SUMMARY
To analyze cellular reprogramming at the single-cell level, mass cytometry was used to simultaneously measure markers of pluripotency, differentiation, cell-cycle status, and cellular signaling throughout the reprogramming process. Time-resolved progression analysis of the resulting data sets was used to construct a continuous molecular roadmap for three independent reprogramming systems. Although these systems varied substantially in Oct4, Sox2, Klf4, and c-Myc stoichiometry, they presented a common set of reprogramming landmarks. Early in the reprogramming process, Oct4highKlf4high cells transitioned to a CD73highCD104highCD54low partially reprogrammed state. Ki67low cells from this intermediate population reverted to a MEF-like phenotype, but Ki67high cells advanced through the M-E-T and then bifurcated into two distinct populations: an ESC-like NanoghighSox2highCD54high population and a mesendoderm-like NanoglowSox2lowLin28high CD24highPDGFR-αhigh population. The methods developed here for time-resolved, single-cell progression analysis may be used for the study of additional complex and dynamic systems, such as cancer progression and embryonic development.
INTRODUCTION
Reprogramming somatic cells to a pluripotent state by forced expression of transcription factors is a dynamic process. How a somatic cell successfully undergoes this transition is poorly understood because low efficiencies, long latency times, and asynchronous progression impede molecular analysis (Hanna et al., 2009; Wernig et al., 2008). Characterization of bulk populations over time has given insight into how entire reprogramming populations progress (Li et al., 2010; Mikkelsen et al., 2008; Samavarchi-Tehrani et al., 2010; Soufi et al., 2012), but as most cells undergoing this process fail to reprogram, bulk analyses of such processes are necessarily biased toward measurement of unproductive reprogramming events.
To address these concerns, several groups have sought to identify and characterize productive reprogramming populations. An early role for transgene stoichiometry was deduced from transgene integrations in induced pluripotent stem cells (iPSCs) and by sorting fibroblasts according to transgene expression levels (Papapetrou et al., 2009; Wernig et al., 2008). Sox2low, Oct4high, Klf4high was found to be an optimal combination and was further verified with polycistronic constructs expressing different transgene stoichiometries (Carey et al., 2009). Single-cell time-lapse imaging analysis revealed an early proliferation phenotype (Koche et al., 2011; Smith et al., 2010). Early work suggested the progression of reprogramming states with sequential acquisition of the pluripotency markers alkaline phosphatase, SSEA1, Nanog, and Oct4 (Stadtfeld et al., 2008). Additionally, repression of the fibroblast marker Thy1 and loss of retroviral expression was observed to occur early in the process. Characterization of these states suggested two waves of reprogramming occur with the first being mediated by c-Myc and Klf4 and the second by Oct4, Sox2, and Klf4 (Polo et al., 2012).
Stable partially reprogrammed lines have also been isolated and characterized (Chen et al., 2013; Ichida et al., 2009; Meissner et al., 2007; Mikkelsen et al., 2008; Polo et al., 2012; Sridharan et al., 2009; Theunissen et al., 2011; Wernig et al., 2008). These partially reprogrammed cells arise late in the process but prior to the acquisition of pluripotency and can be derived from multiple reprogramming populations including fibroblasts, neural stem cells, and B cells (Mikkelsen et al., 2008; Theunissen et al., 2011; Wernig et al., 2008). Morphologically they resemble iPSCs, but have not acquired pluripotency as shown by their inability to form teratomas and dependence on the reprogramming transgenes (Wernig et al., 2008). Although the majority of these cells do not acquire pluripotency under standard conditions, they can be pushed to a pluripotent state with chemical treatment of 5-aza-cytidine and vitamin C or by overexpression of Nanog, suggesting that they resemble an intermediate state where roadblocks inhibit pluripotency acquisition (Mikkelsen et al., 2008; Theunissen et al., 2011).
Although characterization of enriched intermediates has been useful, analysis is still highly reliant on bulk populations where heterogeneity is still prevalent. Recently, Buganim et al. (2012) attempted to address this by conducting single-cell mRNA analysis to identify an early stochastic phase of reprogramming followed by a late deterministic phase correlated with Sox2 expression. Despite the importance of the findings in this study, its conclusions may have been limited by the relatively small sample size of 96 cells that were assayed at each time point, coupled with low reprogramming efficiencies where only two in 100 cells may successfully reprogram. To this end, we have characterized the reprogramming process by single-cell mass cytometry, a flow cytometry technique that uses rare earth metal isotopes for antibody labeling and detection (Bandura et al., 2009). Mass cytometry produces results that are essentially identical to conventional fluorescent flow cytometry (Bendall et al., 2011), but allows over 40 different parameters to be measured simultaneously at ~500 cells per second. Using mass cytometry, we have analyzed three different reprogramming lines during the first 3–4 weeks of reprogramming. Time-resolved, high-dimensional progression analysis of the mass cytometry data sets facilitated construction of continuous molecular maps of reprogramming, which should serve as valuable resources for the continued development of improved iPSC reprogramming protocols.
