Abstract
Background
Diarrheal diseases can be caused by viral, bacterial and parasitic infections. This paper provides a preliminary image of diarrhea with regards to etiology and epidemiologic factors in Tunisian children less than five years of age.
Methods
Overall, 124 diarrhoeal stools were collected from patients suffering from acute diarrhea and 54 stool samples from healthy children. All stools were examined for the presence of enteric pathogens.
Results
In diarrheagenic children, 107 pathogenic bacteria were isolated (12 Salmonella spp. (9.7%) and 95 diarrheagenic Escherichia coli strains (76.6%): 29 enteroaggregative E.coli (EAEC) (23.4%), 15 enteroinvasive E.coli (EIEC) (12.1%), 17 enteropathogenic E.coli (EPEC) (13.7%), 26 enterotoxigenic E.coli (ETEC) (21%) and 2 enterohemoragic E.coli (EHEC) (1.6%). However, in the control group, 23 pathogenic E.coli strains were isolated (42.6%): 8 EAEC (14.8%), 12 EIEC (22.2%) and 3 EPEC (5.5%). Among diarrheagenic E.coli (DEC), only ETEC strains were significantly recovered from diarrheagenic children than from healthy controls (P < 0.0003). Group A rotavirus was identified in 33.9% (n=42) of diarrheagenic children and in 11.1% among the control group (n=6). Concerning norovirus, 8.9% (n=11) of the samples collected from diarrheagenic children and 9.2% (n=5) from the control group were positive. The prevalence of rotaviruses and Salmonella spp were also significantly higher in patients with diarrhea than in controls (P = 0.002 and P < 0.019, respectively). Finally, enteropathogenic parasites (Entamoeba coli and cryptosporidium Oocystes) were isolated from 4.8% and 9.2% of diarrheagenic and control children, respectively.
Conclusion
These results provide baseline data about the relative importance of different enteropathogens in Tunisian children.
Keywords: Enteric pathogens, Escherichia coli, Diarrhea, Children, Diagnosis, Tunisia
Introduction
Acute diarrhea is still a relevant health problem, particularly in early infancy, in both developing and developed countries. Furthermore, acute gastroenteritis is the major cause of morbidity and mortality worldwide (1). Diarrheal diseases cause over two million deaths every year, most of them affecting children less than 5 years of age living in less developed countries (www.who.int., 2005).
A wide range of enteropathogens cause diarrhea in children such as viruses, bacteria and parasites (2, 3, 4). The data on rotavirus and norovirus gastroenteritis published by the United States Centers for Disease Control and Prevention, showed that rotavirus infections were estimated to cause 25 million clinical visits, 2 million hospitalizations and 453,000 deaths worldwide each year in children younger than 5 years (5). However, it was estimated that norovirus caused 900,000 clinical visits and 64,000 hospitalizations among children in developed countries and up to 200,000 deaths of children younger than 5 years in developing countries each year (6).
Among the bacterial pathogens, Salmonella, Shigella and some Escherichia coli (E.coli) strains can cause severe and life-threatening diarrhea. Diarrheagenic E.coli (DEC) strains can be currently classified into five major categories on the basis of distinct epidemiological and clinical features, specific virulence determinants, and association with certain serotypes (7) : (i) enteroaggregative E.coli (EAEC), (ii) enteroinvasive E.coli (EIEC), (iii) enterohemorrhagic E.coli (EHEC), (iv) enteropathogenic E.coli (EPEC), and (v) enterotoxigenic E.coli (ETEC). Other additional categories have been also described as DECstrains (7); however their clinical significance, epidemiology and pathogenesis have not been well understood yet.
In North Africa, several studies about the role of DEC in diarrhea have been carried out especially in Egypt. However, little is known about the role of DEC in diarrheal diseases in the Maghreb area. During the last two decades, only one study has been conducted in Tunis (North of Tunisia), and highlighted an important rate of ETEC in children and adults with or without diarrhea, suggesting an endemic status in our country (8).
The aim of this prospective study was to determine the etiology of infantile diarrhea including viruses, bacteria and parasites and to examine the virulence markers of pathogenic E.coli strains.
Materials and Methods
Subjects and fecal specimens
From December 2008 to November 2009, 124 stool samples were collected from non-hospitalized children (median age: 21 months; range: 3 month to 60 months) who consulted the CNSS polyclinic and suffered from acute diarrhea and gastro-enteric symptoms. A control group consisting of 54 healthy children representing the same age group (median age: 20 months; range: 4 month to 60 months) was also tested for the presence of enteric pathogens. Children with chronic diarrhea were excluded from this study.
Diarrhea was characterized by watery stools with or without blood and mucus. Children patients had no history of travelling outside Tunisia and did not take antibiotics in the week preceding the sampling. Stools were classified as bloody when blood was visible upon examination of the samples macroscopically. Other clinical symptoms, including fever, vomiting, nausea, abdominal pain or dehydration were obtained through questionnaires reported by the physicians.
The samples were immediately transported to the laboratory. Specimens which were not analyzed for more than four hours were rejected. Patients and healthy persons, who mostly were on a low income, were informed by doctors about the study and agreed to participate and to inform doctors of any information needed. This study did not require approval by the ethic committee.
Bacteriological Methods Phenotypic procedures
All fecal specimens were cultured on sorbitol MacConkey agar (Oxoid) and were incubated for 24 hours at 37°C. Colonies suspected to be E.coli (3-5 colonies from each culture plate) were selected. These colonies were further studied by other characteristic: enzymes specific for E.coli strains (Tryptophanase that cleaves Tryptophan into pyruvate, indol, and ammonia; by using reagents (Kovac’s and DMCA). Identification was confirmed with the Api 20E system (bioMérieux, Marcy-l’étoile, France).
