Abstract
Ethanol toxicity in yeast Saccharomyces cerevisiae limits titer and productivity in the industrial production of transportation bioethanol. We show that strengthening the opposing potassium and proton electrochemical membrane gradients is a mechanism that enhances general resistance to multiple alcohols. Elevation of extracellular potassium and pH physically bolster these gradients, increasing tolerance to higher alcohols and ethanol fermentation in commercial and laboratory strains (including a xylose-fermenting strain) under industrial-like conditions. Production per cell remains largely unchanged with improvements deriving from heightened population viability. Likewise, up-regulation of the potassium and proton pumps in the laboratory strain enhances performance to levels exceeding industrial strains. Although genetically complex, alcohol tolerance can thus be dominated by a single cellular process, one controlled by a major physicochemical component but amenable to biological augmentation.
The increased use of renewable transportation fuels such as bioethanol is an accepted strategy to combat global climate change (1). However, the toxicity of ethanol and other alcohols to the yeast Saccharomyces cerevisiae is a primary factor limiting titer and productivity in industrial production (2, 3). Ethanol tolerance is a complex phenotype: studies have shown that no single genetic modification is capable of eliciting greater resistance at high ethanol levels (4–7).
As toxicity may arise from chemical perturbation of the plasma membrane, we surmised that ionic composition of the culture medium could play a role in exacerbating or ameliorating the extent of this destabilization (8–10). Therefore, we supplemented various salts to high cell density and high glucose cultures mimicking industrial fermentation to ascertain their consequences on ethanol production.
Monopotassium phosphate (K-Pi) added to standard yeast synthetic complete (YSC) medium induced the greatest improvement (fig. S1), an effect that we dissected into components deriving from elevated potassium (K+) and pH. Specifically, when the pH of cultures containing elevated potassium chloride (KCl) was manually adjusted with potassium hydroxide (KOH) throughout the course of fermentation to match that of cultures containing elevated K-Pi, ethanol titers were statistically indistinguishable (p = 0.09 from two sample t-test; p ≤ 7.6×10−3 for other pairs) from one another (fig. S2, S3). We also determined that KCl elicited a statistically higher improvement than sodium chloride (NaCl), and that supplementation with NaCl and sodium hydroxide, or with monosodium phosphate, demonstrated a distinguishable boost over NaCl alone (p ≤ 2×10−4 from pair-wise t-tests). Thus, the greatest improvements in ethanol production derive specifically from the increase in K+ concentration and reduction in acidity of the fermentation medium.
Over the course of a 3-day culture, supplementation with KCl and KOH enhanced ethanol titer and volumetric productivity (grams of ethanol per volume per hour), two key benchmarks of fermentative performance (Fig. 1A). Additionally, compared with equimolar KCl or matched pH alone, the combination of K+ supplementation and acidity reduction enabled the complete utilization of glucose and decreases in the synthesis of acetic acid and glycerol, two undesired byproducts of fermentation (fig. S4A–D). Ethanol titers of 115–120 g/L have been reported previously from S288C (11), the inbred laboratory strain used here that is known for its low ethanol resistance (5, 12, 13). However, these studies were typically conducted using chemically undefined (“rich”) media. The 128±0.7 g/L (SD) concentrations observed here were achieved using a purely synthetic formulation, allowing us to identify, and precisely control, the environmental components that impact ethanol tolerance.
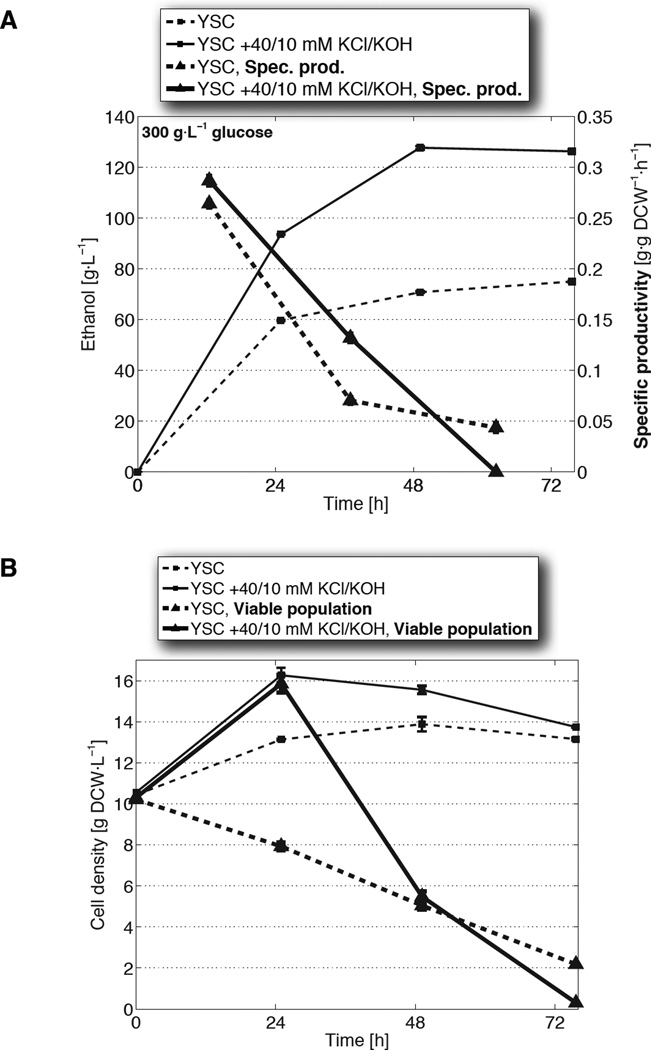
Fig. 1. Elevated extracellular potassium and pH enhance ethanol tolerance and production under high glucose and high cell density conditions.
