of Sciences of the United States of America. 2014;111:1072–1077. doi: 10.1073/pnas.1318685111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Rathinam VA, et al. TRIF Licenses Caspase-11-Dependent NLRP3 Inflammasome Activation by Gram-Negative Bacteria. Cell. 2012;150:606–619. doi: 10.1016/j.cell.2012.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Broz P, Ohlson MB, Monack DM. Innate immune response to Salmonella typhimurium, a model enteric pathogen. Gut Microbes. 2012;3:62–70. doi: 10.4161/gmic.19141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Gurung P, et al. Toll or Interleukin-1 Receptor (TIR) Domain-containing Adaptor Inducing Interferon-beta (TRIF)-mediated Caspase-11 Protease Production Integrates Toll-like Receptor 4 (TLR4) Protein- and Nlrp3 Inflammasome-mediated Host Defense against Enteropathogens. The Journal of biological chemistry. 2012;287:34474–34483. doi: 10.1074/jbc.M112.401406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Broz P, Monack DM. Noncanonical inflammasomes: caspase-11 activation and effector mechanisms. PLoS pathogens. 2013;9:e1003144. doi: 10.1371/journal.ppat.1003144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Lupfer CR, et al. Reactive Oxygen Species Regulate Caspase-11 Expression and Activation of the Non-canonical NLRP3 Inflammasome during Enteric Pathogen Infection. PLoS pathogens. 2014;10:e1004410. doi: 10.1371/journal.ppat.1004410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Casson CN, et al. Caspase-11 activation in response to bacterial secretion systems that access the host cytosol. PLoS pathogens. 2013;9:e1003400. doi: 10.1371/journal.ppat.1003400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Kayagaki N, et al. Noncanonical inflammasome activation by intracellular LPS independent of TLR4. Science. 2013;341:1246–1249. doi: 10.1126/science.1240248. [DOI] [PubMed] [Google Scholar]
- 111.Hagar JA, Powell DA, Aachoui Y, Ernst RK, Miao EA. Cytoplasmic LPS activates caspase-11: implications in TLR4-independent endotoxic shock. Science. 2013;341:1250–1253. doi: 10.1126/science.1240988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Shi J, et al. Inflammatory caspases are innate immune receptors for intracellular LPS. Nature. 2014 doi: 10.1038/nature13683. [DOI] [PubMed] [Google Scholar]
- 113.Kajiwara Y, et al. A critical role for human caspase-4 in endotoxin sensitivity. Journal of immunology. 2014;193:335–343. doi: 10.4049/jimmunol.1303424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Wang S, Miura M, Jung YK, Zhu H, Li E, Yuan J. Murine caspase-11, an ICE-interacting protease, is essential for the activation of ICE. Cell. 1998;92:501–509. doi: 10.1016/s0092-8674(00)80943-5. [DOI] [PubMed] [Google Scholar]
- 115.Demon D, Kuchmiy A, Fossoul A, Zhu Q, Kanneganti TD, Lamkanfi M. Caspase-11 is expressed in the colonic mucosa and protects against dextran sodium sulfate-induced colitis. Mucosal immunology. 2014;7:1480–1491. doi: 10.1038/mi.2014.36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Williams TM, et al. Caspase-11 Attenuates Gastrointestinal Inflammation and Experimental Colitis Pathogenesis. American journal of physiology Gastrointestinal and liver physiology. 2014 doi: 10.1152/ajpgi.00234.2014. ajpgi 00234 02014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Oficjalska K, et al. Protective Role for Caspase-11 during Acute Experimental Murine Colitis. Journal of immunology. 2014 doi: 10.4049/jimmunol.1400501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Aachoui Y, et al. Caspase-11 Protects Against Bacteria That Escape the Vacuole. Science. 2013 doi: 10.1126/science.1230751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Meunier E, et al. Caspase-11 activation requires lysis of pathogen-containing vacuoles by IFN-induced GTPases. Nature. 2014;509:366–370. doi: 10.1038/nature13157. [DOI] [PubMed] [Google Scholar]