Samples were subcultred on Hektoen agar (Oxoid) and on other media (SS agar (Oxoid)) for the identification of Salmonella and Shigella spp. followed by an overnight incubation at 37°C. Colonies morphologically resembling Shigella and Salmonella spp. were further identified based on appropriate biochemical reactions using the API 20E system and the results were confirmed by slide agglutination testing using commercially available antisera (Biorad, France).
Molecular procedure for the identification of diarrheagenic E. coli
For DNA extraction, all E.coli strains were grown in enrichment broth. Genomic DNA was prepared from each isolate by boiling (9) and used as target for polymerase chain reaction (PCR).
Thus, the DNA templates were subjected to multiplex and monoplex PCRs with specific primers for the detection of the following virulence markers: bfpA (BFP1, BFP2) structural gene for the bundle-forming pilus of EPEC (10), eae (eae1, eae2) (attaching and effacing lesions of EPEC (10, 11), shiga toxins and their variants stx, stx1 (STX1f, STX1r) andstx2 (STX2f, STX2r) of EHEC (12), elt (heat-labile enterotoxin) (LTL, LTR) and/or est (heat-stable enterotoxin) (AL65, AL125) (enterotoxins of ETEC) (13, 14), ipaH (ipaIII, ipaIV) invasion-adhesion locus of the invasion plasmid found in EIEC (15) and aggR (aggRks1, aggRkas2) transcriptional activator of AAF I and AAF II of EAEC (16). The sequences of primers selected for use in the amplification method matched the sequences of the corresponding genes of EPEC, EHEC, ETEC, EIEC and EAEC.
The minimum criteria for determining diarrheagenic E.coli were defined as follows: the presence of est and/or elt for ETEC strains, the presence of bfpA and eae for typical EPEC strains whereas the only presence of eae gene confirmed the detection of atypical EPEC strains, the presence of shiga toxins genes and their variants (stx) for STEC strains, the presence of ipaH for EIEC and the presence of aggR for EAEC. Differentiation between EIEC and Shigella spp for strains possessing ipaH gene was also achieved by agglutination with Shigella specific antisera (Bio-Rad laboratories).
To further investigate the toxin type in diarrheagenic EHEC strains, we conducted two other monoplex PCRs. The first monoplex PCR was performed to detect shiga toxins 1 gene (stx1) (STX1f, STX1r) and the second monoplex PCR to detect all shiga toxins 2 gene variants (stx2) (STX2f, STX2r)) (Table 1).
Table 1.
Primers used in multiplex and monoplex PCRs
| Designation | Primer sequence (5′-3′) | Target gene | Amplicon size (bp) |
|---|---|---|---|
| AL65 | 5’-TTAATAGCACCCGGTACAAGCAGG-3’ | est | 147 |
| AL125 | 5’- 5’-CCTGACTCTTCAAAAGAGAAAATTAC-3’ | est | |
| LTL | 5’-TCTCTATGTGCATACGGAGC–3’ | elt | 322 |
| LTR | 5’-CCATACTGATTGCCGCAAT-3’ | elt | 322 |
| eae1 | 5’CTGAACGGCGATTACGCGAA 3’ | eae | 917 |
| eae2 | 5’CCAGACGATACGATCCAG3’ | eae | 917 |
| VTcom-u | 5’gACCgAAATAATTTATATgTg3’ | stx | 518 |
| VTcom-d | 5’TgATgATggCAATTCAgTAT3’ | stx | 518 |
| ipaIII | 5’gTTCCTTgACCgCCTTTCCgATACCgTC3’ | ipaH | 916 |
| ipaIV | 5’gCCggTCAgCCACCCTCTgAgAgTAC3’ | IpaH | 916 |
| aggRks1 | 5’gTATACACAAAAgAAggAAgC3’ | aggR | 254 |
| aggRkas2 | 5’ACAgAATCgTCAgCATCAgC3’ | aggR | 254 |
| BF1 | 5’AATggTgCTTgCgCTTgCTgC3’ | bfpA | 326 |
| BF2 | 5’gCCgCTTTATCCAACCTggTA3’ | bfpA | 326 |
| STX1f | 5’ATAAATCgCCATTCgTTgACTAC3’ | stx1 | 180 |
| STX1r | 5’AgAACgCCCACTgAgATCATC3’ | stx1 | 180 |
| STX2f | 5’ggCACTgTCTgAAACTgCTCC3’ | stx2 | 225 |
| STX2r | 5’TCgCCAgTTATCTgACATTCTg3’ | stx2 | 225 |
All reactions were performed in a final volume of 34 μl containing 5μl of template DNA, 0.2 mM dNTPs, 2 Mm MgCl2, 5.0 units of Taq polymerase, 50 ng of each primer and deionized water to make up the volume. All assays used the same cycling parameters. Primers and cycling parameters used in multiplex and monoplex PCRs were summarized in (Table 1, 2, 3).
Table 2.
Cycling parameters used in multiplex and monoplex PCRs
| Primers | PCR programme | ||
|---|---|---|---|
| Multiplex | STX | Enzyme activation | |
| PCR I | eae | 5min at 94°C | |
| ipaH | 40 Cycles | ||
| Denaturation: 30s at 94°C Primer annealing30s | |||
| 62°C | |||
| DNA extension: 1min at 72°C Final elongation | |||
| 5min at 72°C | |||
| Multiplex | bfpA | Enzyme activation | |
| PCR II | aggR | 5min at 94°C | |
| 40 Cycles | |||
| Denaturation: 30s at 94°C Primer annealing30s | |||
| 62°C | |||
| DNA extension: 1min at 72°C Final elongation | |||
| 5min at 72°C | |||
| Multiplex | Est | Enzyme activation | |
| PCR III | Elt | 5min at 94°C | |
| 40 Cycles | |||
| Denaturation: 30s at 94°C Primer annealing30s | |||
| 62°C | |||
| DNA extension: 1min at 72°C Final elongation | |||
| 5min at 72°C | |||
| Monoplex | STX1 | Enzyme activation | |
| PCR I | 5min at 94°C | ||
| 40 Cycles | |||
| Denaturation: 30s at 94°C Primer annealing30s | |||
| 62°C | |||
| DNA extension: 1min at 72°C Final elongation | |||
| 5min at 72°C | |||
| Monoplex | STX2 | Enzyme activation | |
| PCR II | 5min at 94°C | ||
| 40 Cycles | |||
| Denaturation: 30s at 94°C Primer annealing30s | |||
| 62°C | |||
| DNA extension: 1min at 72°C Final elongation | |||
| 5min at 72°C |
Table 3.
