Abstract
Streptococcus pneumoniae (pneumococcus) is a potential cause of bacterial endophthalmitis in humans that can result in ocular morbidity. We sought to identify pneumococcal genes that are differentially expressed during growth in the vitreous humor of the eye in an experimental endophthalmitis model. Microarray analysis was used to identify genes that were differentially expressed when pneumococci replicated in the vitreous of rabbit eyes as compared with bacteria grown in vitro in Todd Hewitt medium. Array results were verified by quantitative real-time PCR analysis of representative genes. Select genes potentially playing a role in virulence during endophthalmitis were deleted and mutants were tested for reduced eye pathogenesis and altered adhesion to host cells. Array analysis identified 134 genes that were differentially expressed during endophthalmitis. 112 genes demonstrated increased expression during growth in the eye whereas 22 were down-regulated. Real-time analysis verified increased expression of neuraminidase A (SP1693), neuraminidase B (SP1687), and serine protease (SP1954), and decreased expression of RlrA (SP0461) and choline transporter (SP1861). Mutation of neuraminidases A and B had no major effect on pathogenesis. Loss of SP1954 led to increased adherence to host cells. S. pneumoniae enhances and represses expression of a variety of genes during endophthalmitis. While some of these genes reflect changes in metabolic requirements, some appear to play a role in immune evasion and pathogenesis in the eye.
Keywords: pneumococcus, bacterial endophthalmitis, gene expression analysis, microarray
Introduction
Streptococcus pneumoniae (pneumococcus) is a human pathogen responsible for diseases ranging from otitis media and sinusitis to bacteremia and meningitis. Aside from these most prevalent manifestations of pneumococcal disease, the pneumococcus is also a common cause of bacterial endophthalmitis, usually following ocular surgery or trauma [1-4]. Recent information on the occurrence of endophthalmitis implicates the pneumococcus as the third most frequently isolated bacterial species after Staphylococcus aureus and coagulase-negative staphylococci, and the second most isolated species from ocular infections as a whole [5]. Pneumococcal endophthalmitis is a devastating infection, often resulting in evisceration or a poor visual outcome [1, 3, 4, 6].
Two pneumococcal factors that have been identified as important in the pathogenesis of endophthalmitis are pneumolysin, a cholesterol-dependent cytolysin, and the polysaccharide capsule. Vitreal injection of purified pneumolysin was found to induce symptoms of endophthalmitis in rats similar to those caused by whole bacteria [7]. Likewise, deletion of the gene encoding pneumolysin caused a decrease in the severity of endophthalmitis in the early stages of the disease. The in vivo pathogenesis of the pneumolysin-negative strain, however, resembled the parent strain at later stages in the disease [8]. The bacterial polysaccharide capsule was also determined to be important not only for the severity of endophthalmitis, but also for pneumococcal survival in the rabbit vitreous [9]. Despite the importance of these findings regarding pneumococcal pathogenesis in the vitreous, pneumolysin and capsule are not the only factors involved endophthalmitis because deletion of the genes encoding these factors does not result in complete abrogation of the disease [8, 9].
In the current genomics era, microarray technology has allowed us to characterize global transcriptome changes of bacteria in response to various stimuli and even during different stages of infection. Specifically for pneumococcus, bacterial microarray studies have identified differential gene expression involved in colonization [10], phase variation [11], competence [8, 12], metal transport [13-15], host cell interactions [16-20], and invasive disease [17, 21]. Also, numerous studies utilizing microarrays have identified pneumococcus-induced transcriptional effects on host cells [22-25]. The data from these studies have provided invaluable insight into the molecular mechanisms of pneumococcal-host cell interactions.
A genome-wide analysis of pneumococcal factors involved in eye infections is currently lacking. In this study, we characterized the global gene expression profile of S. pneumoniae during experimental endophthalmitis in a rabbit model. Utilizing S. pneumoniae TIGR4 microarray analysis, we identified genes differentially expressed during bacterial replication in the eye that may be contributing to the ocular damage seen during these types of infections.
Methods
Bacterial strains and growth conditions
S. pneumoniae TIGR4 (#BAA-334, American Type Culture Collection, Manassas, VA) and D39 (#6302, American Type Culture Collection) were maintained in Todd Hewitt broth containing 0.5% yeast extract (THY) and 20% glycerol at -80°C. Bacterial colonies were routinely isolated from frozen stocks on blood agar base containing 5% sheep erythrocytes (BAP) and incubation at 37°C and 5% CO2. Several isolated colonies were used to inoculate THY, and each resulting culture was incubated at 37°C and 5% CO2 for 15 hours. Each culture was then diluted 1:100 in THY and was incubated under the same conditions to an optical density (A600) of 0.3, which corresponded to approximately 108 colony-forming units (CFU) per mL as determined by growth curves. Bacterial cultures were then used for in vitro RNA isolation or rabbit vitreous infections.
Mutant construction
Deletion mutants of nanA gene in strain D39 were performed by splice overlap extension (SOEing) PCR method as previously described[26]. Briefly, 500 base pair regions flanking upstream and downstream of target genes were amplified by PCR and spliced to an antibiotic cassette. PCR was then used to amplify the final product using outer primers for each construct and this product was transformed into S. pneumoniae D39 by conventional methods. Deletion mutants (ΔNanA) were selected for by growth on BAP supplemented with erythromycin (0.5μg/ml_). The nanB gene was interrupted by insertion-duplication mutagenesis using the suicide vector pEVP3 kindly provided by Dr. Ed Swiatlo. A 500 nucleotide segment of the nanB gene near the start codon was amplified by PCR primers containing BamHI restriction sites. This product was digested and ligated into pEVP3 and subsequently transformed into E. coli DH5α. Positive clones were selected by growing on Luria-Bertani agar plates containing chloramphenicol (170μg/ml_). The knockout plasmid (pJAT81) was purified by GenElute™ Plasmid Miniprep Kit (Sigma-Aldrich, St Louis, MO) and used to transform S. pneumoniae D39. Insertion mutants were isolated by plating transformations on chloramphenicol plates (5μg/mL). Mutants were verified by PCR. Double mutants were created by transforming the ΔNanB strain with chromosomal DNA from ΔNanA and selection by growth on BAP containing erythromycin and chloramphenicol. Mutations were verified by PCR. All primers used for creating mutants are listed in Table 1.
Table 1.
Oigonucleotide primers used in this Study. All primers are 5′-3′.
| Primer | Sequence | |
|---|---|---|
| Mutagenesis Primers | ||
| NanAKO1 | TTGCCGTATATATGTTACTGAC | |
| NanAKO2 | GTTTGCTTCTAAGTCTTATTTCCTTATTCCAGAAAATGC | |
| NanAKO3 | GAGTCGCTTTTGTAAATTTGGCTAAAGTGCATTTTCTGG | |
| NanAKO4 | TCCTATCCAAGTTCCCCATTC | |
| NanBKO1 | CCGCGGGATCCCTGTTGTAGGCATTAGTC | |
| NanBKO2 | CCGCGGGATCCACCTTCACGATTAACAGCTCC | |
| ErmF | GGAAATAAGACTTAGAAGCAAAC | |
| ErmR | CCAAA T TT ACAAAAGCGACTC | |
| qRT-PCR Primers | Product length (bp) | |
| SP0346-F | GACCGCGGGCGCCATCAAC | 230 |
| SP0346-R | GAAGGAAGATCCATCCGACCTGTCC | |
| SP0461-F | AGGAGTTATCGACCAGCATCGGC | 230 |
| SP0461-R | TCAATGGCTCTGTCAAATGCTCCTCTG | |
| SP1219-F | GCCGTTCGTGGTATGAGCCG | 232 |
| SP1219-R | TAAGACCGGCCAGCAAGCCA | |
| SP1954-F | TTCGGTCGGAGGCATAGATCGCT | 230 |
| SP1954-R | TCGCACCAGAAACT T TAGGAGCAGC | |
| SP1693-F | CCAGATGCCAAGGCCCCAGC | 244 |
| SP1693-R | GGCGCACTCGGCCT T TAGGT | |
| SP1687-F | GCAGAAGCACAAATGGTTGAACTGAG | 249 |
| SP1687-R | GCCTCCCTTGCGGCCACTTC | |
| SP1861-F | GCCGAGTATGGGCATCGCTTACC | 240 |
| SP1861-R | ACGGTCCGCCAACTTCAAGGC | |
| SP1954-F | TTCGGTCGGAGGCATAGATCGCT | 230 |
| SP1954-R | TCGCACCAGAAACTTTAGGAGCAGC |
Rabbit vitreous infections
The use of an animal model was necessary (rather than growing bacteria in vitreous humor in vitro, for instance) because the vitreous cavity undergoes a dynamic movement of fluid and turnover of components including salts, various ions, and hyaluronic acid in the living animal. These changes could not be accurately recreated in vitro. The rabbit has long been a model species for bacterial endophthalmitis for a number of important reasons including size of the eye and similarities to pathology seen in with human infections. New Zealand white rabbits, 2.0-3.0 kg and either sex (Charles River Laboratories, Wilmington, MA), were used in these studies that adhered to the Association for Research in Vision and Ophthalmology Statement for the Use of Animals in Ophthalmic and Vision Research and the tenets of the Declaration of Helsinki. Rabbits were anesthetized by subcutaneous injection of a mixture of xylazine (50 mg/kg; Lloyd Laboratories, Shenandoah, IA) and ketamine hydrochloride (5 mg/kg; Butler Animal Health Supply, Dublin, OH). Proparacaine hydrochloride (Akorn, Inc., Buffalo Grove, IL) was topically applied to each eye. A 30-gauge needle was used to inoculate bacteria into the vitreous humor of each eye. Bacterial cultures were diluted such that each vitreous humor was infected with approximately 102CFU/10 μL. Twenty hours after infection, rabbits were killed by an intravenous overdose of pentobarbital sodium (100 mg/mL; Sigma-Aldrich, St. Louis, MO). Vitreous was aspirated from eyes with a 22-gauge needle, and was used for the determination of bacterial CFU and bacterial RNA isolation. For subsequent experiments involving the analysis of nanA and nanB deletions in the pathogenesis of endophthalmitis, rabbits were examined with the aid of a slit lamp biomicroscope as previously described [27] and by electroretinography (ERG) with a handheld multispecies ERG unit (Ocuscience LLC, Kansas City, MO) 24 and 48 hours after infection, and then euthanized for the determination of bacterial CFU present in the vitreous.
