Abstract
Abnormal activation of insulin-like growth factor (IGF)-Akt signaling is implicated in the development of various diseases, including heart failure. However, the molecular mechanisms that regulate activation of this signaling pathway are not completely understood. Here we show that sirtuin 6 (SIRT6), a nuclear histone deacetylase, functions at the level of chromatin to directly attenuate IGF-Akt signaling. SIRT6-deficient mice developed cardiac hypertrophy and heart failure, whereas SIRT6 transgenic mice were protected from hypertrophic stimuli, indicating that SIRT6 acts as a negative regulator of cardiac hypertrophy. SIRT6-deficient mouse hearts showed hyperactivation of IGF signaling–related genes and their downstream targets. Mechanistically, SIRT6 binds to and suppresses the promoter of IGF signaling–related genes by interacting with c-Jun and deacetylating histone 3 at Lys9 (H3K9). We also found reduced SIRT6 expression in human failing hearts. These findings disclose a new link between SIRT6 and IGF-Akt signaling and implicate SIRT6 in the development of cardiac hypertrophy and failure.
Heart failure is the leading contributor of human morbidity and mortality in the developed world1. Although a number of risk factors have been recognized for heart failure, the molecular mechanisms contributing to the initiation of heart failure are incompletely understood2. Recent studies have demonstrated that heart failure can be prevented or reverted, at least in experimental models, by caloric restriction, a dietary regimen that limits calorie intake3–5.
The health benefits of calorie restriction are thought to be mediated by a family of NAD-dependent deacetylases called sirtuins. The mammalian genome encodes seven ubiquitously expressed sirtuin isoforms (SIRT1–SIRT7), which are emerging as key regulators of a myriad of biological functions, ranging from cell growth to lifespan extension6. Among the sirtuins, SIRT6, a chromatin-associated deacetylase, is considered to have a leading role in regulating genomic stability, cellular metabolism, stress response and aging7–12. SIRT6 deacetylates H3K9 and acts as a transcriptional co-repressor of NF-κB and hypoxia inducible factor-1α (HIF-1α) target genes in a tissue- and context-dependent manner8,10,13. SIRT6-deficient mice show severe hypoglycemia and a multisystemic accelerated aging phenotype and die at around 26 d after birth12. The hypoglycemia of SIRT6-deficient mice seems to result from increased activity of HIF-1α target genes and increased glucose uptake by multiple organs13,14. However, neither correction of hypoglycemia by feeding mice with glucose nor reduced NF-κB signaling could completely rescue the lethality of SIRT6-deficient mice, suggesting that additional mechanisms must contribute to the mutant phenotype of SIRT6-deficient mice8,14.
Mammalian aging and aging-associated diseases have been shown to be associated with dysregulation of IGF-Akt signaling15. In the heart, although short-term activation of IGF-Akt signaling promotes physiologic growth, sustained hyperactivation leads to the development of pathologic hypertrophy and heart failure16,17. Although much is known about the positive activators of this pathway, very little is known about the endogenous negative regulators, which might have potential to block the IGF-Akt pathway and protect the heart from developing heart failure.
In this study, we show that SIRT6 directly controls IGF-Akt signaling at the level of chromatin through c-Jun, a stress-responsive transcription factor, and deacetylation of H3K9. Our findings also demonstrate that SIRT6 deficiency induces hyperactivation of IGF-Akt signaling, which culminates in the development of cardiac hypertrophy and heart failure in mice.
RESULTS
Loss of SIRT6 induces cardiac hypertrophy and failure
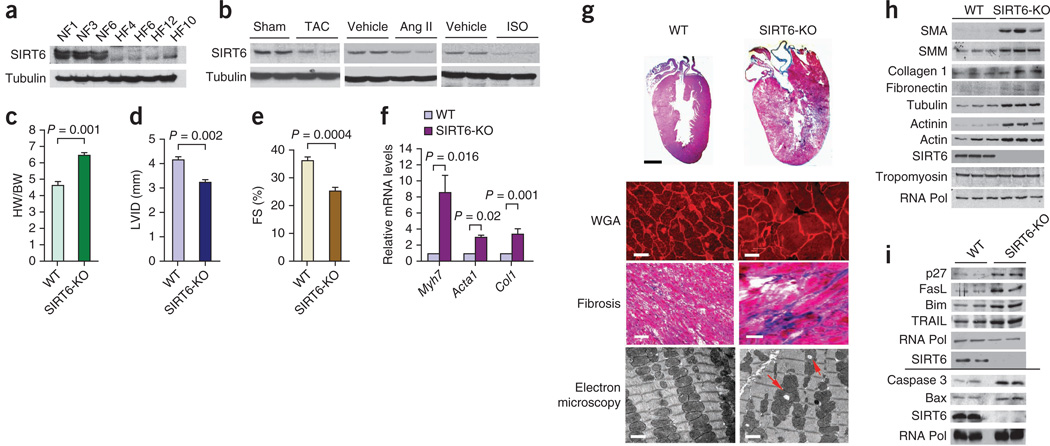
To study the role of SIRT6 in the development of heart failure, we first analyzed SIRT6 expression in failing human hearts and in mouse hearts in which we had induced hypertrophy by either surgically creating transverse aortic constriction (TAC) or infusing the hypertrophic agonists isoproterenol (ISO) or angiotensin-II (Ang-II). In both human and mouse hearts, SIRT6 amounts were markedly reduced compared to control hearts (Fig. 1a,b and Supplementary Table 1), suggesting that SIRT6 deficiency is associated with the development of cardiac hypertrophy and failure. To test whether SIRT6 deficiency contributes to heart disease, we analyzed hearts of SIRT6 whole-body knockout mice. SIRT6 knockout mice on an inbred genetic (129sv) background die at around 26 d of age12, whereas SIRT6 knockout mice on crossbred genetic backgrounds survive for up to 1 year14. We therefore analyzed crossbred SIRT6 knockout mice(129sv/C57BL6) for this study. These crossbred SIRT6 knockout mice spontaneously developed concentric cardiac hypertrophy at around 8–12 weeks of age with a significantly increased heart weight to body weight (HW/BW) ratio, reduced left ventricular internal diameter and reduced fractional shortening (Fig. 1c–e). SIRT6 knockout hearts also showed increased expression of cardiac fetal genes, increased cardiomyocyte size and interstitial fibrosis (Fig. 1f,g). These hearts also showed severe degenerative changes, including aggregation and vacuolization of mitochondria (Fig. 1g). SIRT6 knockout hearts also expressed higher levels of aging-associated cytoskeleton proteins18 such as actin, actinin and tubulin; fibrotic markers such as α-smooth muscle actin, myosin, collagen-1 and fibronectin; and apoptotic markers such as caspase 3, Bax, TRAIL, Bim, FasL and p27 (Fig. 1h,i), which are signs of a failing heart.
Figure 1.
SIRT6 deficiency causes cardiac hypertrophy and degenerative changes in the heart. (a) Western blot showing SIRT6 expression in human heart samples from representative normal control hearts (NF1, NF3, NF6; n = 8) and failing hearts (HF4, HF6, HF12, HF10; n = 24) (Supplementary Table 1). Tubulin was used as a loading control. (b) Representative western blots showing SIRT6 expression in heart samples from mice subjected to TAC or chronic infusion of ISO or angiotensin II (Ang II). n = 8 mice per group. (c) HW/BW ratio of wild-type (WT) and SIRT6 knockout (SIRT6-KO) mice at 8 weeks of age. n = 13 mice per group. Data are presented as means ± s.d. (d,e) Left ventricular internal diameter (LVID; n = 7 mice per group) and fractional shortening (FS; n = 10 mice per group) of WT and SIRT6 knockout mice at 8 weeks of age as determined by echocardiography. Data are presented as means ± s.d. (f) mRNA levels of the indicated genes in heart samples of WT and SIRT6 knockout mice. n = 6 mice per group. Data are presented as the mean ± s.d. For c–f, Student’s t test was used to calculate the P values. (g) Whole-heart sections stained with H&E showing concentric hypertrophy in SIRT6 knockout hearts (top row; scale bar, 2 mm); left ventricular muscle sections stained with wheat germ agglutinin (WGA) to demarcate cell boundaries (second row; scale bars, 10 µm) or Masson’s trichrome to detect fibrosis (third row; scale bars, 40 µm); and electron micrographs showing vacuolization (red arrows) and degeneration of mitochondria in SIRT6 knockout hearts (bottom row; scale bars, 1 µm). (h,i) Representative western blots showing the expression of the indicated fibrosis markers, cytoskeletal proteins and apoptotic markers in WT and SIRT6 knockout hearts. n = 6 per group. The line between the blots indicates two different gels. RNA Pol, RNA polymerase; SMA, α-smooth muscle actin; SMM, smooth muscle myosin.
To test whether a partial reduction in SIRT6 expression, as seen in human failing hearts (Supplementary Fig. 1a), could be sufficient to induce heart failure, we analyzed the hearts of SIRT6+/− heterozygous mice. Young SIRT6+/− mice showed no noticeable cardiac defects; however, at around 6 months of age, they spontaneously developed characteristics of pathologic cardiac hypertrophy, such as increased HW/BW and heart weight to tibia length (HW/TL) ratios, increased cardiomyocyte size and interstitial fibrosis (Supplementary Fig. 1b–d), suggesting that reduced expression of SIRT6 can contribute to the development of cardiac hypertrophy.
