Abstract
Background
We previously reported that specialized and housekeeping genes are differentially regulated via de novo recruitment and pause-release of RNA polymerase II (pol II), respectively, during cardiac hypertrophy. However, the significance of this finding remains to be examined. Therefore, the purpose of this study was to determine the mechanisms that differentially regulate these gene groups and exploit them for therapeutic targeting.
Methods and Results
Here we show that general transcription factor IIB (TFIIB) and cyclin-dependent kinase 9 are upregulated during hypertrophy, both targeted by miR-1, and play preferential roles in regulating those two groups of genes. Chromatin immunoprecipitation-sequencing reveals that TFIIB is constitutively bound to all paused, housekeeping, promoters, whereas, de novo recruitment of TFIIB and pol II is required for specialized genes that are induced during hypertrophy. We exploited this dichotomy to acutely inhibit induction of the latter set, which encompasses cardiomyopathy, immune reaction, and extracellular matrix genes, using locked nucleic acid (LNA)-modified antisense TFIIB oligonucleotide treatment. This resulted in suppression of all specialized genes, while sparing the housekeeping ones, and, thus, attenuated pathological hypertrophy.
Conclusions
The data for the first time reveal distinct general transcription factor IIB dynamics that regulate specialized vs. housekeeping genes during cardiac hypertrophy. Thus, by acutely targeting TFIIB we were able to selectively inhibit the former set of genes and ameliorate pressure overload hypertrophy. We also demonstrate the feasibility of acutely and reversibly targeting cardiac mRNA for therapeutic purposes using LNA-modified antisense oligonucleotides.
Keywords: transcription, hypertrophy, general transcription factor IIB, chromatin immunoprecipitation
Regulation of gene expression is fundamental to organogenesis and pathogenesis, including cardiac hypertrophy. During hypertrophy there are two distinct transcriptional events that occur. One, there is a generalized increase in total RNA synthesis that underlies the 30–50% increase in cell volume1–3. Two, superimposed on this, there is a more substantial increase in the expression of specific genes, such as atrial natriuretic factor (Nppa) and alpha skeletal actin (Acta1)4, 5. The latter mode of transcription has been studied in more detail as reports show that it involves regulation by specific transcription factors and enhancers6, 7. This ultimately requires TFIIB for the recruitment of pol II, promoter assembly of the pre-initiation complex, and transcriptional activation8. Subsequently, phosphorylation of pol II’s C-terminal domain by cyclin-dependent kinase 9 (Cdk9) enhances promoter-proximal clearance of any paused pol II9.
We have recently reported the detection of two mutually exclusive modes of transcriptional regulation during cardiac hypertrophy10. One involves an incremental increase (30–50%) in the elongational activity of preassembled, promoter-paused, pol II that involved ~25% of expressed genes, which are predominantly essential/housekeeping genes (e.g. RNA synthesis and splicing). Another involved a more robust activation, via de novo pol II recruitment, encompassing ~4% of specialized genes (e.g. contractile, immune reaction, and extracellular matrix genes). These results were the first to demonstrate that promoter-paused pol II plays a key role in incrementally and synchronously increasing housekeeping genes, proportionate to the increase in heart size. While we know that recruitment of pol II to any promoter requires TFIIB, it is still unclear how TFIIB is regulated, whether it remains attached to pol II-paused promoters, and if the release of paused pol II is dependent on TFIIB availability. To further dissect the transcriptional regulation during cardiac hypertrophy, we performed genome-wide TFIIB chromatin immunoprecipitation-deep sequencing (ChIP-Seq). In this study, we describe the dichotomy in TFIIB dynamics between housekeeping, pol II-paused, genes, and hypertrophy-induced genes, and how it can be exploited to selectively and dose-dependently inhibit the pathological hypertrophy-induced genes, which include cardiomyopathy, extracellular matrix, and immune reaction genes, in the heart.
Methods
Animals
C57Bl/6 mice and Sprague Dawley rats were used in this study in accordance with US National Institute of Health Guidelines for the Care and Use of Laboratory Animals (No. 85–23). All animal protocols were approved by the Institutional Animal Care and Use Committee at the Rutgers-New Jersey Medical School.
Cell cultures and treatments
Cardiac myocytes were prepared as previously described11. Briefly, hearts were isolated from 1–2 day old Sprague-Dawley rats. After dissociation the cells were subjected to Percoll gradient centrifugation followed by differential pre-plating to enrich for cardiac myocytes and deplete non-myocytes. Myocytes were then plated in DMEM-F12 with 10% fetal bovine serum (FBS) without antibiotics. Twenty-four hours after plating, the medium iof changed and the cells are infected with recombinant adenoviruses at a multiplicity of infection (moi) of 10–20 particles / cell.
Construction of adenoviruses
Recombinant adenoviruses were constructed, propagated and titered, as previously described by Dr. Frank Graham12. Briefly, pBHGloxΔE1,3Cre (Microbix), including the ΔE1 adenoviral genome, is co-transfected with the pDC shuttle vector containing the gene of interest, into 293 cells using Lipofectamine (Invitrogen). Through homologous recombination, the test genes integrate into the E1-deleted adenoviral genome. The viruses were propagated on 293 cells and purified using CsCl2 banding followed by dialysis against 20 mM Tris buffered saline with 2% glycerol. Titering is performed on 293 cells overlaid with Dulbeco’s Modified Eagle’s Medium (DMEM) plus 5% equine serum and 0.5% agarose.
DNA constructs cloned into recombinant adenovirus
MiR-1: the stem-loop of miR-1-2 was cloned into recombinant adenovirus under control of CMV promoter. MiR-1 eraser (antimiR-1): two antisense repeats of the mature miR-1 sequence was synthesized as a double stranded oligonucleotide and cloned into recombinant adenovirus under the control of a U6 promoter. MiR-133a-1: the stem-loop of miR-133a-1 was cloned into recombinant adenovirus under control of CMV and U6 promoter. MiR-SC (scrambled control): the stem-loop expressing a scrambled sequence (GAACCGAGCCCACCAGCGAGC) was cloned into recombinant adenovirus under CMV and used as control for all adenoviral microRNA constructs. TFIIB accession # NM_001514 (Origene) and CDK9 accession # NM_001261.2 (Origene) human cDNA with or without the 3’UTR containing the miR-1 target site were cloned into adenovirus. Short hairpin RNA (shRNA) TFIIB: a hairpin-forming oligo corresponding to bases 827–846 of mus musculus TFIIB (GenBank accession number NM_145546) was cloned into adenovirus as described previously. CMV-GFP-IRES-Luc: MSCV-IRES-Luciferase plasmid was purchased from Addgene (plasmid 18760). The plasmid was digested with EcoR1/ Sal1 and the fragment containing IRES-Luciferase sequence was cloned into 3’ region of pDC316-GFP. The construct was cloned into adenovirus.
Transverse Aortic Constriction
Twelve week-old C57Bl/6 mice are anesthetized intraperitoneally with a mixture of ketamine (65 mg/kg), xylazine (13 mg/kg), and acepromazine (2 mg/kg). The adequacy of the anesthetic is confirmed by the loss of tongue retraction reflex. The transverse thoracic aorta between the innominate artery and the left common carotid artery was dissected free and a 7-0 braided polyester suture was tied around the aorta against a 28-gauge needle with the aid of an operating microscope. The needle was removed, the chest closed, and the mice were extubated and allowed to recover in a Thermocare unit (temperature 88°F or 31°C; humidity 30–50%; oxygen 1–2 ml/min, low flow range). Postoperative Buprenorphine (0.01–0.05 mg/Kg) was administered subcutaneously every 12 h, as needed. The sham operation involved the same procedure, except the aorta was not constricted.
ChIP-Seq of Sham, TAC-induced Hypertrophy and Neonatal hearts
The hearts were isolated from 12–13 wk old C57Bl/6 mice four days after transverse aortic constriction (TAC, n=3) or a sham operation (n=2), or from normal mice (n=1), and from 2 litters of 1day old C57Bl/6 neonate pubs. The hearts from the TAC, sham/normal, and neonatal hearts were pooled and subjected to GTF2B ChIP-Seq (Active Motif, Inc). Immunoprecipitation was performed using anti-GTF2B antibody (Santa cruz Biotechnology, sc-225), followed by high throughput Illumina sequencing using Illumina HiSeq. Sequencing of Input DNA taken before immunoprecipitation served as control for normalization and eliminating background noise.