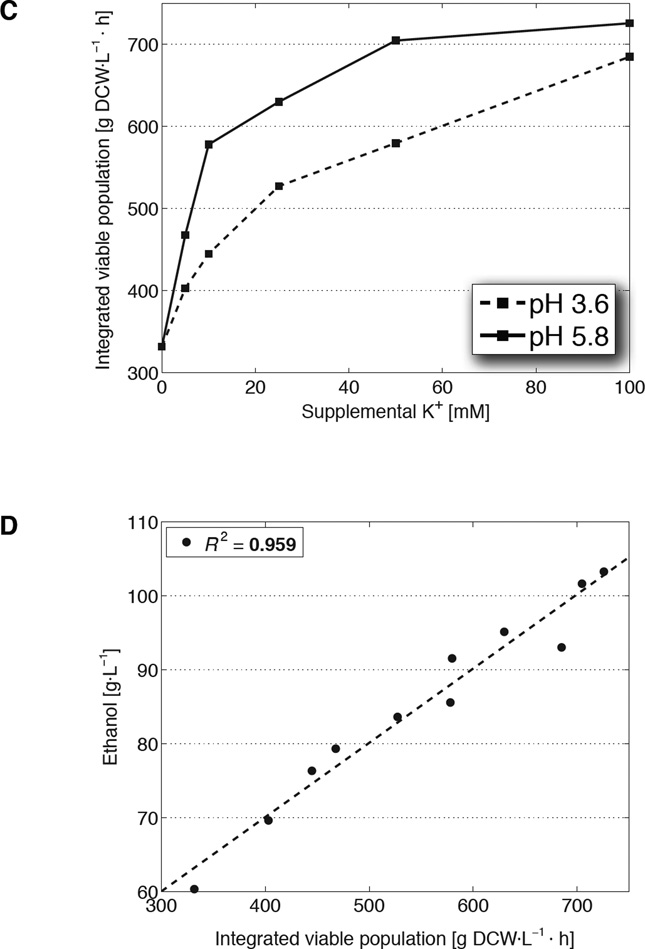
(A) Ethanol titers (squares) and per-cell rates of production (triangles) from fermentations in unmodified synthetic complete medium (YSC; dashed) or YSC supplemented with 40 mM KCl and 10 mM KOH (solid). Specific productivities are calculated from the mean viable population (thick lines from B) during each 24 h period. (B) Cell densities (dry cell weight/DCW; thin squares) and the underlying viable populations (thick triangles) from the fermentations in A. Data are mean ± SD from 3 biological replicates. (C) Net fermentation viability, expressed as time integrals of the viable cell population, as a function of potassium added to YSC in the form of KCl (pH 3.6), or 5 mM KOH + KCl (pH 5.8). (D) Time integral values from C regressed against their final ethanol titers.
The boost in ethanol production from KCl and KOH supplementation did not arise simply from an increase in cell number, but from an increase in cell tolerance. Specifically, the 80±1.3% (SD) jump in titer (Fig. 1A) was accompanied only by an 11±4.6% (SD) average higher cell density (Fig. 1B); therefore, cell growth alone could not explain the rise in output. This discrepancy, however, was resolved when we directly assessed fractions of cells remaining alive throughout fermentation (fig. S5A) and discovered that the addition of KCl and KOH enhanced overall population viability (Fig. 1B, fig. S5B). This enhancement, furthermore, occurred despite the increase in toxicity imposed by higher accumulations of ethanol.
When we calculated specific productivities — rates of ethanol increase normalized by the live, rather than total, cell population — the values from KCl and KOH supplementation differed from the control by an average of 11±7.5% (SD) (Fig. 1A). That these differences account for a minor portion of the increase in titer suggests that elevated K+ and pH acts primarily not by affecting per-cell output, but by boosting tolerance and the overall viable cell population. Additionally, these effects are observed fully in fermentations conducted in anaerobic bioreactors, demonstrating that these tolerance improvements do not depend on oxygen availability and can scale to higher-volume environments (fig. S6).