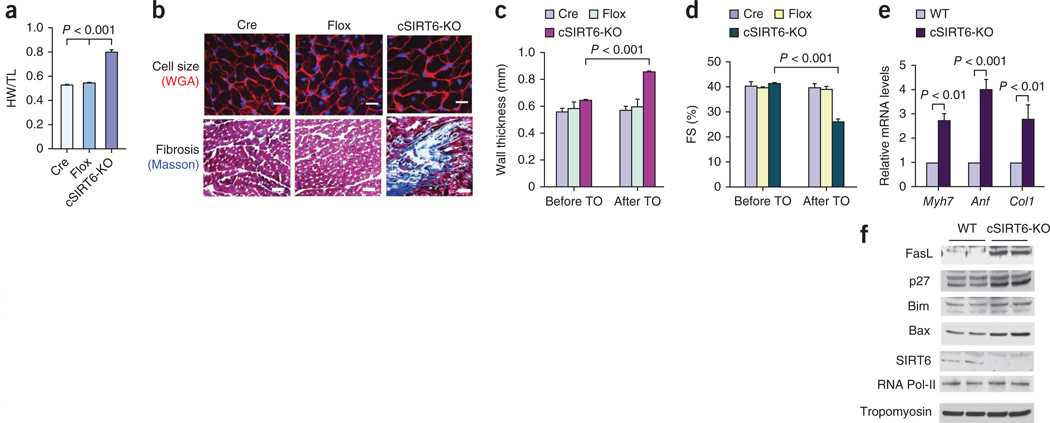
To study the cardiac-specific effects of SIRT6, we used a tamoxifen-inducible α-myosin heavy chain (α-MHC)-Cre system to delete SIRT6 in the hearts of adult mice (Supplementary Fig. 2). Cardiac-specific deletion of SIRT6 resulted in a significantly increased HW/TL ratio, increased left ventricular wall thickness, increased cardiomyocyte cross-sectional area and induction of interstitial fibrosis (Fig. 2a–c). Echocardiography of these mice revealed significantly reduced cardiac contractile function (Fig. 2d). In addition, cardiac-specific SIRT6-deleted hearts had increased expression of fetal genes and cell death markers (Fig. 2e,f). We obtained similar results by analyzing hearts of mice in which SIRT6 was deleted both in the skeletal and cardiac muscles by use of the MCK-Cre system (N.R.S., D.B.L. and M.P.G., unpublished data). These data demonstrate that SIRT6 is an endogenous negative regulator of cardiac hypertrophy.
Figure 2.
Cardiac-specific deletion of SIRT6 causes cardiac hypertrophy and fibrosis. (a) HW/TL ratio of control mice (Cre, α-MHC–Cre; Flox, SIRT6flox/flox) and SIRT6flox/flox–α-MHC–Cre mice injected with tamoxifen to generate a cardiac-specific SIRT6 deletion (cSIRT6-KO). Data are presented as the mean ± s.d. n = 9–15 mice per group. Student’s t test was used to calculate the P value. (b) Heart sections of cardiac-specific SIRT6 knockout and control mice stained with WGA to demarcate cell boundaries (top; scale bars, 10 µm) or with Masson’s trichrome to detect fibrosis (bottom; scale bars, 40 µm). (c,d) Left ventricular wall thickness and fractional shortening of control and cardiac-specific SIRT6 knockout mice before and after tamoxifen (TO) injection. n = 5 mice per group. (e) mRNA levels of the indicated genes in heart samples of control and cardiac-specific SIRT6 knockout mice. Data are presented as the mean ± s.d. n = 4 per group. (f) Western blots showing the expression of the indicated cell death markers in the same hearts as in e. For c–e, Student’s t test was used to calculate the P value.
SIRT6 activation blocks development of cardiac hypertrophy
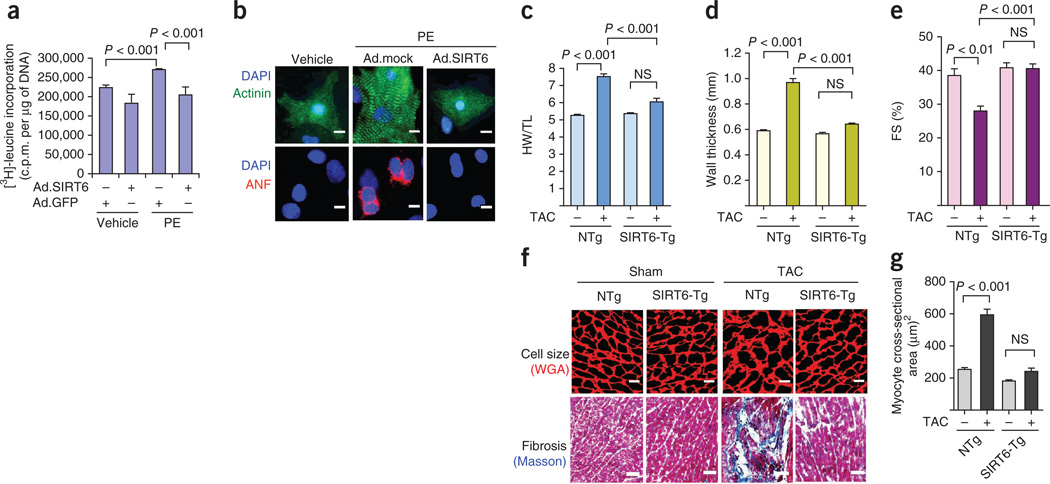
We first examined the effect of SIRT6 overexpression in an in vitro model of cardiomyocyte hypertrophy. We infected neonatal rat cardiomyocytes with either a control vector or a vector expressing SIRT6 (Supplementary Fig. 3a,b). SIRT6-overexpressing cells had a dampened hypertrophic response to phenylephrine treatment based on analysis of protein synthesis, atrial natriuretic factor (ANF) release from nuclei and sarcomere organization, which are hallmarks of cardiomyocyte hypertrophy (Fig. 3a,b). These results indicate an antihypertrophic effect of SIRT6 in cardiomyocytes.
Figure 3.
SIRT6 overexpression blocks the cardiac hypertrophic response. (a) [3H]-leucine incorporation into total cellular protein normalized to the DNA content of neonatal rat cardiomyocytes infected with WT SIRT6 adenovirus (Ad.SIRT6) or control adenovirus (Ad.GFP) and then treated with phenylephrine (PE) (20 µm) for 48 h. Data are presented as the mean ± s.d. n = 5 independent experiments. C.p.m. counts per minute. (b) Sarcomere organization, as determined by immunostaining cells for sarcomeric α-actinin (green), and ANF release (red), as determined by staining cells with an ANF-specific antibody, in cardiomyocytes infected with the indicated adenoviruses (Ad.mock indicates empty vector) and stimulated with phenylephrine. DAPI stain marks the position of nuclei. Scale bars, 10 µm. (c) HW/TL ratios in nontransgenic (NTg) and SIRT6 transgenic (SIRT6-Tg) mice subjected to sham or to TAC treatment for 4 weeks. Data are presented as the mean ± s.d. n = 8 mice per group. (d,e) Left ventricular wall thickness and fractional shortening, as measured by echocardiography, of the same hearts as in c. n = 6 mice per group. For c–e, one-way analysis of variance (ANOVA) was applied to calculate the P value. (f) Heart sections of nontransgenic and SIRT6 transgenic mice subjected to sham or TAC treatment and stained with WGA to demarcate cell boundaries (top; scale bars, 10 µm) or Masson’s trichrome to detect fibrosis (bottom; scale bars, 40 µm). (g) Quantification of myocyte cross-sectional area in nontransgenic and SIRT6 transgenic mice subjected to sham or to TAC treatment. Data are presented as the mean ± s.d. n = 5 mice per group. Student’s t test was used to calculate the P value. NS, not significant.
To evaluate the antihypertrophic effects of SIRT6 in vivo, we generated cardiac-specific SIRT6 transgenic mice (Supplementary Fig. 3c). At baseline, SIRT6 transgenic mice had no noticeable cardiac abnormalities. We induced cardiac hypertrophy in these mice by either surgically creating TAC or infusing ISO into the abdomen. Nontransgenic mice subjected to TAC for 4 weeks developed massive cardiac hypertrophy, as indicated by an increased HW/TL ratio, increased left ventricular wall thickness and decreased fractional shortening (Fig. 3c–e). These changes were accompanied by an increased cardiomyocyte cross-sectional area and induction of fibrosis (Fig. 3f,g). Notably, the hearts of SIRT6 transgenic mice did not undergo similar pathological growth after being subjected to TAC (Fig. 3c–g). We found similar cardioprotective effects of SIRT6 overexpression in the ISO model of cardiac hypertrophy (Supplementary Fig. 3d–g). These data indicate that SIRT6 overexpression is capable of blocking the development of pathologic cardiac hypertrophy.