RESULTS
Single-Cell Analysis of Cellular Reprogramming by Mass Cytometry
Three cellular reprogramming systems were used for this study: (1) Oct4-GFP primary mouse embryonic fibroblasts (MEFs) that express GFP from the endogenous Oct4 locus (Lengner et al., 2007), (2) Nanog-Neo secondary MEFs that express the neomycin resistance gene from the endogenous Nanog locus (Wernig et al., 2008), and (3) Nanog-GFP secondary MEFs that expresses GFP from the endogenous Nanog locus (Wernig et al., 2008). To obtain a comprehensive view of cellular reprogramming in the three MEF systems, cell samples were collected on every other day of the reprogramming time course. The reprogramming regimen consisted of Dox induction for 16 days followed by Dox withdrawal and the addition of LIF (Figure 1A). Additional samples were collected on days 1 and 17, as these days represent points of significant transition in the reprogramming process, immediately after Dox induction and Dox withdrawal, respectively. Samples were collected over 20 days for the primary Oct4-GFP MEFs, and the time course was extended to 30 days for the secondary Nanog-Neo and Nanog-GFP MEFs because the reprogramming process was found to be slower in these systems.
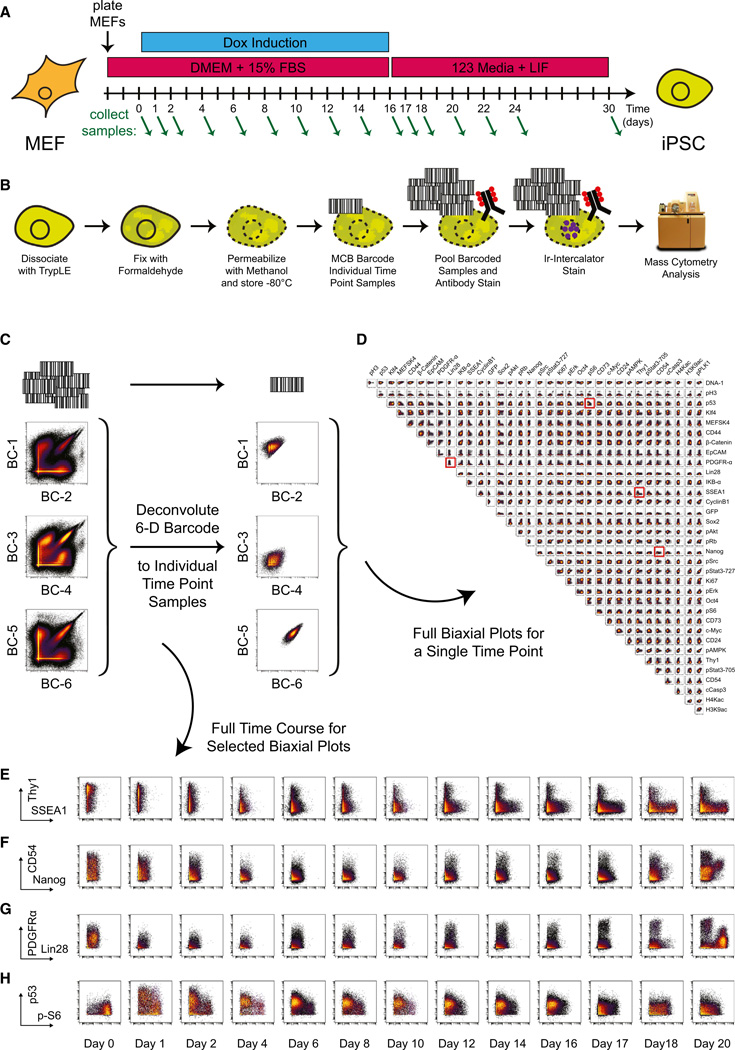
Figure 1. Time Course Analysis of iPS Cell Reprogramming by Mass Cytometry.
(A) Tissue culture conditions for time course analysis of iPS cell reprogramming. Green arrows indicate sample collection time points.
(B) Schematic of sample collection and processing for mass cytometric analysis.
(C) Cell multiplexing and deconvolution with a 6-metal MCB scheme.
(D) All pairwise biaxial plots for antibody-measured parameters from a single deconvoluted cell sample: Oct4-GFP primary MEF infection, day 16. Plots highlighted in red are shown over the full reprogramming time course in (E)–(H).
(E–H) Biaxial plots covering the full reprogramming time course for the following markers: (E) Thy1 × SSEA1, (F) CD54 × Nanog, (G) PDGFRα × Lin28, and (H) p53 × phospho-S6.
See also Figure S1 and Table S1.