Because it is the viable cell population that is actively fermenting, final titers are governed both by the number of live cells and the length of time over which cells maintain viability against rising ethanol toxicity. The time integral of the viable cell population captures these two aspects and quantifies the overall impact of tolerance on ethanol production (fig. S5B, S5C). Indeed, varying the concentration of supplemental KCl resulted in a dose-dependent increase of these time integrals; moreover, when these same additions were done at a higher pH, integrated viable population values were shifted upward correspondingly (Fig. 1C, fig. S7A). A strong linear relationship exists between the time integrals of viable cell density and final titers (R2 = 0.96; p = 1.5×10−7), demonstrating that the ability to endure toxicity is a principal determinant of ethanol output and the primary trait strengthened by KCl and KOH supplementation (Fig. 1D, fig. S7B).
The enhancements conferred by elevated K+ and reduced acidity transcend genetic background and are elicited universally among a random sampling of industrial yeast strains. Those used in the production of biofuel ethanol in Brazil (PE-2) and the United States (Lasaffre Ethanol Red), and of sake wine in Japan (Kyokai No. 7), are typically the result of genetic selection efforts designed to isolate superior ethanol phenotypes (13–15). Consequently, all demonstrate distinctly higher ethanol output than S288C (10±1% – 30±1.2% (SD)) when grown in unmodified medium (Fig. 2A). However, when subjected to KCl and KOH supplementation, all strains responded with enhancements in tolerance that enabled the complete consumption of glucose (fig. S8) and titers of 116±0.9 – 127±1.6 g/L (SD). Under these conditions, S288C performed indistinguishably from the two industrial bioethanol strains (p ≥ 0.08 from pair-wise t-tests). Thus, a strain traditionally deemed ethanol sensitive is inherently capable — without genetic modification — of superior tolerance, indicating that K+ supplementation and acidity reduction drive a process that can supersede advantages conferred by genetic adaptation.
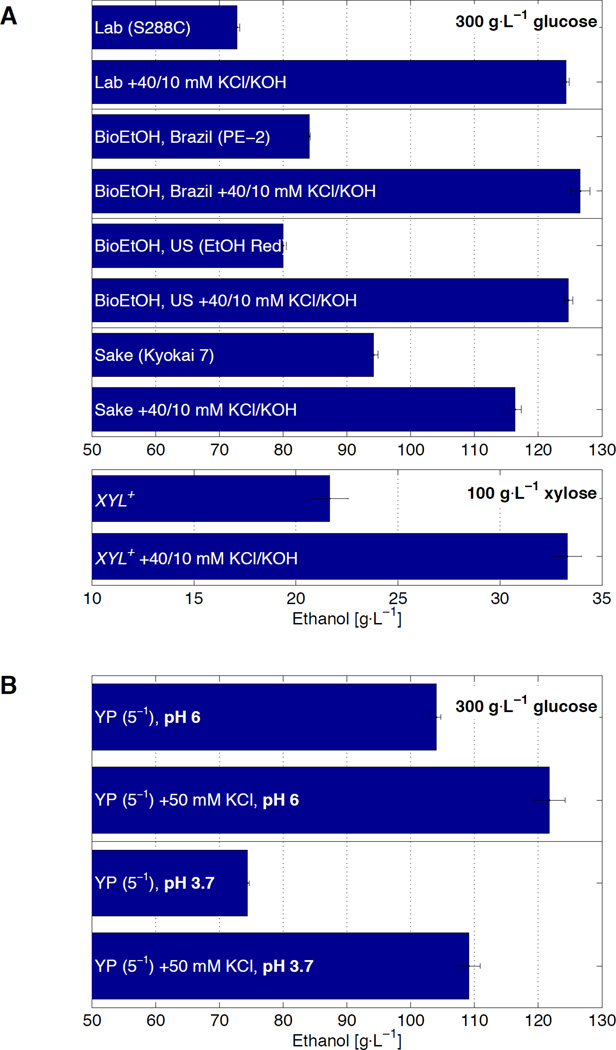
Fig. 2. Elevated potassium and pH are sufficient to enhance tolerance independently of strain genetics, sugar substrate, and alcohol species.