SIRT6 antagonizes IGF signaling
SIRT6 knockout mice on the 129Sv genetic background develop severe hypoglycemia and hypoinsulinemia, which is mediated through alterations in HIF-1α activity13,14. To test whether such metabolic changes contribute to the cardiac phenotype we observed in SIRT6-deficient mice, we analyzed myocardial glucose uptake, serum insulin and blood glucose concentrations and the expression of HIF-1α target glycolytic genes, previously shown to be regulated by SIRT6. We found no change in glucose uptake in the hearts of SIRT6 knockout mice (Supplementary Fig. 4a). We also noted no statistically significant variation in blood glucose or insulin concentrations in these mice (Supplementary Fig. 4b,c). Western blot analysis revealed that expression of the HIF-1α target genes PFK1, TPI1 and GAPDH were unchanged in SIRT6 knockout hearts (Supplementary Fig. 4d). In addition, we found no noticeable changes in the expression of mitochondrial superoxide dismutase 2 (MnSOD) and glutathione S-transferase (GST), which have previously been shown to be targeted by the transcription factor NF-κB (Supplementary Fig. 4d). These findings indicate that the development of heart failure in SIRT6 knockout mice is not mediated by alterations in either systemic glucose homeostasis or activation of HIF-1α or NF-κB signaling in the heart.
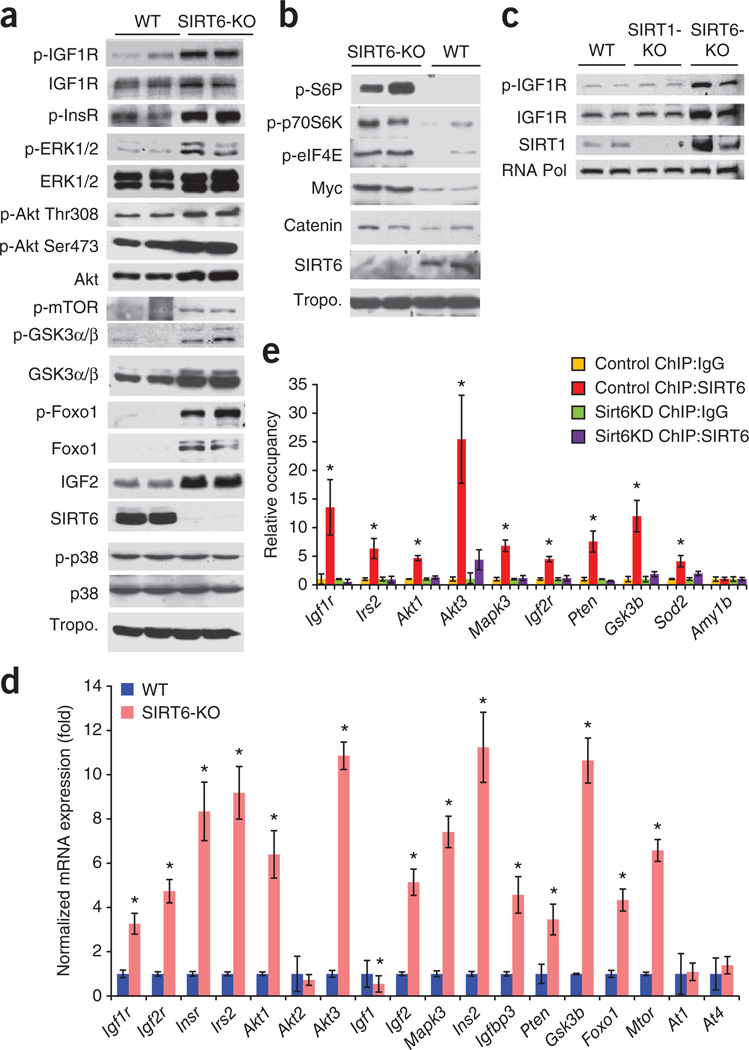
We next tested whether effects on IGF-Akt signaling might underlie the cardiac phenotype of SIRT6 knockout mice. SIRT6 knockout hearts had substantially increased amounts of proteins (and/or phosphoproteins) related to this pathway, including IGF1 receptor (IGF1R), insulin receptor (InsR), extracellular signal-regulated kinase 1 (ERK1)/ERK2, Akt, forkhead box O1 (Foxo1), glycogen synthase kinase 3 (GSK3) and IGF2 (Fig. 4a and Supplementary Fig. 4e). The amount of membrane-bound IGF2 was also increased in SIRT6 knockout hearts (Supplementary Fig. 4f). We also noted that failing human hearts had higher amounts of IGF1R and phosphorylated Akt expression compared to controls; similarly, hypertrophic mouse hearts in the TAC and ISO models had increased expression of these proteins compared to control hearts (Supplementary Fig. 4g,h). Additionally, SIRT6 knockout hearts showed increased expression of transcription and translation factors acting downstream of IGF signaling, including Myc, β-catenin, eukaryotic translation initiation factor 4E (eIF4E), S6P and p70S6K (Fig. 4b). However, SIRT6 knockout hearts did not show increased expression of p38 or tropomyosin, which served as loading controls (Fig. 4a,b). Conversely, the expression and phosphorylation of IGF signaling–related proteins were attenuated in SIRT6-overexpressing transgenic hearts (Supplementary Fig. 5a). These data indicate that induction of cardiac hypertrophy in SIRT6-deficient hearts might be related to hyperactivation of IGF-Akt signaling–related proteins.
Figure 4.
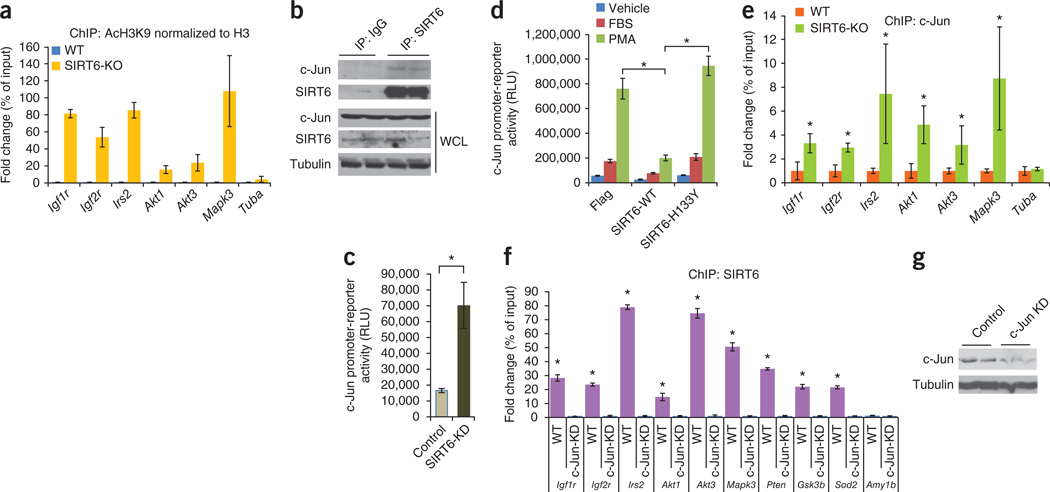
SIRT6 is a negative regulator of IGF signaling. (a) Representative western blots showing increased expression and phosphorylation of IGF signaling–related proteins in SIRT6 knockout hearts compared to WT control hearts. The expression of p38 and tropomyosin (tropo.) were not changed in these hearts and were therefore used as loading controls. n = 6 mice per group. The antibodies used for detecting ERK and GSK3 recognized the ERK1/2 and GSK3α/β isoforms, respectively. The ‘p-’ prefix indicates the phosphorylated form. (b) Representative western blots showing increased expression of transcription and translation factors in SIRT6 knockout hearts. n = 6 mice per group. (c) Representative western blots showing expression of IGF1R, p-IGF1R and SIRT1 in heart lysates of WT, SIRT1 knockout and SIRT6 knockout mice. n = 3 mice per group. (d) Real-time PCR analysis of IGF signaling–related genes in heart samples of WT and SIRT6 knockout mice. n = 6 mice per group. (e) ChIP analysis to detect SIRT6 binding to the promoters of the indicated genes performed with a SIRT6-specific antibody or IgG control antibody in WT or SIRT6 knockdown (Sirt6KD) cardiomyocytes. The occupancy of SIRT6 to promoters is shown relative to background signals with IgG control antibody. n = 4 independent experiments. Data are presented as the mean ± s.d. Student’s t test was applied to calculate the P value. *P < 0.001.