International reference of E.coli strains used as control for PCRs amplifications
| Strain | International Designation | Positive gene(s) |
|---|---|---|
| ETEC | H10407 | elt |
| ETEC | Jep5683 | est |
| E.coli strain(negative control) | HB 101 | No virulence gene |
| EHEC | EDL933 (O157:H7) | stx |
| EPEC | EPEC2348/69 (O127:H6) | eae and bfpA |
| EIEC | EIEC 11741 | ipaH |
| EAEC17-2 | EAEC17-2 | aggR |
A negative control lacking the DNA template was included in each experiment to exclude the possibility of reagent contamination. Multiplex PCR assays were performed with each reference strain of diarrheagenic E. coli from pure culture. We used non-pathogenic Escherichia coli strain HB 101 as a negative control. Positive controls were as follows: enterotoxigenic Escherichia coli ETEC H10407 for the elt gene, Jep5683 for the est gene, EDL933 (O157:H7) for the stx gene, EPEC2348/69 (O127:H6) for the eae and bfpA genes, EIEC 11741 for the ipaH gene and finally, EAEC 17-2 for the aggRgene. These strains were kindly provided by Professor Ridha Ben Aissa (Institut Pasteur Tunis).
Next, in order to test the reproducibility of these results and equally the reproducibility of mPCR method, the mPCR assays were repeated at least twice with all tested strains whereby the DNA template of strain that was freshly prepared for each repeat. Finally, the specificity of the mPCR assay was also tested with each control reference E. coli strain listed above.
Combining the molecular results of all of these primers permitted an easier detection of the five categories of diarrheagenic E. coli and also the E.coli strains that expressed unusual virulence factors (intermediate E. coli pathotypes).
The amplified DNA products were electrophoresed in 2% or 1% (for the multiplex PCR I) agarose gels containing ethidium bromide (Agarose LE; Roche Applied Science, Penzberg, Germany) and were then visualized under UV light. A 100-pb DNA ladder was used as a molecular mass marker.
Virological methods
The stool samples were routinely screened for the presence of VP6 group A rotavirus antigen by enzyme immunoassay using a commercial ELISA kit (Rotavirus ELISA kit, Premier™ Rotaclone®). All rotavirus positive samples were confirmed by RT-PCR and genetically characterized. The viral RNA was extracted as previously described (17). Rotavirus G and P genotyping was performed using semi-nested type specific multiplex RT-PCRs that could detect seven G-types and six P-types, as previously described (17).
Human noroviruses were detected by RT-PCR using several sets of primers in separate reactions. Primer set JV12 and JV13 (Vinjé and Koopmans, 1996) was used to amplify a fragment of the RNA polymerase genes. Primer set G1SKF and G1SKR and primer set G2SKF and G2SKR (Kojima et al., 2002) were used to detect a fragment of the capsid genes of genogroup I (GGI) and genogroup II (GGII) noroviruses, respectively. RT-PCRs were performed using a Qiagen One Step RT-PCR kit (Qiagen, Hilden, Germany) according to the manufacturer’s instructions.
All PCR products were examined by gel electrophoresis in 2% agarose gels containing 10 μg/ml ethidium bromide, and amplicons were viewed with UV light.
The P and G types were determined by the size of the amplicons and the genotyping of noroviruses was performed by direct sequencing of the PCR products with the same primers used for amplification by using an ABI Prism BigDye Terminator cycle sequencing ready reaction kit and a 3100 DNA sequencing system (both from Applera Corporation, Forster City, CA).
Sequence analysis was conducted using Fasta software, version 3, available from the European Bioinformatics Institute.
Parasitological methods
Parasitological examination was based on direct examination of fresh stool followed by a concentration method and completed by a staining method: MIF coloration (Merthiolate-iode-formol).
Statistical analyses
The chi-square test was used to determine the statistical significance of the data. A P value of <0.05 was considered as significant.
Results
Bacterial causes of gastroenteritis
Among the 124 diarrheic children that have visited the CNSS polyclinic during the study period, 107 enteropathogenic bacteria (86.3%) were isolated. These pathogens were classified into two groups: 12 strains of Salmonella spp. (9.7%) and 95 strains of diarrheagenic E. coli (76.6%). For the healthy group, 23 diarrheagenic E. coli (42.6%) from 54 children were isolated. No Shigella strains were isolated from any of the children tested (diarrheic and healthy children). Rates of detection of bacterial pathogens in patients and healthy groups are shown in Table 4.
Table 4.
Distribution of enteropathogens identified from stool samples of 178 children: 124 patients and 54 controls

Pathotype identification
Table 2 recapitulates the different mPCRs used in order to detect the five different categories of diarrheagenic E.coli. PCR products were derived from the pure culture of reference strains of non pathogenic E.coli, EHEC EDL933, EPEC 2348/69, ETEC 10407, ETEC Jep 5683, EIEC 11741 and EAEC 17-2, respectively.
Therefore, when the multiplex PCR was positive for EHEC screened from stool samples, two specific monoplex PCRs were run (separately with primers specific for the shiga toxins1 (stx1 gene), and shiga toxins 2 variants (stx2 gene) to determine whether the strain possessed the stx1 and/or stx2.