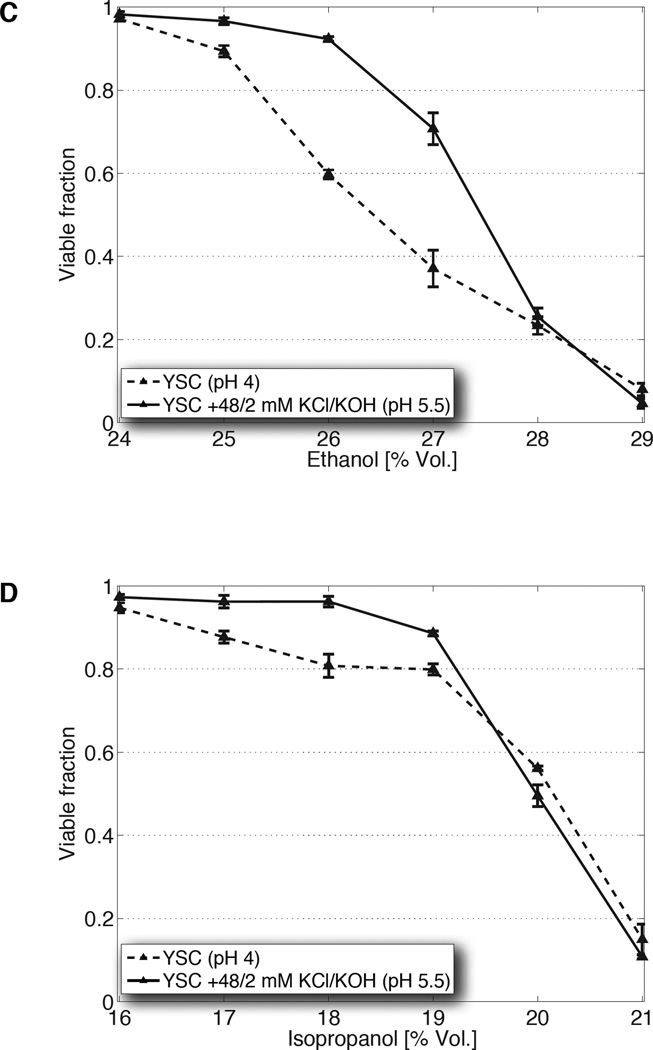
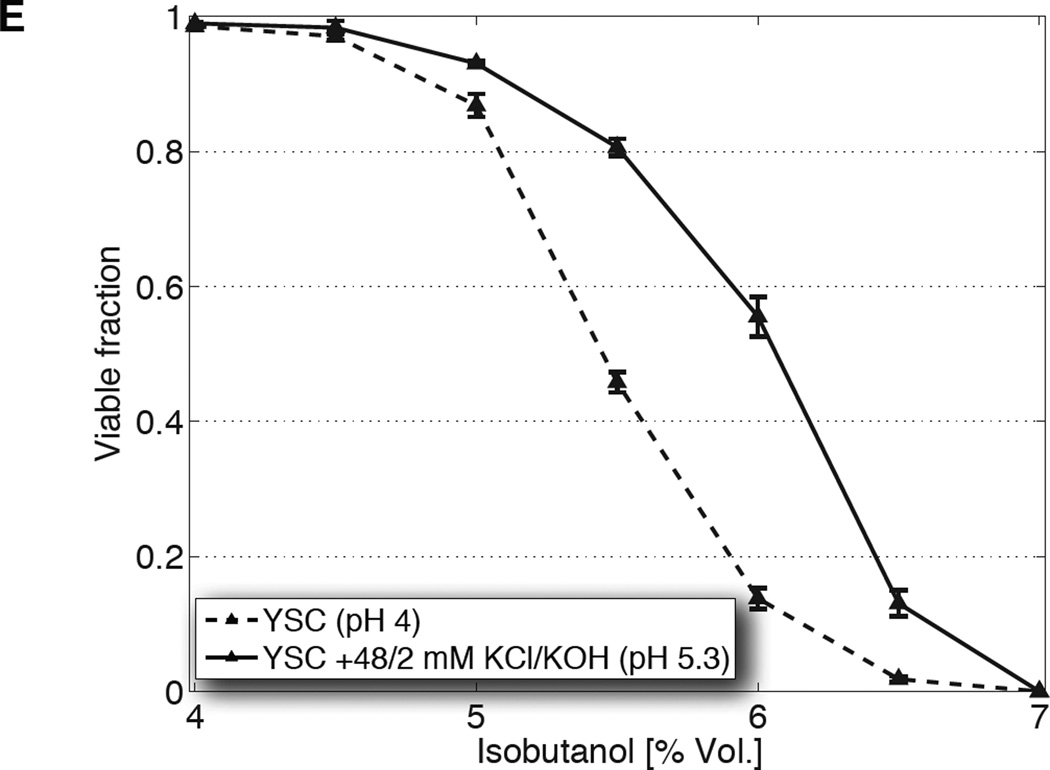
(A) Ethanol titers from glucose fermentation (top) of one laboratory (S288C) and three industrial (PE-2, Ethanol Red, Kyokai 7) yeast strains, or from xylose fermentation (bottom) of an engineered xylose strain, in unmodified YSC or YSC supplemented with 40 mM KCl and 10 mM KOH. (B) Titers from S288C cultured in 20% yeast extract-peptone medium (YP) or that supplemented with potassium, at pH 6 and 3.7. (C) Population fractions of S288C after transfer from overnight growth in unmodified YSC (dashed), or that supplemented with 48 mM KCl and 2 mM KOH (solid), into media containing the indicated concentrations of ethanol. (D, E) Same as C, but with step increases of isopropanol or isobutanol, respectively. All data are mean ± SD from 3 biological replicates.
