Abstract
Amelogenin proteins are critical to the formation of enamel in teeth and may have roles in controlling growth and regulating microstructures of the intricately woven hydroxyapatite (HAP). Leucine-rich amelogenin protein (LRAP) is a 59-residue splice variant of amelogenin and contains the N- and C-terminal charged regions of the full-length protein thought to control crystal growth. Although the quaternary structure of full-length amelogenin in solution has been well studied and can consist of self-assemblies of monomers called nanospheres, there is limited information on the quaternary structure of LRAP. Here, sedimentation velocity analytical ultracentrifugation (SV) and small angle neutron scattering (SANS) were used to study the tertiary and quaternary structure of LRAP at various pH values, ionic strengths, and concentrations. We found that the monomer is the dominant species of phosphorylated LRAP (LRAP(+P)) over a range of solution conditions (pH 2.7 to 4.1, pH 4.5 to 8, 50 mmol/L(mM) to 200 mM NaCl, 0.065 to 2 mg/mL). The monomer is also the dominant species for unphosphorylated LRAP (LRAP(−P)) at pH 7.4 and for LRAP(+P) in the presence of 2.5 mM calcium at pH 7.4. LRAP aggregates in a narrow pH range near the isoelectric point of pH 4.1. SV and SANS show that the LRAP monomer has a radius of ~2.0 nm and an asymmetric structure, and solution NMR studies indicate that the monomer is largely unstructured. This work provides new insights into the secondary, tertiary, and quaternary structure of LRAP in solution and provides evidence that the monomeric species may be an important functional form of some amelogenins.
INTRODUCTION
The amelogenin proteins are necessary for the formation of tooth enamel, representing 90% of the proteins present in enamel fluid.(Termine et al., 1980; Gibson et al., 2001) Humans with mutations in the AMELX gene and knock-out mice engineered to have AMELX null mutations have highly defective and disorganized enamel structure.(Gibson, 2011) Amelogenin proteins are thought to function as a matrix to guide the mineralization of HAP extracellularly because amelogenin nanospheres have been observed along HAP crystallites in immature enamel.(Fincham et al., 1995) In-vitro studies have suggested amelogenin has roles in initiating nucleation,(Tarasevich et al., 2007) controlling growth,(Iijima and Moradian-Oldak, 2004) and affecting the spacing of crystallites.(Moradian-Oldak, Tan, and Fincham, 1998a)
Leucine-rich amelogenin protein (LRAP) is a 59-residue splice variant of amelogenin (Figure 1).(Gibson et al., 1991) Because LRAP appears within the enamel fluid with amelogenin, it has been thought that the protein may have a role in enamel formation.(Fincham et al., 1999) In-vivo studies have shown that LRAP is localized within the extracellular matrix of growing enamel (Gibson et al., 1995) and in-vitro studies have shown that LRAP can control HAP crystal formation(Le Norcy et al., 2011) suggesting that LRAP, like amelogenin, may have an extracellular matrix function in controlling enamel crystal growth. More recent studies have provided evidence that LRAP may promote enamel growth by acting as a cell signaling molecule, affecting ameloblast differentiation and protein expression. For example, LRAP partially rescued the null amelogenin mouse phenotype,(Gibson et al., 2009; 2011) increased enamel growth in tooth explants (Ravindranath et al., 2007) and promoted the differentiation of human enamel organ epithelial cells.(Le et al., 2007) LRAP that was overexpressed in transgenic murine models affected ameloblast differentiation and upregulated amelogenin, MMP-20, and SATB1 proteins.(Stahl et al., 2013)
Figure 1.

Primary amino acid sequence of murine amelogenin with the basic and acidic residues colored blue and red, respectively. The splice variant LRAP is composed of the N-terminal 33 and C-terminal 26 residues, shaded yellow. Both full-length amelogenin and LRAP are post-translationally modified by side chain phosphorylation of S16, colored green. The serine at position 16 was either phosphorylated (LRAP(+P)) or non-phosphorylated (LRAP(−P)).
Although LRAP has a role in enamel formation, recent studies have shown that LRAP can also function as a cell signaling protein to promote differentiation of mesenchymal cells.(Veis et al., 2000; Warotayanont et al., 2008) LRAP has been found to promote osteogenesis of rat muscle fibroblasts,(Veis et al., 2000) cementoblasts, (Boabaid et al., 2004) and mesenchymal stem cells.(Warotayanont et al., 2009; Wen et al., 2011) A porcine enamel matrix derivative has been shown to have therapeutic applications in promoting the regeneration of cementum and bone in periodontal tissue(Hammarstrom, 1997; Heijl et al., 1997; Sculean et al., 2008) and LRAP has been shown to be the main factor within enamel matrix derivatives to promote osteogenesis.(Amin et al., 2012)
In spite of the importance of LRAP as an enamel former, osteogenic protein, or regenerative agent, very little is known about its tertiary and quaternary structure. It will be difficult to fully understand LRAP’s biological function or therapeutic potential without a better grasp of its structure. The dominant quaternary structure of full-length amelogenin in solution is the ‘nanosphere,” aggregates of amelogenin monomers that are 20 nm to 60 nm in diameter.(Moradian-Oldak, 2001) The nanospheres are considered to be self-assemblies because they are believed to have a hierarchical structure consisting of highly ordered oligomers.(Fang et al., 2011) Nanospheres are present in solutions with pH values greater than 6.0 and salt concentrations ranging from 50 mmol/L (mM) to 200 mM.(Moradian-Oldak et al., 1994; Moradian-Oldak, Leung, et al., 1998) Oligomers have also been detected in solutions at pH 5.5 ranging in size from dimers to octamers depending on the protein concentration.(Bromley et al., 2011)
Recently we found that phosphorylated LRAP exists as a monomer in 150 mM NaCl, pH 7.4 solutions.(Tarasevich et al., 2013) Since this quaternary form is in contrast to the nanospheres observed for full-length amelogenin at pH 7.4, we wanted to further investigate the quaternary structure of LRAP over a range of solution conditions including those found in developing enamel.(Sasaki et al., 1991; Robinson et al., 1995) LRAP was studied in solutions ranging from pH 3.0 to 8.0, 50 to 200 mM NaCl, and concentrations from 0.065 to 2 mg/mL. SV was used to determine the distribution of protein species present in the solutions.(Lebowitz et al., 2002) SV has several advantages over other analysis techniques, such as dynamic light scattering (DLS), because it uses a centrifugal field to separate species from monomers to large aggregates based on the species’ masses and shapes, whereas DLS relies on a mathematical separation. Although DLS is well suited for the measurement of sizes in monodisperse systems, it is not as accurate in characterizing polydisperse systems. Light scattering intensity has a size dependence of radius to the sixth power (r6) so that large structures can dominate scattering and prevent the detection of smaller structures. (Filipe et al., 2010) Also, DLS was not able to resolve particle populations that differed in size by a factor of 2 or less.(Filipe et al., 2010) In contrast, SV can directly quantify the proportion of different species in solution on a weight basis, since SV typically uses UV absorbance to monitor sample separation. SV and SANS were also used to determine the size and shape of the LRAP monomers and solution NMR was used to study LRAP secondary structure.
MATERIALS AND METHODS†
Materials
Deuterium oxide (99.9%) was purchased from Cambridge Isotopes Laboratories and used as received. All other chemicals were reagent grade and obtained from Sigma-Aldrich.
Solid-phase LRAP(+P) and LRAP(−P) synthesis
Murine LRAP with phosphoserine (LRAP(+P)) and normal serine (LRAP(−P)) at position 16 (Figure 1) was synthesized using solid-phase methods by the Protein Chemistry Technology Center, University of Texas (Dallas, TX) for SV, SANS, and zeta potential experiments. Each sample was purified by reverse phase HPLC using buffer A, 0.1% trifluoroacetic acid in water; buffer B, 0.1% trifluoroacetic acid in acetonitrile. LRAP eluted at 54% B. Mass spectroscopy was used to characterize the purity and molecular weight of the proteins. After purification, proteins were lyophilized for storage until ready for use.
Recombinant LRAP(−P) (rLRAP(−P)) synthesis
To obtain uniformly 13C- and 15N-labeled LRAP samples for solution NMR studies it was necessary to use recombinant methods. RLRAP(−P) containing a 12-residue (MRGSHHHHHHGS-) N-terminal tag was synthesized for the studies conducted in acetic acid at pH 2.8, as described previously.(Buchko et al., 2010) For the studies conducted in SCP solution at pH 7.4, rLRAP(−P) was synthesized with only a 4-residue (GPGS-) N-terminal tag as described in the supplementary section.
LRAP solution formation
A stock solution of LRAP was dissolved in Millipore purified water at 5–10 mg/ml and stored at 4°C overnight. The solution was centrifuged at 11,000 rpm for 30 minutes and the upper fraction decanted. The concentration of the stock solution was determined by measurement of the UV absorbance at 275 nm using an extinction coefficient of 15,470 (M−1 cm−1). Saturated calcium phosphate (SCP) solutions were prepared containing various NaCl concentrations (50 to 300 mM) and pH values (5.0 to 8.0). They were prepared by adding HAP powder to the NaCl solution, adjusting the pH, stirring for several days, and filtering out any particles as described previously.(Shaw et al., 2004) A SCP solution is being used because it has a small amount of dissolved calcium and phosphate in the solution making it more relevant to the in-vivo enamel forming environment. The amounts of calcium and phosphate do not exceed the saturation limit so there is no calcium phosphate precipitation. The LRAP stock solution was diluted into the various SCP solutions at the appropriate pH to obtain solutions containing 50, 150, and 200 mM NaCl. The solution pH values were initially around pH 3.0 (due to formic acid in the lyophilizing solution) and were readjusted to values ranging from pH 3.0 to pH 8.0 using dilute KOH and HCl. The LRAP solution pH values were stable and did not change over time periods up to 1 month. The final concentrations of the LRAP solutions were 0.065 to 2 mg/ml.