ChIP-Seq analysis (Active Motif Inc)
A. Sequence analysis: 50-nt of the sequencing reads (tags, >10 million) with no more than 2 mismatches were aligned to the genome using BWA algorithm. B. Determination of Fragment density: Tags are extended (150–250 bp) at their 3’end in silico. The genome is divided into 32 nt bins and the density of the fragments (extended Tags) in each bin is determined. The results of this is stored in a BAR (binary analysis results) file. C. Peak Finding (intervals): intervals are defined genomic regions that represent the fragment density peaks. It must have 3 consecutive bins with fragment densities greater than the threshold (usually 10–20). D. Active regions: For the purpose of comparison between samples, regions with overlapping intervals are grouped into active regions. F. Annotation: The locations and proximities to gene annotations of intervals and active regions are defined and compiled in Excel spreadsheets, which include average and peak fragment densities.
ChIP-Seq of Cultured Neonatal Myocytes
Rat neonatal cardiac myocytes were cultured as described above. Sixteen hours after plating, the cells were treated with recombinant adenoviruses expressing with the precursor of miR-1 or a scrambled control for 24hrs. The myocytes were the fixed in freshly prepared 1% formaldehyde solution (11% of 37% formaldehyde, 0.1M NaCl, 1mM EDTA pH 8, 50mM HEPES pH7.9) for 15 minutes (min) at room temperature. The reaction was stopped with 1/20 volume of 2.5M Glycine (5 min at room temperature), the cells collected and washed with 10mL of chilled PBS-Igepal solution (1× PBS, 0.5% Igepal), pelleted (800 xg), and resuspended in PBS-Igepal plus 1mM PMSF. The DNA was analyzed by anti-pol II ChIP-Seq as described above (Active Motif Inc.).
Normalization of the ChIP-Seq data (Active Motif Inc)
“a. Tag Normalization: the tag number of all samples is reduced (by random sampling) to the number of tags present in the smallest sample. b. Input File Analysis: The signal map of the Input/IgG control file is essentially analyzed as an additional “sample”. In this case, the strongest Input/IgG control peaks (which must represent false positives) are determined by simple thresholding of the BAR file, and only Input/IgG control peaks that overlap with Intervals in the ChIP/IP data are used in the analysis. By doing so, the output Active Region table (see Section IV.4, below) will show for each region the corresponding fragment density in the Input/IgG control sample, thus allowing for the identification of possible false positive ChIP/IP peaks.”
Quantitative Polymerase Chain Reaction (qPCR)
Total RNA was reversed transcribed to cDNA using High Capacity cDNA Reverse Transcription Kit or TaqMan MicroRNA Reverse Transcription Kit for microRNAs (applied Biosciences) as per manufacturers protocol. Quantitative PCR was performed using TaqMan gene expression assays (primer/probe sets) on Applied Biosystems 7500 Real-Time PCR system for the following genes; 18S (Mm03928990_g1), GAPDH (Mm99999915_g1), CDK9 (Mm01731275_m1), TFIIB primer 1 (exon boundary 3–4, assay location 283) (Mm01323559_g1), TFIIB primer 2 (exon boundary 5–6, assay location 566) (Mm03047554_g1), ANF (Mm01255747_g1), ANKRD1 (Mm01204056_g1), VDAC1 (Mm00834272_m1), Ppargc1a (Mm01208835_m1), Pnn (Mm00447098_m1), Calm1 (Mm01336281_g1), Mapk1 (Mm00442479_m1), Acta1 (Mm00808218_g1), Sec23a (Mm00486004_m1), Trappc6b (Mm01605498_g1), Col1a1 (Mm00801666_g1), Alb (Mm00802090_m1), Akt (Mm01331626_m1), U6 (001973), miR-1 (002222) and miR-21 (000397).
In vivo LNA anti-TFIIB Oligo injections
In vivo LNA-modified anti-TFIIB and scrambled oligonucleotides were purchased from Exiqon. The antisense TFIIB oligo is a 16mer complementary to bp 567–582 (NM_145546), 5’- +A*+T*+T+C*G*A*G*A*C*A*C*A*+G*+C*+A*+C -3’, * = phosphorothioate bond, + = LNA-modified. Mice (C57/blk, 12wk old male, 24–26gms) were subjected to TAC or sham operations, 1day post TAC mice were randomly divided into two groups, one group received in vivo LNA anti TFIIB oligo (15mg/Kg, bolos, intravenous via tail-vein), while the other received in vivo LNA Scrambled oligo in similar does and route After the indicated time periods the mice underwent echocardiography to measure cardiac function after which were saced, and heart and liver extracted for analysis.
Western Blotting
Ten microgram of protein samples, each, were subjected to electrophoresis on 4–15% SDS-PAGE (Criterion gels; Bio-Rad). The antibodies used for Western blot analysis included anti-ANKRD1 (Santa Cruz), anti-CDK9 (Santa Cruz), anti-Myosin, slow (Sigma), anti-TFIIB (Millipore), anti-GAPDH (Chemicon), anti-VDAC1 (Genscript), Anti-GFP (BD Biosciences), Anti-Luciferase (Novus Biologicals), anti-Nelf-A (SantaCruz). The signal was detected and quantified by the Odyssey Imaging System (LI-COR).
Immunohistochemistry
Hearts extracted were immediately fixed in 10% neutral buffered formalin for 24hrs before paraffin embedding and sectioning (6-µM thickness). Picric acid Sirius red staining was performed to measure Interstitial Fibrosis, for cell size the sections were stained with Wheat germ agglutinin (WGA).
Statistics
Calculation of significance between 2 groups was performed using an unpaired, 2-tailed Student t test (excel software). P <0.05 was considered significant.
Results
TFIIB and Cdk9 are posttranscriptionally upregulated during cardiac growth
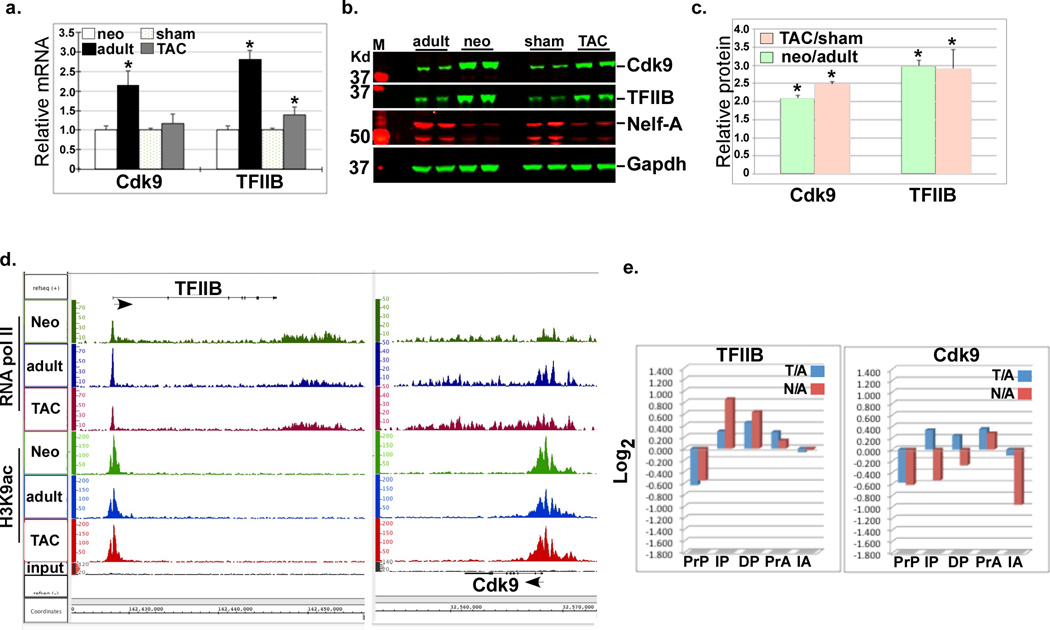
The mechanisms underlying the incremental increase in gene expression that parallels the increase in cardiac myocyte size and mass and its distinction from robust increases in the expression of specific genes during cardiac hypertrophy are not fully elucidated. We have recently reported that these two distinct transcriptional changes are regulated by the incremental release of promoter-paused RNA pol II and de novo recruitment of pol II, respectively10. TFIIB is a key limiting protein that is required for the recruitment of RNA pol II to promoters, while Cdk9 is a component of the positive transcription elongation factor b (P-TEFb) that enhances promoter clearance of this polymerase and has been shown to play a key role in hypertrophic growth13. We predicted that, in addition to Cdk9, TFIIB plays a major role in regulating transcription during hypertrophy via variations in its protein levels. Therefore, we determined the expression pattern of both genes by measuring both the mRNA and protein levels of these genes in 1 day-old neonatal and 12 week-old adult hearts before and after transverse aortic constriction (TAC). The results show that the mRNA levels of both genes were marginally increased in the TAC hearts, whereas in the neonatal heart they were 2–3 folds higher (Fig. 1a). On the other hand, the proteins of both genes were 2–3 fold higher in both the neonatal and TAC hearts (Fig. 1b–c). Thus, both TFIIB and Cdk9 proteins are higher during physiological and pathological cardiac growth.