Data were analyzed using the Statistical Analysis System (SAS) program for computers (Cary, North Carolina, USA). Clinical scores were analyzed using a non-parametric one-way analysis of variance. Bacterial CFU recovery and percent loss of retinal function were analyzed using the general linear models procedure of least squares means. P < 0.05 was considered significant.
Bacterial RNA isolation
S. pneumoniae was harvested from logarithmic phase cultures in vitro or rabbit vitreous in vivo. Cultures or freshly aspirated vitreous were immediately added to RNAprotect Bacteria reagent according to manufacturer instructions (Qiagen, Valencia, CA). Bacteria were collected by centrifugation at 16,000 × g for 5 minutes and suspended in phenol:chloroform saturated solution pH 4.7 for 5 minutes at 65°C. An equal volume of NAES (50 mM C2H3NaO2 pH 5.0, 10 mM EDTA, and 1% SDS) maintained at 65°C was added and the mixture was incubated for 5 minutes at 65°C, then on ice for 2 minutes. The mixture was centrifuged for 2 minutes at 16,000 × g and 4°C, and the upper aqueous phase was collected. A second phenol:chloroform extraction of this aqueous phase was performed, and the RNA was precipitated with 0.1 volume 3 M C2H3NaO2 and 2.5 volumes 100% ethanol for 24 hours at -20°C. The RNA was treated with RNase inhibitor and DNase by standard methods and was further purified by chromatography using RNeasy Mini Kit (Qiagen). If a sample had a 260/230 value of approximately less than 1.0, it was purified prior to priming. The volume of the RNA was adjusted to 100μl prior to addition of 350μl of buffer RLT, mixed, and followed by 250μl of 100% ethanol. The mixture was pipette mixed before loading onto an RNeasy MinElute spin column, centrifuged for 15 seconds at 10,000 xg, washed with 500μl of buffer RPE (previously combined with ethanol in the kit-specified volume), and subjected to a final wash for 2 minutes with 500μl of 80% ethanol. The column was then dried by centrifugation. The purified RNA was eluted with 14μl of nuclease-free water, and quantified once again.
TIGR4 microarray analysis
Total bacterial RNA was isolated using Qiagen RNeasy Mini Kit (Qiagen), quantified on a NanoDrop ND-1000 and genomic integrity was verified using an Agilent Lab-On-A-Chip system. Two μg of total RNA was denatured for 10 min in the presence 6μg of random hexamers (Invitrogen, Carlsbad, CA), and 20U of RNase Inhibitor (Invitrogen), in a total volume of 18.5μl. The RNA was cooled on dry ice-ethanol, spun down, and then converted to aminoallyl cDNA using SuperScriptlll reverse transcriptase (Invitrogen) for 16 hr overnight at 42°C. Microarray experiments were performed as described previously [17]. Whole genome S. pneumoniae oligonucleotide microarrays (version 7) obtained from the PFGRC at the J. Craig Venter Institute (http://pfgrc.jcvi.org/) were used for microarray experiments. The microarray data was analyzed by first filtering out those spots with a signal-to-noise ratio less than 1.5 or signal reading less than 20 in both Cy3 and Cy5 channels. The quantile normalization (DNAMR R-package) on background-subtracted intensity was then applied to each microarray to correct the intensity bias, and only those genes where more than half of their corresponding spots passed the filtering criteria were reported in further analysis as a log2 ratio in comparison to the reference control sample. The average of log2 ratios from three independent biological replicates was calculated and the significantly differential expressed gene was selected to satisfy at least one of the following two conditions: 1) each of three replicates has log2 ratio greater than 2, a 4-fold change of the expression level; 2) two of three replicates have log2 ratio greater than 1.5 while coefficient of variation is less than 0.25.
Quantitative RT-PCR
RNA isolated from in vitro and in vivo grown pneumococci was quantitated using a Nanodrop ND-1000. Due to the low yield of bacterial RNA isolated from vitreous samples, RNA from 4 rabbit eyes (2 rabbits) was pooled and amplified using MessageAmp ll-Bacteria RNA Prokaryotic RNA Amplification Kit (Ambion, Austin, TX) per manufacturer's instructions. One microgram of total RNA from both in vitro and vitreous samples was used as starting material for amplification kit. After amplification, both samples were again quantitated by Nanodrop and 1 μg from each sample was used to synthesize cDNA using SuperScript III First-Strand Synthesis SuperMix for qRT-PCR kit (Invitrogen). One hundred ng of cDNA was used as template for real-time PCR using SYBR GreenER qPCR SuperMix for ABI PRISM (Invitrogen) per manufacturer's instructions. Primers were used at a final concentration of 200nM per reaction and sequences are listed in Table 1.
Adhesion assay
Adhesion and invasion assays were performed in the retinal pigmented epithelial cell line, ARPE-19 (ATCC, CRL-2302). ARPE-19 cells were seeded to 80-90% confluence in a 24-well plate (Invitrogen). Cells were incubated with approximately 5 × 104 CFU/mL bacteria at 37°C in 5% CO2 following a 5 min centrifugation at 2000 rpm. After 30 minutes, cells were washed three times with D-PBS with calcium and magnesium (Thermo Scientific), trypsinized, and 10 μl were plated on blood agar and incubated at 37°C overnight.
Statistical analysis
Analysis of variance for all rabbit endophthalmitis experiments were performed using the SAS system GLM procedure. The least squares means were determined with a Bonferroni adjustment for multiple comparisons. For adhesion assays, values from 3 or more experiments were compared and 2-tailed Student's t-test was used to test for statistical significance. A P value less than 0.05 was considered significant.
Results
Microarray analysis of bacterial gene expression during growth in rabbit vitreous
The global transcriptional response of S. pneumoniae during experimental endophthalmitis in rabbits was compared to that of bacteria grown in vitro. Bacteria were isolated either from the vitreous humor of New Zealand white rabbits 20 h post-inoculation with S. pneumoniae TIGR4 or from THY media containing similar bacterial titers as that achieved in the eye (106 CFU/mL). Bacterial RNA was purified, reverse transcribed and used for hybridization to TIGR4 microarrays. We identified 134 genes that were differentially expressed in the eye compared to growth in culture. These genes are listed in Table 2. One hundred twelve of these genes demonstrated > 3 fold increased expression in the eye compared to bacteria grown in vitro and 22 genes were decreased in their expression (shown in bold). Functions of these genes included metabolism, transport, transcription, cell structure, and hypothetical. Interestingly, we identified numerous groups of contiguous genes displaying similar differential expression patterns (boxed loci in Table 2) that may indicate Operons important during eye infection. Of particular interest was the up-regulation of genes encoding neuraminidases A (NanA) and B (NanB), components of the PTS system, the transcriptional regulator PlcR, and many ABC transporters. Down-regulated loci of interest included genes encoding the transcriptional regulator RlrA, the entire pilus locus, two choline transporters, and all of the type 4 capsule genes (data not shown).
Table 2.