Zeta Potential
The zeta potential of LRAP solutions at concentrations of 0.5 mg/mL, 50 and 150 mM NaCl SCP, and various pH values from 3.0 to 8.0 were determined using a ZetaPALS instrument (Brookhaven Instruments, Holtsville, NY). The protein electrophoretic mobility was measured by detecting the phase shift in the scattered light as the protein moves under an applied field. The protein solution was placed into a cuvette and an Uzgiris cell electrode was inserted into the solution. The Uzgiris cell suppressed electro-osmosis effects. The platinum electrodes were conditioned by 10 runs (30 cycles/run) of a 1 M NaCl solution with both voltage and frequency set to auto. The resulting PtCl2 passivating layer reduced chlorine and protein interactions with the charged electrode. The electrode was operated at 2.5 V and 2.5 Hz for the protein solutions and the data was collected over five runs at 10–20 cycles per run.
Sedimentation Velocity (SV)
Samples were analyzed within 2–3 days after sample preparation and were placed into an AN-60Ti analytical rotor, loaded into a Beckman-Coulter ProteomeLab XL-I analytical ultracentrifuge at 20 °C, and scanned at 280 nm or 230 nm as described previously.(Tarasevich et al., 2013) The data were analyzed using the c(s) method and SEDFIT analysis program (version 11.3) developed previously.(Schuck, 2000) The raw data scans (~37,000 data points) were directly fitted to derive the distribution of sedimentation coefficients, while modeling the influence of diffusion on the data in order to enhance the resolution. This method assigns a diffusion coefficient to each value of sedimentation coefficient based on an assumption that all species have the same overall hydrodynamic shape. That hydrodynamic shape is defined by the f/f0 ratio, where f is the frictional coefficient of the macromolecule and f0 is the frictional coefficient of an anhydrous sphere with the same volume as the macromolecule. The f/f0 values were varied to find the best overall fit of the data for the entire sample. Maximum entropy regularization probabilities of 0.683 (1 σ) were used and time-independent noise was removed.
The partial specific volume (v̄) for LRAP(+P) and LRAP(−P) at 20 °C were calculated as 0.7435 and 0.7476 ml/g, respectively, using the program SEDNTERP (version 1.09).(Laue et al., 1992) Solvent densities and viscosities at 20 °C for the SCP buffer were calculated using SEDNTERP as 1.00442 g/ml and 1.0165 cp respectively. SEDNTERP was also used to convert raw sedimentation coefficients to standardized s20,w values and to calculate f/f0 ratios from the measured sedimentation coefficients.
Small angle neutron scattering (SANS)
LRAP(+P) solutions were studied by SANS to determine the size and shape of the LRAP monomers. LRAP(+P) (1 mg/mL) was prepared in 150 mM NaCl SCP (pH 7.4) or 2% acetic acid (pH 2.7) in 99.9% D2O and was measured in a quartz cuvette with a 2 mm path length on the NG7 30 meter SANS instrument (Glinka et al., 1998) at the NIST Center for Neutron Research (NCNR) in Gaithersburg, MD. The 2% acetic acid sample was also measured at a LRAP(+P) concentration of 2 mg/mL. Preliminary experiments showed that the SANS data for pH 7.4 samples was affected by the presence of oligomers derived from aggregates of LRAP(+P) formed at the isoelectric point. At oligomer concentrations of 5% or greater, the oligomer species dominated the scattering and it was not possible to obtain both a size distribution from the fit and extract size and shape data for the monomer. Methods were developed to reduce the concentration of oligomers in the solutions to 1–2% to allow characterization of the monomers. This was achieved by reducing the protein concentration and adding LRAP stock solution to buffer containing enough base to obtain a final pH of 7.4, avoiding the pH range where LRAP has a low charge and aggregates (pH 4.2–4.5).
The neutron wavelength, λ, was 6 Å, with a wavelength spread, Δλ/λ, of 0.15. Scattered neutrons were detected with a 64 x 64 cm two-dimensional position-sensitive detector with 128 x 128 pixels at a resolution of 0.5 cm/pixel. Sample-to-detector distances of 5 m and 1.5 m were used. The data were reduced using the IGOR program with SANS macro routines developed at the NCNR.(Kline, 2006) Raw counts were normalized to a common monitor count and corrected for empty cell counts, ambient room background counts, and non-uniform detector response. Data were placed on an absolute scale by normalizing the scattered intensity to the incident beam flux. Next, the data were radially averaged to produce scattered intensity, I(q), versus q curves, where q = 4πsin(θ)/λ and 2θ is the scattering angle. The scattered intensities were then further corrected for buffer scattering and incoherent scattering from hydrogen in the samples. The q-range covered by the data after background subtraction was 0.012 Å−1 ≤ q ≤ 0.3 Å−1 for the 2 mg/mL samples and 0.03 Å−1 ≤ q ≤ 0.3 Å−1 for the 1 mg/mL samples.
Initial data analysis was performed using the Guinier approximation,
| (1) |
on the low-q portions of the data to obtain initial values for the radius of gyration, Rg, and the forward scattering intensity, I(0), of the samples. This analysis is valid only in the region where qRg ≤ 1. Standard Kratky analysis was performed on each data set by plotting I(q)•q2 versus q to get a qualitative idea of the degree of protein folding. Both the Guinier and Kratky analyses were performed using the NCNR IGOR SANS macro routines.(Kline, 2006)
GNOM was used to determine the distance distribution functions, P(r), versus r.(Semenyuk and Svergun, 1991) The value of the maximum diameter of the particle, Dmax, was determined empirically by examining the quality of the fit to the experimental data for a range of Dmax values. Since the SANS data were obtained on an absolute scale, the molecular weight, Mw, of the proteins was estimated from the forward scattering intensity, I(0), using the relation:
| (2) |
where Δρ = ρ − ρs is the contrast, or the difference between the scattering length density of the molecule (ρ) and the solvent (ρs), n is the number density of molecules, and V is the molecular volume. The number density can be written as n = cNA /Mw, where c is the concentration, and NA is Avogadro’s number. The volume can be written as V = Mw/(NAd), where d is the mass density. Now, Equation 2 can be rewritten as:
| (3) |
The only unknown parameter in Equation 3 is Mw, since all other parameters can be measured or calculated. I(0) values were taken from the GNOM analysis of the data since this analysis uses all of the data rather than a limited number of points in the Guinier region. The concentration can be directly measured during sample preparation and Δρ can be calculated from the chemical composition of the sample and solvent. The mass density, d, is taken as the inverse of the partial specific volume. It is important to note that I(0) must be on an absolute scale, usually in cm−1, in order to obtain accurate Mw values from equations (2) or (3).
Molecular Modeling
Model SANS curves were calculated for the 10 lowest energy Rosetta model structures of LRAP(+P) derived previously (Tarasevich et al., 2013) using the program XTAL2SAS.(Heidorn and Trewhella, 1988; Krueger et al., 1998) The model SANS curves were compared to the SANS data from 2 mg/mL LRAP(+P) in 2 % acetic acid and scored for quality using the χ2 equation:
| (4) |
where Iexp(q) is the experimentally determined SANS intensity curve, Icalc(q) is the calculated intensity curve from the model structure, and σexp(q) is the q-dependent variance. The sum was taken over 60 data points (N = 60). The lowest energy structure with the model SANS curve that best fit the data was used as a starting point for further structure modeling.
The SASSIE program(Curtis et al., 2012) was used to generate an ensemble of structures from this best-fit starting structure for comparison to the SANS data. A variety of structures were generated by randomly varying backbone dihedral angles within the regions of the protein that are disordered in the Rosetta model structures, i.e., residues 1 to 22, residues 33 to 43, residues 48 to 49 and residues 57 to 59. After each randomly-chosen angle was rotated by a given value, the final value was checked to determine if it was energetically probable, based on an energy function using the Charmm-22 all-atom protein force-field parameters.(MacKerell et al., 1998) Each new configuration was checked for overlap of α-carbon atoms using a distance criterion of 3 Å. If both checks were met, the new structure was accepted and Rg was calculated. All structures were energy minimized using the program NAMD(Phillips et al., 2005) prior to the calculation of model SANS curves.
Examination of a plot of χ2 versus Rg provides an idea of how well the individual structures in the ensemble fit the data. The best (lowest χ2) and worst (highest χ2) fit model SANS curves are noted, along with the average model SANS curve from the entire ensemble of accepted structures. These curves were plotted along with the experimental SANS data to aid in the visualization of the quality of the fits to the data.
Nuclear Magnetic Resonance (NMR) spectroscopy
Varian Inova-500 and -750 spectrometers equipped with an HCN-probe and pulse field gradients were used to collect NMR data at 20°C on the double-labeled (13C-, 15N-) rLRAP(−P) sample (2 mg/ml or ~0.3 mM) in 2% acetic acid (pH 2.8) and in 150 mM NaCl SCP solutions (pH 7.4). Three-dimensional HNCACB and CCC-TOCSY data were collected to assign the amide cross peaks in the two-dimensional 1H-15N HSQC spectrum and to acquire 13Cα and 13Cβ chemical shifts. All NMR data were processed using Felix2007 (MSI, San Diego, CA) software and analyzed with the program Sparky (v3.115) (Goddard). The 1H, 13C, and 15N chemical shifts were referenced using indirect methods.(Wishart et al., 1995) The random coil values used in the 13Cα and 13Cβ chemical shift plots were from CNS (cns_solve_1.1).