These multiplex PCRs showed positive results for diarrheagenic E.coli strains and negative results for non diarrheagenic E.coli strains (Table 4, Fig. 1). The prevalence of diarrheagenic E. coli in both groups was respectively 76.6% and 42.6%. No significant differences in the prevalence of isolation of diarrheagenic E. coli were noted in both diarrheic and healthy groups.
Fig. 1.
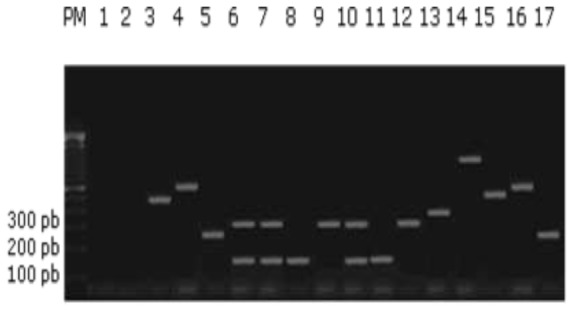
Agarose gel electrophoresis of multiplex PCRs (mPCR 1, 2, and 3) amplification of laboratory strains isolated from diarrheagenic children (Only positive results of each multiplex PCR were present in this Figure) PM; Lanes: 1, negative control: mixture control; 2, non pathogenic E.coli HB101; 3, Strain 103 (EHEC); 4, Strain 113 (EIEC) ; 5, Strain 102 (EAEC); 6,Strain 119 (ETEC); 7, Strain 42 (ETEC); 8, Strain 21 (ETEC); 9, Strain 51 (ETEC); 10, Strain 5 (ETEC); 11, Strain 22 (ETEC); 12, Strain 88 (ETEC); 13, Strain 103 (EPEC); 14, Strain 103 (EPEC); 15, Strain 104 (EHEC); 16, Strain 30 (EIEC); 17, Strain 35 (EAEC)
Among the group with diarrhea the PCR assays detected 29 (23.4%) EAEC isolates (aggR positive gene), 15 (12.1%) EIEC isolates (ipaH positive gene), 17 (13.7%) EPEC isolates among which 11.3% were typical and 2.4% were atypical EPEC strains, 26 (21%) ETEC isolates (elt and/or est positive gene) and 2 (1.6%) EHEC isolates (stx positive gene) (Table 4, Fig. 1). The prevalence of EAEC, EIEC and typical EPEC strains in the healthy group was 14.8% (8 strains), 22.2% (12 strains), and 5.5% (3 strains) respectively. No EHEC, ETEC and atypical EPEC strains were isolated from any of the healthy children tested (Table 4).
Concerning the ETEC isolated strains, 10 of 26 (8.1%) possessed both heat-labile toxin gene and heat-stable toxin gene, 5 of 26 (4%) had only the heat-labile toxin gene and 11 of 26 (8.9%) presented only the heat-stable toxin gene (Table 4).
Interestingly, we detected in patients a collection of diarrheagenic E.coli (n=6) (4.8%) that expressed unusual virulence factor profiles apparently representing intermediate pathotypes: 2 ETEC/EPEC strains (est+, elt+, eae+), 2 ETEC/EIEC strains (est+, ipaH+), 1 ETEC/EAEC strain (est+, aggR+), and 1 ETEC/EPEC strain (est+, bfpA+, eae+). These intermediate pathotypes were absent in the healthy control group (Table 4).
Regarding the two monoplex PCRs that aimed to a further investigation of shiga toxin type in EHEC strains, we noted that the two EHEC strains presented only the shiga toxins 1 target gene.
Twelve Salmonella spp. (9.7%) were isolated from the patient group and none from the healthy group (Table 4) while no Shigella spp. were isolated from any of the children tested (patient and healthy groups).
Viral causes of gastroenteritis
Among the GEA-group (n=124), 53 (42.7%) presented a viral infection. Group A rotaviruses and noroviruses were identified as causative agents in 42 (33.9%) and 11 (8.9%) of the fecal samples, respectively. From the healthy group, we isolated 6 (11.1%) rotaviruses and 5 (9.2%) noroviruses. Mixed infections with the two viruses were detected in 2 stool samples (3.7%).
In diarrheal children, rotavirus genotype combination G2P [4] was predominant with 12.9% (n=16). This was followed by G3P [8] 10.5% (n=13), G1P [8] 6.4% (n=8), and G4P [8] 2.4% (n=3). Among two P [8] rotavirus strains (1.6%), the G type remained nontypeable. In the healthy group, we isolated only genotype G1P [8]. The genotyping of noroviruses revealed that, in diarrheal patients, we detected only genogroup II (GGII) 8.1% (n=10) with GGII.b/II.3 being the predominant genotype. As for children in the control group, the genotype GGI.b/I.6 of genogroup I (GGI) was the most common genotype isolated. It has to be noted that no mixed infections with GGI and GGII strains were observed.
Parasitologic detection
Parasitologic study showed that only 6 (4.8%) of 124 diarrheal samples were positive for parasites (Entamoeba coli (n=2), Cryptosporidium Oocystes (n=2), Giardia lamblia (n=1)and Blastocystis hominis (n=1))
Mixed infections with ≤ 4 different organisms isolated from a single stool sample were observed in the majority of the specimen. A variety of combinations was detected in the diarrheagenic group especially with ETEC pathotype where the most frequent combination was ETEC and other E.coli pathotypes (n=14). ETEC and rotavirus (n=7) were also observed.
Seasonal distribution of enteropathogens
We did not observe any particular temporal distribution of different enteropathogens. In fact, the highest prevalence of bacterial agents was observed in autumn (n=36) and summer (n=34), but bacterial infection can be also detected in winter. Viral infections were detected in autumn and in winter showing a decrease in the frequency of their isolation in summer and spring. However, the six cases of parasitic infections were detected only in autumn.