ORFs differentially expressed during pneumococcal endophthalmitis compared to growth in THY medium.a
| Locusb | Annotation | Gene | Exp 1 | Exp 2 | Exp 3 | Average | CVc |
|---|---|---|---|---|---|---|---|
| SP0061 | PTS system, IIB component | 4.91 | 7.03 | 7.00 | 6.31 | 0.193 | |
| SP0062 | PTS system, IIC component | 5.69 | 7.25 | 6.42 | 6.46 | 0.121 | |
| SP0063 | PTS system, IID component | 4.62 | 5.42 | 5.58 | 5.21 | 0.099 | |
| SP0064 | PTS system, IIA component | 4.68 | 5.56 | 5.38 | 5.21 | 0.090 | |
| SP0065 | sugar isomerase domain protein | agaA | 5.41 | 6.49 | 5.61 | 5.83 | 0.098 |
| SP0066 | aldose-1-epimerase (mutarotase) | galM | 5.82 | 6.24 | 5.91 | 5.99 | 0.037 |
| SP0067 | hypothetical protein | 5.69 | 6.79 | 5.57 | 6.02 | 0.111 | |
| SP0068 | hypothetical protein | 5.68 | 6.54 | 6.15 | 6.13 | 0.071 | |
| SP0069 | choline binding protein I | cbpl | 5.31 | 5.88 | 6.49 | 5.89 | 0.100 |
| SP0091 | ABC transporter, permease protein | 3.17 | 3.64 | 7.21 | 4.67 | 0.473 | |
| SP0092 | ABC transporter, substrate-binding protein | 4.22 | 4.41 | 6.32 | 4.98 | 0.233 | |
| SP0109 | bacteriocin, putative | 2.29 | 2.69 | 3.36 | 2.78 | 0.194 | |
| SP0148 | ABC transporter, substrate-binding protein | -2.42 | -2.13 | -2.20 | -2.25 | 0.068 | |
| SP0162 | hypothetical protein | 4.35 | 6.56 | 4.21 | 5.04 | 0.262 | |
| SP0164 | hypothetical protein | 6.97 | 7.63 | 6.70 | 7.10 | 0.067 | |
| SP0165 | flavoprotein | 7.27 | 8.17 | 6.08 | 7.17 | 0.147 | |
| SP0166 | pyridoxal-dependent decarboxylase, Orn-Lys-Arg family | 5.94 | 6.30 | 5.67 | 5.97 | 0.053 | |
| SP0167 | hypothetical protein | 6.19 | 7.49 | 5.57 | 6.42 | 0.153 | |
| SP0168 | macrolide efflux protein, putative | 5.95 | 7.17 | 5.15 | 6.09 | 0.167 | |
| SP0169 | lactose phosphotransferase system repressor, degenerate | 5.22 | 6.18 | 4.07 | 5.16 | 0.205 | |
| SP0170 | hypothetical protein | 5.42 | 6.86 | 5.59 | 5.96 | 0.132 | |
| SP0171 | ROK family protein | 4.97 | 5.79 | 4.05 | 4.94 | 0.176 | |
| SP0172 | hypothetical protein | 4.72 | 5.17 | 4.12 | 4.67 | 0.113 | |
| SP0213 | 30S ribosomal protein S19 | rpsS | -2.33 | -1.96 | -1.83 | -2.04 | 0.128 |
| SP0285 | alcohol dehydrogenase, zinc-containing | 2.32 | 2.80 | 1.97 | 2.36 | 0.177 | |
| SP0317 | 2-dehydro-3-deoxyphosphogluconate aldolase/4-hydroxy-2-oxoglutarate aldolase | 6.71 | 6.70 | 7.26 | 6.89 | 0.047 | |
| SP0318 | carbohydrate kinase, PfkB family | 7.28 | 5.98 | 5.18 | 6.15 | 0.172 | |
| SP0319 | Hypothetical protein | 7.18 | 5.71 | 5.97 | 6.29 | 0.124 | |
| SP0320 | oxidoreductase, short chain dehydrogenase-reductase family | 7.40 | 5.73 | 6.31 | 6.48 | 0.131 | |
| SP0321 | PTS system, IIA component | 8.29 | 5.84 | 6.94 | 7.02 | 0.175 | |
| SP0322 | glucuronyl hydrolase | 8.41 | 7.99 | 6.77 | 7.72 | 0.110 | |
| SP0323 | PTS system, IIB component | 8.85 | 6.79 | 6.77 | 7.47 | 0.160 | |
| SP0324 | PTS system, IIC component | 9.16 | 7.05 | 6.94 | 7.72 | 0.162 | |
| SP0325 | PTS system, IID component | 9.72 | 8.30 | 7.42 | 8.48 | 0.137 | |
| SP0326 | preprotein translocase, YajC subunit | yajC-1 | 9.71 | 8.29 | 7.14 | 8.38 | 0.154 |
| SP0327 | hypothetical protein, interruption | 2.99 | 2.96 | 4.78 | 3.58 | 0.292 | |
| SP0338 | ATP-dependent Clp protease, ATP-binding subunit, putative | 4.19 | 3.37 | 8.22 | 5.26 | 0.493 | |
| SP0368 | cell wall surface anchor family protein, authentic frameshift | 2.83 | 3.93 | 2.82 | 3.20 | 0.199 | |
| SP0461 | transcriptional regulator, putative | rlrA | -3.94 | -3.64 | -2.77 | -3.45 | 0.176 |
| SP0462 | cell wall surface anchor family protein | -5.46 | -5.65 | -3.79 | -4.97 | 0.206 | |
| SP0463 | cell wall surface anchor family protein | -5.05 | -4.86 | -3.58 | -4.49 | 0.178 | |
| SP0464 | cell wall surface anchor family protein | -5.12 | -4.94 | -3.37 | -4.48 | 0.214 | |
| SP0467 | sortase, putative | -3.67 | -3.67 | -2.73 | -3.36 | 0.161 | |
| SP0468 | sortase, putative | -3.61 | -4.02 | -2.87 | -3.50 | 0.166 | |
| SP0586 | 5,10-methylenetetrahydrofolate reductase, putative | 5.63 | 5.47 | 3.79 | 4.96 | 0.206 | |
| SP0647 | PTS system, IIC component, putative | 4.36 | 5.66 | 4.39 | 4.80 | 0.154 | |
| SP0648 | beta-galactosidase | bgaA | 5.69 | 6.05 | 4.53 | 5.43 | 0.146 |
| SP0703 | hypothetical protein | 2.04 | 1.74 | 2.49 | 2.09 | 0.181 | |
| SP0730 | pyruvate oxidase | spxB | -2.57 | -2.32 | -1.70 | -2.19 | 0.204 |
| SP0731 | conserved domain protein | -3.07 | -2.76 | -2.44 | -2.76 | 0.115 | |
| SP0758 | PTS system, IIABC components | 1.94 | 2.58 | 1.72 | 2.08 | 0.215 | |
| SP0867 | ABC transporter, ATP-binding protein | -2.75 | -2.54 | -2.09 | -2.46 | 0.136 | |
| SP0868 | conserved hypothetical protein | -3.15 | -3.22 | -2.77 | -3.05 | 0.080 | |
| SP0869 | aminotransferase, class-V | -3.19 | -3.60 | -2.71 | -3.16 | 0.140 | |
| SP0870 | hypothetical protein | -3.73 | -3.85 | -3.27 | -3.62 | 0.084 | |
| SP0871 | conserved hypothetical protein | -3.43 | -3.58 | -3.10 | -3.37 | 0.072 | |
| SP0875 | lactose phosphotransferase system repressor | lacR-1 | 3.97 | 4.74 | 4.59 | 4.43 | 0.092 |
| SP0877 | PTS system, fructose-specific enzyme NBC component (FruA) | 2.27 | 2.04 | 4.32 | 2.88 | 0.437 | |
| SP0958 | hypothetical protein | 2.91 | 2.03 | 4.34 | 3.09 | 0.378 | |
| SP1002 | zinc-binding lipoprotein | adcAII | 3.01 | 4.86 | 4.79 | 4.22 | 0.248 |
| SP1003 | conserved hypothetical protein | phtD | 3.48 | 6.06 | 4.36 | 4.63 | 0.284 |
| SP1010 | large conductance mechanosensitive channel protein MscL | mscL | -2.55 | -2.33 | -2.14 | -2.34 | 0.087 |
| SP1058 | hypothetical protein | 7.59 | 8.30 | 7.30 | 7.73 | 0.067 | |
| SP1059 | hypothetical protein | 8.16 | 8.53 | 7.81 | 8.17 | 0.044 | |
| SP1060 | hypothetical protein | 7.43 | 8.55 | 7.53 | 7.84 | 0.079 | |
| SP1061 | protein kinase, putative | 7.82 | 9.31 | 6.28 | 7.80 | 0.194 | |
| SP1062 | ABC transporter, ATP-binding protein | 8.00 | 7.43 | 6.82 | 7.42 | 0.080 | |
| SP1063 | ABC-2 transporter, permease protein, putative | 7.07 | 8.73 | 7.18 | 7.66 | 0.121 | |
| SP1122 | glucose-1-phosphate adenylyltransferase | glgC | 3.69 | 3.93 | 2.79 | 3.47 | 0.173 |
| SP1123 | glycogen biosynthesis protein | glgD | 3.52 | 3.65 | 2.32 | 3.16 | 0.231 |
| SP1124 | glycogen synthase | glgA | 3.61 | 4.05 | 2.93 | 3.53 | 0.159 |
| SP1175 | conserved domain protein | 2.30 | 4.66 | 2.28 | 3.08 | 0.443 | |
| SP1197 | conserved hypothetical protein | 2.31 | 2.94 | 4.92 | 3.39 | 0.402 | |
| SP1339 | hypothetical protein | 3.07 | 3.53 | 2.46 | 3.02 | 0.177 | |
| SP1344 | hypothetical protein | 3.89 | 5.07 | 2.95 | 3.97 | 0.268 | |