RESULTS
Zeta potential of LRAP (+P) as a function of pH
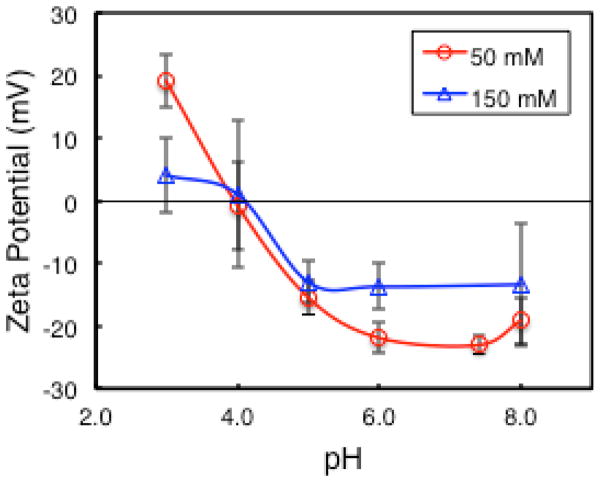
Figure 2 shows the zeta potential of LRAP(+P) solutions at 0.5 mg/mL in SCP solution as a function of pH and ionic strength. The zeta potential is positive at low pH and negative at high pH with an isoelectric point around pH 4.1. The zeta potential decreases with increasing ionic strength from 50 mM to 150 mM NaCl.
Figure 2.
Zeta potential of LRAP (+P) in SCP solution at 50 and 150 mM NaCl as a function of pH. Data points are connected by smoothed lines.
SV studies of quaternary structures
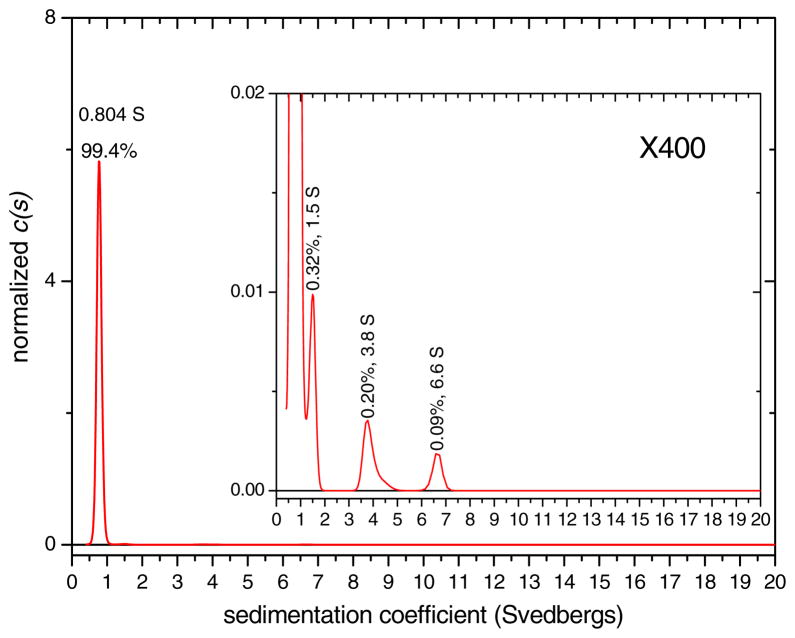
Standardized sedimentation coefficient (s20,w) distributions were obtained for LRAP(+P) in SCP solution at 150 mM NaCl, pH 7.4 as a function of LRAP concentration. The solutions were formed at 2 mg/mL LRAP(+P) and then diluted in SCP solution to 1.0 and 0.5 mg/mL. The species distribution for the 2 mg/mL solution is shown in Figure 3, distributions for 0.5 and 1.0 mg/mL LRAP(+P) are shown in Figure S1, and the sedimentation coefficients (s20,w) and weight % species for all concentrations are summarized in Table 1. The size distributions are dominated by one major peak for all three concentrations. The sedimentation oefficient, together with the best-fit frictional coefficient ratio relative to an anhydrous sphere (f/f0 ratio), imply this major peak has a mass of ~ 6.3 kD, consistent with a monomeric species. The assignment of the major component to a monomer was also confirmed by time derivative analysis.(Philo, 2006) Repetitions of samples showed that the sedimentation coefficient values for the monomers are reproducible with errors of 1.4% and 2.4% for the 1 and 2 mg/mL solutions, respectively (Table S1).
Figure 3.
Normalized sedimentation coefficient distribution for LRAP(+P) in 150 mM NaCl, pH 7.4 SCP solution at 2 mg/mL with a 400x expansion in the inset. Peaks are labeled with % amount and sedimentation coefficient, s20,w, value in svedbergs (S). The main peak was determined to represent a monomer and the s20,w value of the monomer peak is the average over the peak.
Table 1.
Sedimentation coefficient (s20,w) and % species for LRAP(+P) in SCP solution, 150 mm NaCl, pH 7.4, as a function of LRAP concentration.
| [LRAP(+P)] (mg/mL) | [NaCl] (mM) | pH | Monomer1 s20,w (Svedbergs) | Monomer1 w% | ~1.1–1.6 S peaks % | >1.6 S peaks % |
|---|---|---|---|---|---|---|
| 0.5 | 150 | 7.4 | 0.781 | 100 | 0 | 0 |
| 1 | 150 | 7.4 | 0.782 | 99.8 | 0 | 0.2 |
| 2 | 150 | 7.4 | 0.804 | 99.4 | 0.32 | 0.28 |
The monomer peak at these high concentrations represent monomers in weak, rapid reversible association with monomers or other species
The 0.5 mg/mL solution is 100% monomer and the solutions at higher concentration have a very small percentage of larger species (0.2% and 0.6% for the 1.0 mg/mL and 2.0 mg/mL solutions, respectively). The peaks at 1.5 S, 3.6 S, and 6.6 S for the 2.0 mg/mL solution and 5.3 S, 10.5 S, and 13.5 S for the 1.0 mg/mL solution are suggestive of oligomers with a range of sizes. The oligomer species are reported as two classes in Table 1, small oligomers from 1.1 S to 1.6 S, and oligomers larger than 1.6 S. The peak positions and concentration of the oligomers vary between repetitions of the same sample as shown in Figure S2. Also, there were differences in oligomer concentration and distribution with changes in the sample preparation method. As discussed in detail later, we believe that the oligomers are derived from larger aggregates formed at the isoelectric point of LRAP. The oligomers, therefore, are metastable, nonequilibrium species, originating from the sample preparation method. The peak positions for the smallest minor components are in the range of 1.1 S to 1.6 S, consistent with species in the size range of dimers to tetramers.
Although the sedimentation coefficient for pure monomer should decrease with increasing protein concentration by at least 0.9% per mg/mL due to excluded volume effects,(Laue et al., 1992) in this case there is no significant change in sedimentation coefficient with increasing protein concentration (Table 1 and Table S1). Therefore, the sedimentation coefficient values are somewhat larger than expected at the higher protein concentrations, and this difference appears to be larger than can be accounted for by experimental error. This suggests that the sample is exhibiting weak, rapidly reversible, self-association reactions on a time scale too short to result in any resolved oligomer peaks. We believe the peak we have been describing as a “monomer” at concentrations above 0.5 mg/mL, therefore, represents a dynamic mixture of monomers and a few percent or less of short-lived dimers or other larger species. In summary, there are two kinds of structures in the LRAP solutions: a large concentration of monomers in rapid, reversible association with other monomers and a small concentration of oligomers that are metastable.
In addition to the pH 7.4, 150 mM NaCl SCP conditions, we also studied LRAP(+P) structure using sedimentation velocity under a range of solution conditions from 50 to 200 mM NaCl and pH values of 5.8 to 7.4. The results are summarized in Table 2. The data show that the monomer is the dominant species over all of the solution conditions and pH values studied. The monomer percentage was as low as 84.4% for the 50 mM NaCl, pH 5.8 LRAP(+P) solution and as high as 97.9% for the 200 mM NaCl, pH 8.0 LRAP(+P) solutions.
Table 2.
Sedimentation velocity data for LRAP(+P) in SCP solution at various NaCl concentrations and pH values.
| [LRAP(+P)] (mg/mL) | [NaCl] (mM) | pH | Monomer1 % | ~1.1–1.6 S peaks % | >1.6 S peaks % |
|---|---|---|---|---|---|
| 1.0 | 50 | 5.8 | 84.4 | 6.99 | 6.61 |
| 0.78 | 50 | 7.4 | 93.6 | 3.29 | 2.91 |
| 0.78 | 50 | 8.0 | 92.3 | 4.72 | 2.98 |
| 1.0 | 150 | 5.8 | 90.9 | 7.10 | 2.00 |
| 1.0 | 150 | 7.4 | 93.9 | 5.80 | 0.30 |
| 0.92 | 150 | 8.0 | 92.6 | 7.16 | 0.24 |
| 1.0 | 200 | 5.8 | 94.5 | 0 | 5.6 |
| 1.0 | 200 | 7.4 | 97.7 | 0 | 2.3 |
| 0.92 | 200 | 8.0 | 97.9 | 0 | 2.1 |
The monomer peak at these high concentrations represent monomers in weak, rapid reversible association with monomers or other species
Although monomers are the dominant species over most of the pH range, we observed large structures by DLS (~1400 nm diameter) and visible cloudiness in 1 mg/mL solutions in the pH range 4.2 to 4.4. Since our LRAP solutions are below the solubility limit for this pH range,(Tan et al., 1998) we believe the structures are LRAP(+P) aggregates formed at low charge near the isoelectric point of LRAP(+P) as determined by the zeta potential measurements shown in Figure 2. Previous dynamic light scattering (DLS) studies also showed aggregation of 2 mg/mL LRAP in this pH range.(Le Norcy et al., 2011) As the pH is adjusted above pH 4.4, the visible cloudiness disappears, the solutions become clear, and monomers become the dominant species. Above the isoelectric point, therefore, LRAP develops a negative charge which promotes dissolution of the aggregates to monomers and a small fraction of oligomers as small as dimers and trimers. Since the oligomers do not dissolve completely to the equilibrium monomeric species in the time course of the experiments, they represent metastable, nonequilibrium species. The % oligomeric species are highest at pH 5.8 and there was a general trend that the % oligomers decrease with increasing salt concentration from 50 mM to 200 mM NaCl. These trends may represent trends in the degree of dissolution of the aggregates formed near the isoelectric point.