Enteropathogen identification according to sex and age groups
A higher rate of infection among male patients was found for all enteropathogens tested. Infections with enteropathogens were identified in 54.8% of males (n=68) and 45.2% of females (n=56) suffering from gastroenteritis. This one-year surveillance also showed that 50% of cases with gastroenteritis were found in 12-35 months age group (n=35 of male and n=27 of female children).
Discussion
This study, which covered a one-year period, is the first one in our region and the second in Tunisia that has estimated the prevalence of enteric pathogens associated with acute gastroenteritis in outpatient children in Sousse, Tunisia.
Diarrhea in developing countries is caused by an increasingly long list of viral, bacterial, and parasitic pathogens with rotavirus, diarrheagenic E. coli, Shigella spp. and Salmonella spp. (18) being the most prevalent.
In the present study, ETEC were the most common cause of diarrhea in outpatient children followed by rotaviruses and Salmonella (Table 4). These microorganisms were significantly recovered from children with diarrhea than from healthy controls (P < 0.0003, P = 0.002 and P < 0.019, respectively). However, the prevalence of diarrheagenic E.coli was not significantly higher in the patient group than in controls (76.6% versus 42.6%) (P> 0.05).
ETEC and EAEC were the most prevalent pathogens isolated from children with diarrhea. In fact, ETEC pathotype is recognized as one of the main important diarrheagenic E.coli in developing countries. High rates of ETEC have been previously reported in North Africa especially in Egypt where a strong association with diarrhea has been shown (19). However, many other studies have not reported any significant association of ETEC with diarrhea; in fact ETEC strains were found among healthy children (8, 20, 21). These discrepancies have been explained by the geographical and temporal variation of ETEC rates in childhood (22).
Among the 26 ETEC isolates, the percentage of strains possessing est gene only, elt gene only and both elt and est genes were respectively 42.3% (n=11), 19.2% (n=5), and 38.5% (n=10); thus they were different from those previously reported in north Africa (19, 8). In fact Al-Gallas et al. have reported a percentage of 22% of ETEC strains in patients (16% of strains possess est gene only, 3.7% possess elt gene only and 2% possess both elt and est genes) and a percentage of 35% in controls (32.5% of strains possess est gene only and 2.6% possess elt gene only) (8). This wide variation in the frequency of toxin phenotypic subgroups among ETEC isolates was also noted in most developing world. Knowledge of the distribution of ETEC toxin phenotypic subgroups in a population may be useful to assess endemic disease incidence, as LT-ETEC is thought to be less likely to cause disease than ST or ST and LT ETEC (22). Thus, in this study, the high rate of ETEC strains harboring est gene (with or without elt gene) (80.8%) (n=21) may explain why ETEC strains are isolated only in patients with diarrhea but not in healthy controls (Table 4). Other virulence factors (adhesive factors (CFA) in ETEC strains) that were not characterized in this study have been reported to play an effective role in the pathogenesis of ETEC diarrhea and therefore should be explored (7, 19, 22). Co-infections with other pathogens in children with diarrhea are frequently observed with this pathotype. Indeed, 14 co-infections with other E.coli pathotypes, 7 co-infections with rotaviruses and one infection with norovirus have been detected.
In the present study, the identification of EAEC strains was only based on the detection of AggR gene by PCR. This test has been shown to lack sensitivity in comparison to the hep-2 cell adherence test considered as the “gold standard” (7) but the latter requires specialized facilities and is time consuming. Yet, the present study increased the knowledge of the role of EAEC in acute diarrhea in children in the region of Sousse. In fact, EAEC was the most common pathotype among DEC isolated from children (30.5%) (n=29). However, in accord with many previous reports from different geographic areas, among the 95 DEC strains no significant differences between patients and controls (P> 0.05) were found. This lack of association between both groups reflects the high frequency of carrier state among the studied child population (14.8%) (n=8), as it has been reported previously in many less developed countries (23, 24), suggesting their role as enteropathogenic bacteria with an endemic status. Moreover, we have found a high percentage of EAEC in wastewater as well as in entrance and in exit points even after bacteriological wastewater assessment. These environmental findings could explain the important prevalence of EAEC strains in patients and in controls (25). In this way, as it is the first study to report a high rate of EAEC isolates from Tunisia, a further characterization of these strains is required for a better understanding of their role in diarrheal diseases in this country. Co-infections especially with rotaviruses were frequently detected (n=8).
Besides ETEC and EAEC, enteropathogenic E.coli (EPEC) is the leading cause of infantile diarrhea (26). EPEC infection is primarily a disease of infants younger than 2 years of age in developing world. EPEC classification is currently based on the presence of specific genes. The eae (intimin) and the bfpA (bundle-forming-pilus) genes have both been used for the identification of EPEC and for the subdivision of this group of bacteria into typical (eae+, bfpA+) and atypical (eae+, bfpA-) strains. For many years typical EPEC strains have been considered to be the leading cause of infantile diarrhea in developing countries. However current data suggest that atypical EPEC strains are emerging enteropathogens that have been detected worldwide either in developing or in developed countries. As reviewed by Levine and Edelman (27), numerous case-control studies in many countries have found out that EPEC is more frequently isolated from children with diarrhea than from healthy controls (7). In the present study, the prevalence of isolation of EPEC strains from children with diarrhea was not significantly higher than that from the controls (13.7% versus 5.5%) (P> 0.05). 14 EPEC strains (11.3%) isolated possess both eae gene and also structural gene encoding the bundle-forming pilus bfp and so are classified as typical EPEC strains whereas only three strains (2.4%) were atypical EPEC strains. The prevalence of atypical EPEC strains is still low compared with previous reports in Tunisia and elsewhere (8).