| SP1345 | hypothetical protein | 3.49 | 3.78 | 2.41 | 3.23 | 0.224 | |
| SP1435 | ABC transporter, ATP-binding protein | 5.67 | 6.44 | 4.78 | 5.63 | 0.148 | |
| SP1437 | conserved domain protein | 5.53 | 5.19 | 5.70 | 5.47 | 0.048 | |
| SP1438 | ABC transporter, ATP-binding protein | 2.75 | 4.66 | 5.06 | 4.16 | 0.297 | |
| SP1471 | oxidoreductase, putative | 1.68 | 2.44 | 2.74 | 2.29 | 0.237 | |
| SP1472 | oxidoreductase, putative | 2.57 | 4.10 | 3.18 | 3.28 | 0.234 | |
| SP1588 | oxidoreductase, pyridine nucleotide-disulfide, class I | -3.47 | -2.94 | -2.68 | -3.03 | 0.134 | |
| SP1675 | ROK family protein | 2.48 | 2.55 | 2.09 | 2.37 | 0.105 | |
| SP1676 | N-acetylneuraminate lyase, putative | 1.87 | 2.75 | 1.81 | 2.14 | 0.247 | |
| SP1677 | hypothetical protein | 2.41 | 2.82 | 2.20 | 2.48 | 0.127 | |
| SP1679 | hypothetical protein | 3.22 | 4.15 | 3.78 | 3.72 | 0.126 | |
| SP1681 | sugar ABC transporter, permease protein | 3.15 | 4.82 | 2.97 | 3.65 | 0.279 | |
| SP1682 | sugar ABC transporter, permease protein | 3.15 | 4.09 | 3.57 | 3.60 | 0.130 | |
| SP1683 | sugar ABC transporter, sugar-binding protein | 4.73 | 6.52 | 3.59 | 4.95 | 0.298 | |
| SP1684 | PTS system, glucose-specific NBC component | 3.83 | 4.87 | 3.06 | 3.92 | 0.232 | |
| SP1685 | N-acetylmannosamine-6-phosphate 2-epimerase | 2.24 | 2.79 | 4.31 | 3.11 | 0.344 | |
| SP1686 | hypothetical protein | 8.42 | 9.18 | 7.50 | 8.37 | 0.100 | |
| SP1687 | neuraminidase B | NanB | 8.59 | 9.03 | 8.56 | 8.73 | 0.030 |
| SP1688 | ABC transporter, permease protein | 8.47 | 8.27 | 7.60 | 8.11 | 0.056 | |
| SP1689 | ABC transporter, permease protein | 8.05 | 8.93 | 7.04 | 8.01 | 0.118 | |
| SP1690 | ABC transporter, substrate-binding protein | 8.47 | 9.13 | 8.53 | 8.71 | 0.042 | |
| SP1693 | neuraminidase A, authentic frameshift | nanA | 6.57 | 6.09 | 4.56 | 5.74 | 0.183 |
| SP1694 | hypothetical protein | 2.82 | 2.79 | 4.48 | 3.36 | 0.288 | |
| SP1778 | aquaporin | -2.58 | -2.91 | -2.74 | -2.74 | 0.061 | |
| SP1793 | hypothetical protein | 5.40 | 5.49 | 4.36 | 5.08 | 0.124 | |
| SP1809 | transcriptional regulator | 2.57 | 2.75 | 2.56 | 2.63 | 0.040 | |
| SP1860 | choline transporter | proWX | -1.81 | -2.80 | -2.37 | -2.33 | 0.213 |
| SP1861 | choline transporter | proV | -2.34 | -2.98 | -2.42 | -2.58 | 0.134 |
| SP1862 | hypothetical protein | -2.51 | -2.74 | -2.19 | -2.48 | 0.113 | |
| SP1869 | iron-compound ABC transporter, permerase protein | 2.76 | 2.28 | 4.42 | 3.15 | 0.356 | |
| SP1871 | iron-compound ABC transporter, ATP-binding protein | 3.02 | 2.51 | 5.20 | 3.58 | 0.399 | |
| SP1872 | iron-compound ABC transporter | 3.13 | 3.20 | 4.37 | 3.57 | 0.196 | |
| SP1946 | transcriptional activator | PlcR | 2.17 | 2.85 | 2.37 | 2.46 | 0.142 |
| SP1947 | hypothetical protein | 3.98 | 4.67 | 7.96 | 5.54 | 0.385 | |
| SP1948 | conserved domain protein | 7.21 | 7.72 | 7.26 | 7.40 | 0.038 | |
| SP1949 | hypothetical protein | 7.50 | 8.20 | 7.58 | 7.76 | 0.050 | |
| SP1950 | bacteriocin formation protein, putative | 6.28 | 6.35 | 6.42 | 6.35 | 0.011 | |
| SP1951 | conserved hypothetical protein | 6.28 | 6.64 | 7.43 | 6.78 | 0.087 | |
| SP1952 | hypothetical protein | 6.95 | 6.79 | 7.07 | 6.94 | 0.020 | |
| SP1953 | toxin secretion ABC transporter, ATP-binding-permease protein | 7.12 | 8.35 | 7.42 | 7.63 | 0.085 | |
| SP1954 | serine protease, subtilase family | 8.25 | 7.53 | 7.97 | 7.92 | 0.046 | |
| SP1955 | hypothetical protein | 5.20 | 6.81 | 5.59 | 5.87 | 0.143 | |
| SP1956 | hypothetical protein | 5.26 | 6.74 | 5.15 | 5.71 | 0.156 | |
| SP1957 | ABC transporter, ATP-binding protein | 4.85 | 6.33 | 4.73 | 5.30 | 0.168 | |
| SP1958 | hypothetical protein | 4.15 | 4.52 | 4.24 | 4.30 | 0.045 | |
| SP2023 | Phosphotransferase system system, cellobiose-specific IIB component | 4.57 | 3.50 | 2.02 | 3.36 | 0.382 | |
| SP2024 | PTS system, IIA component | 4.89 | 3.99 | 2.08 | 3.65 | 0.393 | |
| SP2026 | alcohol dehydrogenase, iron-containing | 3.24 | 3.86 | 4.22 | 3.77 | 0.132 | |
| SP2058 | queuine tRNA-ribosyltransferase | tgt | -2.01 | -2.59 | -1.60 | -2.06 | 0.242 |
| SP2108 | maltose-maltodextrin ABC transporter, maltose-maltodextrin-binding protein | malX | 3.03 | 3.18 | 2.68 | 2.96 | 0.086 |
| SP2127 | transketolase, C-terminal subunit | 4.88 | 5.32 | 5.26 | 5.15 | 0.046 | |
| SP2128 | transketolase, N-terminal subunit | 5.98 | 6.35 | 6.44 | 6.26 | 0.039 | |
| SP2129 | PTS system, IIC component, putative | 4.48 | 5.27 | 5.34 | 5.03 | 0.096 | |
| SP2130 | PTS system, MB component, putative | 5.40 | 5.66 | 6.60 | 5.89 | 0.108 | |
| SP2131 | transcriptional regulator, BglG family | 3.23 | 4.87 | 6.07 | 4.73 | 0.302 | |
| SP2148 | arginine deiminase | acrA | 4.13 | 4.90 | 7.14 | 5.39 | 0.291 |
| SP2150 | ornithine transcarbamylase | argF | 6.66 | 7.56 | 7.94 | 7.39 | 0.090 |
| SP2152 | conserved hypothetical protein | 5.96 | 5.95 | 5.88 | 5.93 | 0.007 | |
| SP2153 | peptidase, M20-M25-M40 family | 5.29 | 5.95 | 5.35 | 5.53 | 0.066 |
Values are displayed as log2 ratios and expression levels for down-regulated genes are shown in bold. Boxed loci indicate groups of consecutive genes that display similar differential expression.
Locus designation based on TIGR4 genome
Coefficient of variation as described in Methods
Real-time PCR verification of array results
To verify the microarray results, we selected several genes that demonstrated differential gene expression and analyzed their transcription levels by quantitative realtime polymerase chain reaction (qRT-PCR) (Figure 1). RNA samples used for qRT-PCR were purified from vitreous and THY-grown bacteria isolated independently from samples used for microarray analysis. The genes SP1693, SP1687, and SP1954 served as representative up-regulated genes and SP0346, SP0461, and SP1861 served as representative down-regulated genes. SP0346 did not reach the stringent requirements of the genes listed in Table 2; however, it demonstrated decreased expression in all three independent experiments (average log2 expression of -1.52 ± 0.29), as did all genes of the capsule synthesis Cps operon, and was therefore selected as a representative down-regulated gene of interest. Results from qRT-PCR mirrored the transcription trends of the microarray data with SP1687, SP1693, and SP1954 demonstrating 72.7-fold, 4.3-fold, and 13.4-fold increased expression, respectively, and SP0346, SP0461, and SP1861 demonstrating -4.3-fold, -22.3-fold and -23.9-fold decreased expression, respectively.