Figure 1. TFIIB and Cdk9 are posttranscriptionally upregulated during cardiac growth.
a. RNA was extracted from the neonatal, adult, and TAC hearts and subjected to qPCR for the indicated genes. The results were averaged and plotted as relative mRNA change (n=3). Error bars represent standard error of the mean (SEM) and * is p < 0.05 v. sham-operated or normal heart. b. Protein was extracted from the same heart tissue as in (a.) and analyzed by Western blots for the indicated antibodies. c. The Western blot signals for the different proteins were quantified, averaged, and plotted. Error bars represent standard error of the mean (SEM) and * is p < 0.05 v. sham-operated or normal adult (n=3, 2/3 samples shown in the blot). d. IGB images showing the RNA pol II density (y-axis) aligned across the Gtf2b (TFIIB) and Cdk9 genes (x-axis) in the neonatal, adult, and TAC hearts. Arrows positioned at the gene’s start site point to direction of transcription. e. Pol II (P) and H3K9ac (A) densities for the TFIIB and Cdk9 genes were plotted as the Log2 values of TAC/adult (T/A) and neonatal/adult (N/A) of the pol II in promoter proximal (PrP, −300 to +300), the in-gene (IP, +300 to end), and the downstream (DP, end to +5000), and of the H3K9ac in promoter proximal (PrA, −1000 to +1000), the in-gene (IA, +1000 to end) region, were plotted.
To determine whether these increases were a result of transcriptional vs. posttranscriptional mechanisms we explored the dataset generated by our anti-pol II and anti-histone H3 K9-acetyl (H3K9ac) ChIP-Seq analysis in the neonatal, adult, and hypertrophied hearts (Accession: GSE50637). Pol II and H3k9ac density distributions for the TFIIB gene (Ggtf2b) reveal reduced TSS pol II pausing and increased H3K9ac in the growing hearts, with a modest (1.25 and 1.8 fold) increase in in-gene pol II density in the TAC and neonatal vs. adult heart, respectively (Fig. 1d–e). While Cdk9 exhibited a similar pattern in the TAC heart, both pol II and in-gene H3K9ac densities appeared to be reduced in the neonatal heart, signifying reduced transcription (Fig. 1d–e). These data, compared with the increase in the mRNA and protein levels, support a role for both enhanced promoter clearance of pol II (i.e. reduced pausing) and potentially posttranscriptional regulation of TFIIB and Cdk9 during cardiac hypertrophy.
MiR-1 targets transcription factor IIB (TFIIB) and cyclin-dependent kinase 9 (Cdk9) mRNA
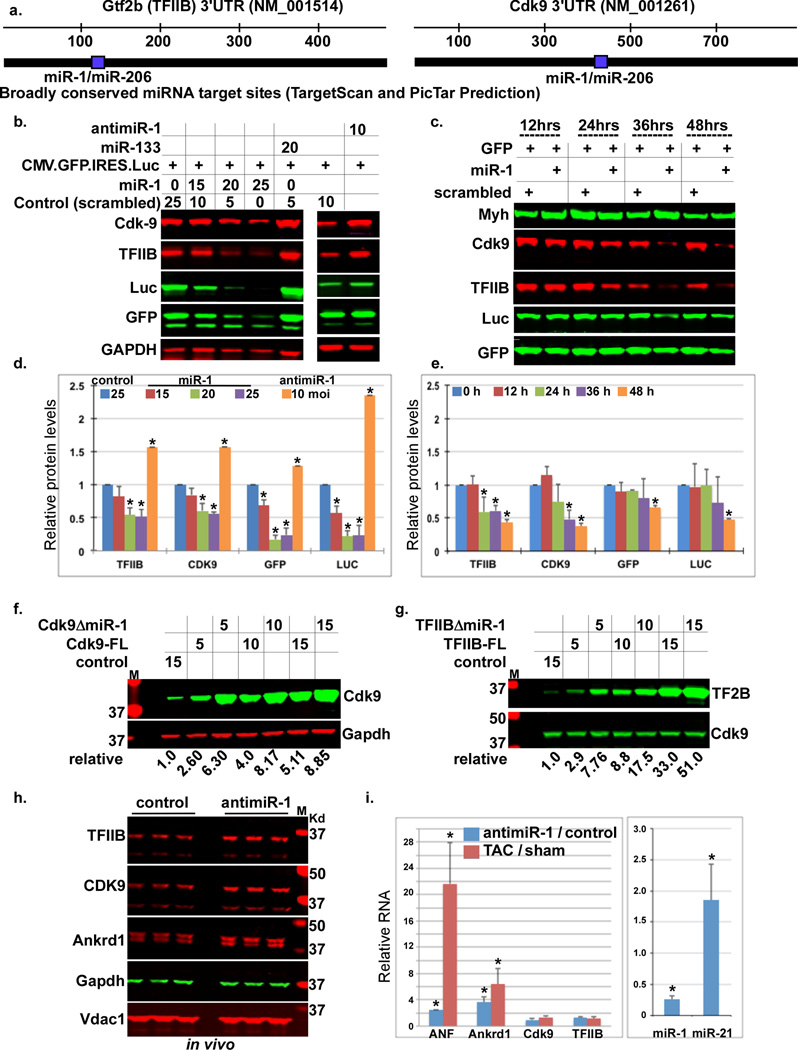
We then examined whether TFIIB and/or Cdk9 are predicted targets of any miRNA. Noteworthy, we found that both TargetScan14–16 and PicTar17, 18, miRNA target prediction software, predict miR-1 as the only, broadly conserved among vertebrates, targeting miRNA of both TFIIB and Cdk9 genes (Fig. 2a). In concordance with a role in regulating these genes during hypertrophic growth, mature miR-1 is significantly higher in the adult vs. neonatal heart, and is downregulated within 24 h of TAC19. To validate these targets, we overexpressed pre-miR-1 in cardiac myocytes before adding a cytomegalovirus (CMV) promoter-driven green florescence protein (GFP)- internal ribosomal entry sequence (IRES)-luciferase (CMV-GFP-Luc) reporter vector used to monitor basic transcription from a constitutive promoter (no miR-1 targeting site is included in this vector). As seen in Figure 2b and quantified in 2d, both TFIIB and Cdk9 proteins were reduced by exogenous miR-1 but not by miR-133, and, conversely, both were increased by an antimiR-1 eraser (Fig. 2b, 2d). The change in TFIIB and Cdk9 expression was paralleled by a similar change in the expression of the GFP and luciferase reporter genes. A time course analysis, in which miR-1 and CMV-GFP-Luc vectors were added simultaneously, revealed that the downregulation of endogenous TFIIB and Cdk9 preceded the downregulation of reporter genes (Fig. 2c, 2e). These results suggest that miR-1 has an effect on basal (constitutive) gene expression, plausibly, through inhibition of TFIIB and/or Cdk9.