EIEC strains were shown to be capable of causing diarrhea in children. This pathotype can be difficult to distinguish from Shigella spp. and from other E. coli strains, including non-pathogenic strains. The ipaH PCR was reported to be effective in identifying EIEC strains from other E. coli pathotypes but other techniques like DNA probes could be used in conjunction with molecular method to characterize better EIEC strains (28). In Sousse, 15 EIEC (12.1%) in children with diarrhea were isolated. But, interestingly, the prevalence of this pathotype in children older than 1 year of age was significantly lower than from children younger than 1 year of age (P < 0.05). To sum up, it has been reported that the prevalence of EIEC pathotype vary in different geographical areas (23, 26, 29, 30).
In contrast to the relatively high prevalence of EIEC in this study in both diarrheagenic and control group, EHEC strains were detected at a low prevalence in diarrheagenic children (1.6%) and were not isolated in control group.
This result assert with the low prevalence of EHEC infection in many developing countries (23, 31-33). Interestingly, 6 intermediate E.coli strains (4.8%) that presented specific virulence genes of more than one specific E. coli pathotype were also found. These intermediate E.coli strains that expressed unusual virulence factor profiles can apparently be explained by the extraordinary plasticity of the E. coli genome. These intermediate pathotypes were also described in a previous report in Tunisia (8).
For rotaviruses, all fecal positive specimens contained only one type of rotavirus strain. The predominant single G/P combination was G2P [4] (12.9%), followed by G3P [8] (10.5%), G1P [8] (6.4%), G4P [8] (2.4%). This variability in the genotype profiles has been shown in previous studies to be dependent on a given place and a period of time (34-36).
For noroviruses, strains classified as GGII.b/II.3 were almost the only ones detected in diarrheagenic children while, in the control group, GGI.b/I.6 was the predominant circulating genotype. Another study in Tunisia (37) demonstrated the predominance of genogroup II and showed that recombinant GGII.b strains were the second most frequently detected noroviruses in Tunisian children. The natural emergent recombinant GGII.b strains were detected for the first time in France in August 2000 (38) and then spread throughout France and all over Europe in the following seasons, with outbreaks and sporadic cases reported in Spain, Sweden, and Hungary (39-41). They were also reported in other parts of the world, such as in Japan in the 2003-2004 seasons (42) and India in the 2005-2007 seasons (43). This study has limitations on account of analysis restricted to a small size of samples and a short period of collection, it is indicative of the notable role of noroviruses in causing gastroenteritis and the need for longitudinal studies on surveillance of norovirus diseases and strains in Tunisia. Earlier reports have described GGII.b strains as naturally occurring recombinant norovirus strains with different capsid types (44). This requires more work towards a complete characterization of RNA polymerase and capsid genes for a better understanding of the mechanism involved in the recombination and pathogenicity of noroviruses in Tunisia.
In this study, mixed infections with more than one pathogen were frequently detected with the majority of E.coli pathotype, particularly bacterial co-infections. Viral co-infections were also observed (n=7).
Regarding the epidemiology of enteric infections, all detected enteropathogens occurred most frequently in children aged between one and three years (n=62) (data not shown). This distribution can be explained by the interruption of breast-feeding after the age of one year and the exposure of these children to environmental contamination (food and water contamination) (25). Therefore, improvement in the quality of the water distribution network might help reduce the burden on the healthcare system by reducing the number of infected children.
The peak of viral infections occurred during the winter months, while bacterial infections were often predominant during the summer or warm months (data not shown). These tendencies agree with the findings of other studies (45-48).
Conclusion
An understanding of the relative contribution of enteric pathogens in gastroenteritis is essential for the implementation of appropriate public health measures controlling these diseases.
The findings of this study should urge us to implement management guidelines for E.coli associated diarrhea in Tunisia. In particular, laboratory diagnostic methods have to take in account ETEC and EAEC as potential pathogens that should be correctly identified.
Ethical considerations
Ethical issues (Including plagiarism, Informed Consent, misconduct, data fabrication and/or falsification, double publication and/or submission, redundancy, etc) have been completely observed by the authors.
Acknowledgments
This work was supported by the Laboratory of Infectious Diseases and Biological Agents, Faculty of Pharmacy, TU-5000 Monastir, Tunisia (LR99ES27) and also by the Laboratory of Hygiene, 5 Rue de Kairouan 4000 Sousse, Tunisia. The viral study was supported by the AUF Project (code 2092 RR823). We would like to gratefully thank the technical assistance from the National Reference Center for Enteric Viruses, CHU Dijon, France, and Nedra Kerkeni for editorial assistance. The authors declare that there is no conflict of interests.