These adjustments to the medium, furthermore, enhance fermentation from xylose, an important hemicellulosic sugar that cannot be consumed by standard strains of S. cerevisiae. In an engineered strain (16), 22±0.9 g/L (SD) ethanol was produced from unmodified medium containing 100 g/L xylose (Fig. 2A). When fermented with the addition of KCl and KOH, we observed a 54±5.7% (SD) increase in titer, commensurate with the complete assimilation of xylose (fig. S8). Thus, K+ supplementation and acidity reduction enhance tolerance in a manner impartial to the type of substrate.
The improvements conferred by elevated K+ and pH generalize beyond synthetic media to chemically undefined broths, provided that such formulations do not already saturate for these effects. For example, in yeast extract-peptone (YP) medium (~pH 6 and unknown concentrations of individual nutrients (17)), cells ferment all sugar such that no margin is available for improvement (fig. S9B). However, the impact of specific supplements can be assessed if the YP components are made limiting. Indeed, when YP was decreased to 30% or 3% while maintaining the same glucose concentration, supplementation with K+ improved ethanol output whereas additives shown to be fermentation-neutral (from fig. S1) did not (fig. S9A). Using YP diluted to 20%, titers of 104±0.8 g/L (SD) are produced, while the addition of K+ enhanced output 17±2.5% (SD) (Fig. 2B). When pH was reduced from 6 to 3.7, production was concomitantly reduced 28±0.8% (SD). However, the subsequent addition of K+ compensated for this decrease, restoring titers 47±2.5% to 109±1.8 g/L (SD). Thus, in media with undefined composition, extracellular K+ and pH are also sufficient to quantitatively modulate ethanol performance.
To isolate the effects of KCl and KOH supplementation on tolerance from other fermentation variables (e.g., decreasing turgor pressure from glucose consumption), we subjected yeast to non-physiological step increases in ethanol concentration and quantified population fractions surviving after 80 min, a period much shorter than the length of fermentation but adequate for cell viability to be impacted. In medium containing a subsistence amount of glucose that minimizes newly produced ethanol, elevated K+ and pH enhanced viability in shocks up to 27% (vol/vol) when compared to cells stressed in unmodified conditions (Fig. 2C). Analogous experiments performed using high glucose (mimicking the osmotic conditions of high gravity fermentation) and heightened K-Pi yielded a similar result, albeit at a lower range of ethanol concentrations (fig. S10A). These results indicate that the impact of elevated K+ and reduced acidity is relatively immediate, does not require adaptation to ethanol accumulation over the course of days, and is capable of overcoming the combined stress of high sugar and ethanol (2).
The boost in tolerance conferred by heightened K+ and pH extends to higher alcohols capable of serving as unmodified substitutes for gasoline. Although at lower concentrations when compared to ethanol (reflecting their increased toxicity), we observed that viability is similarly enhanced when cells are shocked using step increases of isopropanol and isobutanol (Fig. 2D, 2E, fig. S10B). That the improvements are not unique to ethanol suggest that these adjustments to the medium augment a more general cellular process involved in alcohol resistance or membrane integrity.
Given the dominant effects exerted by extracellular K+ and pH, we hypothesized that K+ and proton (H+) regulation may be a primary mechanism mediating the tolerance phenotype. Because opposing gradients of K+ and H+ ions are generated across the yeast plasma membrane by the K+ importer TRK1 and H+ exporter PMA1 (18, 19), we surmised that genetic modifications of these ATP-dependent pumps, designed to perturb or strengthen these gradients, would produce corresponding effects to fermentative performance. Indeed, genetic deletions that debilitate K+ import or H+ export weakened ethanol output (fig. S11). Likewise, hyper-activation of TRK1, accomplished by deletions of PPZ1 and PPZ2 (19), generated an increase, yielding an 18±1.6% (SD) improvement in titer over the wild type when cultured in unmodified medium (Fig. 3, fig. S12A). Given the electroneutral co-dependence of the K+ and H+ gradients (18), we, furthermore, reasoned that up-regulation of PMA1 alongside augmented TRK1 would further enhance the ethanol phenotype; indeed, the combination increased titers by 27±2.2% (SD) over the wild type. These improvements in output mirrored enhancements in net fermentation viability (fig. S12B), affirming the coupled nature of production and tolerance. Incidentally, over-expression of PMA1 without hyper-activation of TRK1 did not enhance ethanol performance (Fig. 3), supporting the proposed notion that K+ uptake creates the dominant electromotive force and H+ efflux acts primarily as a response current (19).