Previous studies have observed LRAP “nanospheres” adsorbed onto surfaces and imaged by TEM(Le Norcy et al., 2011) and AFM. (Habelitz et al., 2006; Tarasevich et al., 2010) Our SV studies at similar solution conditions have found that the solutions contain primarily monomers with up to 16% oligomers. Our studies, therefore, suggest that the “nanospheres” seen previously on surfaces may be the same as the structures we observe in solutions that we are calling “nonequilibrium oligomers.” The monomers we observe as the majority species may also have adsorbed onto the surfaces previously studied but may be difficult to detect experimentally.
We also found that the monomer is the dominate species in solutions containing LRAP(+P) in 2.5 mM CaCl2, pH 7.4 solutions and LRAP(−P) in 150 mM NaCl SCP, pH 7.4 solutions as shown in Table S2. Previous studies used small angle x-ray scattering (SAXS) to determine that LRAP(+P) under similar solution conditions (2 mg/ml, 2.5 mM CaCl2, pH 7.4) had a radius of gyration, Rg, of 4.4 nm.(Le Norcy et al., 2012) They interpreted this result to suggest that LRAP formed particles or aggregates under this solution condition. Since we found 94% monomers and 6% oligomers in our samples at a similar solution condition (Table S2), another way to interpret the SAXS data is that the 4.4 nm diameter represents the average size over the monomers and oligomers present in solution. SAXS is a scattering technique that measures the average size over the intensity weighted distribution of species in contrast to SV which determines a species distribution. Our experience with SANS, another scattering technique, is that if we had 5–10% oligomers in our solutions, we obtained average sizes of around 4–8 nm Rg, even though SV of the same sample showed we had 90–95% monomers at 2 nm Rh. The presence of oligomers mixed with monomers, therefore, can shift up the average size of species studied by scattering techniques such as SAXS and SANS. If we were careful about making our solutions and reduced the % oligomers down to 1–2%, we obtained SANS sizes of around 2 nm Rg, consistent with the SV size we found for the LRAP monomer.
SV studies of monomer tertiary structure
Since the monomer is the dominant quaternary structure of LRAP observed over a range of protein concentrations, pH values, salt concentrations, and phosphorylation state, we decided to further study the tertiary structure of the LRAP monomer by determining its hydrodynamic radius (Rh) and f/f0 ratio, both of which were determined from the observed sedimentation coefficient and the known monomer mass. The f/f0 ratio is the frictional coefficient of the macromolecule relative to the frictional coefficient of an anhydrous sphere and is a function of the shape and hydration of the species. In the absence of hydration, an f/f0 ratio of 1 indicates a spherical shape, while an f/f0 ratio greater than 1 suggests an asymmetric shape. We obtained sedimentation coefficient distributions and f/f0 ratios of LRAP as a function of solution pH at solution concentrations of 0.065 mg/mL. A low protein concentration was used to decrease the extent of self-association of LRAP monomers which would affect the measured s20,w values. The f/f0 ratio was then calculated from the measured sedimentation coefficient of the monomer peak and the known monomer mass.
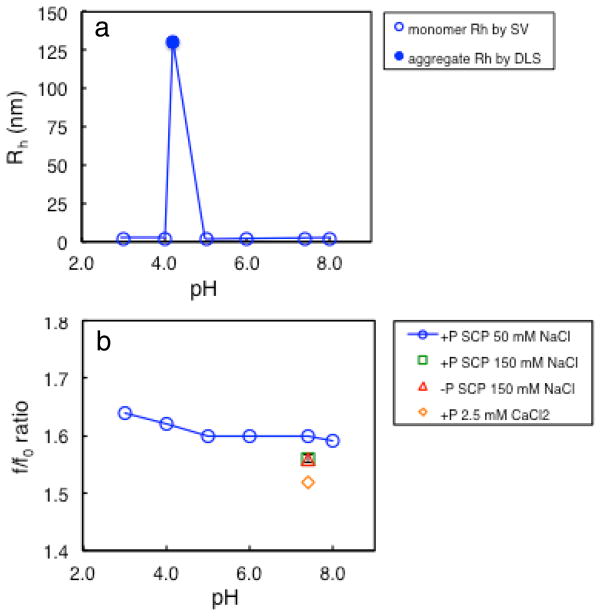
Table 3 is a summary of sedimentation coefficient values, Rh values, and f/f0 ratios for the monomer ordered by pH and Table S3 shows a summary of percent species for the various solution conditions. Repetitions of LRAP(+P) in 150 mM NaCl, SCP, pH 7.4 sample resulted in percent errors of ~0.5% for the sedimentation coefficient, Rh, and f/f0 ratio values. The hydrodynamic radius, Rh, of LRAP(+P) aggregates at pH 4.2 was also determined by DLS. Table 1 and the plot in Figure 4a show that the solutions contain ~2 nm radius monomers over the entire pH range except near the isoelectric point where 130 nm aggregates are formed. Figure 4b shows the f/f0 ratio plotted as a function of solution pH. The f/f0 ratio for SCP solutions at 50 mM NaCl is highest at pH 3 (1.64) and decreases slightly with increasing pH. LRAP has f/f0 ratios ranging from 1.52 to 1.64 over all of the solution conditions used, indicating that LRAP has a more extended or flexible structure than globular proteins which have f/f0 ratios of 1.2 to 1.3.(Schuck, 2000)
Table 3.
Sedimentation velocity results including hydrodynamic radius (Rh) and f/f0 ratios for the LRAP monomer in dilute solutions (0.065 mg/mL) as a function of pH, and DLS results for the hydrodynamic radius of LRAP aggregates at pH 4.2.
| LRAP | Buffer type | [NaCl] (mM) | [CaCl2] (mM) | pH | Monomer s20,w (Svedbergs) | Rh (nm) | f/f0 ratio |
|---|---|---|---|---|---|---|---|
| +P | SCP | 50 | - | 3.0 | 0.740 | 2.09 | 1.64 |
| +P | SCP | 50 | - | 4.0 | 0.749 | 2.06 | 1.62 |
| +P | SCP | 50 | - | 4.2 | 130* | ||
| +P | SCP | 50 | - | 5.0 | 0.760 | 2.03 | 1.60 |
| +P | SCP | 50 | - | 6.0 | 0.760 | 2.03 | 1.60 |
| +P | SCP | 50 | - | 7.4 | 0.761 | 2.03 | 1.60 |
| +P | SCP | 50 | - | 8.0 | 0.765 | 2.02 | 1.59 |
| +P | SCP | 150 | - | 7.4 | 0.781 | 1.98 | 1.56 |
| −P | SCP | 150 | - | 7.4 | 0.758 | 1.98 | 1.56 |
| +P | CaCl2 | 3.1 | 2.5 | 7.4 | 0.799 | 1.93 | 1.52 |
Rh of aggregates determined by DLS
Figure 4.
a) Rh for LRAP(+P) species in 0.065 mg/ml 50 mM NaCl SCP solutions including Rh of monomers determined by SV (open blue circles) and Rh of aggregates at pH 4.2 determined by DLS (filled blue circle), b) SV-determined f/f0 ratios for monomers in 0.065 mg/mL LRAP solutions including LRAP(+P) in 50 mM NaCl SCP (blue circles), LRAP(+P) in 150 mM NaCl SCP at pH 7.4 (green square), LRAP(−P) in 150 mM NaCl SCP at pH 7.4 (red triangle), and LRAP(+P) in 2.5 mM CaCl2 at pH 7.4 (orange diamond).
Table 3 shows that the Rh also decreases slightly for LRAP(+P) at 50 mM NaCl with increasing pH, from 2.09 at pH 3 to 2.02 at pH 8. Increasing the NaCl concentration to 150 mM at pH 7.4 results in a smaller Rh and f/f0 ratio suggesting a more compact structure. LRAP(+P) and LRAP(−P) has the same size and shape in 150 mM, pH 7.4 SCP solutions suggesting that phosphorylation of S16 has little effect on the protein’s tertiary structure at that pH. LRAP(+P) solutions containing 2.5 mM CaCl2 at pH 7.4 have the smallest Rh (1.93 nm) and smallest f/f0 ratio (1.52), indicating that the monomer collapses in the presence of calcium. A previous small angle X-ray scattering (SAXS) study also showed that LRAP is more compact in the presence of calcium.(Le Norcy et al., 2012)
SANS studies of tertiary structure
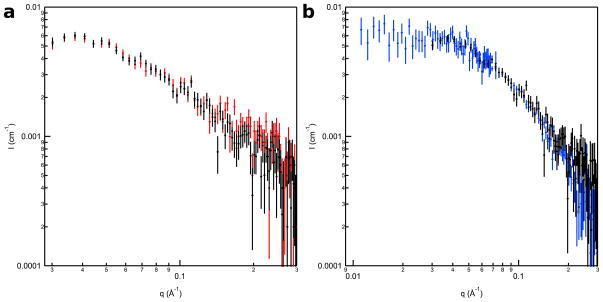
The I(q) vs. q SANS curves are shown in Figure 5 and the corresponding Guinier, Kratky, and P(r) versus r plots are provided in the supplementary section (Figures S3–S5). Figure 5a shows the SANS data for 1 mg/mL LRAP(+P) in 2% acetic acid (pH 2.7) and in 150 mM NaCl, SCP solution (pH 7.4) where the two curves are identical within experimental error. Figure 5b shows the SANS data for LRAP(+P) in 2% acetic acid at 1 and 2 mg/mL where the curves are also identical within experimental error. Table 4 lists the radius of gyration, Rg, and I(0) values determined for these samples from the SANS data and those calculated from the LRAP amino acid sequence.
Figure 5.