References
- Musher DM, Musher BL (2004). Contagious acute gastrointestinal infections. N Engl J Med, 351: 2417–2427. [DOI] [PubMed] [Google Scholar]
- Baudry B, Savarino SJ, Vial P, Kaper JB, Levine MM (1990). A sensitive and specific DNA probe to identify enteroaggregative Escherichia coli, a recently discovered diarrheal pathogen. J Infect Dis, 161: 1249–1251. [DOI] [PubMed] [Google Scholar]
- Jertborn M, Svennerhalm AM (1991). Enterotoxtin-producing bacteria isolated from Swedish travelers with diarrhea. Scand J Infect Dis, 23: 473–479. [DOI] [PubMed] [Google Scholar]
- Behiry IK, Abada EA, Ahmed EA, Labeeb RS (2011). Enteropathogenic Escherichia coli Associated with Diarrhea in Children in Cairo, Egypt. Scientific World J, 11: 2613–2619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tate JE, Burton AH, Boschi-Pinto C, Steele AD, Duque J, Parashar UD (2012). 2008 estimate of worldwide rotavirus-associated mortality in children younger than 5 years before the introduction of universal rotavirus vaccination programmes: a systematic review and meta-analysis. Lancet Infect Dis, 12(2): 136–141. [DOI] [PubMed] [Google Scholar]
- Patel MM, Widdowson MA, Glass RI, Akazawa K, Vinjé J, Parashar UD (2008). Systematic literature review of role of noroviruses in sporadic gastroenteritis. Emerg Infect Dis, 14: 1224–1231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nataro JP, Kaper JB (1998). Diarrheagenic Escherichia coli. Clin Microbiol Rev, 11: 142–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Gallas N, Bahri O, Bouratbeen A, Ben Hassen A, Ben Aissa R (2007). Etiology of acute diarrhea in children and adults in Tunis, Tunisia, with emphasis on diarrheagenic Escherichia coli, phenotyping and molecular epidemiology. Am J Trop Med Hyg, 3: 571–582. [PubMed] [Google Scholar]
- Agarwal A, Makker A, Goel SK (2002). Application of the PCR technique for a rapid, specific and sensitive detection of Salmonella Spp. in foods. Mol Cell Probes, 16(4): 243–250. [DOI] [PubMed] [Google Scholar]
- Gunzburg ST, Tornieporth NG, Riley LW (1995). Identification of enteropathogenic Escherichia coli by PCR-based detection of the bundle-forming pillus gene. J Clin Microbiol, 33: 1375–1377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paton AW, Paton JC (1998). Detection and characterization of shigatoxigenic Escherichia coli by using multiplex PCR assays for stx1, stx2, eaeA, enterohemorrhagic E. coli hly, rfb o111, and rfb o157. J Clin Microbiol, 36: 598–602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamasaki S, Lin Z, Shirai H, Terai A, Oku Y, Ito Het al. (1996). Typing of verotoxins by DNA colony hybridization with poly and oligonucleotide probe a based-enzyme-linked immunosorbent assay and polymerase chain reaction. Microbiol Immunol, 40(5): 345–352. [DOI] [PubMed] [Google Scholar]
- Hornes E, Wasteson Y, Olsvik O (1991). Detection of Escherichia coli heat-stable enterotoxin genes in pig stool specimens by immobilized colorimetric, nested polymerase chain reaction. J Clin Microbiol, 29(11): 2375–2379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamanai-Shacoori Z, Jolivet-Gougeon A (1994). Detection of enterotoxigenic Escherichia coli in watter by polymerase chain reaction amplification and hybridization. Can J Microbiol, 40: 243–249. [DOI] [PubMed] [Google Scholar]
- Sethabutr O, Venkatesan M, Murphy GS, Eampokalap B, Hogoe CW, Echeverria P (1993). Detection of shigellae and enteroinvasive Escherichia coli by amplification of the invasion plasmid antigen H DNA sequence in patients with dysentery. J Infect Dis, 167: 458–461. [DOI] [PubMed] [Google Scholar]
- Ratchtrachenchai O, Subpasu AS, Ito K (1997). Investigation on enteroaggregative Escherichia coli infection by multiplex PCR. Bull Dep Med Sci, 39: 211–220. [Google Scholar]
- Hassine-Zaafrane M, Sdiri-Loulizi K, Ben Salem I, Kaplon J, Ayouni S, Ambert-Balay Ket al. 2011. The molecular epidemiology of circulating rotaviruses: three-year surveillance in the region of Monastir, Tunisia. BMC Infect Dis, 11: 266doi:10.1186/1471-2334-11-266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor DN, Echeverria P (1993). Diarrheal disease: current concepts and future challenges. Molecular biological approaches to the epidemiology of diarrheal diseases in developing countries. Trans. R. Soc. Trop Med Hyg, 87(Suppl 3): 3–5. [DOI] [PubMed] [Google Scholar]
- Shaheen HI, Khalil SB, Rao MR, Elyazeed RA, Wierzba TF, Peruski LF (2004). Phenotypic Profiles of Enterotoxigenic Escherichia coli Associated with Early Childhood Diarrhea in Rural Egypt. J Clin Microbiol, 42(12): 5588–5595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolk M, Ohad E, Shafran R, Safir S, Cohen Y, Wiklund G, Svennerholm AM (1995). Epidemiological aspects of enterotoxigenic Escherichia coli diarrhoea in infants in the Jerusalem area. Public Health Rev, 23: 25–33. [PubMed] [Google Scholar]
- Viboud GI, Jouve MJ, Binsztein N, Vergara M, Rivas M, Quiroga M (1999). Prospective cohort study of enterotoxigenic Escherichia coli infections in Argentinean children. J Clin Microbiol, 37: 2829–2833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qadri F, Svennerholm AM, Faruque ASG, Bradley-Sack R (2005). Enterotoxigenic Escherichia coli in Developing Countries: Epidemiology, Microbiology, Clinical Features, Treatment, and Prevention. Clin Microbiol Rev, 18(3): 465–483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okeke IN, Lamikanra A, Steinrück H, Kaper JB (2000). Characterization of Escherichia coli Strains from Cases of Childhood Diarrhea in Provincial Southwestern Nigeria. J Clin Microbiol, 38(1): 7–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scaletsky IC, Fabbricotti SH, Carvalho RL, Nunes CR, Maranhao HS, Morais MBet al. (2002). Diffusely adherent Escherichia coli as a cause of acute diarrhea in young children in northeast Brazil: a case-control study. J Clin Microbiol, 40(2): 645–648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben Salem I, Ouardani I, Hassine M, Aouni M (2011). Bacteriological and physico-chemical assessment of wastewater in different region of Tunisia: impact on human health. BMC Research Notes, 4: 144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albert MJ, Faruque AS, Faruque SM, Sack RB, Mahalanabis D (1999). Case-Control Study of Enteropathogens Associated with Childhood Diarrhea in Dhaka, Bangladesh. J Clin Microbiol, 37(11): 3458–3464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine MM, Edelman R (1984). Enteropathogenic Escherichia coli of classic serotypes associated with infant diarrhea: epidemiology and pathogenesis. Epidemiol Rev, 6: 31–51. [DOI] [PubMed] [Google Scholar]