Because SIRT6-deficient mice develop a multiorgan aging phenotype, we asked whether the effect of SIRT6 on IGF signaling is limited to the heart or is a more widespread phenomenon. We first analyzed the effects of nicotinamide (NAM), a general inhibitor of sirtuins, and trichostatin A (TSA), a class I and II histone deacetylase (HDAC) inhibitor. Treatment with NAM, but not TSA, increased the expression and phosphorylation of IGF1R in three different cells (adult mouse cardiac fibroblasts and 3T3L and Cos7 cells) analyzed, indicative of the involvement of a sirtuin in IGF1R activation (Supplementary Fig. 5b). We next analyzed the effects of SIRT6 deficiency in mouse embryonic fibroblasts (MEFs). As in the heart, SIRT6 knockout MEFs showed hyperactivation of IGF signaling–related proteins (Supplementary Fig. 5c). SIRT6 knockdown in 293T cells using retroviral vectors targeting SIRT6 had a similar effect (Supplementary Fig. 5d). To confirm these findings, we performed a reconstitution experiment in which we re-expressed SIRT6 in SIRT6 knockout cells. Re-expression of wild-type SIRT6, but not a SIRT6 mutant lacking deacetylase activity, attenuated IGF1R activation in SIRT6 knockout MEFs (Supplementary Fig. 5e), indicating that SIRT6-dependent regulation of IGF signaling is not limited to the heart. We also tested whether deficiency of other sirtuins present in the nucleus, such as SIRT1, can activate IGF1R. However, IGF1R activation occurred in SIRT6 knockout but not in SIRT1 knockout hearts (Fig. 4c). We have recently shown that Akt membrane localization and activation are under the control of SIRT1 (ref. 19). SIRT6 knockout hearts, in which Akt was activated, showed markedly increased expression of SIRT1 (Fig. 4c), which might be needed for mediating Akt activation in these hearts. Taken together, these findings indicate that SIRT6 has a central role in regulating IGF-Akt signaling.
SIRT6 blocks IGF signaling by deacetylating H3K9
Because SIRT6 is a chromatin-associated deacetylase, we posited that it could directly influence the expression of IGF signaling–related genes by modifying chromatin. To test this hypothesis, we used available microarray data from the skeletal muscle of SIRT6 knockout mice13. The results of a clustering analysis using these data suggested that SIRT6 deficiency substantially increased the expression of key IGF signaling–related genes, including Igf2, Igf2r, Foxo1, Igf1r, Mapk3, Pten, Gsk3, Irs2, Akt3 and Ins2 (Supplementary Fig. 6a), whereas the expression of other such genes, including Ins1 and Igf1, was down regulated (Supplementary Fig. 6b). To test whether similar changes also take place in the heart, we performed real-time PCR. In SIRT6 knockout hearts, the mRNA levels of key IGF signaling–related genes, including Igf1r, Igf2r, Insr, Irs2, Akt1, Akt3, Igf2, Mapk3, Ins2, Igfbp3, Pten, Gsk3b, Foxo1 and Mtor, were significantly upregulated, whereas Igf1 was downregulated (Fig. 4d). The expression of Akt2, an IGF signaling–related gene, and At1 and At4, which are unrelated to IGF signaling, were unchanged (Fig. 4d), indicating that SIRT6 deficiency specifically induces certain IGF signaling genes rather than nonspecifically activating the cellular transcription machinery. By chromatin immunoprecipitation (ChIP), we found that SIRT6 binds to promoters of IGF signaling–related genes in cardiomyocytes, and knockdown of SIRT6 reduced its binding to these promoters (Fig. 4e and Supplementary Fig. 6c), indicating the specificity of SIRT6 binding to these genes.
SIRT6 deacetylates H3K9 and thereby regulates gene expression by modifying chromatin structure10. We therefore asked whether SIRT6 deficiency results in increased H3K9 acetylation in the promoters of upregulated IGF signaling–related genes. ChIP analysis revealed increased acetylation of H3K9 in SIRT6-deficient cells compared to wild-type control cells (Fig. 5a and Supplementary Fig. 7a). However, acetylation of H3K14 was not increased, which served as a negative control (Supplementary Fig. 7b,c). These data demonstrate that SIRT6 directly suppresses the expression of key IGF signaling–related genes by deacetylating H3K9 at their promoters.
Figure 5.
SIRT6 is a co-repressor of c-Jun transcriptional activity. (a) ChIP analysis to detect H3K9 acetylation at the promoters of the indicated IGF signaling–related genes in WT and SIRT6 knockout heart samples. Antibodies to acetylated H3K9 (AcH3K9) and to H3 were used, and acetylated H3K9 levels are shown relative to total H3 levels. n = 4 independent experiments. Data are presented as the mean ± s.d. (b) SIRT6 binding to c-Jun, as determined by western blots of SIRT6 or control IgG antibody immunoprecipitates from rat cardiomyocytes. WCL, whole-cell lysate. (c) Luciferase activity in cell extracts from control or SIRT6 knockdown (SIRT6-KD) 293T cells transfected with a c-Jun–dependent luciferase reporter plasmid. n = 5 independent experiments. Data are presented as the mean ± s.d. RLU, relative light units. Student’s t test was applied to calculate the P value. *P < 0.001. (d) Luciferase activity in rat cardiomyocytes cotransfected with the c-Jun–dependent luciferase reporter plasmid and plasmids expressing WT SIRT6 (SIRT6-WT) or mutant SIRT6 (SIRT6-H133Y); the cells were serum starved overnight and induced with FBS or with the c-Jun activator phorbol 12-myristate 13-acetate (PMA). n = 8 independent experiments. Data are presented as the mean ± s.d. ANOVA was applied to calculate the P value. *P < 0.001. (e) ChIP analysis using a c-Jun–specific antibody to detect c-Jun occupancy at promoters of IGF signaling–related genes in WT or SIRT6 knockout mouse heart homogenates. n = 4 independent experiments. Data are presented as the mean ± s.d. *P < 0.001 compared to WT (Student’s t test). (f) ChIP analysis using a SIRT6-specific antibody to detect SIRT6 occupancy at the promoters of the indicated genes in control (WT) and c-Jun knockdown (c-Jun-KD) rat cardiomyocytes. SIRT6 occupancy at the promoters is shown relative to the background signal with IgG control antibody. *P < 0.001 (Student’s t test). n = 4 independent experiments. Data are presented as the mean ± s.d. (g) Western blots showing c-Jun knockdown in rat cardiomyocytes.
SIRT6 suppresses the transcriptional activity of c-Jun
To search for a transcription factor that could be targeted by SIRT6 and that might regulate the expression of IGF signaling–related genes, we performed an in silico analysis and identified an evolutionarily conserved c-Jun binding site in the promoters of IGF signaling–related genes (Supplementary Fig. 8). c-Jun is a stress-responsive subunit of the AP-1 transcription factor20. Supporting the potential role of c-Jun in SIRT6 function, we found marked changes in the expression levels of well-known AP-1 target genes in SIRT6 knockout skeletal muscle based on the available microarray data13 (Supplementary Fig. 9a). We found that SIRT6 was bound to AP-1 target genes by ChIP analysis (Supplementary Fig. 9b) and that SIRT6 physically interacts with c-Jun in cardiomyocytes (Fig. 5b and Supplementary Fig. 10a), consistent with the idea that SIRT6 is a regulator of c-Jun–dependent gene transcription. We next tested the effect of SIRT6 on the transcriptional activity of c-Jun, as assessed by a luciferase reporter assay using a reporter construct carrying multiple c-Jun binding sites. This assay indicated that SIRT6 knockdown enhanced the transcriptional activity of endogenous c-Jun (Fig. 5c and Supplementary Fig. 10b). Moreover, wild-type SIRT6, but not a catalytic mutant (H133Y SIRT6), attenuated the transcriptional activity of c-Jun (Fig. 5d and Supplementary Fig. 10c), suggesting that the deacetylase activity of SIRT6 is needed for attenuating c-Jun–dependent gene transcription. As assessed by ChIP analysis using a c-Jun–specific antibody, c-Jun was bound to the promoters of IGF signaling–related genes and loss of SIRT6 enhanced the binding of c-Jun to target promoters (Fig. 5e and Supplementary Fig. 10d,e), indicating that the enhanced transcriptional activity of c-Jun in SIRT6-deficient cells contributes to the increased expression of IGF signaling–related genes.
We next asked whether loss of c-Jun affects the occupancy of SIRT6 at promoters of IGF signaling–related genes. Knockdown of c-Jun significantly reduced SIRT6 occupancy at these promoters (Fig. 5f,g and Supplementary Fig. 10f), indicating that SIRT6 is specifically recruited to these promoters by forming a complex with c-Jun.