Figure 2. TFIIB and Cdk9 are uniquely targeted by miR-1.
a. TargetScan and Pictar miR-1 predicted targeting sites in the 3’UTR of Gtf2b and Cdk9. b. Cardiac myocytes were infected with the specified multiplicity of infection (moi) of adenoviral (Ad) vectors Ad.miR-1, Ad.miR-133, Ad.antimiR-1, or a control virus for 12 h before adding Ad.CMV.GFP.IRES.Luc. Twenty-four hours later, protein was extracted and analyzed by Western blots. c. Cardiac myocytes were simultaneously infected with the indicated Ad vectors. After the specified intervals, protein was extracted and analyzed by Western blots. d. The Western blot signals in (b.), and 2 other blots, for the different proteins, were quantified, averaged, and plotted (n=3). Error bars represent standard error of the mean (SEM) and * is p < 0.05 v. control. e. The Western blot signals in (c.), and 2 other blots, for the different proteins, were quantified, averaged, and plotted (n=3). Error bars represent standard error of the mean (SEM) and * is p < 0.05 v. control. f.–g. The full-length (FL) cDNA or mutants lacking the miR-1 target site (ΔmiR-1) of f. Cdk9 or g. TFIIB were designed, cloned, and equal doses expressed in growth-arrested cardiac myocytes. Protein was extracted, analyzed by Western blots, the signal quantified and normalized, and listed below the blots (n=2). h–i. LNA-modified antimiR-1 (15 mg/Kg), or a nonsense control, was injected into mice via the tail vein. h. Hearts were isolated after 7 days and analyzed by Western blots for the indicated proteins (n=3). i. The same set of hearts was analyzed by qPCR for the indicated miRNA and genes and the results plotted as fold change of antimiR-1/control hearts. Error bars represent standard error of the mean (SEM) and * is p < 0.05 v. control.
To determine whether the predicted miR-1 target site within the 3’UTRs of Cdk9 or TFIIB affects their translation levels, we engineered expression vectors of TFIIB and Cdk9 cDNA that either encompassed [full-length (FL)] or lacked (ΔmiR-1) the miR-1 target site within the 3’UTR. These vectors were delivered to myocytes cultured in serum-free conditions that promote higher miR-1 expression (Supplementary Figure 1S); no exogenous miR-1 was added. A dose response shows that the constructs lacking miR-1 recognition site expressed ~ 2.5 fold higher protein levels (Fig. 2f–g). This difference diminished with increasing concentrations of the constructs, plausibly due to a “sponging” effect. The data suggest that miR-1 directly suppresses TFIIB and Cdk9 and as a consequence also inhibits constitutive gene expression.
MiR-1 is downregulated during cardiac hypertrophy19. Therefore, it remained necessary to determine the relevance of this reduction on the levels of TFIIB and Cdk9 in vivo. The results show that intravenous delivery of locked nucleic acid (LNA)-modified antisense miR-1 oligonucleotides resulted in ~70% knockdown of endogenous miR-1 in the heart accompanied by an increase in both TFIIB and Cdk9 proteins within one week (Fig. 2h–i); confirming that downregulation of miR-1 is indeed sufficient for increasing the expression of these targets. This, however, was not sufficient for inducing fulminant hypertrophy, although, it did induce significant increases in miR-21, ANF, and Ankrd1, albeit to a much lower extent than observed with TAC (Fig. 2h–i). This shows that downregulation of miR-1 is sufficient for inducing upregulation of Cdk9 and TFIIB, which, however, is not sufficient for inducing cardiac hypertrophy.
TFIIB and pol II are constitutively bound to paused promoters but recruited de novo to hypertrophy-induced promoters, which is inhibited by miR-1
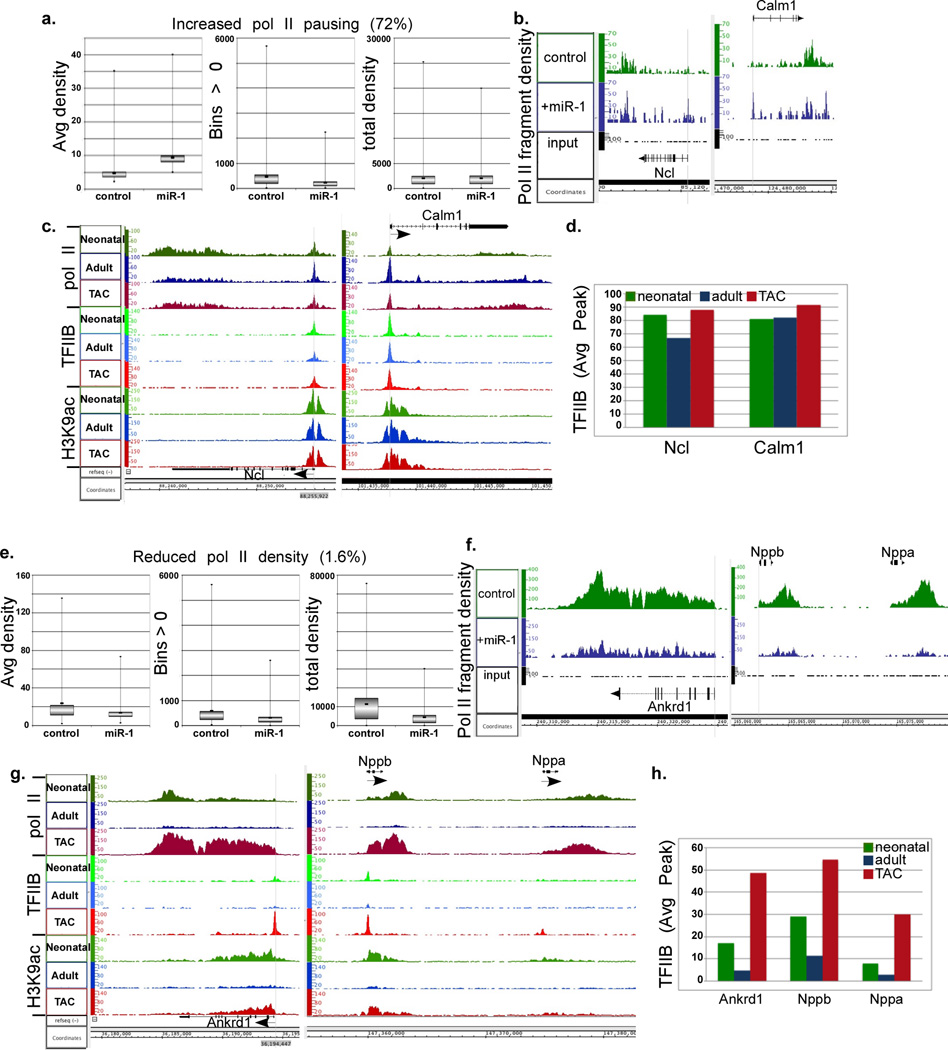
Since miR-1 proved to be a direct inhibitor of TFIIB and Cdk9, we predicted that it would dampen general transcription by inhibiting pol II recruitment and induce its pausing. To test this, we supplied cultured neonatal rat myocytes with exogenous miR-1 for 24 h before extracting the DNA for pol II ChIP-Seq analysis. The results show two major patterns of miR-1-induced changes in pol II densities. In one group, the average pol II density increased with miR-1, while the number of bins (32 nt) was reduced and total pol II density (average density × bins) remained unchanged (Fig. 3a). The aligned sequence data of representative genes (Ncl and Calm1) are displayed in Figure 3b (Fig. 3b). As reflected in those graphs, pol II peaks were generally higher, in particular at the TSS, reflecting an increase in pol II pausing. Importantly, total bound pol II was unchanged; suggesting that pol II recruitment is unaffected in this case. This group includes 72% of genes that are enriched in housekeeping (e.g. ribosomal and proteasomal) and signaling pathway genes [DAVID v6.720–22 (Table 1)]. Notably, we have previously reported that a corresponding set of genes exhibited promoter-proximal pol II pausing in the adult vs. neonatal heart that was incrementally released during hypertrophy, and was, likewise, independent of an increase in de novo pol II recruitment (Fig. 3c)10. The second major group of genes that is regulated by miR-1 is one that characterized by reduced average density, bin numbers, and total pol II densities, reflecting reduced pol II recruitment to genes (Fig. 3e). Shown, as examples of this group, are Ankrd1, Nppa, and Nppb genes, exhibiting reduced pol II density across the gene, however, pol II promoter clearance and advancement were not impacted (Fig. 3f). This group includes 1.6% of genes that encompass those that are involved in hypertrophic and dilated cardiomyopathy pathways [DAVID v6.720–22 (Table 2)]. These represent a similar set of genes that are induced during hypertrophy by de novo recruitment of pol II and does not exhibit promoter-proximal pol II pausing (Fig. 3g)10.
Figure 3. Constitutive vs. induced binding of TFIIB and pol II to housekeeping and specialized promoters during hypertrophy, respectively, and inhibition of the latter by miR-1.