- Hien BTT, Scheutz F, Cam PD, Serichantalergs O, Huong TT, Thu TM, Dalsgaard A (2008). Diarrheagenic Escherichia coli and Shigella Strains Isolated from Children in a Hospital Case-Control Study in Hanoi, Vietnam. J Clin Microbiol, 46: 996–1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baqui AH, Sack RB, Black RE, Haider K, Hossain A, Alim ARMAet al. (1992). Enteropathogens associated with acute and persistent diarrhea in Bangladeshi children less than 5 years of age. J Infect Dis, 166: 792–796. [DOI] [PubMed] [Google Scholar]
- Echeverria P, Hoge CW, Bodhidatta L, Tungtaem C, Herrmann J, Imlarp Set al. (1994). Etiology of diarrhea in a rural community in western Thailand: importance of enteric viruses and enterovirulent Escherichia coli. J Infect Dis, 169(4): 916–919. [DOI] [PubMed] [Google Scholar]
- Strockbine NA, Faruque SM, Kay BA, Haider K, Alam K, Alam ANet al. (1992). DNA probe analysis of diarrhoeagenic Escherichia coli: detection of EAF-positive isolates of traditional enteropathogenic E. coli serotypes among Bangladeshi paediatric diarrhoea patients. Mol Cell Probes, 6: 93–99. [DOI] [PubMed] [Google Scholar]
- Albert MJ, Faruque SM, Faruque AS, Neogi PK, Ansaruzzaman M, Bhuiyan NAet al. (1995). Controlled study of Escherichia coli diarrheal infections in Bangladeshi children. J Clin Microbiol, 33(4): 973–977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pershing D (2007). Automated molecular dignosistic. Gen Eng Biotech. 27: 35–39. [Google Scholar]
- Naficy AB, Abu-Elyazeed R, Holmes JL, Rao MR, Savarino SJ, Kim Yet al. (1999). Epidemiology of rotavirus diarrhea in Egyptian children and implications for disease control. Am J Epidemiol, 150(7): 770–777. [DOI] [PubMed] [Google Scholar]
- Khalili B, Cuevas LE, Reisi N, Dove W, Cunliffe NA, Hart CA (2004). Epidemiology of rotavirus diarrhoea in Iranian children. J Med Virol, 73(2): 309–312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamel AH, Ali MA, El Nady HG, de Rougemont A, Pothier P, Belliot G (2009). Predominance and circulation of enteric viruses in the region of Greater Cairo, Egypt. J Clin Microbiol, 47(4): 1037–1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sdiri-Loulizi K, Ambert-Balay K, Gharbi-Khelifi H, Sakly N, Hassine M, Chouchane Set al. (2009). Molecular Epidemiology of Norovirus Gastroenteritis Investigated Using Samples Collected from Children in Tunisia during a Four-Year Period: Detection of the Norovirus Variant GGII.4 Hunter as Early as January 2003. J Clin Microbiol, 47(2): 421–429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ambert-Balay K, Bon F, Le Guyader F, Pothier P, Kohli E (2005). Characterization of new recombinant noroviruses. J Clin Microbiol, 43: 5179–5186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buesa J, Collado B, Lopez-Andujar P, Abu-Mallouh R, Diaz JR, Diaz AGet al. (2002). Molecular epidemiology of caliciviruses causing outbreaks and sporadic cases of acute gastroenteritis in Spain. J Clin Microbiol, 40: 2854–2859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindell AT, Grillner L, Svensson L, Wirgart BZ (2005). Molecular epidemiology of norovirus infections in Stockholm, Sweden, during the years 2000 to 2003: association of the GGIIb genetic cluster with infection in children. J Clin Microbiol, 43: 1086–1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reuter G, Krisztalovics K, Vennema H, Koopmans M, Szucs G (2005). Evidence of the etiological predominance of norovirus in gastroenteritis outbreaks emerging new-variant and recombinant noroviruses in Hungary. J Med Virol, 76: 598–607. [DOI] [PubMed] [Google Scholar]
- Phan TG, Kuroiwa T, Kaneshi K, Ueda Y, Nakaya S, Nishimura Set al. (2006). Changing distribution of norovirus genotypes and genetic analysis of recombinant GIIb among infants and children with diarrhea in Japan. J Med Virol, 78: 971–978. [DOI] [PubMed] [Google Scholar]
- Chhabra P, Chitambar SD (2008). Norovirus genotype IIb associated acute gastroenteritis in India. J Clin Virol, 42: 429–32. [DOI] [PubMed] [Google Scholar]
- Reuter G, Vennema H, Koopmans M, Szvcs G (2006). Epidemic spread of recombinant noroviruses with four capsid types in Hungary. J Clin Virol, 35: 84–8. [DOI] [PubMed] [Google Scholar]
- Gomes TA, Rassi V, MacDonald KL, Ramos SR, Trabulsi LR, Vieira MAet al. (1991). Enteropathogens associated with acute diarrheal disease in urban infants in Sao Paulo, Brazil. J Infect Dis, 164: 331–7. [DOI] [PubMed] [Google Scholar]
- Levine MM, Ferreccio C, Prado V, Cayazzo M, Abrego P, Martinez Jet al. (1993). Epidemiologic studies of Escherichia coli diarrheal infections in a low socioeconomic level peri-urban community in Santiago, Chile. Am J Epidemiol, 138: 849–69. [DOI] [PubMed] [Google Scholar]
- Fang ZY, Yang H, Zhang J, Li YF, Hou AC, Ma Let al. (2000). Child rotavirus infection in association with acute gastroenteritis in two Chinese sentinel hospitals. Pediatr Int, 42: 401–405. [DOI] [PubMed] [Google Scholar]
- Maneekarn N, Ushijima H (2000). Epidemiology of rotavirus infection in Thailand. Pediatr Int, 42: 415–421. [DOI] [PubMed] [Google Scholar]