- 120.Knodler LA, et al. Noncanonical Inflammasome Activation of Caspase-4/Caspase-11 Mediates Epithelial Defenses against Enteric Bacterial Pathogens. Cell host & microbe. 2014;16:249–256. doi: 10.1016/j.chom.2014.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Pilla DM, et al. Guanylate binding proteins promote caspase-11-dependent pyroptosis in response to cytoplasmic LPS. Proceedings of the National Academy of Sciences of the United States of America. 2014;111:6046–6051. doi: 10.1073/pnas.1321700111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Akhter A, et al. Caspase-11 promotes the fusion of phagosomes harboring pathogenic bacteria with lysosomes by modulating actin polymerization. Immunity. 2012;37:35–47. doi: 10.1016/j.immuni.2012.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Shenoy AR, et al. GBP5 promotes NLRP3 inflammasome assembly and immunity in mammals. Science. 2012;336:481–485. doi: 10.1126/science.1217141. [DOI] [PubMed] [Google Scholar]
- 124.Poyet JL, Srinivasula SM, Tnani M, Razmara M, Fernandes-Alnemri T, Alnemri ES. Identification of Ipaf, a human caspase-1-activating protein related to Apaf-1. The Journal of biological chemistry. 2001;276:28309–28313. doi: 10.1074/jbc.C100250200. [DOI] [PubMed] [Google Scholar]
- 125.Franchi L, et al. Cytosolic flagellin requires Ipaf for activation of caspase-1 and interleukin 1beta in salmonella-infected macrophages. Nature immunology. 2006;7:576–582. doi: 10.1038/ni1346. [DOI] [PubMed] [Google Scholar]
- 126.Miao EA, et al. Cytoplasmic flagellin activates caspase-1 and secretion of interleukin 1beta via Ipaf. Nature immunology. 2006;7:569–575. doi: 10.1038/ni1344. [DOI] [PubMed] [Google Scholar]
- 127.Miao EA, et al. Innate immune detection of the type III secretion apparatus through the NLRC4 inflammasome. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:3076–3080. doi: 10.1073/pnas.0913087107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Endrizzi MG, Hadinoto V, Growney JD, Miller W, Dietrich WF. Genomic sequence analysis of the mouse Naip gene array. Genome research. 2000;10:1095–1102. doi: 10.1101/gr.10.8.1095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Kofoed EM, Vance RE. Innate immune recognition of bacterial ligands by NAIPs determines inflammasome specificity. Nature. 2011;477:592–595. doi: 10.1038/nature10394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Zhao Y, et al. The NLRC4 inflammasome receptors for bacterial flagellin and type III secretion apparatus. Nature. 2011;477:596–600. doi: 10.1038/nature10510. [DOI] [PubMed] [Google Scholar]
- 131.Yang J, Zhao Y, Shi J, Shao F. Human NAIP and mouse NAIP1 recognize bacterial type III secretion needle protein for inflammasome activation. Proceedings of the National Academy of Sciences of the United States of America. 2013;110:14408–14413. doi: 10.1073/pnas.1306376110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Rayamajhi M, Zak DE, Chavarria-Smith J, Vance RE, Miao EA. Cutting Edge: Mouse NAIP1 Detects the Type III Secretion System Needle Protein. Journal of immunology. 2013;191:3986–3989. doi: 10.4049/jimmunol.1301549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Tenthorey JL, Kofoed EM, Daugherty MD, Malik HS, Vance RE. Molecular basis for specific recognition of bacterial ligands by NAIP/NLRC4 inflammasomes. Molecular cell. 2014;54:17–29. doi: 10.1016/j.molcel.2014.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Miao EA, et al. Caspase-1-induced pyroptosis is an innate immune effector mechanism against intracellular bacteria. Nature immunology. 2010;11:1136–1142. doi: 10.1038/ni.1960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Franchi L, et al. NLRC4-driven production of IL-1beta discriminates between pathogenic and commensal bacteria and promotes host intestinal defense. Nature immunology. 2012;13:449–456. doi: 10.1038/ni.2263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Kupz A, et al. NLRC4 inflammasomes in dendritic cells regulate noncognate effector function by memory CD8(+) T cells. Nature immunology. 2012;13:162–169. doi: 10.1038/ni.2195. [DOI] [PubMed] [Google Scholar]