Figure 1.
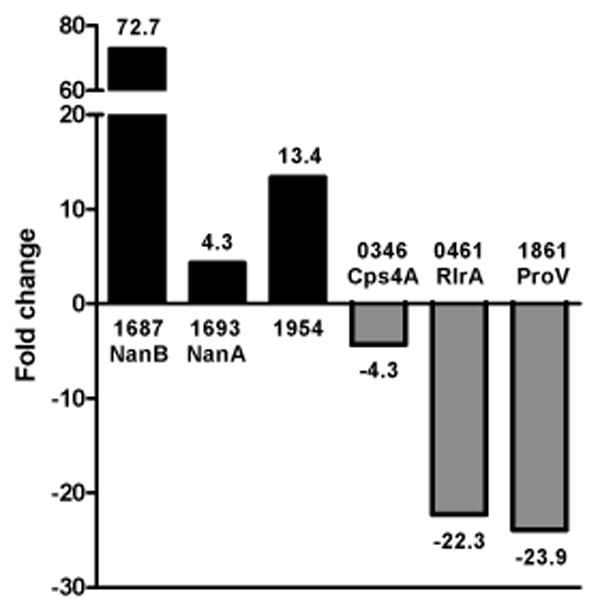
Validation of microarray results by qRT-PCR. Transcription of representative genes (x axis) displaying differential expression was analyzed by qRT-PCR of pooled bacterial RNA isolated independently from samples used for array experiments. Genes are listed by their TIGR4 accession number (SP) and gene name. Scale of the fold-change (y axis) is not continuous due to highly induced expression of SP1687 (NanB) and is indicated by the broken bar.
Assessment of neuraminidase contribution to pathogenesis
Microarrays and real-time PCR showed that the genes encoding NanA and NanB were upregulated in rabbit vitreous. Both neuraminidases are known to play a role in bacterial survival within the host [28], therefore, these proteins were chosen for analysis as possible virulence factors during endophthalmitis. In an effort to determine the contribution of these neuraminidases to virulence, we created isogenic mutants of S. pneumoniae D39 lacking either NanA, NanB, or both neuraminidases and tested their virulence in a rabbit model of endophthalmitis. Loss of NanA, NanB, or both did not result in any significant decreases in the severity of endophthalmitis 24 or 48 hours after infection (Figure 2A). In fact, eyes infected with D39ΔnanA had significantly higher clinical scores than those infected with the parent strain or the other two mutant strains (P < 0.001). Rabbits infected with D39ΔnanA were euthanized immediately after the 24 hour examination for humane reasons, therefore no data were obtained for this group 48 hours after infection. Bacterial recovery from the vitreous humor of infected rabbits was not significantly different between any group at either time point (Figure 2B). Examination of the loss of retinal function by electroretinography yielded variable results. Eyes infected with D39ΔnanA retained more retinal function than those infected with D39 24 hours after infection, and eyes infected with D39ΔnanB retained more function than those infected with D39ΔnanAB 48 hours after infection (Figure 2C).
Figure 2.
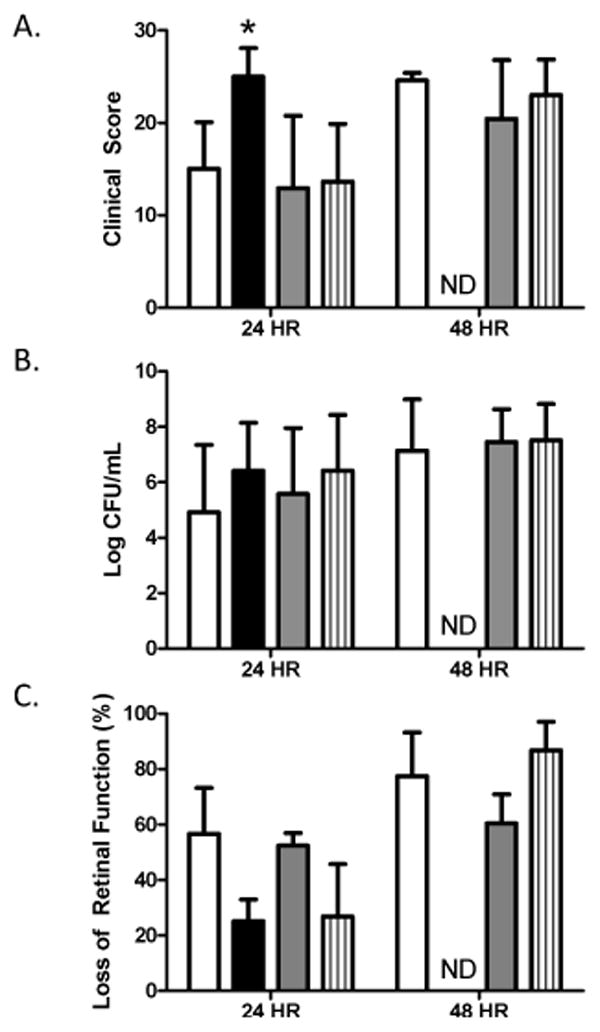
Effect of neuraminidases during pneumococcal endophthalmitis. Rabbit eyes were infected with S. pneumoniae D39 (open bars), D39ΔnanA (solid bars), D39ΔnanB (gray bars), and D39ΔnanAB (striped bars). A) Clinical severity scores of eyes; n = 8, 8, 8, and 10 for D39, D39ΔnanA, D39ΔnanB, and D39ΔnanAB, respectively, at 24 hours. N = 8, 4, and 10 for D39, D39ΔnanB, and D39ΔnanAB, respectively, at 48 hours. B) Bacterial recovery from the vitreous humor; n = 8, 4, 8, and 9 for D39, D39ΔnanA, D39ΔnanB, and D39ΔnanAB, respectively, at 24 hours. N = 8, 4, and 10 for D39, D39ΔnanB, and D39ΔnanAB, respectively, at 48 hours. C) Percent loss of retinal function; n = 4 per group per time point. Asterisks indicate statistical significance (P<0.05). ND = not done.
Adhesion of wild type and mutant strains to retinal cells
Mutants lacking representative genes found to be differentially expressed during endophthalmitis were tested for their ability to adhere to retinal pigmented epithelial cells (ARPE-19) in an in vitro adherence assay. None of the mutants tested (ΔSpxB, ΔAdcAII, ΔPhtD, ΔSP1954, or ΔCbpl) demonstrated reduced adherence to ARPE-19 cells compared to the wild type parental strain T4R (Figure 3). Interestingly, the Δ1954 mutant demonstrated significantly increased binding to ARPE-19 cells compared to T4R.
Figure 3.
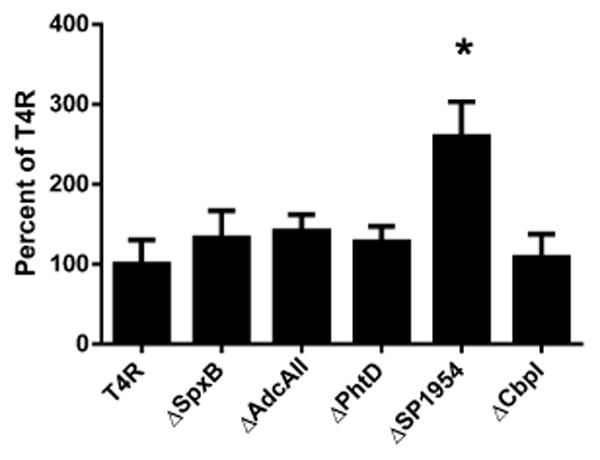
Bacterial adhesion to ARPE-19 cells. Adhesion of T4R or isogenic mutants ΔSpxB, ΔAdcAII, ΔPhtD, ΔSP1954, or ΔCbpl was assessed after 30 min incubation with ARPE-19 cells. Colony forming units (CFU's) are expressed as percent adhesion compared to wild type T4R. Asterisk indicates statistical significance as determined by Student's t-test comparison of each mutant to T4R. (p<0.01).
Discussion
Examining bacterial gene expression during an active infection is key to unraveling the mechanisms by which a pathogen causes a specific form of disease. These types of studies also have potential to unmask previously unidentified targets for prevention and therapeutics. The majority of pneumococcal transcriptome studies have logically focused on the most common forms of invasive disease such as pneumonia, sepsis, and meningitis. However, less common types of infections still occur and the mechanisms involved are not well understood. To elucidate the mechanisms involved in bacterial survival and virulence during ocular infections, we utilized microarray analysis to assess transcriptional responses of pneumococcus during experimental endophthalmitis. Our results demonstrate that S. pneumoniae differentially expresses a large number of genes during growth in the vitreous, many of which are known to be important for pneumococcal virulence. Little is known about how the pneumococcus adapts to growth in the eye and what factors may be expressed. The importance of pneumolysin and the polysaccharide capsule has been demonstrated in endophthalmitis models [9, 29]. Immunization against pneumolysin was found to protect rabbit retinas from complete destruction following vitreal infection with S. pneumoniae [29]. However, in the current study, we did not identify a consistent up or down regulation of pneumolysin.