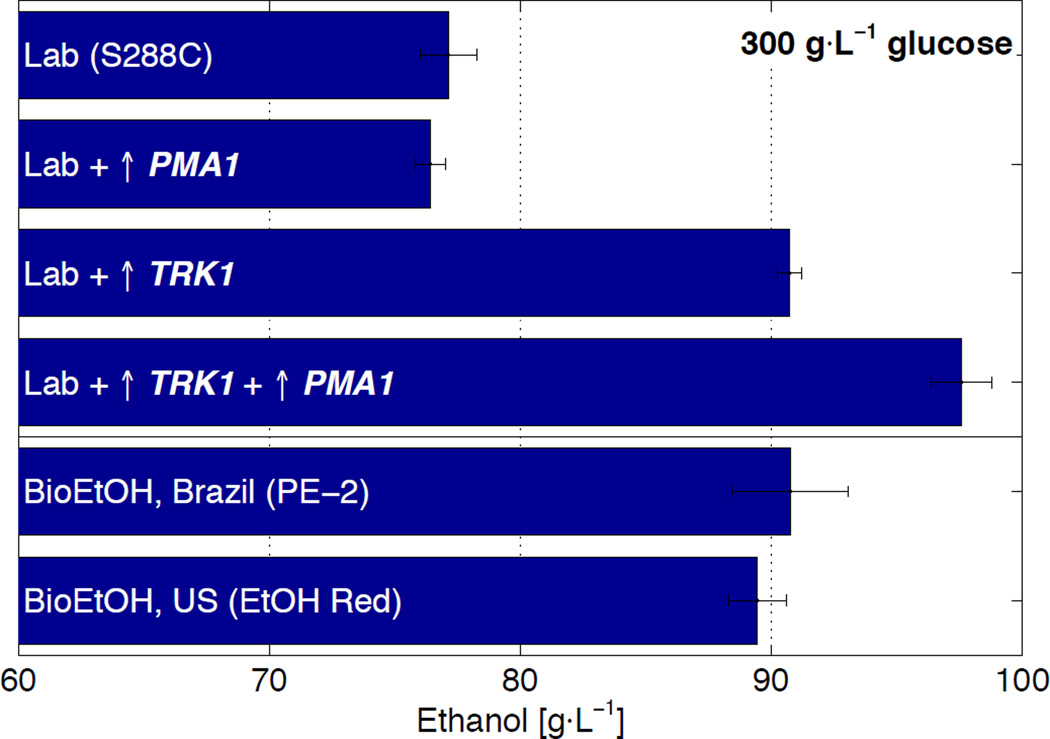
Fig. 3. Genetic augmentation of the plasma membrane potassium (TRK1) and proton (PMA1) pumps increase ethanol production to levels exceeding industrial strains.
Ethanol titers from a wild type laboratory strain (S288C) transformed with empty over-expression plasmid, S288C transformed with a plasmid over-expressing PMA1, S288C containing hyper-activated TRK1 (via deletions of PPZ1 and PPZ2) and transformed with empty over-expression plasmid, the TRK1 hyper-activated strain transformed with a plasmid over-expressing PMA1, and bioethanol production strains from Brazil (PE-2) and the US (Ethanol Red), all cultured in unmodified YSC lacking uracil. Data are mean ± SD from 3 biological replicates.
Genetic augmentation of the K+ and H+ transporters increased ethanol titer of the laboratory strain to that surpassing the two bioethanol production strains (Fig. 3). These specific genetic modifications are, therefore, sufficient to create a superior phenotype previously available only through selection. That hyper-activation of TRK1 alone is sufficient to match the titers of PE-2 and Ethanol Red, combined with the observation that these industrial strains respond to KCl and KOH supplementation, lends support to the possibility that polymorphisms enhancing the K+ and H+ gradients may be responsible for intrinsically higher ethanol tolerances (20).
Collectively, our results suggest a toxicity model where alcohols attack viability not at threshold concentrations that solubilize lipid bilayers, but at lower concentrations that increase permeability of the plasma membrane and dissipate the cell’s ionic membrane gradients. That genetically unchanged cells can be made to tolerate higher ethanol concentrations by modulating extracellular K+ and pH indicates that many observed tolerance thresholds (e.g., the sub-100 g/L titers from unmodified medium) represent a physiological, rather than chemical, limit. Ethanol has been known to decrease intracellular pH in a dose-dependent fashion, demonstrating that its amphipathicity permeabilizes the plasma membrane to H+ (and, potentially, other ions) (8, 21). Furthermore, that the coupled K+ and H+ gradients comprise a dominant portion of the yeast electrical membrane potential, used to power many of the cell’s exchange processes with the environment, hints that the cessation of nutrient and waste transport due to gradient dissipation may be a primary mode of cell death (18, 21–25).