- 137.Miao EA, Ernst RK, Dors M, Mao DP, Aderem A. Pseudomonas aeruginosa activates caspase 1 through Ipaf. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:2562–2567. doi: 10.1073/pnas.0712183105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Sutterwala FS, Mijares LA, Li L, Ogura Y, Kazmierczak BI, Flavell RA. Immune recognition of Pseudomonas aeruginosa mediated by the IPAF/NLRC4 inflammasome. The Journal of experimental medicine. 2007;204:3235–3245. doi: 10.1084/jem.20071239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Suzuki S, et al. Shigella type III secretion protein MxiI is recognized by Naip2 to induce Nlrc4 inflammasome activation independently of Pkcdelta. PLoS pathogens. 2014;10:e1003926. doi: 10.1371/journal.ppat.1003926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Suzuki S, et al. Shigella IpaH7.8 E3 ubiquitin ligase targets glomulin and activates inflammasomes to demolish macrophages. Proceedings of the National Academy of Sciences of the United States of America. 2014;111:E4254–4263. doi: 10.1073/pnas.1324021111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Liu Z, et al. Role of inflammasomes in host defense against Citrobacter rodentium infection. The Journal of biological chemistry. 2012;287:16955–16964. doi: 10.1074/jbc.M112.358705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Nordlander S, Pott J, Maloy KJ. NLRC4 expression in intestinal epithelial cells mediates protection against an enteric pathogen. Mucosal immunology. 2014;7:775–785. doi: 10.1038/mi.2013.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Amer A, et al. Regulation of Legionella phagosome maturation and infection through flagellin and host Ipaf. The Journal of biological chemistry. 2006;281:35217–35223. doi: 10.1074/jbc.M604933200. [DOI] [PubMed] [Google Scholar]
- 144.Lightfield KL, et al. Critical function for Naip5 in inflammasome activation by a conserved carboxy-terminal domain of flagellin. Nature immunology. 2008;9:1171–1178. doi: 10.1038/ni.1646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Wu J, Fernandes-Alnemri T, Alnemri ES. Involvement of the AIM2, NLRC4, and NLRP3 inflammasomes in caspase-1 activation by Listeria monocytogenes. J Clin Immunol. 2010;30:693–702. doi: 10.1007/s10875-010-9425-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Cai S, Batra S, Wakamatsu N, Pacher P, Jeyaseelan S. NLRC4 inflammasome-mediated production of IL-1beta modulates mucosal immunity in the lung against gram-negative bacterial infection. Journal of immunology. 2012;188:5623–5635. doi: 10.4049/jimmunol.1200195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Brodsky IE, et al. A Yersinia effector protein promotes virulence by preventing inflammasome recognition of the type III secretion system. Cell host & microbe. 2010;7:376–387. doi: 10.1016/j.chom.2010.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Tomalka J, et al. A novel role for the NLRC4 inflammasome in mucosal defenses against the fungal pathogen Candida albicans. PLoS pathogens. 2011;7:e1002379. doi: 10.1371/journal.ppat.1002379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Cohen TS, Prince AS. Activation of inflammasome signaling mediates pathology of acute P. aeruginosa pneumonia. The Journal of clinical investigation. 2013;123:1630–1637. doi: 10.1172/JCI66142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Faure E, et al. Pseudomonas aeruginosa type-3 secretion system dampens host defense by exploiting the NLRC4-coupled inflammasome. American journal of respiratory and critical care medicine. 2014;189:799–811. doi: 10.1164/rccm.201307-1358OC. [DOI] [PubMed] [Google Scholar]
- 151.Mariathasan S, et al. Differential activation of the inflammasome by caspase-1 adaptors ASC and Ipaf. Nature. 2004;430:213–218. doi: 10.1038/nature02664. [DOI] [PubMed] [Google Scholar]