SANS I(q) vs. q data for a) 1 mg/mL LRAP(+P) in 2% acetic acid (pH 2.7) (black) and 150 mM NaCl, SCP (pH 7.4) (red) and b) 2 mg/mL (blue) and 1 mg/mL (black) LRAP(+P) in 2% acetic acid (pH 2.7) with the 2 mg/mL data normalized to the 1 mg/mL data to allow better comparison of the curves.
Table 4.
Rg calculations for LRAP(+P) based on P(r) analysis of SANS data with Dmax = 70 Å ± 5 Å.
| Condition | pH | [LRAP(+P)] mg/mL | Rg (nm) | Measured I(0) (cm−1) | Calculated I(0) (cm−1)* | Apparent Mw (kDa) |
|---|---|---|---|---|---|---|
| 2% (v/v) AA | 2.7 | 2.0 | 2.06 ± 0.01 | 0.0114 ± 0.0004 | 0.0152 | 5.3 ± 0.2 |
| 2% (v/v) AA | 2.7 | 1.0 | 2.05 ± 0.06 | 0.0067 ± 0.0003 | 0.0076 | 6.2 ± 0.2 |
| SCP | 7.4 | 1.0 | 2.06 ± 0.06 | 0.0067 ± 0.0002 | 0.0076 | 6.2 ± 0.2 |
I(0) was calculated using Equation 3 with d = 1.34 g/cm3 and assuming the LRAP monomer was measured in 100 % D2O SCP buffer. The LRAP sequence was taken from the Rosetta model PDB file. The monomer Mw = 6.9 kDa, assuming 90 % of exchangeable H are exchanged for D.
Since all of the SANS curves are essentially identical, we chose to perform structural modeling on the 2 mg/mL LRAP(+P), 2% acetic acid (pH 2.7) data set because it contained the best signal-to-noise and the widest q-range. The lowest energy Rosetta model structure that best fit the SANS data had a χ2 value of 1.6. Using this Rosetta model structure as the starting point, an ensemble of 9427 accepted, energy-minimized structures was generated with SASSIE. Model SANS curves were calculated for each of the structures in the ensemble and compared to the data. Figure 6a shows the original 2 mg/mL 2% acetic acid, pH 2.7 SANS data along with model SANS traces for the average and best-fit SASSIE structures. Both model traces lie within the experimental limits of the original data showing that the SASSIE analyses generated structures that fit the SANS data well. Figure 6b is a plot of χ2 versus Rg that includes the SASSIE-derived structures along with the 10 lowest energy Rosetta structures. Clearly, the SASSIE ensemble provides a broader distribution of structures that fit the SANS data than the 10 best Rosetta model structures. The distribution of SASSIE structures indicates that LRAP(+P) can adopt a range of conformations dominated by extended structures with Rg of 1.5 – 2.5 nm.
Figure 6.
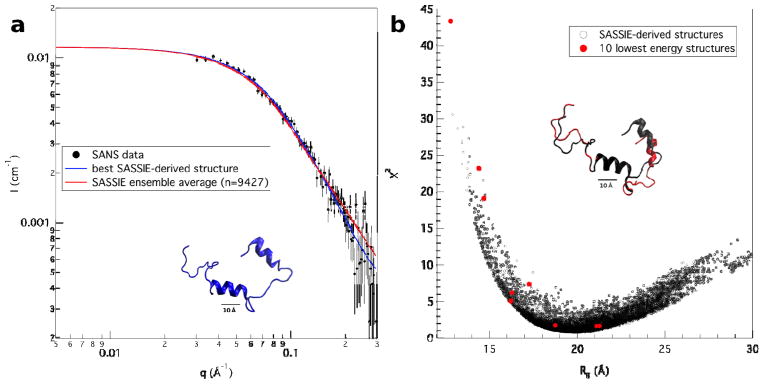
a) The SANS data from 2 mg/mL LRAP(+P) in 2% acetic acid (pH 2.7) along with the corresponding average (red) and best-fit (blue) model trace from the SASSIE calculations. The best-fit (blue) LRAP(+P) structure is shown under the traces. b) A χ2 versus Rg plot for the ensemble of 9427 SASSIE structures derived from the SANS data for 2 mg/mL LRAP(+P) in 2% acetic acid (pH 2.7). The corresponding points for the 10 lowest energy Rosetta model structures are shown in red. The best-fit (black) SASSIE structure and best-fit (red) lowest energy Rosetta structure are superimposed above the traces.
NMR studies of the secondary structure of rLRAP(−P)
Studies of the secondary structure of full-length amelogenin have been performed using NMR at low pH (~3–4) where amelogenin exists as a monomer. (Buchko et al., 2008; Delak et al., 2009) It is not possible to obtain structural information of the amelogenin monomer within the oligomer or nanosphere at physiological conditions (pH 5.8 – 8.0) using current NMR methods because of severe spectral line broadening. Our discovery that LRAP is a monomer under most solution conditions provides an opportunity to study the secondary structure of an amelogenin under physiological conditions using NMR. We were limited to studying LRAP with unphosphorylated S16 (LRAP(−P) because NMR studies required recombinant synthesis to uniformly label LRAP with 15N and 13C. It is not possible to recombinantly synthesize LRAP and enzymatically phosphorylate only one serine residue at S16. Attempts were made to obtain HSQC spectra on unlabeled LRAP (+P) but the concentration of naturally abundant 15N and 13C wasn’t enough to give good spectra.
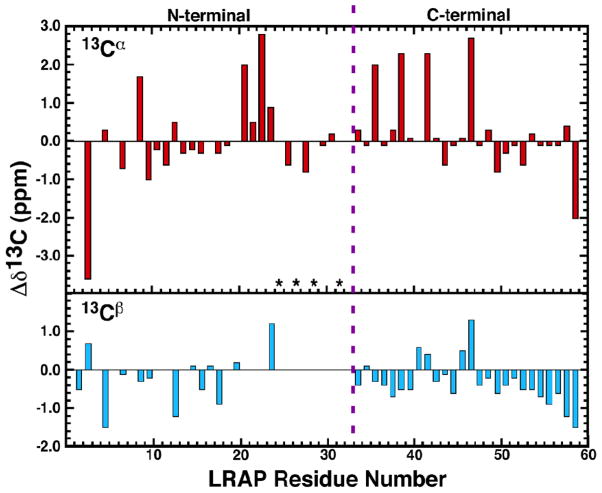
Regions of secondary structure in a protein may be identified from the chemical shifts of the 13Cα and 13Cβ side chain atoms of amino acid residues. Relative to random coil chemical shift values (Δδ13C = δRandom coil-δObserved), these carbon chemical shifts are associated with α-helical (negative 13Cα, positive 13Cβ) or β-strand (positive 13Cα, negative 13Cβ) secondary structure. The deviations of 13Cα and 13Cβ for LRAP(−P) in 150 mM NaCl, SCP buffer, pH 7.4 is shown in Figure 7. Most of the 13Cα and 13Cβ chemical shifts for LRAP(−P) do not deviate significantly from random coil values, with most differences less then 1 ppm. Furthermore, there are no consecutive runs for four or more residues of corresponding positive/negative or negative/positive deviations. Similar observations were made for the deviations of the 13Cα and 13Cβ of LRAP(−P) in 2% acetic acid (pH 2.8) (Figure S6). The solution NMR data for LRAP(−P), therefore, is consistent with random coil structures in the C-terminal and N-terminal regions. It was not possible to obtain 13Cβ data in the K24-I30 region where a canonical α-helix has been observed by solid state NMR (SSNMR) and Rosetta simulations.(Masica et al., 2011) The NMR data suggests a largely disordered structure for LRAP (−P), similar to the disordered, extended conformation reported for full-length porcine amelogenin at pH 3.8 in solution.(Delak et al., 2009) Circular dichroism (CD) spectra obtained for the phosphorylated LRAP (+P) in SCP at pH 7.4 and in solutions with 3 mM CaCl2 (Figure S7) show that LRAP (+P) also has a largely disordered secondary structure.
Figure 7.
Analysis of the observed 13Cα and 13Cβ chemical shift deviations from random coil values for LRAP(−P) in 150 mM NaCl, SCP solution, pH 7.4 where Δδ13C = δRandom coil-δObserved. Red = 13Cα, cyan = 13Cβ. Asterisks indicate residues where it was not possible to obtain unambiguous 13Cβ assignments. The dashed purple line separated the residues present in the N- and C-terminal ends of full-length murine amelogenin. Random coil values obtained from cns_solve_1.1.
DISCUSSION
LRAP quaternary structure
Although numerous studies have shown that full-length amelogenin can self-assemble into quaternary structures called nanospheres, (Fincham et al., 1995; Moradian-Oldak, Tan, and Fincham, 1998b) there has been little research on the quaternary structures of LRAP. Our data shows that LRAP(+P) is primarily a monomer over a wide range of protein concentrations, salt concentrations, pH values, and in the presence of calcium. The only solution condition where monomers are not the dominant species is near the isoelectric point where LRAP(+P) has a low charge and aggregates due to the loss of electrostatic repulsive forces. LRAP(+P), therefore, is a monomer at pH 3, aggregates at low charge in a pH range of 4.2 – 4.5, then forms monomers at pH values from 4.5 to 8.0, as shown in Figure 4a. This behavior is in sharp contrast to full length amelogenin which can form a range of quaternary structures as a function of pH. Amelogenin exists as monomers at pH 3, oligomers at pH 5.5, and nanospheres, self-assemblies of oligomers, at pH 6.8 and higher. (Bromley et al., 2011)
Our data shows that LRAP(−P) has the same monomeric quaternary structure, size, and shape as LRAP(+P) at pH 7.4 in solution. Since the solution structures of LRAP(−P) and LRAP(+P) appear similar, further discussion will refer to LRAP in a general way. Although monomers are the dominant species in LRAP solutions at pH values higher than the isoelectric point, the solutions also contain a relatively low concentration of oligomeric species (0 – 16%). Our studies suggest that the oligomers are derived from the larger aggregates that are formed near the isoelectric point as the solution pH is increased from the starting pH of ~3.0. As the pH is increased above the isoelectric point, LRAP develops a negative charge which promotes dissolution of the aggregates to monomers and a small fraction of oligomers as small as dimers and trimers. When we change the way the solutions are made and dilute LRAP into basic buffer, avoiding the low charge pH regime, we find that the oligomer concentrations are greatly reduced.