While capsule is known to be important for preventing phagocytosis of the pneumococcus by professional phagocytes, it is also differentially expressed depending on location within the host. However, in fluid microenvironments, phagocytosis is less efficient (as seen in the cerebrospinal fluid [30]), so the expression of capsule will be less essential than at a site such as a mucosal surface [31]. Capsule production is one factor associated with phase variation in pneumococci, with the bacteria expressing lower levels of capsule in the nasopharynx, when attachment to host cells is critical, and higher levels during replication in the blood to enhance immune evasion [32]. These two phenotypic variants have traditionally been referred to as “transparent” and “opaque”, respectively. At the time point selected for our microarray analysis, there was a decrease in expression of the entire type 4 capsule locus. We also saw increased transcription of a large group of genes previously identified to be overexpressed by pneumococci of the transparent type [11].
Our study found all the genes between SP1675 and SP1694 to be increased in expression in at least 1 array experiment. Three of the genes in this region encode neuraminidases NanA (SP1693) and NanB (SP1688) as well as an N-acetylneuraminate lyase, indicating considerable capacity for sialic acid cleavage during growth in the eye. Additionally, three genes in this region (SP1681-1683) have recently been shown to code for an ABC transporter of sialic acid (satABC) [33], which aids in colonization. An additional ABC transporter also exists in this overexpressed region (SP1688-SP1690) along with a predicted N-acetylmannosamine-6-phosphate 2-epimerase (SP1685). The gene for beta-galactosidase (SP0648) was also increased in expression during endophthalmitis providing further opportunity for sequential cleavage of N-terminal glycans as previously described [34]. These results indicate that while growing within the eye, the pneumococcus may be utilizing its ability to cleave terminal oligosaccharides from host glycoproteins as an energy source or to enhance attachment to host cells. Also, NanA and beta-galactosidase have been shown to be induced when pneumococci are in contact with A549 epithelial cells or macrophages indicating these genes may be part of an early response triggered by host cells [16, 18]. Increased expression of NanA, NanB, N-acetyl neuraminate lyase, and beta-galactosidase along with decreased expression of capsule genes was also identified in pneumococcal conjunctivitis strains, indicating potential expression patterns relating to the eye infections [20].
Loss of either or both neuraminidases had essentially no effect on either bacterial replication or damage to the eye. This finding was surprising and indicates that the pneumococcus could possess additional factors that provide a redundancy of function either in growth and survival in the ocular environment, or in pathogenesis. The current study and previous studies utilizing the rabbit as a model for pneumococcal endophthalmitis have shown that S. pneumoniae can grow to high numbers in the vitreous humor [9, 29, 35]. One of the eyes in the current study yielded approximately 3 × 109 CFU/mL 48 hours after infection, which is near the upper limit of growth for S. pneumoniae in rich medium in vitro. This observation suggests that the rabbit vitreous humor is an enriched environment for S. pneumoniae to grow. The vitreous humor contains glucose which decreases in concentration following bacterial endophthalmitis [36]. Given that S. pneumoniae possesses a number of sugar transport genes that were increased in expression in our model, it is likely that the ability of this bacterium to thrive in a sugar-rich environment, despite the absence of the neuraminidases, would contribute to its pathogenicity due to enhanced growth.
A variety of ABC sugar transporters as well as components of phosphotransferase systems (PTS) for sorbose (SP0061-0064), N-acetyl glucosamine (putative, SP0321-0325), and the pentitol family (SP2131-SP2129) [33] and genes involved in carbohydrate metabolism demonstrated differential gene expression during pneumococcal endophthalmitis. Many of these genes have not previously been shown to be differentially expressed during in vivo growth in other studies and may be important for survival of the pneumococcus during eye infection. However, differences in sugar concentrations between our in vitro medium (THY) and the vitreous could be responsible for some of these changes.
We identified 4 transcriptional regulators that were differentially expressed during pneumococcal endophthalmitis: RlrA (SP0461), SP1809, PlcR (SP1946), and SP2131. SP1809 has not been well characterized but has been shown to be induced in response to competence factor [37] and SP2131 is a transcriptional regulator for the genes of the pentitol family PTS, previously mentioned. RlrA is known to regulate expression of a 14-kb pathogenicity island (rlrA islet) which includes rlrA itself, as well as 6 other genes encoding the three structural proteins of the pilus and three sortases involved in linking pilus to the pneumococcal cell wall [38-40]. RlrA has been shown to be important during colonization and lung infection and its effect on pilus expression allows for increased invasion of host cells [19, 41]. Interestingly, our results indicate this entire locus is down-regulated during eye infection. This could be due to a number of factors since the rlrA islet is regulated by complex networks including two-component systems and metalloregulators [19].
The transcriptional regulator PlcR has been well characterized in Bacillus species where it is known to affect expression of a multitude of proteins, many of which are involved in virulence [42-45]. PlcR (SP1946) as well as 12 consecutive genes (SP1947-SP1958) downstream of PlcR, but transcribed in opposite direction, were all increased in expression during endophthalmitis. The function of many of these genes is not known but within this group were genes encoding a putative bacteriocin protein (SP1950), a toxin secretion ABC transporter (SP1953), and a serine protease (SP1954). Interestingly, loss of SP1954 led to a significant increase in adhesion to ARPE cells. It is unclear how loss of a protease would enhance adhesion. However, we are investigating this mechanism. Increased transcription of two other groups of genes (SP1058-SP1063 and SP0164-SP0172), both of which are immediately downstream of putative PlcR genes (SP1057 and SP0163), was also seen during endophthalmitis. Interestingly, all of these genomic regions were previously identified as being overexpressed during interaction of S. pneumoniae with THP-1 macrophages [18]. Macrophages and neutrophils are known to be present in the eye during the time frame selected for these studies [9]. Interaction of pneumococci with these cells could potentially trigger a PlcR-dependent transcriptional response which enhances survival of the bacteria during growth within the eye.
Other genes of interest displaying differential gene expression in smaller Operons may be important for metal availability during growth in the eye. These included genes of iron ABC transporter piu Operon (SP1869-SP1872) and an operon containing AdcAII (SP1002) and PhtD (SP1003). The genes of the piu operon encode components of one of three ABC transporters the pneumococcus possesses for acquiring iron [46]. Increased expression of the operon during endophthalmitis may indicate low availability of this metal during growth in the eye. Likewise, AdcAII and PhtD are surface proteins known to bind zinc regulate zinc homeostasis [47-52]. Antibodies against PhtD have been shown to protect mice against systemic pneumococcal disease [53].
Changes in oxidative metabolism were also identified during growth in the eye with the bacteria overexpressing the gene for pyruvate oxidase (SpxB, SP0730), which is responsible for production of large amounts of hydrogen peroxide by the bacteria [54]. Two putative oxidoreductases (SP1471, SP1472) were increased in expression with an additional oxidoreductase (SP1588) demonstrating reduced expression during growth in the eye. Decreased expression during endophthalmitis was also seen for two putative choline transporters (SP1860, SP1861) although the function of these proteins has not been determined [55].
The mechanisms of pneumococcal eye infection are poorly understood when compared to what is known about other presentations of disease. This study sheds light on some of the potential genetic alterations the bacterium employs during growth within the eye. While some of these transcriptional changes may correspond to restructuring of catabolic pathways during adaptation to nutrient availability, many likely serve to protect the bacterium from host responses and some may even promote tissue damage and invasion. Also, the transcriptional responses identified in this report may be similar to those seen during other forms of eye infection. Understanding these mechanisms will provide new insight into strategies of prevention and treatment of bacterial eye infections.
Acknowledgments
We thank Elaine Tuomanen and The Institute for Genomic Research (TIGR) for supplying the TIGR4 microarrays. We also thank Ed Swiatlo for providing us with the suicide vector pEVP3 used in making the ΔNanB insertion-duplication mutant. This work was supported in part by NIH R01 27913 to ET, American Lebanese Syrian Associated Charities, Mississippi State University College of Arts and Sciences and the Department of Biological Sciences.
Footnotes
Competing interests. None to disclose.
Authors' contributions. JT performed RNA amplifications, real-time qPCR, bacterial mutagenesis and mutant screening, adhesion assays, and drafted the manuscript. NT and MM infected rabbits and prepared RNA for arrays. MS and MM performed the endophthalmitis experiments with neuraminidase knockouts. GR and YW performed RNA quality control, microarray hybridizations, and data analysis. ST assisted with plating and enumeration of bacteria in various experiments. LM and MM provided substantial contributions to conception and design of the study as well as interpretation of data and drafting of the manuscript.