- 152.Akhter A, et al. Caspase-7 activation by the Nlrc4/Ipaf inflammasome restricts Legionella pneumophila infection. PLoS pathogens. 2009;5:e1000361. doi: 10.1371/journal.ppat.1000361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Qu Y, et al. Phosphorylation of NLRC4 is critical for inflammasome activation. Nature. 2012 doi: 10.1038/nature11429. [DOI] [PubMed] [Google Scholar]
- 154.Man SM, et al. Actin polymerization as a key innate immune effector mechanism to control Salmonella infection. Proceedings of the National Academy of Sciences of the United States of America. 2014 doi: 10.1073/pnas.1419925111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Chen KW, et al. The neutrophil NLRC4 inflammasome selectively promotes IL-1beta maturation without pyroptosis during acute Salmonella challenge. Cell reports. 2014;8:570–582. doi: 10.1016/j.celrep.2014.06.028. [DOI] [PubMed] [Google Scholar]
- 156.Perez-Lopez A, Rosales-Reyes R, Alpuche-Aranda CM, Ortiz-Navarrete V. Salmonella Downregulates Nod-like Receptor Family CARD Domain Containing Protein 4 Expression To Promote Its Survival in B Cells by Preventing Inflammasome Activation and Cell Death. Journal of immunology. 2013;190:1201–1209. doi: 10.4049/jimmunol.1200415. [DOI] [PubMed] [Google Scholar]
- 157.Sellin ME, et al. Epithelium-Intrinsic NAIP/NLRC4 Inflammasome Drives Infected Enterocyte Expulsion to Restrict Salmonella Replication in the Intestinal Mucosa. Cell host & microbe. 2014;16:237–248. doi: 10.1016/j.chom.2014.07.001. [DOI] [PubMed] [Google Scholar]
- 158.Hu B, et al. Inflammation-induced tumorigenesis in the colon is regulated by caspase-1 and NLRC4. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:21635–21640. doi: 10.1073/pnas.1016814108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Ayres JS, Trinidad NJ, Vance RE. Lethal inflammasome activation by a multidrug-resistant pathobiont upon antibiotic disruption of the microbiota. Nature medicine. 2012;18:799–806. doi: 10.1038/nm.2729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.von Moltke J, et al. Rapid induction of inflammatory lipid mediators by the inflammasome in vivo. Nature. 2012;490:107–111. doi: 10.1038/nature11351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Hu Z, et al. Crystal structure of NLRC4 reveals its autoinhibition mechanism. Science. 2013;341:172–175. doi: 10.1126/science.1236381. [DOI] [PubMed] [Google Scholar]
- 162.Romberg N, et al. Mutation of NLRC4 causes a syndrome of enterocolitis and autoinflammation. Nature genetics. 2014;46:1135–1139. doi: 10.1038/ng.3066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Canna SW, et al. An activating NLRC4 inflammasome mutation causes autoinflammation with recurrent macrophage activation syndrome. Nature genetics. 2014;46:1140–1146. doi: 10.1038/ng.3089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Kitamura A, Sasaki Y, Abe T, Kano H, Yasutomo K. An inherited mutation in NLRC4 causes autoinflammation in human and mice. The Journal of experimental medicine. 2014 doi: 10.1084/jem.20141091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Hornung V, et al. AIM2 recognizes cytosolic dsDNA and forms a caspase-1-activating inflammasome with ASC. Nature. 2009;458:514–518. doi: 10.1038/nature07725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Fernandes-Alnemri T, Yu JW, Datta P, Wu J, Alnemri ES. AIM2 activates the inflammasome and cell death in response to cytoplasmic DNA. Nature. 2009;458:509–513. doi: 10.1038/nature07710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Burckstummer T, et al. An orthogonal proteomic-genomic screen identifies AIM2 as a cytoplasmic DNA sensor for the inflammasome. Nature immunology. 2009;10:266–272. doi: 10.1038/ni.1702. [DOI] [PubMed] [Google Scholar]
- 168.Roberts TL, et al. HIN-200 proteins regulate caspase activation in response to foreign cytoplasmic DNA. Science. 2009;323:1057–1060. doi: 10.1126/science.1169841. [DOI] [PubMed] [Google Scholar]