Because the oligomers are present in relatively low concentrations, vary in concentration and size sample to sample, and are not present when the aggregation pH regime is avoided during sample preparation, we suggest that these species represent nonequilibrium structures. They are derived from aggregates formed near the isoelectric point of LRAP that do not dissolve completely to the equilibrium monomeric species at the final pH within the 2–3 day period between sample preparation and the SV measurement. We conclude, therefore, that LRAP does not form stable nanospheres under physiological conditions (pH 5.8 – 8.0). The lack of nanosphere formation for LRAP is consistent with a previous study on LRAP interactions using a yeast two-hybrid assay(Paine and Snead, 1997).
The differences in behavior between LRAP and amelogenin may lead to clues on what protein-protein interactions are important in self-assembly. Table 1 shows that murine LRAP consists of the N-terminal residues M1-P33 and C-terminal residues P155-D180 of murine amelogenin but lacks the large central Y34-Q154 region. This suggests that the missing central region is necessary for nanosphere formation. This central region is very hydrophobic, rich in histidine (H, 13 residues), proline (P, 29 residues), and glutamine (Q, 23 residues). Fluorescence experiments of single tryptophan porcine amelogenin mutants showed that there are protein-protein interactions in the W25 and W45 region that increase as amelogenin forms oligomers at pH 5.5 and nanospheres at pH 8.0.(Bromley et al., 2011) The W45 region is located in the N-terminal side of the central domain that is missing in LRAP. The authors suggest that interactions near W25 and W45 promote oligomer formation at pH 5.5 and that further deprotonation of histidine residues in the central domain as the pH is raised leads to aggregation of the oligomers to form nanospheres. Hydrophobic interactions involving the central domain may also promote oligomer-oligomer binding.
Although the central hydrophobic region is necessary to promote oligomer and nanosphere formation in full length amelogenin, the N and C-terminal domains have been proposed to have important roles in controlling the interactions,(Buchko et al., 2008) size,(Moradian-Oldak, 2000) polydispersity,(Moradian-Oldak, 2000) and hierarchical structure (Fang et al., 2011) of the nanospheres. For example, removal of the N-terminal domain resulted in aggregates ranging in size from 3 to 38 nm radius in contrast to the 20 nm radius monodisperse nanospheres of full-length amelogenin.(Moradian-Oldak, 2000) Removal of the C-terminus resulted in larger aggregates (49 nm radius). Fang et al. proposed that the C-terminus may be important in the time-dependent self-assembly of amelogenin into a double-ring barrel dodecamer as an intermediate to the formation of nanospheres as studied by cryo-TEM(Fang et al., 2011). Amelogenin that was lacking the C-terminus (rM166) did not form the dodecamer oligomer intermediate.
LRAP tertiary structure (size and shape)
The SASSIE simulations show that LRAP has limited tertiary folding and is flexible enough to have a number of possible tertiary conformations. The hydrodynamic radius, Rh, from SV and radius of gyration, Rg, from SANS are both ~2.0 nm. This size is consistent with the smallest LRAP sizes obtained previously by DLS.(Le Norcy et al., 2011) Both SV and SANS show that LRAP is extended with an asymmetric structure. For example, the SV f/f0 ratio of ~1.6 are typical of asymmetric proteins such as antibodies and indicate a significantly more extended or flexible structure than globular proteins with f/f0 ratios of 1.2–1.3.(Schuck, 2000) The SASSIE simulations from the SANS data shown in Figure 6 also indicate that LRAP has an asymmetric structure. The size and shape of LRAP was the same at pH 2.7 and pH 7.4 according to the SANS studies. This suggests that the small increases in the f/f0 ratio from 1.59 at pH 8 to 1.64 at pH 3 may be due to slight increases in hydration and not due to any significant changes in shape.
LRAP secondary structure
Solution NMR studies of LRAP(−P) at low (2.8) and high (7.4) pH are consistent with a predominantly unstructured protein, consistent with circular dichroism studies of LRAP (Le et al., 2006; Chen et al., 2007). SSNMR studies have also shown evidence for random coil structures in the C-terminus and random coil to loose helices in the inner N-terminus of the protein when lyophilized from solution. (Masica et al., 2011; Lu et al., 2013; Tarasevich et al., 2013) Rosetta simulations showed that the C-terminus (48–55) and inner N-terminus(10–20) had regions that were partially helical, sampling both helical and turn-like conformations.(Tarasevich et al., 2013) The solution NMR studies presented here suggest that any loose or partially helical conformations in the terminal regions observed in the solid state or by simulation are likely transient in nature when LRAP is in solution. The Rosetta simulations(Masica et al., 2011) especially overestimate the degree of helical structure in the C and N-terminal regions compared to the experimental studies. The most structured part of LRAP is a stable helix localized in the L23-R31 region as evidenced by both SSNMR experiments and Rosetta simulations.(Masica et al., 2011) This region, however, was experimentally inaccessible by the solution state NMR experiments described here.
Although only LRAP(−P) was studied by solution NMR in this study, CD studies show that LRAP(+P) is also predominantly unstructured. The local secondary structures of both LRAP(+P) and LRAP(−P) in the lyophilized state were also studied by SSNMR measurements of the 13C-15N backbone distance. (Masica et al., 2011; Lu et al., 2013) There were no significant differences in structure between LRAP(+P) and LRAP(−P) in the V19-L23 and K24-S28 regions and moderate differences in the G8-Y12 and K15-V19 regions. Interestingly, CD studies show no evidence of secondary structure for LRAP(+P) in the presence of calcium even though our SV studies and previous SAXS studies(Le Norcy et al., 2012) find that LRAP has a more collapsed tertiary structure in the presence of calcium. This result suggests that calcium does not induce a random coil to α-helix or β-sheet transition for LRAP, consistent with previous studies.(Le et al., 2006)
Physiological significance
The general LRAP secondary, tertiary, and quaternary structures are similar to those of the SIBLING family of biomineralization proteins, known to be involved in the formation of bone and dentin.(George and Veis, 2008) These glycoproteins include bone sialoprotein (BSP), dental phosphophoryn (DPP), dentin matrix protein 1 (DMP1), and dentin sialoprotein (DSP). The SIBLING proteins exist as monomers, have flexible backbones, and have limited global secondary or tertiary structure.(Fisher et al., 2001) There is evidence that the SIBLING proteins can have dual roles in bone and dentin formation - a structural function in guiding crystal nucleation and growth and a cell signaling function.(George and Veis, 2008) For example, BSP has been suggested to be the main nucleator of HAP in bone(Ganss et al., 1999) and promotes osteoblast differentiation.(Gordon et al., 2007) Like the SIBLING proteins, the monomeric, flexible, and relatively unstructured nature of LRAP may be important in its function as a cell signaling molecule, promoting binding interactions with cell surface receptors. In addition, a flexible, monomeric protein conformation may be important in controlling the nucleation and growth of HAP crystals, allowing adaptation to the growing crystal lattice. The SV, SANS, and NMR techniques developed here to elucidate the structure of LRAP can also be applied to studying truncated forms of LRAP formed by cleavage by the enamel protease Mmp-20. (Li et al., 1999; Nagano et al., 2009) LRAP and other amelogenin proteins degrade during the process of enamel formation and the cleavage products may have different biological functions compared to the parent proteins.
SUMMARY
Our studies have shown that analytical techniques such as SV and SANS can provide important insights into the structure and function of biomineralization proteins. We found that LRAP does not form nanospheres like the higher molecular weight amelogenins and exists as a monomer over a range of solution conditions. This result supports the hypothesis that the central hydrophobic domain present in full length amelogenin is critical to self-assembly. The LRAP monomer is extended and flexible with limited tertiary folding and a limited degree of local secondary structure. The quaternary and tertiary structure of LRAP is relatively unchanged as a function of solution pH which may be critical to its function in an enamel environment of changing solution conditions.
Supplementary Material
Acknowledgments
This work was supported by NIH-NIDCR Grant DE-015347 (PNNL) and was performed in part at Pacific Northwest National Laboratory, operated by Battelle for the US-DOE. A portion of the research was performed in the EMSL, a national scientific user facility sponsored by the DOE-OBER at PNNL. The SANS studies benefitted from CCP-SAS software developed by Joseph Curtis through a joint EPSRC (EP/K039121/1) and NSF (CHE-1265821) grant.