Neonatal rat myocytes in complete growth medium were supplemented with exogenous Ad.miR-1 for 24 h or an Ad.control (scrambled), before they were subjected to pol II ChIP-Seq analysis. a. Box plots of the average densities, bin numbers, and total densities of genes with average values of miR-1/control > 1 (cutoff of pausing) combined with a total density of miR-1/control that is >0.7 and <1.6, which are the cutoff values for downregulated and upregulated genes, respectively, that are excluded from this group. b. Two gene representative of group in (a.) are shown on the right as a plot of the binary analysis results (BAR) files aligned in Affimetrix’s Integrated Genome Browser (IGB), which shows the fragments densities of pol II (y-axis) aligned in 32-nt bins along the coordinates (x-axis) for the individual genes listed. c. The IGB images for the same genes in (b.) represented in our neonatal, adult, and hypertrophy pol II and H3K9ac databases. In addition, these were aligned to our recent TFIIB ChIP-Seq data. D. The blot shows the quantitative average TFIIB peak values for the listed genes. e. Box plots of the average densities, bin numbers, and total densities of genes with average values of miR-1/control < 0.7. f. Two genes representative of group in (e.) are shown on the right as IGB images. g.–h. See the description provided for 3c.–d.
Table 1.
Genes that exhibit pol II pausing in the miR-1-treated neonatal myocytes were analyzed for functional categories using Database for Annotation, Visualization and Integrated Discovery (DAVID) v6.7, NIAID/NIH. Paused gene are categorized as those that exhibit an increase in average pol II density but unchanged total pol II density (average density × bins).
| Functional pathway | # of genes | P value |
|---|---|---|
| Ribosome | 44 | 7.0E-23 |
| Renal cell carcinoma | 25 | 8.5E-9 |
| Neurotrophin signaling pathway | 30 | 5.4E-6 |
| Pathways in cancer | 56 | 6.9E-6 |
| Chronic myeloid leukemia | 20 | 5.7E-5 |
| Insulin signaling pathway | 28 | 1.1E-4 |
| Proteasome | 15 | 1.7E-4 |
| Huntington's disease | 35 | 2.3E-4 |
| Cell cycle | 26 | 3.1E-4 |
| Prostate cancer | 20 | 7.3E-4 |
| Non-small cell lung cancer | 14 | 9.7E-4 |
| MAPK signaling pathway | 42 | 1.4E-3 |
| Bladder cancer | 11 | 1.6E-3 |
| Pancreatic cancer | 16 | 1.9E-3 |
| Parkinson's disease | 26 | 1.9E-3 |
| ErbB signaling pathway | 18 | 2.5E-3 |
| Endometrial cancer | 13 | 2.6E-3 |
| Regulation of actin cytoskeleton | 34 | 2.6E-3 |
| Alzheimer's disease | 32 | 3.7E-3 |
| Colorectal cancer | 17 | 3.8E-3 |
| Glioma | 14 | 4.5E-3 |
| Acute myeloid leukemia | 13 | 5.1E-3 |
| Wnt signaling pathway | 25 | 5.3E-3 |
| Focal adhesion | 31 | 6.0E-3 |
| Adipocytokine signaling pathway | 14 | 1.0E-2 |
| Long-term potentiation | 14 | 1.0E-2 |
| Small cell lung cancer | 16 | 1.2E-2 |
| Melanoma | 14 | 1.3E-2 |
| Ubiquitin mediated proteolysis | 21 | 1.6E-2 |
| Oocyte meiosis | 19 | 1.8E-2 |
| Fc gamma R-mediated phagocytosis | 16 | 1.9E-2 |
| Oxidative phosphorylation | 22 | 2.0E-2 |
| Thyroid cancer | 8 | 2.1E-2 |
| Spliceosome | 20 | 2.1E-2 |
| GnRH signaling pathway | 16 | 3.3E-2 |
| Gap junction | 14 | 4.4E-2 |
Table 2.
Genes that exhibit reduced pol II recruitment in the miR-1-treated neonatal myocytes were analyzed for functional categories using Database for Annotation, Visualization and Integrated Discovery (DAVID) v6.7, NIAID/NIH. Genes exhibiting reduced pol II recruitment are characterized by reduced average density, bin numbers, and total pol II density (average density × bins).
| Functional pathway | # of genes | P value |
|---|---|---|
| Dilated cardiomyopathy | 11 | 3.9E-13 |
| Hypertrophic cardiomyopathy (HCM) | 10 | 1.1E-11 |
| Cardiac muscle contraction | 6 | 1.0E-5 |
| Focal adhesion | 7 | 7.8E-5 |
| Arrhythmogenic right ventricular cardiomyopathy (ARVC) RT | 5 | 1.7E-4 |
| Tight junction RT | 5 | 1.5E-3 |
| Viral myocarditis RT | 4 | 4.5E-3 |
| Adherens junction RT | 3 | 3.4E-2 |
| Leukocyte transendothelial migration RT | 3 | 7.4E-2 |
TFIIB is required for the recruitment of pol II to all promoters. However, whether TFIIB remains bound to pol II-paused promoters, or whether it is released with the release of pausing, remains unknown. Therefore, to determine the dynamics of TFIIB on active promoters, we performed a TFIIB ChIP-Seq analysis on neonatal, adult, and TAC-induced hearts and aligned the results with our previous pol II and H3K9ac datasets (Fig. 3c, 3g). The results show that TFIIB is constitutively bound to promoters that have paused pol II, and undergoes incremental changes (e.g. Ncl and Calm1) in inverse relation with changes in pol II pausing, as total gene pol II density remains unchanged (Fig. 3d). This was in stark contrast to genes requiring de novo pol II recruitment, which were associated with an equivalent 5–15 fold increase in TFIIB recruitment (Fig. 3g, 3h). The data confirm that all expressed promoters require TFIIB, albeit with varying binding dynamics - constitutive vs. induced.
Knockdown of TFIIB differentially inhibits genes requiring de novo recruitment of pol II vs. promoter-paused pol II
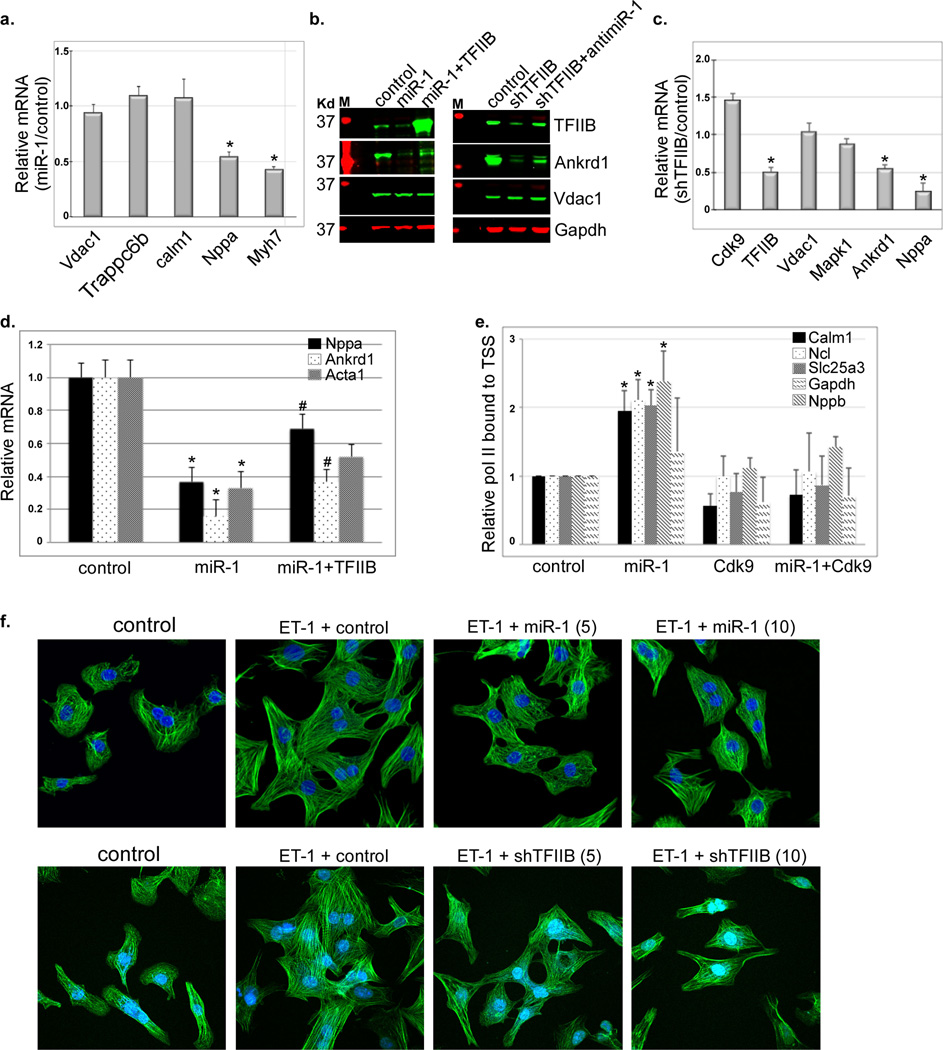
To confirm the transcriptional data, we measured the mRNA output of select genes. In concordance, there was no significant reduction in Calm1, Trappc6b, or Vdac1 mRNA versus 45% and 55% reductions in Nppa and Myh7 mRNA, respectively, in the presence of excess miR-1 (Fig. 4a). Similarly, there was no detectable change in Vdac1 protein, in contrast to an 80% reduction of Ankrd1 protein. Knockdown of TFIIB mimicked the effect of miR-1 overexpression in downregulating Ankrd1 protein and mRNA by 50%, and Nppa mRNA by ~ 70%, and, likewise, had little or no effect on Cdk9, Vdac1, or Mapk1 mRNA levels (Fig. 4c). Replenishing TFIIB could only marginally rescue inhibition of Ankrd1 and Nppa by miR-1 overexpression (Fig. 4b, 4d), indicating that miR-1 effects are mediated though multiple targets. In contrast, ChIP-qPCR shows that overexpression of Cdk9 was sufficient for the rescue of miR-1-induced promoter pausing of calm1, Ncl, Slc25a3, and Gapdh (Fig. 4e). Either overexpression of miR-1 or TFIIB knockdown partly inhibited endothelin-induced myocyte hypertrophy (Fig. 4f), suggesting that the increase in cell size is not only dependent on both the increase in housekeeping genes but also the increase in specialized genes [e.g growth factors and extracellular matrix genes (Supplementary Fig. 2S)].