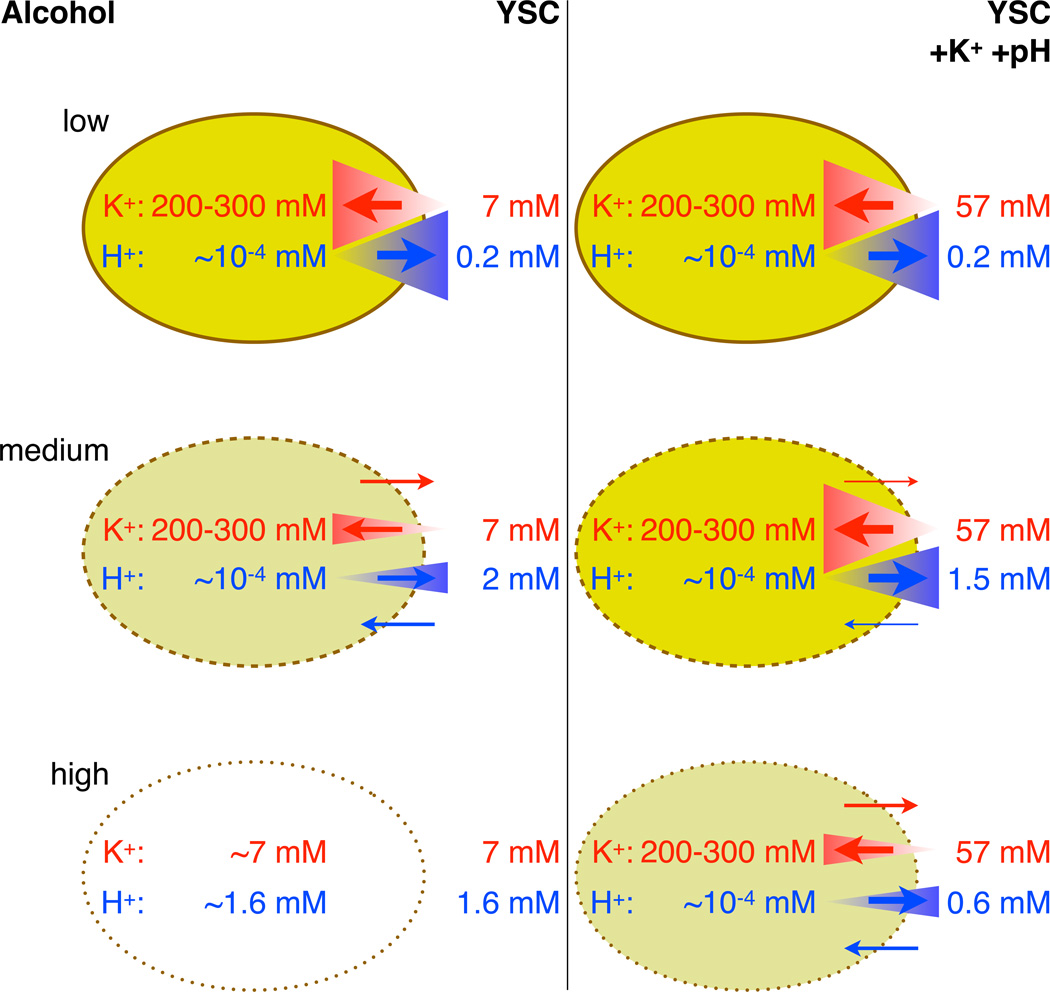
These ionic gradients are likely disrupted with increasing strength as ethanol accumulates during fermentation, requiring the cell to expend escalating amounts of energy to reestablish the steep separation of charges. Conditions that bolster the cell’s efforts to maintain high concentrations of intracellular K+ and low intracellular H+ (estimated to be 200–300 mM and pH 7, respectively (23, 24)) thus likely enhance tolerance by raising the threshold to which alcohols will collapse these gradients (Fig. 4). Physical reinforcements in the form of K+ supplementation and acidity reduction generate the greatest improvements: not only do higher concentrations of extracellular K+ facilitate import (by potentially reducing the gradient from 36- to 4-fold, using the above estimates), and lower concentrations of extracellular H+ facilitate export, the corresponding rates of ion leakage are decreased due to reduced concentration differences across the membrane. Likewise, genetic modifications strengthening the ion pump activities responsible for establishing the K+ and H+ gradients increase performance without alteration of the medium. However, that KCl and KOH supplementation confers larger improvements suggests that tolerance may contain a considerable physically driven component that ultimately constrains biological augmentation.
Fig. 4. Potential biophysical mechanism depicting how elevated potassium and pH counteract rising alcohol toxicity.
In the absence of stress (top row), the opposing potassium (K+) and proton (H+) pumps maximally maintain steep gradients of K+ and H+ across the plasma membrane. Rising alcohol levels perturb these gradients by permeabilizing the membrane and increasing ion leakage (middle left). Elevated potassium and pH, however, bolster the gradients by slowing rates of ion leakage (due to reduced concentration differences) and allowing transporters to pump against a less precipitous differential (middle right). Therefore, the threshold alcohol concentration that collapses these gradients is raised, allowing cells to maintain viability at higher toxicity levels (bottom row). Red corresponds to K+, blue to H+, size of triangles to concentration gradient steepness, and thickness of arrows to magnitude of ion flux.