Footnotes
Certain commercial equipment, instruments, or materials are identified in this paper to foster understanding. Such identification does not imply recommendation or endorsement by the National Institute of Standards and Technology, nor does it imply that the materials or equipment identified are necessarily the best available for the purpose.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Amin HD, Olsen I, Knowles JC, Donos N. Differential Effect of Amelogenin Peptides on Osteogenic Differentiation In Vitro: Identification of Possible New Drugs for Bone Repair and Regeneration. Tissue Eng Part A. 2012;18:1193–1202. doi: 10.1089/ten.TEA.2011.0375. [DOI] [PubMed] [Google Scholar]
- Boabaid F, Gibson CW, Kuehl MA, Berry JE, Snead ML, Nociti FHJ, Katchburian E, Somerman MJ. Leucine-rich amelogenin peptide: a candidate signaling molecule during cementogenesis. J Periodontol. 2004;75:1126–36. doi: 10.1902/jop.2004.75.8.1126. [DOI] [PubMed] [Google Scholar]
- Bromley KM, Kiss AS, Lokappa SB, Lakshminarayanan R, Fan D, Ndao M, Evans JS, Moradian-Oldak J. Dissecting amelogenin protein nanospheres: characterization of metastable oligomers. J Biol Chem. 2011;286:34643–34653. doi: 10.1074/jbc.M111.250928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchko GW, Tarasevich BJ, Bekhazi J, Snead ML, Shaw WJ. A Solution NMR Investigation into the Early Events of Amelogenin Nanosphere Self-Assembly Initiated with Sodium Chloride or Calcium Chloride †. Biochemistry. 2008;47:13215–13222. doi: 10.1021/bi8018288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchko GW, Tarasevich BJ, Roberts J, Snead ML, Shaw WJ. A solution NMR investigation into the murine amelogenin splice-variant LRAP (Leucine-Rich Amelogenin Protein) Biochimica et Biophysica Acta (BBA) - Proteins & Proteomics. 2010;1804:1768–1774. doi: 10.1016/j.bbapap.2010.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Wang Q, Shen J, Pan H, Wu T. Adsorption of Leucine-Rich Amelogenin Protein on Hydroxyapatite (001) Surface through -COO- Claws. J Phys Chem C. 2007;111:1284–1290. [Google Scholar]
- Curtis Raghunandan, Nanda Krueger. SASSIE: A program to study intrinsically disordered biological molecules and macromolecular ensembles using experimental scattering restraints. Comput Phys Commun. 2012;183:8–8. [Google Scholar]
- Delak K, Harcup C, Lakshminarayanan R, Sun Z, Fan Y, Moradian-Oldak J, Evans JS. The Tooth Enamel Protein, Porcine Amelogenin, Is an Intrinsically Disordered Protein with an Extended Molecular Configuration in the Monomeric Form †. Biochemistry. 2009;48:2272–2281. doi: 10.1021/bi802175a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang PA, Conway JF, Margolis HC, Simmer JP, Beniash E. Hierarchical self-assembly of amelogenin and the regulation of biomineralization at the nanoscale. P Natl Acad Sci Usa. 2011;108:14097–14102. doi: 10.1073/pnas.1106228108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filipe V, Hawe A, Jiskoot W. Critical evaluation of Nanoparticle Tracking Analysis (NTA) by NanoSight for the measurement of nanoparticles and protein aggregates. Pharm Res. 2010;27:796–810. doi: 10.1007/s11095-010-0073-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fincham AG, Moradian-Oldak J, Diekwisch TG, Lyaruu DM, Wright JT, Bringas P, Slavkin HC. Evidence for amelogenin “nanospheres” as functional components of secretory-stage enamel matrix. Journal of Structural Biology. 1995;115:50–59. doi: 10.1006/jsbi.1995.1029. [DOI] [PubMed] [Google Scholar]
- Fincham AG, Moradian-Oldak J, Simmer JP. The Structural Biology of the Developing Dental Enamel Matrix. Journal of Structural Biology. 1999;126:30–30. doi: 10.1006/jsbi.1999.4130. [DOI] [PubMed] [Google Scholar]
- Fisher LW, Torchia DA, Fohr B, Young MF, Fedarko NS. Flexible structures of SIBLING proteins, bone sialoprotein, and osteopontin. Biochemical and Biophysical Research Communications. 2001;280:460–465. doi: 10.1006/bbrc.2000.4146. [DOI] [PubMed] [Google Scholar]
- Ganss B, Kim RH, Sodek J. Bone sialoprotein. Critical Reviews in Oral Biology & Medicine. 1999;10:79–98. doi: 10.1177/10454411990100010401. [DOI] [PubMed] [Google Scholar]
- George A, Veis A. Phosphorylated Proteins and Control over Apatite Nucleation, Crystal Growth, and Inhibition. Chem Rev. 2008;108:4670–4693. doi: 10.1021/cr0782729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson CW. The Amelogenin Proteins and Enamel Development in Humans and Mice. J Oral Biosci. 2011;53:248–256. doi: 10.2330/joralbiosci.53.248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson CW, Golub E, Ding W, Shimokawa H, Young M, Termine J, Rosenbloom J. Identification of the leucine-rich amelogenin peptide (LRAP) as the translation product of an alternatively spliced transcript. Biochemical and Biophysical Research Communications. 1991;174:1306–1312. doi: 10.1016/0006-291x(91)91564-s. [DOI] [PubMed] [Google Scholar]
- Gibson CW, Kucich U, Collier P, Shen G, Decker S, Bashir M, Rosenbloom J. Analysis of amelogenin proteins using monospecific antibodies to defined sequences. Connect Tissue Res. 1995;32:109–14. doi: 10.3109/03008209509013711. [DOI] [PubMed] [Google Scholar]
- Gibson CW, Li Y, Daly B, Suggs C, Yuan ZA, Fong H, Simmons D, Aragon M, Kulkarni AB, Wright JT. The Leucine-Rich Amelogenin Peptide Alters the Amelogenin Null Enamel Phenotype. Cells Tissues Organs. 2009;189:169–174. doi: 10.1159/000151384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson CW, Li Y, Suggs C, Kuehl MA, Pugach MK, Kulkarni AB, Wright JT. Rescue of the murine amelogenin null phenotype with two amelogenin transgenes. European Journal of Oral Sciences. 2011;119(Suppl 1):70–4. doi: 10.1111/j.1600-0722.2011.00882.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson CW, Yuan ZA, Hall B, Longenecker G, Chen E, Thyagarajan T, Sreenath T, Wright JT, Decker S, Piddington R, Harrison G, Kulkarni AB. Amelogenin-deficient mice display an amelogenesis imperfecta phenotype. J Biol Chem. 2001;276:31871–31875. doi: 10.1074/jbc.M104624200. [DOI] [PubMed] [Google Scholar]
- Glinka CJ, Barker JG, Hammouda B, Krueger S, Moyer JJ, Orts WJ. The 30 m Small-Angle Neutron Scattering Instruments at the National Institute of Standards and Technology. J Appl Crystallogr. 1998;31:430–445. [Google Scholar]
- Gordon JAR, Tye CE, Sampaio AV, Underhill TM, Hunter GK, Goldberg HA. Bone sialoprotein expression enhances osteoblast differentiation and matrix mineralization in vitro. Bone. 2007;41:462–473. doi: 10.1016/j.bone.2007.04.191. [DOI] [PubMed] [Google Scholar]
- Habelitz S, DenBesten PK, Marshall SJ, Marshall GW, Li W. Self-assembly and effect on crystal growth of the leucine-rich amelogenin peptide. European Journal of Oral Sciences. 2006;114:315–319. doi: 10.1111/j.1600-0722.2006.00312.x. [DOI] [PubMed] [Google Scholar]
- Hammarstrom L. The role of enamel matrix proteins in the development of cementum and periodontal tissues. Ciba Found Symp. 1997;205:246–55. doi: 10.1002/9780470515303.ch17. discussion 255–60. [DOI] [PubMed] [Google Scholar]
- Heidorn DBD, Trewhella JJ. Comparison of the crystal and solution structures of calmodulin and troponin C. Biochemistry. 1988;27:909–915. doi: 10.1021/bi00403a011. [DOI] [PubMed] [Google Scholar]
- Heijl LL, Heden GG, Svärdström GG, Ostgren AA. Enamel matrix derivative (EMDOGAIN) in the treatment of intrabony periodontal defects. J Clin Periodontol. 1997;24:705–714. doi: 10.1111/j.1600-051x.1997.tb00253.x. [DOI] [PubMed] [Google Scholar]
- Iijima M, Moradian-Oldak J. Control of octacalcium phosphate and apatite crystal growth by amelogenin matrices. J Mater Chem. 2004;14:2189. [Google Scholar]
- Kline SR. Reduction and analysis of SANS and USANS data using IGOR Pro. J Appl Crystallogr. 2006;39:895–900. [Google Scholar]
- Krueger SS, Gorshkova II, Brown JJ, Hoskins JJ, McKenney KHK, Schwarz FPF. Determination of the conformations of cAMP receptor protein and its T127L,S128A mutant with and without cAMP from small angle neutron scattering measurements. J Biol Chem. 1998;273:20001–20006. doi: 10.1074/jbc.273.32.20001. [DOI] [PubMed] [Google Scholar]
- Laue TM, Shah BD, Ridgeway TM, Pelletier SL. Computer-aided interpretation of analytical sedimentation data for proteins. In: Harding SE, Rowe AJ, Horton JC, editors. Analytical Ultracentrifugation in Biochemistry and Polymer Science. Royal Society of Chemistry; Cambridge: 1992. pp. 90–125. [Google Scholar]
- Le Norcy E, Kwak SY, Allaire M, Fratzl P, Yamakoshi Y, Simmer JP, Margolis HC. Effect of phosphorylation on the interaction of calcium with leucine-rich amelogenin peptide. European Journal of Oral Sciences. 2012;119(Suppl 1):97–102. doi: 10.1111/j.1600-0722.2011.00900.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Norcy E, Kwak SY, Wiedemann-Bidlack FB, Beniash E, Yamakoshi Y, Simmer JP, Margolis HC. Leucine-rich amelogenin peptides regulate mineralization in vitro. J Dent Res. 2011;90:1091–1097. doi: 10.1177/0022034511411301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le TQ, Gochin M, Featherstone JDB, Li W, DenBesten PK. Comparative calcium binding of leucine-rich amelogenin peptide and full-length amelogenin. European Journal of Oral Sciences. 2006;114(Suppl 1):320–6. doi: 10.1111/j.1600-0722.2006.00313.x. discussion 327–9, 382. [DOI] [PubMed] [Google Scholar]