Figure 4. Knockdown of TFIIB selectively inhibits genes regulated by de novo recruitment of pol II and inhibits myocyte hypertrophy.
a. RNA extracted from neonatal myocytes supplemented with a control or Ad.miR-1 for 24 h was analyzed by qPCR. The data for 5 genes are averaged and plotted as relative miR-1/control values (n=3). Error bars represent standard error of the mean (SEM) and * is p < 0.05 v. control. b. Protein was extracted from cells treated as in (a.) in the absence or presence of exogenous Ad.TFIIB, or with a shRNA targeting TFIIB (Ad.shTFIIB) in the absence or presence of Ad.antimiR-1, and analyzed by Western blotting. c. RNA extracted from neonatal myocytes supplemented with a control or Ad.shTFIIB for 24 h was analyzed by qPCR (n=3). The data for 6 genes are averaged and plotted as relative shTFIIB/control values. Error bars represent standard error of the mean (SEM) and * is p < 0.05 v. control. d. RNA was extracted from cells treated as in (a.) in the absence or presence of exogenous Ad.TFIIB, and analyzed by qPCR for the specified genes (n=3). e. Myocytes were treated with control, Ad.miR-1, Ad.Cdk9, or Ad.miR-1+Cdk9 for 24. Cells were then fixed and subjected to ChIP-qPCR encompassing the TSS of the specified genes. The results were averaged and plotted as relative bound pol II. * is p < 0.05 v. control. f. Myocytes were treated with control (10 moi), Ad.miR-1 (5 or 10 moi, upper pannels), or Ad.shTFIIB (5 or 10 moi, lower panels) for 24 h before stimulating them with 100 nM endothelin (ET-1). After 24 h the cells were fixed and stained with Dapi and phalloidin.
Acute, systemic, delivery of LNA-modified anti-TFIIB oligonucleotide restricts cardiac hypertrophy
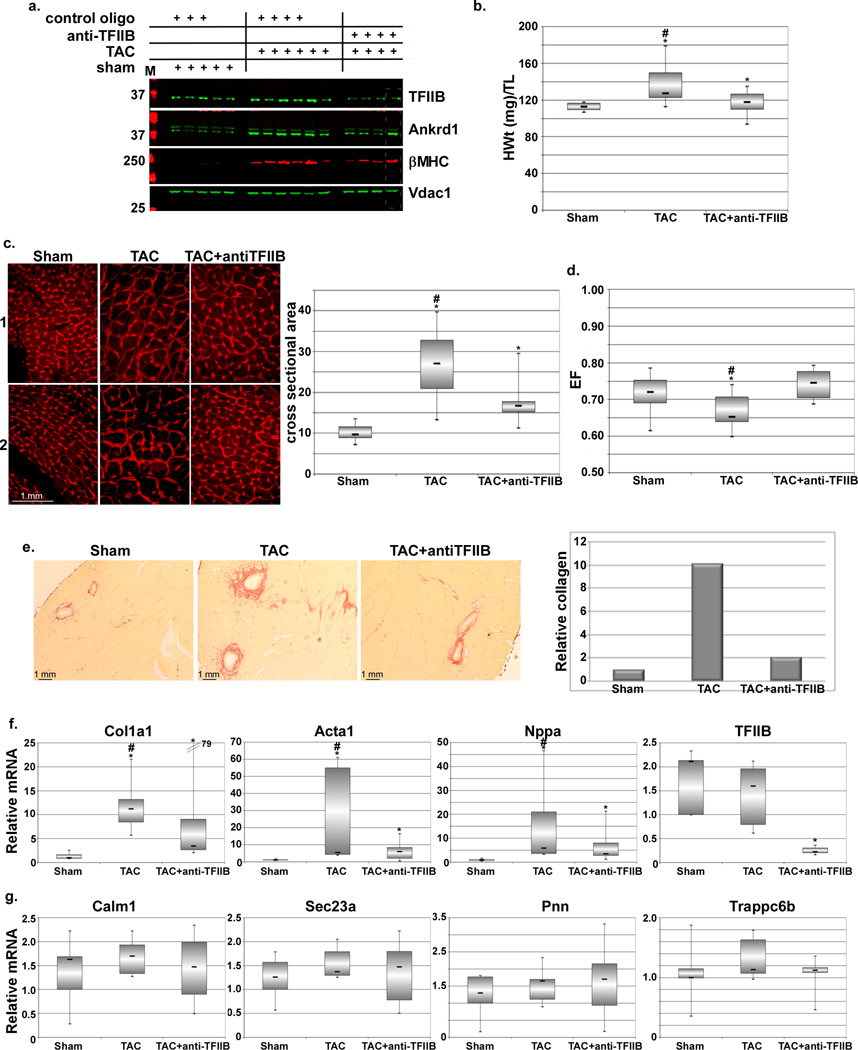
The above results led us to speculate that acute targeting of TFIIB would suppress the expression of cardiomypathy-induced genes while sparing the housekeeping genes, thereby, reducing pathological aspects of hypertrophy. To test this we systemically delivered locked-nucleic acid (LNA)-modified antisense TFIIB oligo 24 h post-TAC in mice. After three weeks, 7 of 7 injected mice exhibited 80–90% suppression by occupancy vs. degradation of TFIIB mRNA in the heart (Fig. 5f and Supplementary Fig. 3S), in addition to other organs (Fig.5f and Supplementary Fig. 3S), whereas only 6 of 7 mice exhibited complete inhibition of hypertrophy-induced increase in TFIIB protein (Fig. 5a and Supplementary Fig. 4S). The latter also correlated with a significant reduction in heart weight (Fig. 5b), myocyte cross sectional area (Fig. 5c) and collagen deposition (Fig. 5e), and improved ejection fraction (Fig. 5d). In addition, gene expression showed a significant reduction in Myh7 and Ankrd1 proteins (Fig. 5a and Supplementary Fig. 3S), and Col1a1, Acta1, and Nppa mRNA (Fig. 5f). This is in contrast to the housekeeping genes, Calm1, Pnn, Sec23a, and Trappc6b, which exhibited no significant change during hypertrophy (Fig. 5g). Similarly, anti-TFIIB had a differential effect on liver genes (Supplementary Fig. 3S). Thus, acute anti-TFIIB treatment reduces the extent of hypertrophy plausibly by reducing, but not completely inhibiting, the extent of pressure overload hypertrophy-induced genes that majorly contribute to pathological hypertrophy. Importantly, this treatment does not affect housekeeping genes.