At each collection time point, cell cultures were dissociated into a single-cell suspension, permeabilized (Krutzik et al., 2005), and then multiplexed by Mass-tag Cell Barcoding (Bodenmiller et al., 2012) before antibody staining (Table S1) and mass cytometry analysis (Bendall et al., 2011) (Figure 1B). A six-metal barcoding scheme was used to encode 20 unique Mass-tag barcodes (Figure 1C) (Zunder et al., 2015), allowing the entire time course for each MEF system to be pooled and stained within a single tube, eliminating tube-to-tube variability in antibody staining and minimizing the effect of variable instrument sensitivity. After mass cytometry measurement of the multiplexed samples, barcode deconvolution was performed to recover individual samples from the pooled data set (Figure 1D).
Over the course of reprogramming, the MEF marker Thy1 decreased and the pluripotency marker SSEA1 increased (Figure 1E), in agreement with previous studies (Stadtfeld et al., 2008). Elevated Nanog expression was not observed until after release from Dox-induced transgene expression on day 16 and correlated with expression of ICAM-1/CD54 (Figure 1F), a marker associated with successful cellular reprogramming (O’Malley et al., 2013). PDGFR-α was expressed by MEFs, depressed during the initial stages of reprogramming, and gradually returned during the later stages of reprogramming. One late-stage cell population expressed PDGFR-α at a higher level than seen in the uninduced MEFs, and another Lin28-expressing population expressed PDGFR-α at an intermediate level (Figure 1G). Total protein levels of the tumor suppressor protein p53, which has been shown to inhibit cellular reprogramming (Hong et al., 2009; Kawamura et al., 2009; Li et al., 2009; Marió n et al., 2009; Utikal et al., 2009), increased rapidly after transgene induction, and phosphorylation levels of the ribosomal protein S6, a downstream marker of mTOR activity and translational control (Dufner and Thomas, 1999) decreased rapidly (Figure 1H), indicating a stress-related response to forced expression of the four transcription factors (Feng et al., 2005).
Hierarchical clustering identified MEF-like markers that decreased over time, transitional markers that appeared at intermediate time points, and ESC-like markers that appeared during the latest stages of reprogramming (Figures S1A–S1D). Replicate analysis (Figure S1E) indicated these observed trends are reproducible. While hierarchical clustering provides a useful overview of the data sets, this approach does not take advantage of the single-cell nature of the data and is not well suited for analysis of low abundance populations. Therefore, additional methods were employed to better exploit the single-cell nature of the three reprogramming time course data sets.
High-Dimensional Analysis of Cellular Reprogramming by Force-Directed Layout of Clustered Cells
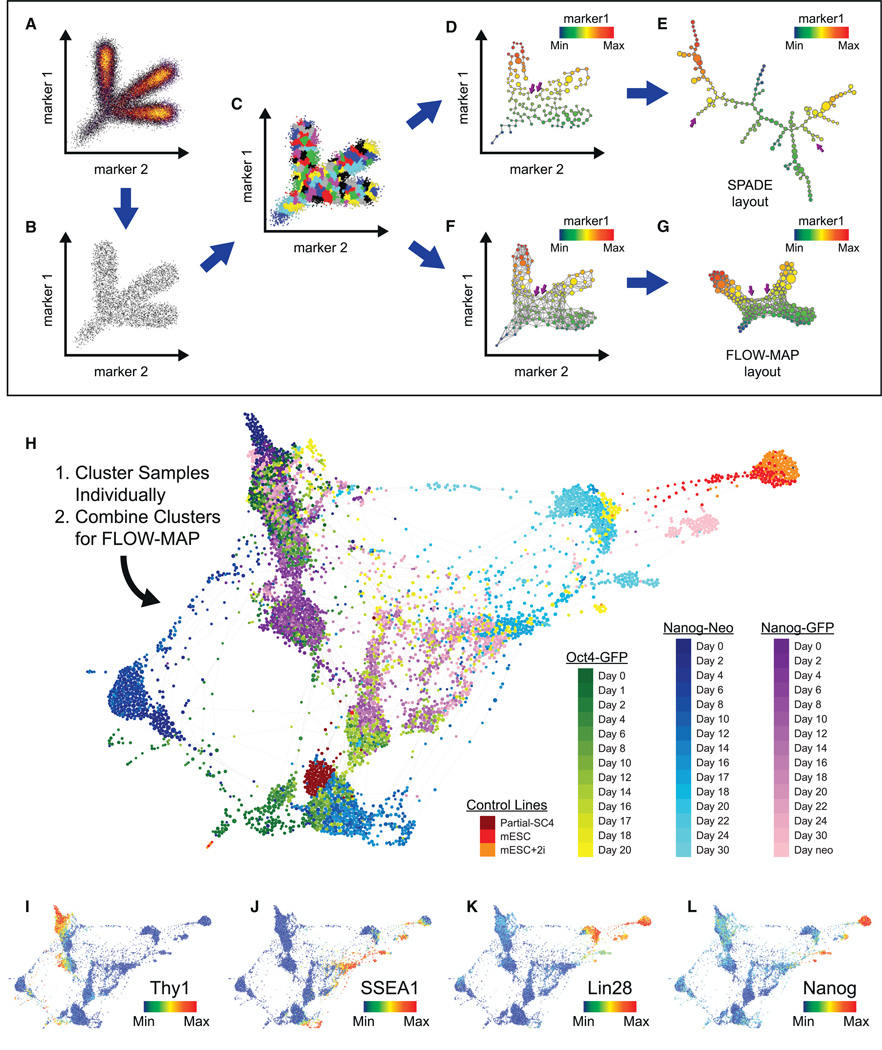