- 169.Jin T, et al. Structures of the HIN domain:DNA complexes reveal ligand binding and activation mechanisms of the AIM2 inflammasome and IFI16 receptor. Immunity. 2012;36:561–571. doi: 10.1016/j.immuni.2012.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Lu A, et al. Unified Polymerization Mechanism for the Assembly of ASC-Dependent Inflammasomes. Cell. 2014;156:1193–1206. doi: 10.1016/j.cell.2014.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Belhocine K, Monack DM. Francisella infection triggers activation of the AIM2 inflammasome in murine dendritic cells. Cellular microbiology. 2012;14:71–80. doi: 10.1111/j.1462-5822.2011.01700.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Jones JW, et al. Absent in melanoma 2 is required for innate immune recognition of Francisella tularensis. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:9771–9776. doi: 10.1073/pnas.1003738107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Fernandes-Alnemri T, et al. The AIM2 inflammasome is critical for innate immunity to Francisella tularensis. Nature immunology. 2010;11:385–393. doi: 10.1038/ni.1859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Rathinam VA, et al. The AIM2 inflammasome is essential for host defense against cytosolic bacteria and DNA viruses. Nature immunology. 2010;11:395–402. doi: 10.1038/ni.1864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Kim S, et al. Listeria monocytogenes is sensed by the NLRP3 and AIM2 inflammasome. Eur J Immunol. 2010;40:1545–1551. doi: 10.1002/eji.201040425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Sauer JD, Witte CE, Zemansky J, Hanson B, Lauer P, Portnoy DA. Listeria monocytogenes triggers AIM2-mediated pyroptosis upon infrequent bacteriolysis in the macrophage cytosol. Cell host & microbe. 2010;7:412–419. doi: 10.1016/j.chom.2010.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Warren SE, et al. Cutting edge: Cytosolic bacterial DNA activates the inflammasome via Aim2. Journal of immunology. 2010;185:818–821. doi: 10.4049/jimmunol.1000724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Tsuchiya K, et al. Involvement of absent in melanoma 2 in inflammasome activation in macrophages infected with Listeria monocytogenes. Journal of immunology. 2010;185:1186–1195. doi: 10.4049/jimmunol.1001058. [DOI] [PubMed] [Google Scholar]
- 179.Fang R, et al. Type I interferon signaling regulates activation of the absent in melanoma 2 inflammasome during Streptococcus pneumoniae infection. Infection and immunity. 2014;82:2310–2317. doi: 10.1128/IAI.01572-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Saiga H, et al. Critical role of AIM2 in Mycobacterium tuberculosis infection. Int Immunol. 2012;24:637–644. doi: 10.1093/intimm/dxs062. [DOI] [PubMed] [Google Scholar]
- 181.Henry T, Brotcke A, Weiss DS, Thompson LJ, Monack DM. Type I interferon signaling is required for activation of the inflammasome during Francisella infection. The Journal of experimental medicine. 2007;204:987–994. doi: 10.1084/jem.20062665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Mariathasan S, Weiss DS, Dixit VM, Monack DM. Innate immunity against Francisella tularensis is dependent on the ASC/caspase-1 axis. The Journal of experimental medicine. 2005;202:1043–1049. doi: 10.1084/jem.20050977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Sampson TR, et al. A CRISPR-Cas system enhances envelope integrity mediating antibiotic resistance and inflammasome evasion. Proceedings of the National Academy of Sciences of the United States of America. 2014;111:11163–11168. doi: 10.1073/pnas.1323025111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Ge J, Gong YN, Xu Y, Shao F. Preventing bacterial DNA release and absent in melanoma 2 inflammasome activation by a Legionella effector functioning in membrane trafficking. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:6193–6198. doi: 10.1073/pnas.1117490109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Khare S, et al. The PYRIN domain-only protein POP3 inhibits ALR inflammasomes and regulates responses to infection with DNA viruses. Nature immunology. 