Inhibition of c-Jun or IGF-Akt signaling blocks cardiac hypertrophy
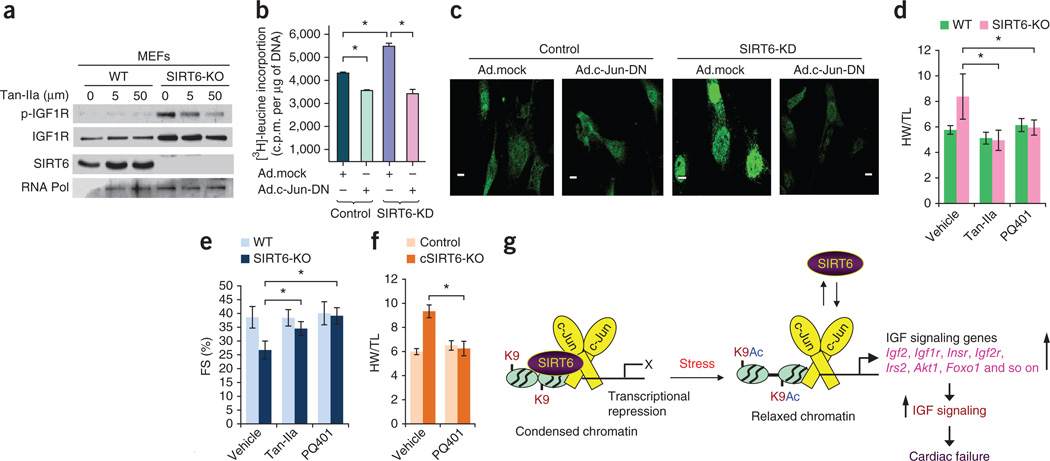
Having shown that SIRT6 deficiency activates c-Jun–dependent transcription, we next asked whether inhibition of c-Jun could block the hypertrophy of SIRT6-deficient cardiomyocytes. Inhibition of c-Jun in SIRT6 knockout MEFs by treating them with an AP-1–specific inhibitor, tanshinone IIa (Tan-IIa)21, for 24 h led to decreased expression and phosphorylation of IGF1R in a concentration-dependent manner (Fig. 6a). The compound did not have this effect in wild-type cells (Fig. 6a). We next inhibited c-Jun function using an adenovirus expressing a dominant-negative version of c-Jun, which, when infected into cardiomyocytes, blocked the activity of endogenous c-Jun (Supplementary Fig. 11a). The hypertrophic response of SIRT6-deficient cardiomyocytes, as measured by [3H]-leucine incorporation into total cellular protein, expression of ANF and sarcomere reorganization, was completely blocked by c-Jun inhibition (Fig. 6b,c and Supplementary Fig. 11b,c). These effects were associated with reduced phosphorylation of IGF1R and reduced mRNA expression of key IGF signaling–related genes, such as Igf2, Igf1r and Irs2 (Supplementary Fig. 11d,e), suggesting that the hypertrophy of SIRT6-deficient cardiomyocytes can be attributed to the increased transcriptional activity of c-Jun. Confirming these findings, we found that Tan-IIa treatment also blocked the hypertrophic response of SIRT6-deficient cardiomyocytes (Supplementary Fig. 12a,b). Administration of Tan-IIa to SIRT6 knockout mice blocked the cardiac hypertrophic response in vivo (Fig. 6d,e and Supplementary Fig. 13a–c), and this effect was associated with inhibition of Igf2 and fetal gene expression (Supplementary Fig. 13d). These data demonstrate that SIRT6 deficiency induces cardiac hypertrophy through hyperactivation of c-Jun.
Figure 6.
Inhibition of c-Jun or IGF signaling blocks hypertrophy of SIRT6-deficient hearts. (a) Western blots showing effects of the indicated concentrations of the AP-1 inhibitor Tan-IIa on IGF1R and p-IGF1R abundance in WT and SIRT6 knockout MEFs. (b) [3H]-leucine incorporation into total cellular protein in control and SIRT6 knockdown neonatal rat cardiomyocytes infected with control (Ad.mock) or c-Jun dominant-negative (Ad.c-Jun-DN) adenovirus vectors. Data are presented as the mean ± s.d. n = 6–8 independent experiments. *P < 0.001 (Student’s t test). (c) Confocal imaging of ANF in the same group of cardiomyocytes as in b. Scale bars, 10 µm. (d,e) HW/TL ratio (d) and fractional shortening (FS) (e) of WT and SIRT6 knockout mice injected with vehicle, the AP-1 inhibitor Tan-IIa or the IGF1R inhibitor PQ401 for 2 weeks. n = 6–9 mice per group. Data are presented as the mean ± s.d. *P < 0.001 (ANOVA). (f) HW/TL ratio of control (Sirt6flox/flox) and cardiac-specific SIRT6 knockout mice injected with vehicle or the IGF1R inhibitor PQ401 for 2 weeks. n = 6 mice per group. Data are presented as the mean ± s.d. *P < 0.001 (Student’s t test). (g) Under normal conditions, SIRT6 inhibits the expression of IGF signaling–related genes by deacetylating histones and repressing c-Jun activity, thereby restraining IGF signaling. Under pathological stress, cardiac SIRT6 expression is reduced, leading to increased acetylation of H3K9 (K9Ac) at the promoters of IGF signaling genes and c-Jun–mediated transcriptional activation. Increased expression of multiple IGF-Akt signaling–related genes leads to the development of cardiac hypertrophy, fibrosis and heart failure.
To obtain evidence that cardiac hypertrophy of SIRT6-deficient cells results from c-Jun–dependent hyperactivation of IGF-Akt signaling, we performed a series of rescue experiments using specific inhibitors of IGF-Akt signaling. Treatment of SIRT6-deficient cardiomyocytes with PQ401, a pharmacological inhibitor of IGF signaling, significantly (P < 0.001) blocked hypertrophy of cardiomyocytes (sarcomeric reorganization, ANF release, protein synthesis and NFAT (nuclear factor of activated T cells) promoter activity) (Supplementary Fig. 12b–e). Knockdown of IGF1R expression using siRNA had similar effects (Supplementary Fig. 14a–e). An Akt inhibitor, Akt-v (triciribine)19, also blocked the hypertrophic response of SIRT6-deficient cardiomyocytes (Supplementary Fig. 12b,d–f). To test whether inhibition of IGF signaling could block the cardiac hypertrophy of SIRT6 knockout mice in vivo, we treated these mice with PQ401 for 14 d. PQ401 treatment of SIRT6 knockout mice prevented the increase in the HW/TL ratio and the cross-sectional area of cardiomyocytes, with significant improvement in contractile function, which was associated with reduced activation of IGF-Akt signaling (Fig. 6d,e and Supplementary Fig. 15a–c). Treatment of cardiac-specific SIRT6-deleted mice with PQ401 had similar effects (Fig. 6f and Supplementary 15d,e). From these studies, we conclude that hyperactivation of IGF-Akt signaling contributes to the development of cardiac hypertrophy and heart failure in SIRT6-deficient mice.
DISCUSSION
In this study, we identify a direct link between two evolutionarily conserved nutrient-sensitive signaling pathways, the insulin/IGF and sirtuin pathways, which are widely implicated in pathophysiological conditions. Our data show that IGF signaling–related genes are negatively regulated by SIRT6 at the level of chromatin. SIRT6 deficiency enhanced H3K9 acetylation and augmented the promoter binding and transcriptional activity of c-Jun, which resulted in hyperactivation of multiple IGF signaling–related genes and the development of cardiac hypertrophy and failure.
Hyperactivation of signaling by insulin and the IGF family of growth factors has been implicated in the pathophysiology of many diseases, including cancer22, diabetes23, obesity23, musculoskeletal disease24 and neurodegenerative disease25. Dysregulation of insulin/IGF signaling has been also reported in human end-stage failing hearts26. In SIRT6-deficient hearts, most of the key components of insulin/IGF signaling pathways, including IGF1R, InsR, IGF2R, IGF2, IRS2, MAPK3 and Akt, were overexpressed, but IGF1 was not. In fact, IGF1 expression was downregulated in SIRT6 knockout hearts, consistent with previous reports in which low circulating IGF1 concentrations were found in SIRT6 knockout mice12–14. We also found increased amounts of membrane-bound IGF2 in SIRT6 knockout hearts, suggesting that membrane-associated IGF2 might drive constant IGF signaling, contributing to the development of cardiac hypertrophy, despite low cardiac expression of IGF1. IGF2 acts on multiple receptors, including, IGF1R, IGF2R and InsR, to promote the development of pathologic cardiac hypertrophy and tumorigenesis27,28. Consistent with our data, transgenic mice overexpressing IGF2 have cardiac structural changes that reflect heart failure29. We found that PTEN mRNA, a suppressor of IGF signaling, was modestly upregulated in SIRT6 knockout cardiomyocytes, raising the question of why increased PTEN expression was unable to block hyperactive IGF signaling in SIRT6 knockout hearts. From these studies, we deduce that SIRT6 has a major role in regulating the expression of IGF signaling–related genes and that dysregulation of this pathway might contribute to the pathophysiology of several diseases, including heart failure.
Mechanistically, SIRT6 blocks the expression of IGF signaling–related genes through its ability to suppress the transcriptional activity of c-Jun (Fig. 6g). c-Jun is a subunit of the transcription factor AP-1, which regulates cell proliferation, differentiation and apoptosis20. c-Jun is involved in the development of the embryonic heart30, whereas in the adult heart, c-Jun activation leads to hypertrophy and subsequent failure31–36. Transgenic mice with whole-body c-Jun overexpression develop tumors of multiple organs leading to death of the mice29. Elevated expression of c-Jun has also been reported during aging37. Growth factors and oxidative stress promote the transcriptional activity of c-Jun, whereas dietary restriction antagonizes it37,38. In our in silico analysis, we found evolutionarily conserved c-Jun binding sites in several insulin/IGF signaling–related gene promoters, suggesting that these genes are probably regulated by a c-Jun–dependent transcription mechanism. Consistent with this, a previous report has shown that c-Jun has a crucial role in the regulation of Igf2 expression6. In addition to IGF signaling–related genes, it is also possible that other c-Jun targets participate in the development of pathologic cardiac hypertrophy in SIRT6 knockout mice. In particular, as we found increased expression of many proapoptotic and prohypertrophic genes in SIRT6 knockout hearts (Trail, Bim, P27, Myc and Ctnnb1 (β-catenin)), a role of these factors in the cardiac hypertrophy of these mice cannot be excluded. However, inhibition of IGF signaling was sufficient to block the cardiac hypertrophic response in SIRT6 knockout mice, suggesting that IGF signaling has a major role in this response. Notably, we did not find upregulation of previously identified HIF-1α–targeted glycolytic genes, suggesting that SIRT6 deficiency–induced cardiac failure is not mediated through HIF-1α.