Spanning-tree progression analysis of density-normalized events (SPADE) has proven useful in identifying relationships between cell types in high-dimensional space, by downsampling to uniform density (Figures 2A and 2B), hierarchical clustering (Figure 2C), and connecting the resulting clusters with a minimum spanning tree (MST) (Figure 2D) that can be visualized in two dimensions (Figure 2E) to reveal high-dimensional relationships between cell types (Bendall et al., 2011; Linderman et al., 2012; Qiu et al., 2011). The MST used by SPADE is susceptible to overfitting the data and is not robust to local variation (Figures 2D and 2E). To improve the ability of the SPADE algorithm to robustly identify high-dimensional relationships between cell types, the MST was replaced with a more highly connected graph structure, where the number of connections between cell clusters is determined by local density (Figure 2F). This new graph structure is then employed to produce a force-directed layout of a weighted graph containing multidimensional agglomeratively clustered points (FLOW-MAP) plot. The FLOW-MAP layout of cell clusters is more reproducible than a MST-derived layout, because the underlying graph structure is highly connected and therefore less susceptible to local edge and cluster variability (Figure 2G).
Figure 2. Force-Directed Layout of MEF Reprogramming SPADE Clusters.
(A–G) Schematic of the SPADE algorithm and modified FLOW-MAP layout. A 2D synthetic example data set (A) is first downsampled to a uniform density in ndimensional space (B), clustered hierarchically, and then upsampled (C), after which the cell clusters are connected by a minimum spanning tree, shown with the true 2D coordinates (D) and minimum spanning tree-derived layout (E), or alternatively connected by a FLOW-MAP graph, shown with the true 2D coordinates (F) and FLOW-MAP-derived layout (G). Two clusters that are close in 2D space, far apart on the minimum spanning tree layout and correctly placed adjacent to one another with the FLOW-MAP layout are indicated by purple arrows.
(H) Combined FLOW-MAP analysis of the full time course data sets for the Oct4-GFP, Nanog-GFP, and Nanog-Neo reprogramming systems. Hierarchical clustering was performed on each sample/time point individually using the following markers: PDGFRα, CD24, CD44, CD54, CD73, EpCAM, H3K9ac, Ki67, Klf4, Lin28, MEFSK4, Nanog, Oct4, SSEA1, Sox2, Thy1, and c-Myc to produce 500 clusters per sample. The cell clusters from each sample were then combined into a single FLOW-MAP graph layout and colored by cell system and time point as indicated.
(I–L) The FLOW-MAP layout from Figure 4H, colored to indicate the protein expression level of the specified markers: (I) Thy1, (J) SSEA1, (K) Lin28, and (L) Nanog.
See also Figure S2.
To facilitate comparison between the three reprogramming systems, each timed sample was clustered individually and then combined into a single FLOW-MAP graph (Figure 2H). Cell clusters from two mESC samples and a partially reprogrammed iPSC line were also included as reference points. As expected, the three MEF systems occupied a similar high-dimensional space before Dox induction, characterized by high expression of MEF markers such as Thy1 (Figure 2I), PDGFRα, and MEFSK4 (Figure S2). Late in the reprogramming process, all three culture systems contained ESC-like cell populations that expressed high levels of non-transgene markers of pluripotency such as SSEA1, Lin28, and Nanog (Figures 2J–2L). These ESC-like populations formed distinct groups in the combined FLOW-MAP layout that may represent metastable pluripotent states (Hayashi et al., 2008) or may be unique stages on the path to a pluripotent “ground state” (Ying et al., 2008). These populations vary mainly between high-level and mid-level expression of pluripotency markers and group closely in the graph with the mESC control samples.