Figure 5. Acute antisense inhibition of TFIIB reduces cardiac hypertrophy-induced gene expression and the increase in heart weight.
Twelve-week old, male, mice were subjected to a sham or transverse aortic constriction (TAC) operation. After 1d the mice were randomly selected for injection with saline or 15 mg/Kg LNA-modified control or antisense TFIIB (anti-TFIIB) oligo, as indicated (n=7 each). a. After 3 weeks the hearts were isolated, protein was extracted and subjected to Western blotting for the specified genes. One experimental set is shown here; the second set is shown in the online data (set = same day surgery for all included mice). b. Box-plot of the heart weights of the mice, adjusted to tibial length. c. Similarly treated heart were isolated, sectioned and stained with wheat germ agglutinin for delineating cross sectional area. Panels 1 and 2 represent sections from 2 different hearts. Cross sectional area of 10 myocyte/section/heart was measured and plotted (graph, right). * is p < 0.05 v. sham, # is p < 0.5 v. TAC+anti-TFIIB. d. All mice were analyzed by echocardiography before sacrifice and the ejection fraction calculated and plotted. * is p < 0.05 v. sham, # is p < 0.5 v. TAC+anti-TFIIB. e. Sectioned hearts were stained with Sirius red for estimating collagen content (n=2). The red-stained collaged was quantified, averaged, and plotted (graph, right). f. Total mRNA was extracted from all hearts and select hypertrophy-related genes were quantified by qPCR. * is p < 0.05 v. sham, # is p < 0.5 v. TAC+anti-TFIIB. g. * is p < 0.05 v. sham, # is p < 0.5 v. TAC+anti-TFIIB. g. Total mRNA was extracted from all hearts and select housekeeping genes were quantified by qPCR. * is p < 0.05 v. sham, # is p < 0.5 v. TAC+anti-TFIIB. g. * is p < 0.05 v. sham, # is p < 0.5 v. TAC+anti-TFIIB.
Discussion
Our previous data show two distinct, mutually exclusive, modes of transcription during cardiac hypertrophy with regards to pol II dynamics10. One involved promoter-proximal pausing of pol II that was incrementally released upon induction of pressure overload, while the other required de novo pol II recruitment. These modes of transcription strictly distinguished two categories of genes. The first includes housekeeping genes (e.g. genes involved in basic transcription, translation, nucleic acid metabolism..etc), which constitutes ~40% of the genes in the heart, while the second includes specialized (~4% of genes in the heart), cardiac-specific (e.g. Acta1, Ankrd1, Xirp2…etc), extracellular matrix (e.g. collagen and fibronectin), and immune response genes (e.g. chemokines and interleukins). Our previous study also reveals that reduced pol II pausing at housekeeping genes is a common feature of physiological, postnatal, cardiac growth, as well as, pathological pressure-overload cardiac hypertrophy. In contrast, most of the specialized genes are either not expressed during postnatal growth (e.g. Acta1, xirp2, chemokines), or are expressed at significantly lower levels than seen in pathological growth (e.g. ankrd1, Nppa, Col1a1, Ctgf). For the complete genome please see our Gene Expression Omnibus ChIP-Seq dataset (Accession: GSE50637). While some of these genes promote adaptation to pressure overload and may counteract cardiac hypertrophy, their excessive upregulation is what may contribute to the pathology23, 24. Thus, for the first time, our data revealed distinct basic transcriptional mechanisms that regulate housekeeping and specialized genes that could be exploited for selective inhibition of the latter group during hypertrophy.
Our objective was to collectively inhibit the upregulation of genes induced by pressure overload in an acute, dose-dependent, and reversible fashion. Therefore, we directed our attention to general transcription factors. TFIIB is a limiting general transcription factor that is required for the recruitment of pol II to promoter start sites, and, accordingly, the transcription of all promoters8. However, the dynamics of TFIIB binding to pol II-paused vs. de novo pol II-recruited promoters has not been determined yet. As demonstrated by our previous ChIP-Seq data10, a comparison between the neonatal, adult, and pressure-overloaded hearts afford us a unique and ideal model for addressing this question. Thus, we performed a TFIIB ChIP-Seq and aligned it with the pol II results. As expected, all pol II-bound promoters were also occupied by TFIIB. However, in contrast to promoters that required robust de novo recruitment of pol II and TFIIB, those that exhibited pol II pause-release displayed constitutive TFIIB binding, with only incremental, changes during hypertrophy. These results led us to speculate that acute and transient inhibition of TFIIB-induced expression during hypertrophy would selectively inhibit the former group of genes. Indeed, short hairpin targeting of TFIIB in neonatal myocytes decreased expression of Myh7 (~55%) and Nppa mRNA (~45%), while having no apparent effect of Vdac or MAPK. This would suggest that the turnover of pol II on the latter promoters is much slower, or that the immediate downstream-accumulated pol II is recycled to the promoter without detaching (gene looping)25.
One of the promising targeting tools that are designed for specific mRNA inhibition/knockdown are modified antisense oligonucleotides, which are widely utilized in cancer clinical trial26–28. In particular, we chose the locked-nucleic acid (LNA)-modified antisense oligo for its superior affinity to complimentary mRNA and, thereby, its high efficiency in inhibiting its target, either by occupancy or by inducing degradation. At a dose of 15 mg/Kg, the oligo inhibited TFIIB by ~80% via mRNA occupancy for up to 3 wks. Notably, hypertrophy-induced upregulation of TFIIB protein was completely inhibited. It should be emphasized here that TFIIB is required for the transcription of all genes, as confirmed by out ChIP-Seq data (Gene Expression Omnibus, GSE56813), however, since the paused promoters have both preassembled TFIIB and pol II, they were not immediately affected by knockdown of TFIIB. One of the drawbacks of this method, as is the case with most therapies, though, is the systemic delivery of the oligo, which would impact other organs. In our study, we tested the effect on liver gene expression, as in the heart, the oligo had no effect on the housekeeping genes (AKT or MAPK), but inhibited the expression of albumin, which is an inducible gene, by ~ 50% (Supplementary Fig. 2S).
While the involvement of RNA pol II in cardiac hypertrophy has been established in earlier studies1, 3, 13, 29, including alpha 1-adrenergic-induced hypertrophy4, the details of its involvement and how, in particular, it regulates the expression of housekeeping genes, and their incremental increase in parallel with cell size, has not been determined. We expect that the mechanism presented in our pressure-induced hypertrophy model, with respect to the dichotomous effect of pol II and TFIID on transcriptional dynamics, to be similar in other forms of hypertrophy. In support, the set of housekeeping genes that exhibit decreased pausing in hypertrophy, are similar to those exhibiting reduced pausing during postnatal growth. Although the epigenetics that potentially regulates pol II assembly on paused vs. inducible promoters has not been delineated, the regulation of pol II pausing by phosphorylation of its c-terminal domain (CTD) with Cdk9 is well established9, and has previously shown to play a major role in the development of cardiac hypertrophy30. In addition, recently pol II CTD has been shown to be modified by the p300 acetylase, which regulates its elongational activity in inducible genes, but has no role on housekeeping genes in human fibroblasts31. Thus, in conclusion, our study provides new mechanistic insights into general transcriptional regulation during cardiac hypertrophy, which offers a potentially new therapeutic target that promises to reduce pathological aspects of cardiac hypertrophy.
Supplementary Material
Acknowledgments
We want to thank Dr. Junichi Sadoshima, Chairman of the Department of Cell Biology and Molecular Medicine, for his support.
Sources of Funding
This work was supported by the National Institutes of Health [HL119726] and the American Heart Association Scientist Development Grants [12SDG8820011] to the corresponding authors.
Footnotes
Disclosures
None.