The expression of IGF2 is also regulated by genomic imprinting28. In the present study, two different findings argue against the loss of genomic imprinting as the cause of Igf2 activation in SIRT6 knockout cells. First, other genes that were upregulated in SIRT6 knockout cells, such as Igf1r, Akt, Gsk3 and Mapk3, are not controlled by genomic imprinting. Second, IGF2 activation was blocked by inhibiting c-Jun. Thus, it seems that activation of c-Jun is the mechanism responsible for SIRT6-mediated activation of IGF signaling–related genes and the development of cardiac hypertrophy.
In many previous studies, lifespan extension in response to caloric restriction has been linked with the activation of sirtuins and a reduction in IGF-Akt signaling4,6. Among the seven members of the sirtuin family, SIRT1, SIRT3 and SIRT6 have been shown to be activated in response to caloric restriction39–41. However, recent studies have shown that SIRT1 deacetylates and activates Akt and thereby promotes cardiac hypertrophy and tumorigenesis in mice42, casting doubt on the idea that SIRT1 promotes longevity. The activation of SIRT3 has been linked to an extension of lifespan in humans28. Although, the underlying mechanism of the antiaging effect of SIRT3 is not known, growing evidence suggests that SIRT3 might promote longevity by facilitating mitochondrial function42 and blocking cellular oxidative stress43. Our data show that the SIRT6 sirtuin directly controls IGF-Akt signaling, supporting the idea that SIRT6 is capable of promoting longevity. Consistent with our findings, it was recently reported that SIRT6-overexpressing transgenic male mice that had an extended lifespan had reduced IGF signaling44. From these studies, it can be concluded that SIRT6 has an important role in controlling pro-growth and pro-aging IGF-Akt signaling in mammals.
Other HDACs, particularly class II HDACs, are capable of blocking the cardiac hypertrophic response. These HDACs function by shuttling in and out of the nucleus as needed to protect the heart from hypertrophic stimuli45. This mechanism does not apply to the negative regulatory effects of SIRT6, as we found no evidence of SIRT6 export from the nucleus during the hypertrophy of cardiomyocytes. Instead, SIRT6 levels were substantially reduced in failing human and mouse hearts in which IGF-Akt signaling was hyperactivated, suggesting that loss of SIRT6 contributes to the development of heart failure. This finding raises the possibility that low levels of SIRT6 expression might be a risk factor for heart failure in humans. In summary, this study demonstrates that the nuclear sirtuin SIRT6 directly controls the expression of insulin/IGF-Akt signaling–related genes at the level of chromatin and, hence, the development of cardiac hypertrophy and heart failure.
ONLINE METHODS
Knockout mice
All animal protocols were reviewed and approved by the University of Chicago Institutional Animal Care and Use Committee. Whole-body SIRT6 knockout mice were obtained from the laboratory of F. Alt. Initially, Sirt6+/− male mice were backcrossed to 129Sv females for several generations to obtain 129Sv Sirt6+/− mice. After that, 129Sv Sirt6+/− males were crossed to C57BL6 females to obtain Sirt6-deficient mice with a 129sv/C57BL6 mixed background. Mice with loxP recombination sites flanking exons 2 and 3 of the SIRT6 gene are described elsewhere9,14. These mice were crossed with mice carrying the α-MHC–MerCreMer transgene (Jackson Laboratory; Tg(Myh6-cre/Esr1*)1Jmk/J). For cardiac specific-deletion of SIRT6, 4-hydroxytamoxifen (4-OHT) was injected into mice positive for the α-MHC–MerCreMer transgene and containing loxP sites flanking the SIRT6 gene at 4 weeks of age. Fifty milligrams of 4-OHT (H-7904, Sigma) was dissolved in 1 ml of ethanol. Approximately 4.0 ml of peanut oil (Sigma) was added to give a final concentration of 10 mg/ml. Before injection, the emulsion was sonicated, and 100 µl containing 1 mg of 4-OHT was injected intraperitoneally (i.p.) daily for 3 consecutive days. PCR genotyping was performed as described9. The cardiac phenotype of the mice was analyzed 3 months after 4-OHT injection. The phenotype of SIRT1 knockout mice has been described in our previous publication42. In all knockout mice experiments, WT littermates were used as controls. For the experiments involving mice with cardiac-restricted SIRT6 deficiency, we used both SIRT6flox/flox alone and α-MHC–Cre alone as controls. We used both sexes for experiments involving whole-body or conditional SIRT6-deficient mice and only male mice for the SIRT1 knockout experiments.
Transgenic mice
Transgenic mice with cardiac-restricted expression of Sirt6 were generated using a cDNA amplified from a plasmid encoding human SIRT6 (Addgene, plasmid 13817), which was subcloned into the α-MHC promoter vector. (a gift from J. Robbins, Cincinnati Children’s Hospital, Cincinnati, Ohio, USA). Transgenic mice expressing SIRT6 were generated in the CD1 mouse strain according to the standard procedure of the University of Chicago Transgenic facility. At 2–3 weeks of age, tail DNA was analyzed to confirm mice positive for the transgene. The following primers were used for genotyping: forward, 5′-CTTCCAGCCCTCTCTTTCTC-3′; reverse, 5′-CGGACGTACTGCTGCGTCTTACA-3′; forward, 5′-TTCACATTGCATGTGTGG-3′; reverse, 5′-TAGCCTGCGTAGTGTTGGTG-3′. The expected band sizes for endogenous and transgenic mice are 423 bp and 700 bp, respectively. The transgenic mice were backcrossed to CD-1 mice to obtain SIRT6 transgenic mice in the CD-1 background. We used females for all transgenic mice experiments. Nontransgenic littermates were used as controls.
Treatment of mice with Tan-IIa and PQ401
Tan-IIa (Sigma, T4952) was dissolved in DMSO at a concentration of 25 mg/ml, which was further diluted with peanut oil and administrated at a dose of 1 mg per kg body weight daily i.p. Similarly, PQ401 (Sigma, P0113) was also diluted with peanut oil and administrated at the dose of 1 mg per kg body weight daily i.p. The hearts of WT and SIRT6 knockout mice were analyzed after 2 weeks of continuous treatment of Tan-IIa or PQ401 treatment.
Cell culture
Cardiomyocyte cell cultures from 2-day-old neonatal rat hearts were prepared as described43,46. Adult mouse cardiac fibroblasts were prepared using standard protocol. U2OS, 3T3L, Cos7 and 293T cells were obtained from American Type Culture Collection and grown in DMEM supplemented with penicillin-streptomycin and 10% FBS. MEFs were prepared from embryonic day 18 embryos of WT 129Sv mice using standard protocols. All experiments using MEFs were performed from passage 2 to 4. Flag-tagged SIRT6 (Flag-SIRT6) and Flag-SIRT6 catalytic mutant constructs were obtained from E. Verdin’s laboratory. Mouse Sirt6-expressing plasmid construct was obtained from F. Van Gool’s laboratory. Adenovirus for recombinant human SIRT6 (1556) and Ad–CMV–c-Jun (dominant negative, DN) (1046) were purchased from Vector Biolabs. Ad.GFP, a vector expressing GFP protein; Ad.mock, empty vector. For all adenovirus infections, viruses were used at a multiplicity of infection of 100. Retroviral vectors synthesizing SIRT6 shRNA or control shRNA were generously provided by K. Chua10. The c-Jun lentiviral vectors synthesizing shRNA (sequence: 3′-CCGGACTCATGCTAACGCAGCAGTTCTCGAGAACTGCTGCGTTAGCATGAGTTTTTTG-5′ or 3′-CCGGGAAGCGCATGAGGAACCGCATCTCGAGATGCGGTTCCTCATGCGCTTCTTTTTG-5′) were purchased from Open Biosystems. For in vitro experiments, we used PQ401 and Tan-IIa at a concentration of 10 µM (unless otherwise indicated) and Akt-v at a concentration of 0.5 µM. We used Stealth siRNA targeting rat Igf1r (sense, 5′-CCUGUGAAAGUGAUGUUCUCCGUUU-3′; antisense, 5′-AAACGGAGAACAUCACUUUCACAGG-3′ (Life Technology, Inc) at a concentration of 200 nM. The methods for inducing hypertrophy of cardiomyocytes, analysis of fetal genes expression, protein synthesis and sarcomeric reorganization and luciferase reporter assays were described in our previous publication43.
Induction of hypertrophy in mice
ISO and Ang II were infused chronically by implanting osmotic minipumps (ALZET, model 2002) into the peritoneal cavity of mice as described in our previous publication43. Pressure overload hypertrophy was induced by TAC of the ascending aorta of mice, as described elsewhere47. The measurement of cardiac hypertrophy, myocyte cross-sectional area, fibrosis and the expression of hypertrophic marker genes were carried out essentially as described earlier43. Transmission electron microscopy was performed with the help of Electron Microscopy Core Facility at University of Chicago as per standard protocols46.