- Le TQ, Zhang Y, Li W, Denbesten PK. The effect of LRAP on enamel organ epithelial cell differentiation. J Dent Res. 2007;86:1095–9. doi: 10.1177/154405910708601114. [DOI] [PubMed] [Google Scholar]
- Lebowitz JJ, Lewis MSM, Schuck PP. Modern analytical ultracentrifugation in protein science: a tutorial review. Protein Science. 2002;11:2067–2079. doi: 10.1110/ps.0207702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W, Machule D, Gao C, Denbesten PK. Activation of recombinant bovine matrix metalloproteinase-20 and its hydrolysis of two amelogenin oligopeptides. European Journal of Oral Sciences. 1999;107:352–9. doi: 10.1046/j.0909-8836.1999.eos107506.x. [DOI] [PubMed] [Google Scholar]
- Lu JX, Xu YS, Shaw WJ. Phosphorylation and Ionic Strength Alter the LRAP–HAP Interface in the N-Terminus. Biochemistry. 2013;52:2196–2205. doi: 10.1021/bi400071a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacKerell AD, Bashford D, Bellott, Dunbrack RL, Evanseck JD, Field MJ, Fischer S, Gao J, Guo H, Ha S, Joseph-McCarthy D, Kuchnir L, Kuczera K, Lau FTK, Mattos C, Michnick S, Ngo T, Nguyen DT, Prodhom B, Reiher WE, Roux B, Schlenkrich M, Smith JC, Stote R, Straub J, Watanabe M, Wiórkiewicz-Kuczera J, Yin D, Karplus M. All-Atom Empirical Potential for Molecular Modeling and Dynamics Studies of Proteins †. J Phys Chem B. 1998;102:3586–3616. doi: 10.1021/jp973084f. [DOI] [PubMed] [Google Scholar]
- Masica DL, Gray JJ, Shaw WJ. Partial High-Resolution Structure of Phosphorylated and Non-phosphorylated Leucine-Rich Amelogenin Protein Adsorbed to Hydroxyapatite. J Phys Chem C Nanomater Interfaces. 2011;115:13775–13785. doi: 10.1021/jp202965h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moradian-Oldak J. Self-Assembly Properties of Recombinant Engineered Amelogenin Proteins Analyzed by Dynamic Light Scattering and Atomic Force Microscopy. Journal of Structural Biology. 2000;131:27–37. doi: 10.1006/jsbi.2000.4237. [DOI] [PubMed] [Google Scholar]
- Moradian-Oldak J. Amelogenins: assembly, processing and control of crystal morphology. Matrix Biol. 2001;20:293–305. doi: 10.1016/s0945-053x(01)00154-8. [DOI] [PubMed] [Google Scholar]
- Moradian-Oldak J, Leung W, Fincham AG. Temperature and pH-dependent supramolecular self-assembly of amelogenin molecules: a dynamic light-scattering analysis. Journal of Structural Biology. 1998;122:320–327. doi: 10.1006/jsbi.1998.4008. [DOI] [PubMed] [Google Scholar]
- Moradian-Oldak J, Simmer JP, Lau EC, Sarte PE, Slavkin HC, Fincham AG. Detection of monodisperse aggregates of a recombinant amelogenin by dynamic light scattering. Biopolymers. 1994;34:1339–1347. doi: 10.1002/bip.360341006. [DOI] [PubMed] [Google Scholar]
- Moradian-Oldak J, Tan J, Fincham AG. Interaction of amelogenin with hydroxyapatite crystals: an adherence effect through amelogenin molecular self-association. Biopolymers. 1998a;46:225–38. doi: 10.1002/(SICI)1097-0282(19981005)46:4<225::AID-BIP4>3.0.CO;2-R. [DOI] [PubMed] [Google Scholar]
- Moradian-Oldak J, Tan J, Fincham AG. Interaction of amelogenin with hydroxyapatite crystals: an adherence effect through amelogenin molecular self-association. Biopolymers. 1998b;46:225–238. doi: 10.1002/(SICI)1097-0282(19981005)46:4<225::AID-BIP4>3.0.CO;2-R. [DOI] [PubMed] [Google Scholar]
- Nagano T, Kakegawa A, Yamakoshi Y, Tsuchiya S, Hu JCC, Gomi K, Arai T, Bartlett JD, Simmer JP. Mmp-20 and Klk4 Cleavage Site Preferences for Amelogenin Sequences. J Dent Res. 2009;88:823–828. doi: 10.1177/0022034509342694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paine ML, Snead ML. Protein interactions during assembly of the enamel organic extracellular matrix. J Bone Miner Res. 1997;12:221–227. doi: 10.1359/jbmr.1997.12.2.221. [DOI] [PubMed] [Google Scholar]
- Phillips JCJ, Braun RR, Wang WW, Gumbart JJ, Tajkhorshid EE, Villa EE, Chipot CC, Skeel RDR, Kalé LL, Schulten KK. Scalable molecular dynamics with NAMD. J Comput Chem. 2005;26:1781–1802. doi: 10.1002/jcc.20289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Philo JS. Improved methods for fitting sedimentation coefficient distributions derived by time-derivative techniques. Anal Biochem. 2006;354:238–246. doi: 10.1016/j.ab.2006.04.053. [DOI] [PubMed] [Google Scholar]
- Ravindranath RMH, Devarajan A, Bringas P., Jr Enamel formation in vitro in mouse molar explants exposed to amelogenin polypeptides. Arch Oral Biol. 2007;52:1161–1171. doi: 10.1016/j.archoralbio.2007.06.008. [DOI] [PubMed] [Google Scholar]
- Robinson C, Kirkham J, Brookes SJ, Bonass WA, Shore RC. The chemistry of enamel development. Int J Dev Biol. 1995;39:145–152. [PubMed] [Google Scholar]
- Sasaki SS, Takagi TT, Suzuki MM. Cyclical changes in pH in bovine developing enamel as sequential bands. Arch Oral Biol. 1991;36:227–231. doi: 10.1016/0003-9969(91)90090-h. [DOI] [PubMed] [Google Scholar]
- Schuck PP. Size-Distribution Analysis of Macromolecules by Sedimentation Velocity Ultracentrifugation and Lamm Equation Modeling. Biophysj. 2000;78:14–14. doi: 10.1016/S0006-3495(00)76713-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sculean AA, Kiss AA, Miliauskaite AA, Schwarz FF, Arweiler NBN, Hannig MM. Ten-year results following treatment of intra-bony defects with enamel matrix proteins and guided tissue regeneration. J Clin Periodontol. 2008;35:817–824. doi: 10.1111/j.1600-051X.2008.01295.x. [DOI] [PubMed] [Google Scholar]
- Semenyuk AV, Svergun DI. GNOM – a program package for small-angle scattering data processing. J Appl Crystallogr. 1991;24:537–540. doi: 10.1107/S0021889812007662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw WJ, Campbell AA, Paine ML, Snead ML. The COOH Terminus of the Amelogenin, LRAP, Is Oriented Next to the Hydroxyapatite Surface. J Biol Chem. 2004;279:40263–40266. doi: 10.1074/jbc.C400322200. [DOI] [PubMed] [Google Scholar]
- Stahl J, Nakano Y, Kim SO, Gibson CW, Le T, Denbesten P. Leucine rich amelogenin peptide alters ameloblast differentiation in vivo. Matrix Biol. 2013;32:432–442. doi: 10.1016/j.matbio.2013.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan J, Leung W, Moradian-Oldak J, Zeichner-David M, Fincham AG. Quantitative Analysis of Amelogenin Solubility. J Dent Res. 1998;77:1388–1396. doi: 10.1177/00220345980770060301. [DOI] [PubMed] [Google Scholar]
- Tarasevich BJ, Howard CJ, Larson JL, Snead ML, Simmer JP, Paine M, Shaw WJ. The nucleation and growth of calcium phosphate by amelogenin. J Cryst Growth. 2007;304:407–415. doi: 10.1016/j.jcrysgro.2007.02.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarasevich BJ, Lea S, Shaw WJ. The leucine rich amelogenin protein (LRAP) adsorbs as monomers or dimers onto surfaces. Journal of Structural Biology. 2010;169:266–276. doi: 10.1016/j.jsb.2009.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarasevich BJ, Perez-Salas U, Masica DL, Philo J, Kienzle P, Krueger S, Majkrzak CF, Gray JL, Shaw WJ. Neutron Reflectometry Studies of the Adsorbed Structure of the Amelogenin, LRAP. J Phys Chem B. 2013;117:3098–3109. doi: 10.1021/jp311936j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Termine JD, Belcourt AB, Christner PJ, Conn KM, Nylen MU. Properties of dissociatively extracted fetal tooth matrix proteins. I. Principal molecular species in developing bovine enamel. J Biol Chem. 1980;255:9760–9768. [PubMed] [Google Scholar]
- Veis A, Tompkins K, Alvares K, Wei K, Wang L, Wang XS, Brownell AG, Jengh SM, Healy KE. Specific amelogenin gene splice products have signaling effects on cells in culture and in implants in vivo. J Biol Chem. 2000;275:41263–41272. doi: 10.1074/jbc.M002308200. [DOI] [PubMed] [Google Scholar]
- Warotayanont R, Frenkel B, Snead ML, Zhou Y. Leucine-rich amelogenin peptide induces osteogenesis by activation of the Wnt pathway. Biochemical and Biophysical Research Communications. 2009;387:558–63. doi: 10.1016/j.bbrc.2009.07.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warotayanont R, Zhu D, Snead ML, Zhou Y. Leucine-rich amelogenin peptide induces osteogenesis in mouse embryonic stem cells. Biochemical and Biophysical Research Communications. 2008;367:1–6. doi: 10.1016/j.bbrc.2007.12.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen X, Cawthorn WP, MacDougald OA, Stupp SI, Snead ML, Zhou Y. The influence of Leucine-rich amelogenin peptide on MSC fate by inducing Wnt10b expression. Biomaterials. 2011;32:6478–6486. doi: 10.1016/j.biomaterials.2011.05.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wishart DS, Bigam CG, Yao J, Abildgaard F, Dyson HJ, Oldfield E, Markley JL, Sykes BD. 1H, 13C and 15N chemical shift referencing in biomolecular NMR. J Biomol NMR. 1995;6:135–140. doi: 10.1007/BF00211777. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.