Time-Resolved Progression Analysis of iPS Cell Reprogramming
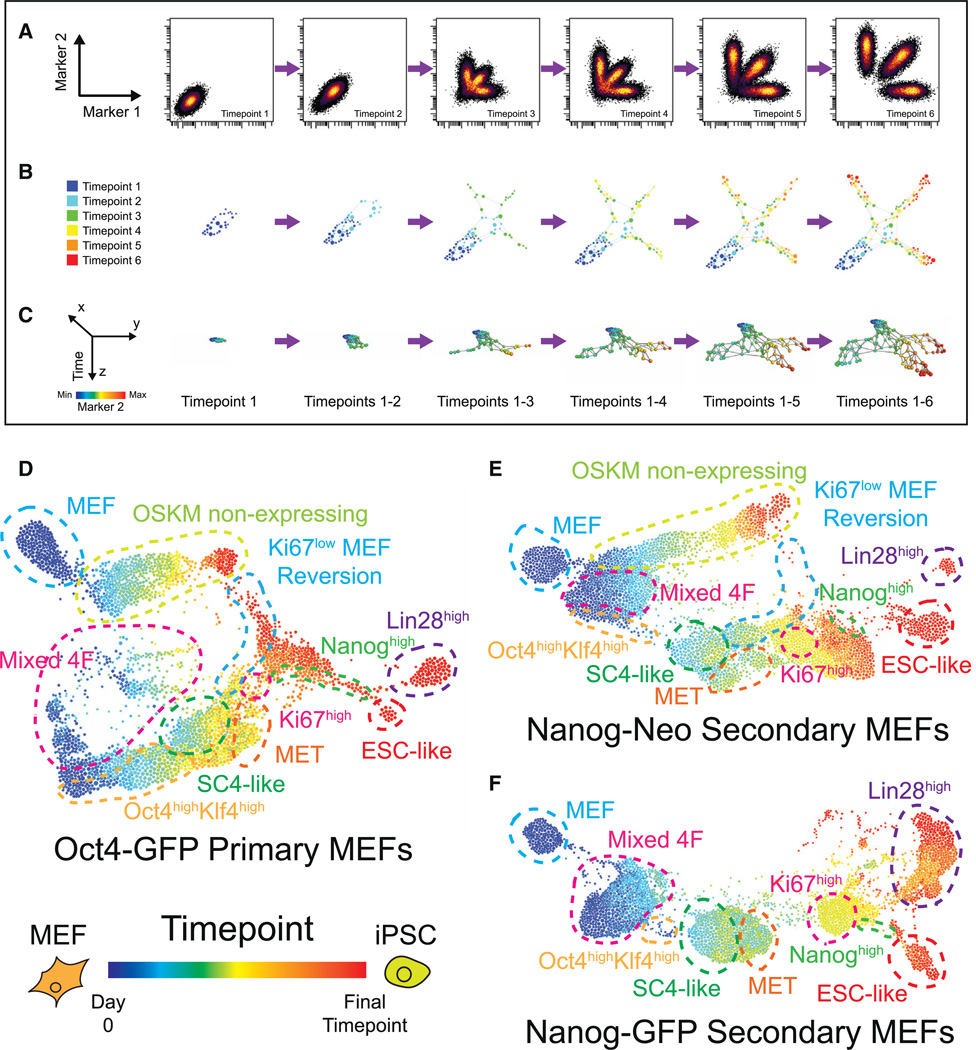
SPADE and the FLOW-MAP algorithm draw connections between populations that are similar in n-dimensional space, but cannot utilize the temporal information present in time course data sets. To exploit this temporal information, the FLOW-MAP algorithm was extended to include multiple graph drawing steps, where cell clusters are added to the graph sequentially for each time point (Figures 3A, 3B, and S3A–S3C). In this scheme, only cell clusters that reside in identical or adjacent time points may be connected in the final FLOW-MAP graph. Display of the resulting x–y layouts with time on the z axis in a 3D environment such as PyMOL (http://www.pymol.org) (Figure 3C) allows simultaneous visualization of experimental time point along with additional markers (Supplemental Files 1, 2, and 3). Time-resolved FLOW-MAP analysis revealed several similarities between the Oct4-GFP, Nanog-Neo, and Nanog-GFP MEF reprogramming systems (Figures 3D–3F), as well as two additional reprogramming time course experiments (Figures S3D and S3E).
Figure 3. Time-Resolved Progression Analysis of iPS Cell Reprogramming.
(A–C) Schematic of the FLOW-MAP algorithm modified for time-ordered data sets. A 2D synthetic example data set (A) is downsampled and clustered individually as in Figures 2A–2C and then clusters from each time point are added to the FLOW-MAP graph sequentially (B), shown here in the final force-directed layout position. 3D representation with the FLOW-MAP layout on the x–y axes and sequential time points on the z axis allows improved visualization of the relationship between marker intensity (synthetic marker 2 is shown) and time (C).