Human heart samples
Human heart samples were obtained from the University of Chicago cardiac transplant program. Informed consent was obtained from all patients participating in this study. All procedures involving human tissue use were approved by the University of Chicago Institutional Review Board. Control samples were obtained intraoperatively from nonfailing hearts undergoing ventricular corrective surgery or from donor dysfunctional hearts. Failing heart specimens were obtained from diseased hearts that were removed during orthotopic heart transplantation.
Antibodies
SIRT6-specific antibody (Sigma-Aldrich, S4322) was used for analyzing endogenous SIRT6 by western blot, ChIP, coimmunoprecipitation and immunostaining in human and mouse samples. SIRT6-specific antibody (Novus Biological, NB100-2522) was used for the analysis of mouse SIRT6 in western blotting. Antibodies to SIRT1 (04-1557,#07-131), c-Jun (06-225), c-Fos (06-341) and p27 (06-445) were purchased from Millipore. Antibodies to H3 (ab1791), acetylated H3K9 (ab10812), Foxo4 (ab12075), Flag tag (ab1162), Foxo1 (ab12161) and Bax (ab7977) were purchased from Abcam. Antibodies to tubulin (sc-32293), actinin (sc-7454), RNA polymerase (sc-5943), FasL (sc-6237), Bim (sc-11425), TRAIL (sc-6079), tropomyosin (sc18174), p38 (sc-7972), IGF2 (sc-5622), Foxo3a (sc-34897), Akt (sc-1618, sc-5298), p-Akt Ser473 (sc-7985-R), ERK1/2 (sc-98), tuberous sclerosis 2 (TSC2) (sc-893), p-TSC2 (sc-32839), Myc (sc-40, sc-41, sc-789) and β-catenin (sc-1496) were purchased from Santa Cruz. Antibodies to SMA (A5228), SMM (M7786), actinin (A7811), vimentin (V6630), c-Myc (C3956) and Flag beads (A2220) were obtained from Sigma-Aldrich. Antibodies to IGF1R (3018), p-IGF1R (3918, 6113, 4568, 3024), p-InsR (3023), p-Akt Ser473 (4060), p-Akt Thr308 (4056, 5106), p–GSK3-β (9322), p–GSK3-α/β (9331), c-Jun (9165), p-S6RP (2211), p38 MAPK (9212), p-p38 (9216), p-mTOR (2971, 2974), mTOR (2983) and p-ERK1/2 (9101) were purchased from Cell Signaling. GSK3-α/β–specific antibody (2199) was obtained from Epitomics.
Immunoblots and ChIP analyses
Immunoprecipitation and immunoblot analyses were done using standard protocols, as described previously43. Chromatin immunoprecipitation was performed using a commercial kit (EZ-ChIP Kit, Millipore) following the manufacturer’s instructions. In vivo ChIP experiments with heart samples were performed using a previously described protocol48. The primer sequences used for ChIP and PCR analysis are given in Supplementary Table 2.
Bioinformatic analyses
The transcription factor binding site analysis was done using TRANSFAC software (Biobase GmbH), and evolutionary conservation analysis was carried out using UCSC Genome Browser. In Supplementary Figure 8, nucleotide sequences in bold denote AP-1/c-Jun consensus binding sites, and the numbers below denote distance (bp) from the transcription start site for the respective mouse gene.
Statistical analyses
All values are expressed as the mean ± s.d. Statistical differences among groups were determined using either Student’s t test (for two groups) or one-way ANOVA (for more than two groups) using Graph-Pad Prism Software.
Supplementary Material
Acknowledgments
We thank F. Alt, Harvard Medical School, for providing SIRT6 knockout mice, E. Verdin, University of California, San Francisco, for providing Flag-SIRT6 wild-type and mutant plasmids, F. VanGool, Institut de Biologie, Universite Libre de Bruxelles, Gosselies, Belgium, for providing mouse-SIRT6 expression plasmid and K. Chua, Stanford University, for providing SIRT6 retroviral vectors and SIRT6 knockout MEFs. The α-MHC promoter vector used to make cardiac-specific SIRT6 transgenic mice was provided by J. Robbins, University of Cincinnati. This study was supported by US National Institutes of Health grants RO1 HL-117041, HL-83423 and 111455 to M.P.G. N.R.S. was supported by a postdoctoral fellowship from the American Heart Association.
Footnotes
Note: Supplementary information is available in the online version of the paper.
AUTHOR CONTRIBUTIONS
N.R.S. and M.P.G. designed the study and wrote the manuscript. N.R.S. performed the majority of experiments. P.V. performed the ChIP experiments. L.Z. analyzed tissue microarray data. G.K. did echocardiography of mice. S.S. analyzed human samples and performed electron microscopy. V.P. performed in silico analysis and identified c-Jun target genes. V.B.P. performed experiments with IGF1R inhibitors. P.V.R. generated transgenic mice and did in vitro hypertrophy experiments. M.G. analyzed cardiac fetal gene program and cell death markers. V.J. provided human cardiac tissue samples during surgery. J.M.C. planned and supervised ChIP experiments. C.-X.D. provided Sirt6loxP mice. D.B.L. provided heart samples from SIRT6-MCK-Cre mice and participated in discussing experiments. R.M. provided microarray data and discussed the whole study multiple times. M.P.G. coordinated with different investigators, supervised the whole study and generated the final draft of the manuscript.
COMPETING FINANCIAL INTERESTS
The authors declare no competing financial interests.
References
- 1.Lloyd-Jones D, et al. Heart disease and stroke statistics—2010 update: a report from the American Heart Association. Circulation. 2010;121:e46–e215. doi: 10.1161/CIRCULATIONAHA.109.192667. [DOI] [PubMed] [Google Scholar]
- 2.Donmez G, Guarente L. Aging and disease: connections to sirtuins. Aging Cell. 2010;9:285–290. doi: 10.1111/j.1474-9726.2010.00548.x. [DOI] [PubMed] [Google Scholar]
- 3.Kahn AJ. Development, aging, and life duration: effects of nutrient restriction. Am. J. Clin. Nutr. 1972;25:822–828. doi: 10.1093/ajcn/25.8.822. [DOI] [PubMed] [Google Scholar]
- 4.Fontana L, Partridge L, Longo VD. Extending healthy life span—from yeast to humans. Science. 2010;328:321–326. doi: 10.1126/science.1172539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Weiss EP, Fontana L. Caloric restriction: powerful protection for the aging heart and vasculature. Am. J. Physiol. Heart Circ. Physiol. 2011;301:H1205–H1219. doi: 10.1152/ajpheart.00685.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Haigis MC, Guarente LP. Mammalian sirtuins—emerging roles in physiology, aging, and calorie restriction. Genes Dev. 2006;20:2913–2921. doi: 10.1101/gad.1467506. [DOI] [PubMed] [Google Scholar]
- 7.Kanfi Y, et al. SIRT6 protects against pathological damage caused by diet-induced obesity. Aging Cell. 2010;9:162–173. doi: 10.1111/j.1474-9726.2009.00544.x. [DOI] [PubMed] [Google Scholar]
- 8.Kawahara TL, et al. SIRT6 links histone H3 lysine 9 deacetylation to NF-κB–dependent gene expression and organismal life span. Cell. 2009;136:62–74. doi: 10.1016/j.cell.2008.10.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kim HS, et al. Hepatic-specific disruption of SIRT6 in mice results in fatty liver formation due to enhanced glycolysis and triglyceride synthesis. Cell Metab. 2010;12:224–236. doi: 10.1016/j.cmet.2010.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Michishita E, et al. SIRT6 is a histone H3 lysine 9 deacetylase that modulates telomeric chromatin. Nature. 2008;452:492–496. doi: 10.1038/nature06736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Michishita E, Park JY, Burneskis JM, Barrett JC, Horikawa I. Evolutionarily conserved and nonconserved cellular localizations and functions of human SIRT proteins. Mol. Biol. Cell. 2005;16:4623–4635. doi: 10.1091/mbc.E05-01-0033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mostoslavsky R, et al. Genomic instability and aging-like phenotype in the absence of mammalian SIRT6. Cell. 2006;124:315–329. doi: 10.1016/j.cell.2005.11.044. [DOI] [PubMed] [Google Scholar]
- 13.Zhong L, et al. The histone deacetylase Sirt6 regulates glucose homeostasis via Hif1α. Cell. 2010;140:280–293. doi: 10.1016/j.cell.2009.12.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Xiao C, et al. SIRT6 deficiency results in severe hypoglycemia by enhancing both basal and insulin-stimulated glucose uptake in mice. J. Biol. Chem. 2010;285:36776–36784. doi: 10.1074/jbc.M110.168039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Holzenberger M, et al. IGF-1 receptor regulates lifespan and resistance to oxidative stress in mice. Nature. 2003;421:182–187. doi: 10.1038/nature01298. [DOI] [PubMed] [Google Scholar]