References
- 1.Miller JJ, Scott IU, Flynn HW, Jr, Smiddy WE, Corey RP, Miller D. Endophthalmitis caused by Streptococcus pneumoniae. Am J Ophthalmol. 2004;138(2):231–6. doi: 10.1016/j.ajo.2004.03.008. [DOI] [PubMed] [Google Scholar]
- 2.Nouri M, Terada H, Alfonso EC, Foster CS, Durand ML, Dohlman CH. Endophthalmitis after keratoprosthesis: incidence, bacterial causes, and risk factors. Arch Ophthalmol. 2001;119(4):484–9. doi: 10.1001/archopht.119.4.484. [DOI] [PubMed] [Google Scholar]
- 3.Recchia FM, Baumal CR, Sivalingam A, Kleiner R, Duker JS, Vrabec TR. Endophthalmitis after pediatric strabismus surgery. Arch Ophthalmol. 2000;118(7):939–44. [PubMed] [Google Scholar]
- 4.Soriano F, Perez-Trallero E, Pallares R, Meseguer MA, Fleites A, Gene A, Gonzalez A, Linares J, Esteban J, Baquero F, Valdes E, Munoz-Almagro C. Streptococcus pneumoniae endophthalmitis: a study of 36 cases with special reference to antibiotic resistance and treatment options. Clin Microbiol Infect. 2006;12(6):519–26. doi: 10.1111/j.1469-0691.2006.01418.x. [DOI] [PubMed] [Google Scholar]
- 5.Bharathi MJ, Ramakrishnan R, Shivakumar C, Meenakshi R, Lionalraj D. Etiology and antibacterial susceptibility pattern of community-acquired bacterial ocular infections in a tertiary eye care hospital in south India. Indian J Ophthalmol. 2010;58(6):497–507. doi: 10.4103/0301-4738.71678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kuriyan AE, Weiss KD, Flynn HW, Jr, Smiddy WE, Berrocal AM, Albini TA, Miller D. Endophthalmitis caused by streptococcal species: clinical settings, microbiology, management, and outcomes. Am J Ophthalmol. 2014;157(4):774–780. el. doi: 10.1016/j.ajo.2013.12.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ng EW, Samiy N, Rubins JB, Cousins FV, Ruoff KL, Baker AS, D'Amico DJ. Implication of pneumolysin as a virulence factor in Streptococcus pneumoniae endophthalmitis. Retina. 1997;17(6):521–9. [PubMed] [Google Scholar]
- 8.Robertson GT, Ng WL, Foley J, Gilmour R, Winkler ME. Global transcriptional analysis of clpP mutations of type 2 Streptococcus pneumoniae and their effects on physiology and virulence. J Bacteriol. 2002;184(13):3508–20. doi: 10.1128/JB.184.13.3508-3520.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sanders M, Norcross E, Robertson ZM, Moore QC, Fratkin J, Marquart ME. The Streptococcus pneumoniae capsule is required for full virulence in pneumococcal endophthalmitis. Invest Ophthalmol Vis Sci. 2010 doi: 10.1167/iovs.10-5513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sebert ME, Palmer LM, Rosenberg M, Weiser JN. Microarray-based identification of htrA, a Streptococcus pneumoniae gene that is regulated by the CiαRH two-component system and contributes to nasopharyngeal colonization. Infect Immun. 2002;70(8):4059–67. doi: 10.1128/IAI.70.8.4059-4067.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.King SJ, Hippe KR, Gould JM, Bae D, Peterson S, Cline RT, Fasching C, Janoff EN, Weiser JN. Phase variable desialylation of host proteins that bind to Streptococcus pneumoniae in vivo and protect the airway. Mol Microbiol. 2004;54(1):159–71. doi: 10.1111/j.1365-2958.2004.04252.x. [DOI] [PubMed] [Google Scholar]
- 12.Dagkessamanskaia A, Moscoso M, Henard V, Guiral S, Overweg K, Reuter M, Martin B, Wells J, Claverys JP. Interconnection of competence, stress and CiaR regulons in Streptococcus pneumoniae: competence triggers stationary phase autolysis of ciαR mutant cells. Mol Microbiol. 2004;51(4):1071–86. doi: 10.1111/j.1365-2958.2003.03892.x. [DOI] [PubMed] [Google Scholar]
- 13.Ulijasz AT, Andes DR, Glasner JD, Weisblum B. Regulation of iron transport in Streptococcus pneumoniae by RitR, an orphan response regulator. J Bacteriol. 2004;186(23):8123–36. doi: 10.1128/JB.186.23.8123-8136.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kloosterman TG, van der Kooi-Pol MM, Bijlsma JJ, Kuipers OP. The novel transcriptional regulator SczA mediates protection against Zn2+ stress by activation of the Zn2+-resistance gene czcD in Streptococcus pneumoniae. Mol Microbiol. 2007;65(4):1049–63. doi: 10.1111/j.1365-2958.2007.05849.x. [DOI] [PubMed] [Google Scholar]
- 15.Rosch JW, Gao G, Ridout G, Wang YD, Tuomanen EI. Role of the manganese efflux system mntE for signalling and pathogenesis in Streptococcus pneumoniae. Mol Microbiol. 2009;72(1):12–25. doi: 10.1111/j.1365-2958.2009.06638.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Song XM, Connor W, Hokamp K, Babiuk LA, Potter AA. Streptococcus pneumoniae early response genes to human lung epithelial cells. BMC Res Notes. 2008;1:64. doi: 10.1186/1756-0500-1-64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Orihuela CJ, Radin JN, Sublett JE, Gao G, Kaushal D, Tuomanen EI. Microarray analysis of pneumococcal gene expression during invasive disease. Infect Immun. 2004;72(10):5582–96. doi: 10.1128/IAI.72.10.5582-5596.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Song XM, Connor W, Hokamp K, Babiuk LA, Potter AA. Transcriptome studies on Streptococcus pneumoniae, illustration of early response genes to THP-1 human macrophages. Genomics. 2009;93(1):72–82. doi: 10.1016/j.ygeno.2008.09.008. [DOI] [PubMed] [Google Scholar]
- 19.Rosch JW, Mann B, Thornton J, Sublett J, Tuomanen E. Convergence of regulatory networks on the pilus locus of Streptococcus pneumoniae. Infect Immun. 2008;76(7):3187–96. doi: 10.1128/IAI.00054-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Williamson YM, Gowrisankar R, Longo DL, Facklam R, Gipson IK, Ades EP, Carlone GM, Sampson JS. Adherence of nontypeable Streptococcus pneumoniae to human conjunctival epithelial cells. Microb Pathog. 2008;44(3):175–85. doi: 10.1016/j.micpath.2007.08.016. [DOI] [PubMed] [Google Scholar]
- 21.Molzen TE, Burghout P, Bootsma HJ, Brandt CT, van der Gaast-de Jongh CE, Eleveld MJ, Verbeek MM, Frimodt-Moller N, Ostergaard C, Hermans PW. Genome-wide identification of Streptococcus pneumoniae genes essential for bacterial replication during experimental meningitis. Infect Immun. 2011;79(1):288–97. doi: 10.1128/IAI.00631-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rogers PD, Thornton J, Barker KS, McDaniel DO, Sacks GS, Swiatlo E, McDaniel LS. Pneumolysin-dependent and -independent gene expression identified by cDNA microarray analysis of THP-1 human mononuclear cells stimulated by Streptococcus pneumoniae. Infect Immun. 2003;71(4):2087–94. doi: 10.1128/IAI.71.4.2087-2094.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bootsma HJ, Egmont-Petersen M, Hermans PW. Analysis of the in vitro transcriptional response of human pharyngeal epithelial cells to adherent Streptococcus pneumoniae: evidence for a distinct response to encapsulated strains. Infect Immun. 2007;75(11):5489–99. doi: 10.1128/IAI.01823-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Joyce EA, Popper SJ, Falkow S. Streptococcus pneumoniae nasopharyngeal colonization induces type l interferons and interferon-induced gene expression. BMC Genomics. 2009;10:404. doi: 10.1186/1471-2164-10-404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Banerjee A, Van Sorge NM, Sheen TR, Uchiyama S, Mitchell TJ, Doran KS. Activation of brain endothelium by pneumococcal neuraminidase NanA promotes bacterial internalization. Cell Microbiol. 2010;12(11):1576–88. doi: 10.1111/j.1462-5822.2010.01490.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ho SN, Hunt HD, Horton RM, Pullen JK, Pease LR. Site-directed mutagenesis by overlap extension using the polymerase chain reaction. Gene. 1989;77(1):51–9. doi: 10.1016/0378-1119(89)90358-2. [DOI] [PubMed] [Google Scholar]