(D–F) Time-resolved FLOW-MAP analysis of the three MEF reprogramming systems: Oct4-GFP primary MEFs (D), Nanog-GFP secondary MEFs (E), and Nanog-Neo secondary MEFs (F). Hierarchical clustering (200 clusters per sample) and FLOW-MAP graph construction were performed using 18 marker expression levels: Oct4, Sox2, Klf4, c-Myc, Nanog, GFP, Lin28, SSEA1, EpCAM, CD54, CD73, Thy1, MEFSK4, PDGFR-α, CD24, CD44, H3K9ac, and Ki67. GFP indicates endogenous Oct4 promoter activity in the Oct4-GFP MEFs, endogenous Nanog promoter activity in the Nanog-GFP MEFs and was not used for clustering the Nanog-Neo MEFs. The resulting FLOW-MAP graphs are colored by time point, and cell populations common among the three reprogramming systems are labeled with colored dashed lines.
See also Figure S3 and Files S1, S2, and S3.
After Dox induction, all three systems contain populations with mixed stoichiometry of the four reprogramming factors, and the Oct4-GFP and Nanog-Neo MEFs contained a sizable proportion of cells that do not express any of the four reprogramming factors at the protein level. The reprogramming factor non-expressing FLOW-MAP branches show increased PDGFR-a and CD54/ICAM-1 expression at later time points, behavior also observed in uninfected mock-reprogramming MEF cultures (Figure S3F). An Oct4highKlf4high population emerged from the mixed stoichiometry population in the FLOW-MAP plot of each of the three reprogramming systems. This population immediately precedes a partially reprogrammed state in the FLOW-MAP plot, which in turn extends to the mesenchymal-epithelial transition (MET). Ki67low cells appeared to revert to a MEF-like state, while proliferative Ki67high cells appeared to transition into a Nanoghigh population. At the end of the reprogramming time course, the Nanoghigh population diverged into two major end-stage populations in the FLOW-MAP plot: one that was ESC-like with high levels of Oct4, Sox2, and Nanog protein expression, and another state that was Lin28high but low in Sox2 and Nanog protein expression, which represents a state immunophenotypically similar to multipotent mesendodermal progenitor cells (Tada et al., 2005; Wang et al., 2012).
Variability in Reprogramming Factor Stoichiometry
Exogenous reprogramming factor expression is the sole driver of cellular reprogramming (Takahashi and Yamanaka, 2006), and the stoichiometry and timing of their expression plays a critical role in the successful transition to a pluripotent state (Carey et al., 2011; Papapetrou et al., 2009). We therefore investigated the differences in transgene stoichiometry between the three reprogramming MEF systems and their functional consequences at the single cell level. The percentage of cells that were positive for Oct4, Sox2, Klf4, and c-Myc during Dox induction provided a global overview of transgene expression at the protein level in the three MEF reprogramming systems (Figure 4A). While the antibodies used for mass cytometry do not distinguish between endogenous and exogenous reprogramming factors, these measurements are likely to be primarily exogenous because the Dox-induced expression levels are substantially higher than endogenous levels, even in the pluripotent state (Figures S4A and S4B), and expression levels fell rapidly after Dox withdrawal on day 16 (Figures 4A, S4A, and S4B).
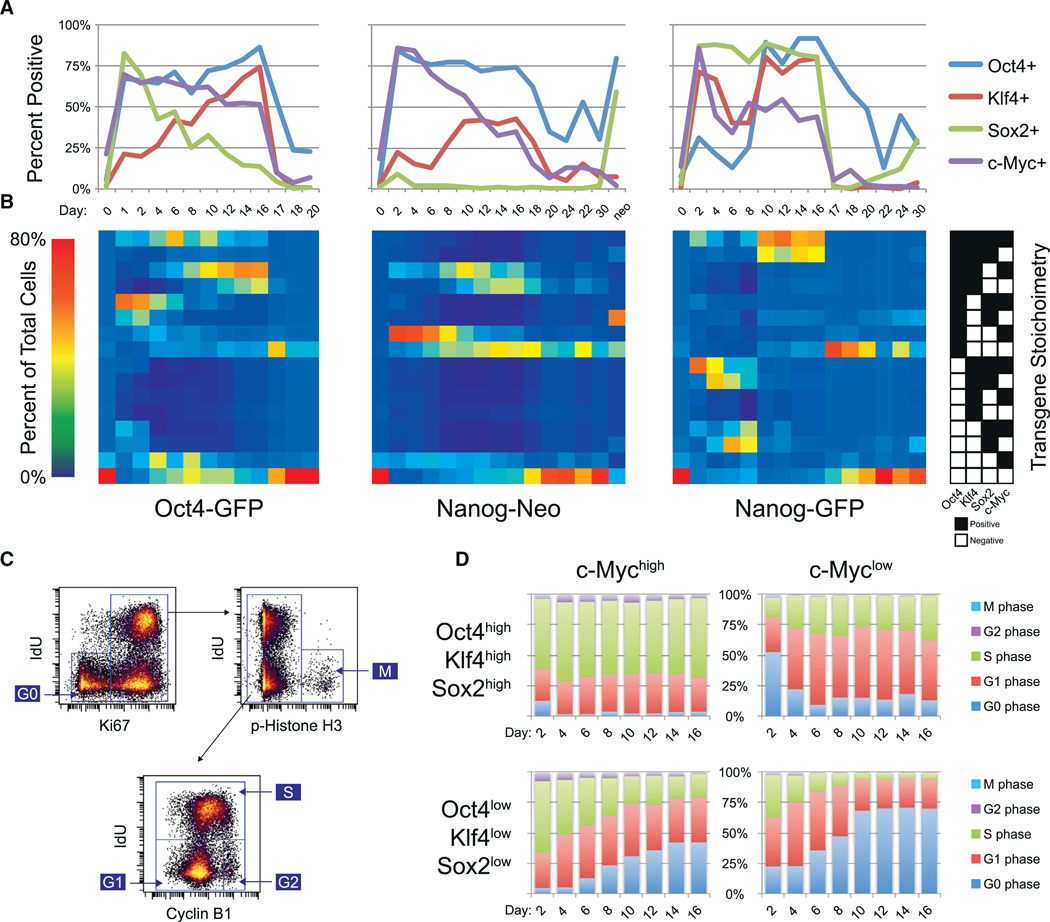
Figure 4. Variability in Transgene Stoichiometry between Reprogramming Systems and within Each System.