Supplementary Material
Acknowledgments
We are grateful to present and former members of the Fink and Stephanopoulos labs for intellectual and technical guidance. Additionally, we thank V. Vyas and D. Pincus for critical commentary on the manuscript, and Phibro for the gift of Lesaffre Ethanol Red. Support was provided by the MIT Energy Initiative and Dept. of Energy grant DESC0008744 to G.S., and National Institutes of Health grant R01-GM035010 to G.R.F. No competing financial interests impinged on this study. F.H.L., G.R.F., and G.S. are listed as co-inventors on a patent application filed by MIT and the Whitehead Institute. F.H.L., G.R.F., and G.S. designed all experiments and collaborated on the manuscript; A.G. conducted all bioreactor studies; F.H.L. performed all other experiments, strain construction, and analysis of data.
Footnotes
Supporting data are available in the supplementary materials.
References and Notes
- 1.Demain AL. J Ind Microbiol Biotechnol. 2009;36:319–332. doi: 10.1007/s10295-008-0521-8. [DOI] [PubMed] [Google Scholar]
- 2.Bai FW, Chen LJ, Zhang Z, Anderson WA, Moo-Young M. J Biotechnol. 2004;110:287–293. doi: 10.1016/j.jbiotec.2004.01.017. [DOI] [PubMed] [Google Scholar]
- 3.Stephanopoulos G. Science. 2007;315:801–804. doi: 10.1126/science.1139612. [DOI] [PubMed] [Google Scholar]
- 4.van Voorst F, Houghton-Larsen J, Jønson L, Kielland-Brandt MC, Brandt A. Yeast. 2006;23:351–359. doi: 10.1002/yea.1359. [DOI] [PubMed] [Google Scholar]
- 5.Swinnen S, et al. Genome Res. 2012;22:975–984. doi: 10.1101/gr.131698.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Stanley D, Bandara A, Fraser S, Chambers PJ, Stanley GA. J Appl Microbiol. 2010;109:13–24. doi: 10.1111/j.1365-2672.2009.04657.x. [DOI] [PubMed] [Google Scholar]
- 7.Alper H, Moxley J, Nevoigt E, Fink GR, Stephanopoulos G. Science. 2006;314:1565–1568. doi: 10.1126/science.1131969. [DOI] [PubMed] [Google Scholar]
- 8.Madeira A, et al. FEMS Yeast Res. 2010;10:252–258. doi: 10.1111/j.1567-1364.2010.00607.x. [DOI] [PubMed] [Google Scholar]
- 9.Dickey AN, Faller R. Biophys J. 2007;92:2366–2376. doi: 10.1529/biophysj.106.097022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chanda J, Bandyopadhyay S. Langmuir. 2006;22:3775–3781. doi: 10.1021/la053398r. [DOI] [PubMed] [Google Scholar]
- 11.Pais TM, et al. PLoS Genet. 2013;9:e1003548. doi: 10.1371/journal.pgen.1003548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lewis JA, Elkon IM, McGee MA, Higbee AJ, Gasch AP. Genetics. 2010;186:1197–1205. doi: 10.1534/genetics.110.121871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Argueso JL, et al. Genome Res. 2009;19:2258–2270. doi: 10.1101/gr.091777.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Akao T, et al. DNA Res. 2011;18:423–434. doi: 10.1093/dnares/dsr029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Katou T, Namise M, Kitagaki H, Akao T, Shimoi H. J. Biosci. Bioeng. 2009;107:383–393. doi: 10.1016/j.jbiosc.2008.12.014. [DOI] [PubMed] [Google Scholar]
- 16.Zhou H, Cheng J-S, Wang BL, Fink GR, Stephanopoulos G. Metab Eng. 2012;14:611–622. doi: 10.1016/j.ymben.2012.07.011. [DOI] [PubMed] [Google Scholar]
- 17.Sherman F. Meth Enzymol. 2002;350:3–41. doi: 10.1016/s0076-6879(02)50954-x. [DOI] [PubMed] [Google Scholar]
- 18.Yenush L, Merchan S, Holmes J, Serrano R. Mol Cell Biol. 2005;25:8683–8692. doi: 10.1128/MCB.25.19.8683-8692.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yenush L, Mulet JM, Ariño J, Serrano R. EMBO J. 2002;21:920–929. doi: 10.1093/emboj/21.5.920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Reisser C, et al. G3 (Bethesda) 2013 doi: 10.1534/g3.113.005884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cartwright CP, et al. Microbiology. 1986;132:369–377. [Google Scholar]
- 22.Madrid R, Gómez MJ, Ramos J, Rodríguez-Navarro A. J Biol Chem. 1998;273:14838–14844. doi: 10.1074/jbc.273.24.14838. [DOI] [PubMed] [Google Scholar]
- 23.Orij R, Brul S, Smits GJ. Biochim Biophys Acta. 2011;1810:933–944. doi: 10.1016/j.bbagen.2011.03.011. [DOI] [PubMed] [Google Scholar]
- 24.Cyert MS, Philpott CC. Genetics. 2013;193:677–713. doi: 10.1534/genetics.112.147207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Horák J. Biochim Biophys Acta. 1997;1331:41–79. doi: 10.1016/s0304-4157(96)00015-9. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.