- 16.Matsui T, et al. Phenotypic spectrum caused by transgenic overexpression of activated Akt in the heart. J. Biol. Chem. 2002;277:22896–22901. doi: 10.1074/jbc.M200347200. [DOI] [PubMed] [Google Scholar]
- 17.Matsui T, Nagoshi T, Rosenzweig A. Akt and PI 3-kinase signaling in cardiomyocyte hypertrophy and survival. Cell Cycle. 2003;2:220–223. [PubMed] [Google Scholar]
- 18.Kostin S, Hein S, Arnon E, Scholz D, Schaper J. The cytoskeleton and related proteins in the human failing heart. Heart Fail. Rev. 2000;5:271–280. doi: 10.1023/A:1009813621103. [DOI] [PubMed] [Google Scholar]
- 19.Sundaresan NR, et al. The deacetylase SIRT1 promotes membrane localization and activation of Akt and PDK1 during tumorigenesis and cardiac hypertrophy. Sci. Signal. 2011;4:ra46–ra58. doi: 10.1126/scisignal.2001465. [DOI] [PubMed] [Google Scholar]
- 20.Hess J, Angel P, Schorpp-Kistner M. AP-1 subunits: quarrel and harmony among siblings. J. Cell Sci. 2004;117:5965–5973. doi: 10.1242/jcs.01589. [DOI] [PubMed] [Google Scholar]
- 21.Chiu YC, et al. Peptidoglycan enhances IL-6 production in human synovial fibroblasts via TLR2 receptor, focal adhesion kinase, Akt, and AP-1–dependent pathway. J. Immunol. 2009;183:2785–2792. doi: 10.4049/jimmunol.0802826. [DOI] [PubMed] [Google Scholar]
- 22.Gallagher EJ, LeRoith D. The proliferating role of insulin and insulin-like growth factors in cancer. Trends Endocrinol. Metab. 2010;21:610–618. doi: 10.1016/j.tem.2010.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gatenby VK, Kearney MT. The role of IGF-1 resistance in obesity and type 2 diabetes-mellitus–related insulin resistance and vascular disease. Expert Opin. Ther. Targets. 2010;14:1333–1342. doi: 10.1517/14728222.2010.528930. [DOI] [PubMed] [Google Scholar]
- 24.Glass DJ. PI3 kinase regulation of skeletal muscle hypertrophy and atrophy. Curr. Top. Microbiol. Immunol. 2010;346:267–278. doi: 10.1007/82_2010_78. [DOI] [PubMed] [Google Scholar]
- 25.Freude S, Schilbach K, Schubert M. The role of IGF-1 receptor and insulin receptor signaling for the pathogenesis of Alzheimer’s disease: from model organisms to human disease. Curr. Alzheimer Res. 2009;6:213–223. doi: 10.2174/156720509788486527. [DOI] [PubMed] [Google Scholar]
- 26.Haq S, et al. Differential activation of signal transduction pathways in human hearts with hypertrophy versus advanced heart failure. Circulation. 2001;103:670–677. doi: 10.1161/01.cir.103.5.670. [DOI] [PubMed] [Google Scholar]
- 27.Chu CH, et al. Activation of insulin-like growth factor II receptor induces mitochondrial-dependent apoptosis through Gαq and downstream calcineurin signaling in myocardial cells. Endocrinology. 2009;150:2723–2731. doi: 10.1210/en.2008-0975. [DOI] [PubMed] [Google Scholar]
- 28.Chao W, D’Amore PA. IGF2: epigenetic regulation and role in development and disease. Cytokine Growth Factor Rev. 2008;19:111–120. doi: 10.1016/j.cytogfr.2008.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zaina S, et al. Shortened life span, bradycardia, and hypotension in mice with targeted expression of an Igf2 transgene in smooth muscle cells. Endocrinology. 2003;144:2695–2703. doi: 10.1210/en.2002-220944. [DOI] [PubMed] [Google Scholar]
- 30.Eferl R, et al. Functions of c-Jun in liver and heart development. J. Cell Biol. 1999;145:1049–1061. doi: 10.1083/jcb.145.5.1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schunkert H, Jahn L, Izumo S, Apstein CS, Lorell BH. Localization and regulation of c-fos and c-jun protooncogene induction by systolic wall stress in normal and hypertrophied rat hearts. Proc. Natl. Acad. Sci. USA. 1991;88:11480–11484. doi: 10.1073/pnas.88.24.11480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Iwaki K, Sukhatme VP, Shubeita HE, Chien KR. α- and β-adrenergic stimulation induces distinct patterns of immediate early gene expression in neonatal rat myocardial cells. fos/jun expression is associated with sarcomere assembly; Egr-1 induction is primarily an α 1–mediated response. J. Biol. Chem. 1990;265:13809–13817. [PubMed] [Google Scholar]
- 33.Takemoto Y, et al. Increased JNK, AP-1 and NF-κB DNA binding activities in isoproterenol-induced cardiac remodeling. J. Mol. Cell. Cardiol. 1999;31:2017–2030. doi: 10.1006/jmcc.1999.1033. [DOI] [PubMed] [Google Scholar]
- 34.Nadruz W, Jr, Corat MA, Marin TM, Guimaraes Pereira GA, Franchini KG. Focal adhesion kinase mediates MEF2 and c-Jun activation by stretch: role in the activation of the cardiac hypertrophic genetic program. Cardiovasc. Res. 2005;68:87–97. doi: 10.1016/j.cardiores.2005.05.011. [DOI] [PubMed] [Google Scholar]
- 35.Reiss K, et al. ANG II receptors, c-myc, and c-jun in myocytes after myocardial infarction and ventricular failure. Am. J. Physiol. 1993;264:H760–H769. doi: 10.1152/ajpheart.1993.264.3.H760. [DOI] [PubMed] [Google Scholar]
- 36.Freire G, Ocampo C, Ilbawi N, Griffin AJ, Gupta M. Overt expression of AP-1 reduces α myosin heavy chain expression and contributes to heart failure from chronic volume overload. J. Mol. Cell Cardiol. 2007;43:465–478. doi: 10.1016/j.yjmcc.2007.07.046. [DOI] [PubMed] [Google Scholar]
- 37.Kim HJ, et al. Modulation of redox-sensitive transcription factors by calorie restriction during aging. Mech. Ageing Dev. 2002;123:1589–1595. doi: 10.1016/s0047-6374(02)00094-5. [DOI] [PubMed] [Google Scholar]
- 38.Jung KJ, et al. Effect of short term calorie restriction on pro-inflammatory NF-κB and AP-1 in aged rat kidney. Inflamm. Res. 2009;58:143–150. doi: 10.1007/s00011-008-7227-2. [DOI] [PubMed] [Google Scholar]
- 39.Cohen HY, et al. Calorie restriction promotes mammalian cell survival by inducing the SIRT1 deacetylase. Science. 2004;305:390–392. doi: 10.1126/science.1099196. [DOI] [PubMed] [Google Scholar]
- 40.Kanfi Y, et al. Regulation of SIRT6 protein levels by nutrient availability. FEBS Lett. 2008;582:543–548. doi: 10.1016/j.febslet.2008.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hirschey MD, et al. SIRT3 regulates mitochondrial fatty-acid oxidation by reversible enzyme deacetylation. Nature. 2010;464:121–125. doi: 10.1038/nature08778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sundaresan NR, et al. The deacetylase SIRT1 promotes membrane localization and activation of Akt and PDK1 during tumorigenesis and cardiac hypertrophy. Sci. Signal. 2011;4:ra46. doi: 10.1126/scisignal.2001465. [DOI] [PubMed] [Google Scholar]
- 43.Sundaresan NR, et al. Sirt3 blocks the cardiac hypertrophic response by augmenting Foxo3a-dependent antioxidant defense mechanisms in mice. J. Clin. Invest. 2009;119:2758–2771. doi: 10.1172/JCI39162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kanfi Y, et al. The sirtuin SIRT6 regulates lifespan in male mice. Nature. 2012;483:218–221. doi: 10.1038/nature10815. [DOI] [PubMed] [Google Scholar]
- 45.Zhang CL, et al. Class II histone deacetylases act as signal-responsive repressors of cardiac hypertrophy. Cell. 2002;110:479–488. doi: 10.1016/s0092-8674(02)00861-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sundaresan NR, Samant SA, Pillai VB, Rajamohan SB, Gupta MP. SIRT3 is a stress-responsive deacetylase in cardiomyocytes that protects cells from stress-mediated cell death by deacetylation of Ku70. Mol. Cell. Biol. 2008;28:6384–6401. doi: 10.1128/MCB.00426-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pillai JB, et al. Activation of SIRT1, a class III histone deacetylase, contributes to fructose feeding-mediated induction of the α-myosin heavy chain expression. Am. J. Physiol. Heart Circ. Physiol. 2008;294:H1388–H1397. doi: 10.1152/ajpheart.01339.2007. [DOI] [PubMed] [Google Scholar]
- 48.Wu CL, Kandarian SC, Jackman RW. Identification of genes that elicit disuse muscle atrophy via the transcription factors p50 and Bcl-3. PLoS ONE. 2011;6:e16171. doi: 10.1371/journal.pone.0016171. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.