(A) Percentage of cells with positive expression for each of the four transgenes over course of reprogramming. Percentages are based on the gates shown in Figures S4A and S4B.
(B) Percentage of cells that fall within each of 16 possible binary combinations of transgene expression over the course of reprogramming. Percentages are based on the gates from Figures S4A and S4B. Each binary combination is indicated by the key on the right, where black boxes denote positive expression above background.
(C) Cell-cycle gating strategy for assignment to G0, G1, S, G2, or M phase using IdU incorporation and the markers Ki67, phospho-Histone H3, and Cyclin B1.
(D) Percentage of cells in each cell-cycle phase for selected c-Mychigh and c-Myclow populations of the time course of Dox induction.
See also Figure S4.
Most of the 16 possible binary transgene combinations were well represented in the Oct4-GFP primary MEFs on day 2, but the Nanog-Neo secondary MEFs display high Oct4 protein expression and low Sox2 protein expression, while the reverse is true for the Nanog-GFP secondary MEFs (Figures 4A, S4A, and S4B), corroborating previous mRNA measurements (Wernig et al., 2008). Early time point replicate analysis indicated that the opposite Oct4/Sox2 stoichiometries of the Nanog-Neo and Nanog-GFP secondary MEFs were reproducible (Figure S4C). The presence of multiple populations with unique transgene stoichiometries is not surprising in the primary Oct4-GFP MEFs, because they were infected with individual randomly integrating lentiviral supernatants, but is surprising in the two secondary MEF systems, because Nanog-GFP and Nanog-Neo MEFs are derived from clonal iPSC lines and are therefore expected to be genetically homogenous. The observed temporal shift of transgene stoichiometries is likely due to increased proliferation of cells that express certain transgene combinations or due to selective lentiviral silencing. To assess selective proliferation rates, the level of IdU incorporation and the protein expression levels of Ki67, phospho-Histone H3, and Cyclin B1 in the reprogramming samples were used to quantify the percentage of cells in G0, G1, S,G2, and M phase of the cell cycle (Figure 4C) (Behbehani et al., 2012).
Expression of the reprogramming factor c-Myc was strongly correlated with cell-cycle progression across all combinations of the remaining three transgenes (Figure 4D). c-Myc has been reported to act as a global amplifier of transcription (Lin et al., 2012) and may play that role during cellular reprogramming as well. We observed a linear relationship between c-Myc protein expression and the reprogramming factors Oct4 and Klf4, but not with Sox2 (Figures S4D and S4E). c-Myc protein expression levels also correlated with markers of proliferation and metabolic activity (Figure S4F), consistent with its role as a driver of cell proliferation. However, this correlation was lost for a set of cell-signaling proteins in the Oct4-GFP primary MEFs, possibly because these cells were virally transduced unlike the secondary MEFs. The innate immune response to retroviral infection has been reported to improve the efficiency of cellular reprogramming by inducing TLR signaling though NF-κB (Lee et al., 2012). Consistent with this model, we observed the Oct4-GFP MEFs reprogrammed faster than the Nanog-Neo and Nanog- GFP secondary MEFs, and protein levels of the NF-κB inhibitor IκBα were anti-correlated with c-Myc in these cells (Figure S4G).