2014;15:343–353. doi: 10.1038/ni.2829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Yin Q, et al. Molecular mechanism for p202-mediated specific inhibition of AIM2 inflammasome activation. Cell reports. 2013;4:327–339. doi: 10.1016/j.celrep.2013.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Sagulenko V, et al. AIM2 and NLRP3 inflammasomes activate both apoptotic and pyroptotic death pathways via ASC. Cell death and differentiation. 2013 doi: 10.1038/cdd.2013.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Pierini R, et al. AIM2/ASC triggers caspase-8-dependent apoptosis in Francisella-infected caspase-1-deficient macrophages. Cell death and differentiation. 2012;19:1709–1721. doi: 10.1038/cdd.2012.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Dombrowski Y, et al. Cytosolic DNA triggers inflammasome activation in keratinocytes in psoriatic lesions. Sci Transl Med. 2011;3:82ra38. doi: 10.1126/scitranslmed.3002001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Dihlmann S, et al. Increased expression and activation of absent in melanoma 2 inflammasome components in lymphocytic infiltrates of abdominal aortic aneurysms. Molecular medicine. 2014;20:230–237. doi: 10.2119/molmed.2013.00162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Javierre BM, et al. Changes in the pattern of DNA methylation associate with twin discordance in systemic lupus erythematosus. Genome research. 2010;20:170–179. doi: 10.1101/gr.100289.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Dihlmann S, et al. Lack of Absent in Melanoma 2 (AIM2) expression in tumor cells is closely associated with poor survival in colorectal cancer patients. Int J Cancer. 2014 doi: 10.1002/ijc.28891. [DOI] [PubMed] [Google Scholar]
- 193.Ponomareva L, et al. AIM2, an IFN-inducible cytosolic DNA sensor, in the development of benign prostate hyperplasia and prostate cancer. Molecular cancer research : MCR. 2013;11:1193–1202. doi: 10.1158/1541-7786.MCR-13-0145. [DOI] [PubMed] [Google Scholar]
- 194.Dihlmann S, et al. Lack of Absent in Melanoma 2 (AIM2) expression in tumor cells is closely associated with poor survival in colorectal cancer patients. International journal of cancer Journal international du cancer. 2014 doi: 10.1002/ijc.28891. [DOI] [PubMed] [Google Scholar]
- 195.Cai X, et al. Prion-like polymerization underlies signal transduction in antiviral immune defense and inflammasome activation. Cell. 2014;156:1207–1222. doi: 10.1016/j.cell.2014.01.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Franklin BS, et al. The adaptor ASC has extracellular and ‘prionoid’ activities that propagate inflammation. Nature immunology. 2014;15:727–737. doi: 10.1038/ni.2913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Baroja-Mazo A, et al. The NLRP3 inflammasome is released as a particulate danger signal that amplifies the inflammatory response. Nature immunology. 2014;15:738–748. doi: 10.1038/ni.2919. [DOI] [PubMed] [Google Scholar]
- 198.Hara H, et al. Phosphorylation of the adaptor ASC acts as a molecular switch that controls the formation of speck-like aggregates and inflammasome activity. Nature immunology. 2013;14:1247–1255. doi: 10.1038/ni.2749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Rodgers MA, et al. The linear ubiquitin assembly complex (LUBAC) is essential for NLRP3 inflammasome activation. The Journal of experimental medicine. 2014;211:1333–1347. doi: 10.1084/jem.20132486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Shi CS, et al. Activation of autophagy by inflammatory signals limits IL-1beta production by targeting ubiquitinated inflammasomes for destruction. Nature immunology. 2012;13:255–263. doi: 10.1038/ni.2215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Yu JW, et al. Cryopyrin and pyrin activate caspase-1, but not NF-kappaB, via ASC oligomerization. Cell death and differentiation. 2006;13:236–249. doi: 10.1038/sj.cdd.4401734. [DOI] [PubMed] [Google Scholar]