- 27.Callegan MC, Booth MC, Jett BD, Gilmore MS. Pathogenesis of gram-positive bacterial endophthalmitis. Infect Immun. 1999;67(7):3348–56. doi: 10.1128/iai.67.7.3348-3356.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Manco S, Hernon F, Yesilkaya H, Paton JC, Andrew PW, Kadioglu A. Pneumococcal neuraminidases A and B both have essential roles during infection of the respiratory tract and sepsis. Infect Immun. 2006;74(7):4014–20. doi: 10.1128/IAI.01237-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sanders ME, Norcross EW, Moore QC, 3rd, Fratkin J, Thompson H, Marquart ME. Immunization with pneumolysin protects against both retinal and global damage caused by Streptococcus pneumoniae endophthalmitis. J Ocul Pharmacol Ther. 2010;26(6):571–7. doi: 10.1089/jop.2010.0077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ernst JD, Decazes JM, Sande MA. Experimental pneumococcal meningitis: role of leukocytes in pathogenesis. Infect Immun. 1983;41(1):275–9. doi: 10.1128/iai.41.1.275-279.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wood WB, Jr, Smith MR, Watson B. Surface Phagocytosis-Its Relation to the Mechanism of Recovery in Pneumococcal Pneumonia. Science. 1946;104(2689):28–9. doi: 10.1126/science.104.2689.28. [DOI] [PubMed] [Google Scholar]
- 32.Weiser JN, Austrian R, Sreenivasan PK, Masure HR. Phase variation in pneumococcal opacity: relationship between colonial morphology and nasopharyngeal colonization. Infect Immun. 1994;62(6):2582–9. doi: 10.1128/iai.62.6.2582-2589.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Marion C, Burnaugh AM, Woodiga SA, King SJ. Sialic acid transport contributes to pneumococcal colonization. Infect Immun. 2011;79(3):1262–9. doi: 10.1128/IAI.00832-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.King SJ, Hippe KR, Weiser JN. Deglycosylation of human glycoconjugates by the sequential activities of exoglycosidases expressed by Streptococcus pneumoniae. Mol Microbiol. 2006;59(3):961–74. doi: 10.1111/j.1365-2958.2005.04984.x. [DOI] [PubMed] [Google Scholar]
- 35.Sanders ME, Norcross EW, Moore QC, Onwubiko C, King LB, Fratkin J, Marquart ME. A comparison of pneumolysin activity and concentration in vitro and in vivo in a rabbit endophthalmitis model. Clin Ophthalmol. 2008;2(4):793–800. doi: 10.2147/opth.s3941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Davey P, Barza M, Peckman C. Spontaneous inhibition of bacterial growth in experimental gram-negative endophthalmitis. Invest Ophthalmol Vis Sci. 1987;28(5):867–73. [PubMed] [Google Scholar]
- 37.Rimini R, Jansson B, Feger G, Roberts TC, de Francesco M, Gozzi A, Faggioni F, Domenici E, Wallace DM, Frandsen N, Polissi A. Global analysis of transcription kinetics during competence development in Streptococcus pneumoniae using high density DNA arrays. Mol Microbiol. 2000;36(6):1279–92. doi: 10.1046/j.1365-2958.2000.01931.x. [DOI] [PubMed] [Google Scholar]
- 38.Hava DL, Hemsley CJ, Camilli A. Transcriptional regulation in the Streptococcus pneumoniae rlrA pathogenicity islet by RlrA. J Bacteriol. 2003;185(2):413–21. doi: 10.1128/JB.185.2.413-421.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.LeMieux J, Hava DL, Basset A, Camilli A. RrgA and RrgB are components of a multisubunit pilus encoded by the Streptococcus pneumoniae rlrA pathogenicity islet. Infect Immun. 2006;74(4):2453–6. doi: 10.1128/IAI.74.4.2453-2456.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.LeMieux J, Woody S, Camilli A. Roles of the sortases of Streptococcus pneumoniae in assembly of the RlrA pilus. J Bacteriol. 2008;190(17):6002–13. doi: 10.1128/JB.00379-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hava DL, Camilli A. Large-scale identification of serotype 4 Streptococcus pneumoniae virulence factors. Mol Microbiol. 2002;45(5):1389–406. [PMC free article] [PubMed] [Google Scholar]
- 42.Gohar M, Okstad OA, Gilois N, Sanchis V, Kolsto AB, Lereclus D. Two-dimensional electrophoresis analysis of the extracellular proteome of Bacillus cereus reveals the importance of the PlcR regulon. Proteomics. 2002;2(6):784–91. doi: 10.1002/1615-9861(200206)2:6<784::AID-PROT784>3.0.CO;2-R. [DOI] [PubMed] [Google Scholar]
- 43.Gominet M, Slamti L, Gilois N, Rose M, Lereclus D. Oligopeptide permease is required for expression of the Bacillus thuringiensis PlcR regulon and for virulence. Mol Microbiol. 2001;40(4):963–75. doi: 10.1046/j.1365-2958.2001.02440.x. [DOI] [PubMed] [Google Scholar]
- 44.Fagerlund A, Brillard J, Furst R, Guinebretiere MH, Granum PE. Toxin production in a rare and genetically remote cluster of strains of the Bacillus cereus group. BMC Microbiol. 2007;7:43. doi: 10.1186/1471-2180-7-43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gohar M, Faegri K, Perchat S, Ravnum S, Okstad OA, Gominet M, Kolsto AB, Lereclus D. The PlcR virulence regulon of Bacillus cereus. PLoS One. 2008;3(7):e2793. doi: 10.1371/journal.pone.0002793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Brown JS, Gilliland SM, Holden DW. A Streptococcus pneumoniae pathogenicity island encoding an ABC transporter involved in iron uptake and virulence. Mol Microbiol. 2001;40(3):572–85. doi: 10.1046/j.1365-2958.2001.02414.x. [DOI] [PubMed] [Google Scholar]
- 47.Loisel E, Jacquamet L, Serre L, Bauvois C, Ferrer JL, Vernet T, Di Guilmi AM, Durmort C. AdcAII, a new pneumococcal Zn-binding protein homologous with ABC transporters: biochemical and structural analysis. J Mol Biol. 2008;381(3):594–606. doi: 10.1016/j.jmb.2008.05.068. [DOI] [PubMed] [Google Scholar]
- 48.Loisel E, Chimalapati S, Bougault C, Imberty A, Gallet B, Di Guilmi AM, Brown J, Vernet T, Durmort C. Biochemical Characterization of the Histidine Triad Protein PhtD as a Cell Surface Zinc-Binding Protein of Pneumococcus. Biochemistry. 2011 doi: 10.1021/bi200012f. [DOI] [PubMed] [Google Scholar]
- 49.Bayle L, Chimalapati S, Schoehn G, Brown J, Vernet T, Durmort C. Zinc uptake by Streptococcus pneumoniae depends on both AdcA and AdcAII and is essential for normal bacterial morphology and virulence. Mol Microbiol. 2011;82(4):904–16. doi: 10.1111/j.1365-2958.2011.07862.x. [DOI] [PubMed] [Google Scholar]
- 50.Bersch B, Bougault C, Roux L, Favier A, Vernet T, Durmort C. New insights into histidine triad proteins: solution structure of a Streptococcus pneumoniae PhtD domain and zinc transfer to AdcAII. PLoS One. 2013;8(11):e81168. doi: 10.1371/journal.pone.0081168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Plumptre CD, Eijkelkamp BA, Morey JR, Behr F, Counago RM, Ogunniyi AD, Kobe B, O'Mara ML, Paton JC, McDevitt CA. AdcA and AdcAII employ distinct zinc acquisition mechanisms and contribute additively to zinc homeostasis in Streptococcus pneumoniae. Mol Microbiol. 2014;91(4):834–51. doi: 10.1111/mmi.12504. [DOI] [PubMed] [Google Scholar]
- 52.Plumptre CD, Hughes CE, Harvey RM, Eijkelkamp BA, McDevitt CA, Paton JC. Overlapping functionality of the Pht proteins in zinc homeostasis of Streptococcus pneumoniae. Infect Immun. 2014;82(10):4315–24. doi: 10.1128/IAI.02155-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Adamou JE, Heinrichs JH, Erwin AL, Walsh W, Gayle T, Dormitzer M, Dagan R, Brewah YA, Barren P, Lathigra R, Langermann S, Koenig S, Johnson S. Identification and characterization of a novel family of pneumococcal proteins that are protective against sepsis. Infect Immun. 2001;69(2):949–58. doi: 10.1128/IAI.69.2.949-958.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Spellerberg B, Cundell DR, Sandros J, Pearce BJ, Idanpaan-Heikkila I, Rosenow C, Masure HR. Pyruvate oxidase, as a determinant of virulence in Streptococcus pneumoniae. Mol Microbiol. 1996;19(4):803–13. doi: 10.1046/j.1365-2958.1996.425954.x. [DOI] [PubMed] [Google Scholar]
- 55.Tettelin H, Nelson KE, Paulsen IT, Eisen JA, Read TD, Peterson S, Heidelberg J, DeBoy RT, Haft DH, Dodson RJ, Durkin AS, Gwinn M, Kolonay JF, Nelson WC, Peterson JD, Umayam LA, White O, Salzberg SL, Lewis MR, Radune D, Holtzapple E, Khouri H, Wolf AM, Utterback TR, Hansen CL, McDonald LA, Feldblyum TV, Angiuoli S, Dickinson T, Hickey EK, Holt IE, Loftus BJ, Yang F, Smith HO, Venter JC, Dougherty BA, Morrison DA, Hollingshead SK, Fraser CM. Complete genome sequence of a virulent isolate of Streptococcus pneumoniae. Science. 2001;293(5529):498–506. doi: 10.1126/science.1061217. [DOI] [PubMed] [Google Scholar]