References
- 1.Abdellatif M, Packer SE, Michael LH, Zhang D, Charng MJ, Schneider MD. A Ras-dependent pathway regulates RNA polymerase II phosphorylation in cardiac myocytes: Implications for cardiac hypertrophy. Mol. Cell. Biol. 1998;18:6729–6736. doi: 10.1128/mcb.18.11.6729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cutilletta AF, Dowell RT, Rudnik M, Arcilla RA, Zak R. Regression of myocardial hypertrophy. I. Experimental model, changes in heart weight, nucleic acids and collagen. J Mol Cell Cardiol. 1975;7:761–780. doi: 10.1016/0022-2828(75)90042-5. [DOI] [PubMed] [Google Scholar]
- 3.Kamala J, Mariappan M, Rajamanickam C, Cutilletta AF. Changes associated with rat heart chromatin during cardiac hypertrophy. Biochem Int. 1986;13:271–286. [PubMed] [Google Scholar]
- 4.Simpson PC, Long CS, Waspe LE, Henrich CJ, Ordahl CP. Transcription of early developmental isogenes in cardiac myocyte hypertrophy. J Mol Cell Cardiol. 1989;21(Suppl 5):79–89. doi: 10.1016/0022-2828(89)90774-8. [DOI] [PubMed] [Google Scholar]
- 5.Feldman AM, Weinberg EO, Ray PE, Lorell BH. Selective changes in cardiac gene expression during compensated hypertrophy and the transition to cardiac decompensation in rats with chronic aortic banding. Circ Res. 1993;73:184–192. doi: 10.1161/01.res.73.1.184. [DOI] [PubMed] [Google Scholar]
- 6.Schneider MD, McLellan WR, Black FM, Parker TG. Growth factors, growth factor response elements, and the cardiac phenotype. Basic Res Cardiol. 1992;87(Suppl 2):33–48. doi: 10.1007/978-3-642-72477-0_4. [DOI] [PubMed] [Google Scholar]
- 7.Paradis P, MacLellan WR, Belaguli NS, Schwartz RJ, Schneider MD. Serum response factor mediates AP-1-dependent induction of the skeletal alpha-actin promoter in ventricular myocytes. J Biol Chem. 1996;271:10827–10833. doi: 10.1074/jbc.271.18.10827. [DOI] [PubMed] [Google Scholar]
- 8.Deng W, Roberts SG. TFIIB and the regulation of transcription by RNA polymerase II. Chromosoma. 2007;116:417–429. doi: 10.1007/s00412-007-0113-9. [DOI] [PubMed] [Google Scholar]
- 9.Lis JT, Mason P, Peng J, Price DH, Werner J. P-TEFb kinase recruitment and function at heat shock loci. Genes Dev. 2000;14:792–803. [PMC free article] [PubMed] [Google Scholar]
- 10.Sayed D, He M, Yang Z, Lin L, Abdellatif M. Transcriptional regulation patterns revealed by high resolution chromatin immunoprecipitation during cardiac hypertrophy. J Biol Chem. 2013;288:2546–2558. doi: 10.1074/jbc.M112.429449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Abdellatif M, Mcllelan WR, Schneider MD. p21 Ras as a governer of global gene expression. JBC. 1994;269:15423–15426. [PubMed] [Google Scholar]
- 12.Graham FL, Prevec L. Methods in Molecular Biology. Vol. 7. Clifton, NJ: The Humana Press Inc.; 1991. [DOI] [PubMed] [Google Scholar]
- 13.Sano M, Abdellatif M, Oh H, Xie M, Bagella L, Giordano A, Michael LH, DeMayo FJ, Schneider MD. Activation and function of cyclin T-Cdk9 (positive transcription elongation factor-b) in cardiac muscle-cell hypertrophy. Nat Med. 2002;8:1310–1317. doi: 10.1038/nm778. [DOI] [PubMed] [Google Scholar]
- 14.Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- 15.Grimson A, Farh KK, Johnston WK, Garrett-Engele P, Lim LP, Bartel DP. MicroRNA targeting specificity in mammals: determinants beyond seed pairing. Mol Cell. 2007;27:91–105. doi: 10.1016/j.molcel.2007.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Friedman RC, Farh KK, Burge CB, Bartel DP. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res. 2009;19:92–105. doi: 10.1101/gr.082701.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Krek A, Grun D, Poy MN, Wolf R, Rosenberg L, Epstein EJ, MacMenamin P, da Piedade I, Gunsalus KC, Stoffel M, Rajewsky N. Combinatorial microRNA target predictions. Nat Genet. 2005;37:495–500. doi: 10.1038/ng1536. [DOI] [PubMed] [Google Scholar]
- 18.Creighton CJ, Nagaraja AK, Hanash SM, Matzuk MM, Gunaratne PH. A bioinformatics tool for linking gene expression profiling results with public databases of microRNA target predictions. RNA. 2008;14:2290–2296. doi: 10.1261/rna.1188208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sayed D, Hong C, Chen IY, Lypowy J, Abdellatif M. MicroRNAs play an essential role in the development of cardiac hypertrophy. Circ Res. 2007;100:416–424. doi: 10.1161/01.RES.0000257913.42552.23. [DOI] [PubMed] [Google Scholar]
- 20.Dennis G, Jr, Sherman BT, Hosack DA, Yang J, Gao W, Lane HC, Lempicki RA. DAVID: Database for Annotation, Visualization, and Integrated Discovery. Genome Biol. 2003;4:3. [PubMed] [Google Scholar]
- 21.Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 22.Jiao X, Sherman BT, Huang da W, Stephens R, Baseler MW, Lane HC, Lempicki RA. DAVID-WS: a stateful web service to facilitate gene/protein list analysis. Bioinformatics. 2012;28:1805–1806. doi: 10.1093/bioinformatics/bts251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chen MM, Lam A, Abraham JA, Schreiner GF, Joly AH. CTGF expression is induced by TGF- beta in cardiac fibroblasts and cardiac myocytes: a potential role in heart fibrosis. J Mol Cell Cardiol. 2000;32:1805–1819. doi: 10.1006/jmcc.2000.1215. [DOI] [PubMed] [Google Scholar]
- 24.Zolk O, Marx M, Jackel E, El-Armouche A, Eschenhagen T. Beta-adrenergic stimulation induces cardiac ankyrin repeat protein expression: involvement of protein kinase A and calmodulin-dependent kinase. Cardiovasc Res. 2003;59:563–572. doi: 10.1016/s0008-6363(03)00476-0. [DOI] [PubMed] [Google Scholar]
- 25.Laine JP, Singh BN, Krishnamurthy S, Hampsey M. A physiological role for gene loops in yeast. Genes Dev. 2009;23:2604–2609. doi: 10.1101/gad.1823609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Visser ME, Witztum JL, Stroes ES, Kastelein JJ. Antisense oligonucleotides for the treatment of dyslipidaemia. Eur Heart J. 2012;33:1451–1458. doi: 10.1093/eurheartj/ehs084. [DOI] [PubMed] [Google Scholar]
- 27.Zhang Y, Castaneda S, Dumble M, Wang M, Mileski M, Qu Z, Kim S, Shi V, Kraft P, Gao Y, Pak J, Sapra P, Bandaru R, Zhao H, Vessella RL, Horak ID, Greenberger LM. Reduced expression of the androgen receptor by third generation of antisense shows antitumor activity in models of prostate cancer. Mol Cancer Ther. 2011;10:2309–2319. doi: 10.1158/1535-7163.MCT-11-0329. [DOI] [PubMed] [Google Scholar]
- 28.Zhang Y, Qu Z, Kim S, Shi V, Liao B, Kraft P, Bandaru R, Wu Y, Greenberger LM, Horak ID. Down-modulation of cancer targets using locked nucleic acid (LNA)-based antisense oligonucleotides without transfection. Gene Ther. 2011;18:326–333. doi: 10.1038/gt.2010.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cutilletta AF, Rudnik M, Zak R. Muscle and non-muscle cell RNA polymerase activity during the development of myocardial hypertrophy. J. of Molecular and Cellular Cardiology. 1978;10:677–687. doi: 10.1016/0022-2828(78)90403-0. [DOI] [PubMed] [Google Scholar]
- 30.Sano M, Schneider MD. Cyclin-dependent kinase-9: an RNAPII kinase at the nexus of cardiac growth and death cascades. Circ Res. 2004;95:867–876. doi: 10.1161/01.RES.0000146675.88354.04. [DOI] [PubMed] [Google Scholar]
- 31.Schroder S, Herker E, Itzen F, He D, Thomas S, Gilchrist DA, Kaehlcke K, Cho S, Pollard KS, Capra JA, Schnolzer M, Cole PA, Geyer M, Bruneau BG, Adelman K, Ott M. Acetylation of RNA polymerase II regulates growth-factor-induced gene transcription in mammalian cells. Mol Cell. 2013;52:314–324. doi: 10.1016/j.molcel.2013.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.