- 202.Seshadri S, Duncan MD, Hart JM, Gavrilin MA, Wewers MD. Pyrin levels in human monocytes and monocyte-derived macrophages regulate IL-1beta processing and release. Journal of immunology. 2007;179:1274–1281. doi: 10.4049/jimmunol.179.2.1274. [DOI] [PubMed] [Google Scholar]
- 203.Xu H, et al. Innate immune sensing of bacterial modifications of Rho GTPases by the Pyrin inflammasome. Nature. 2014;513:237–241. doi: 10.1038/nature13449. [DOI] [PubMed] [Google Scholar]
- 204.Elinav E, et al. NLRP6 inflammasome regulates colonic microbial ecology and risk for colitis. Cell. 2011;145:745–757. doi: 10.1016/j.cell.2011.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Wlodarska M, et al. NLRP6 inflammasome orchestrates the colonic host-microbial interface by regulating goblet cell mucus secretion. Cell. 2014;156:1045–1059. doi: 10.1016/j.cell.2014.01.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Anand PK, et al. NLRP6 negatively regulates innate immunity and host defence against bacterial pathogens. Nature. 2012;488:389–393. doi: 10.1038/nature11250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Vladimer GI, et al. The NLRP12 inflammasome recognizes Yersinia pestis. Immunity. 2012;37:96–107. doi: 10.1016/j.immuni.2012.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Ataide MA, et al. Malaria-induced NLRP12/NLRP3-dependent caspase-1 activation mediates inflammation and hypersensitivity to bacterial superinfection. PLoS pathogens. 2014;10:e1003885. doi: 10.1371/journal.ppat.1003885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Zaki MH, Man SM, Vogel P, Lamkanfi M, Kanneganti TD. Salmonella exploits NLRP12-dependent innate immune signaling to suppress host defenses during infection. Proceedings of the National Academy of Sciences of the United States of America. 2014;111:385–390. doi: 10.1073/pnas.1317643111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Allen IC, et al. Characterization of NLRP12 during the in vivo host immune response to Klebsiella pneumoniae and Mycobacterium tuberculosis. PloS one. 2013;8:e60842. doi: 10.1371/journal.pone.0060842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Zaki MH, et al. The NOD-like receptor NLRP12 attenuates colon inflammation and tumorigenesis. Cancer Cell. 2011;20:649–660. doi: 10.1016/j.ccr.2011.10.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Allen IC, et al. NLRP12 suppresses colon inflammation and tumorigenesis through the negative regulation of noncanonical NF-kappaB signaling. Immunity. 2012;36:742–754. doi: 10.1016/j.immuni.2012.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Unterholzner L, et al. IFI16 is an innate immune sensor for intracellular DNA. Nature immunology. 2010;11:997–1004. doi: 10.1038/ni.1932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Kerur N, et al. IFI16 acts as a nuclear pathogen sensor to induce the inflammasome in response to Kaposi Sarcoma-associated herpesvirus infection. Cell host & microbe. 2011;9:363–375. doi: 10.1016/j.chom.2011.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Ansari MA, et al. Constitutive interferon-inducible protein 16-inflammasome activation during Epstein-Barr virus latency I, II, and III in B and epithelial cells. Journal of virology. 2013;87:8606–8623. doi: 10.1128/JVI.00805-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Johnson KE, Chikoti L, Chandran B. Herpes simplex virus 1 infection induces activation and subsequent inhibition of the IFI16 and NLRP3 inflammasomes. Journal of virology. 2013;87:5005–5018. doi: 10.1128/JVI.00082-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Horan KA, et al. Proteasomal degradation of herpes simplex virus capsids in macrophages releases DNA to the cytosol for recognition by DNA sensors. Journal of immunology. 2013;190:2311–2319. doi: 10.4049/jimmunol.1202749. [DOI] [PMC free article] [PubMed] [Google Scholar]