An Oct4/Klf4-Expressing Partially Reprogrammed State Precedes the MET
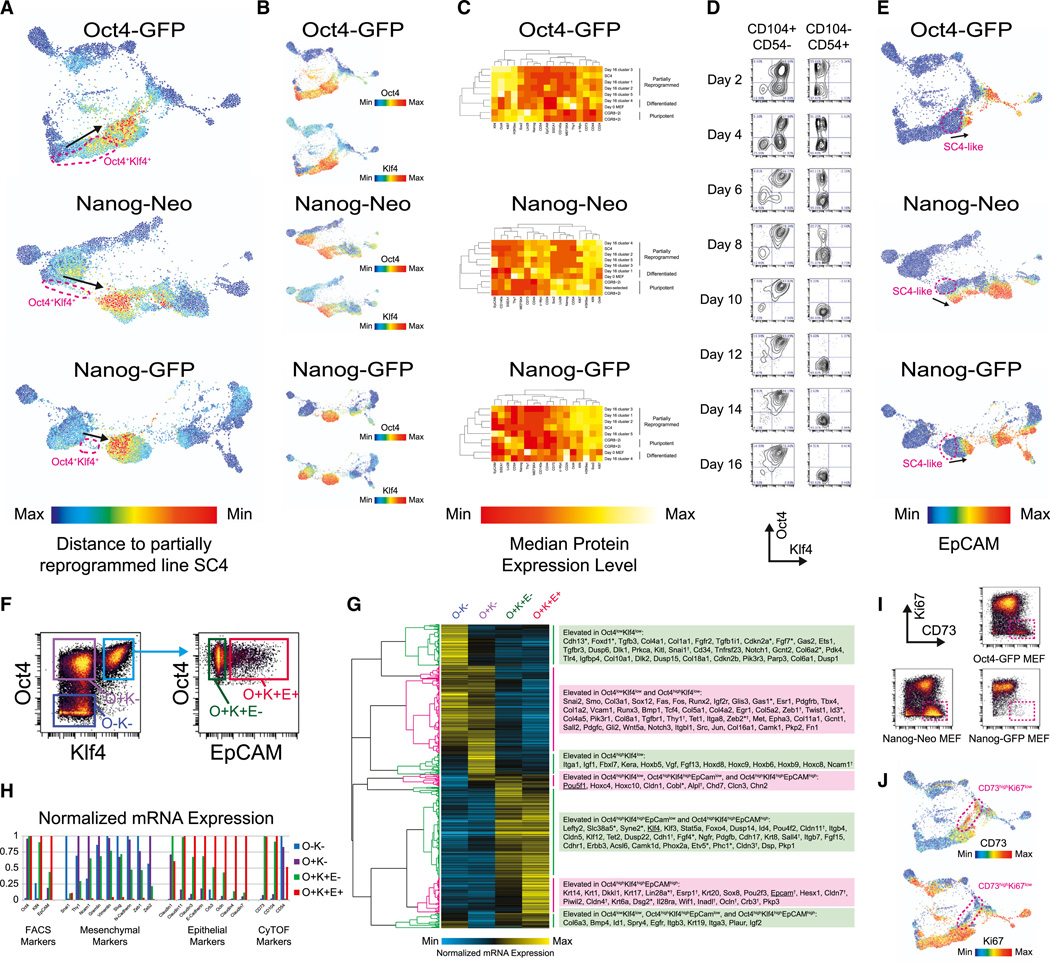
Comparison to the partially reprogrammed iPSC line SC4 (Meissner et al., 2007) revealed that an immunophenotypically similar population of cells was present between days 8 and 16 in all three reprogramming MEF systems (Figure 5A). This SC4-like population is Oct4highKlf4high in all three reprogramming systems and is preceded by an earlier immunophenotypically distinct Oct4highKlf4high transitional population (Figure 5B). 2D cluster analysis of the day 16 samples in combination with MEF, ESC, and SC4 reference samples identified CD73 and CD54/ICAM-1 as the markers that best distinguish this partially reprogrammed population, which is CD73high and CD54/ICAM-1low (Figure 5C). The marker CD73 has previously been used to identify a rare somatic cell type with extensive lineage plasticity that can differentiate to all three germ layers (Roy et al., 2013). Additional experiments identified CD104 as another positive marker for this SC4-like population. Gating by CD104 and CD54/ICAM-1 segregated the reprogramming culture into Klf4high and Klf4low populations as early as 2 days after Dox induction, and at later time points this gating scheme separated Oct4highKlf4high from Oct4lowKlf4low populations (Figure 5D). This SC4-like population precedes the MET, as measured by expression of EpCAM (Figure 5E), an E-cadherin-associated epithelial surface marker (Samavarchi-Tehrani et al., 2010) and therefore represents an earlier stage of the reprogramming process.
Figure 5. A Pre-MET Reprogramming State Marked by Oct4 and Klf4 Expression.
(A) Time-resolved FLOW-MAP plots of similarity to partially reprogrammed iPSCs for the three MEF reprogramming systems. Similarity was calculated by the manhattan distance metric relative to the median expression levels of Oct4, Sox2, Klf4, c-Myc, Nanog, Lin28, SSEA1, EpCAM, CD54, CD73, Thy1, MEFSK4, PDGFR-α, CD24, CD44, H3K9ac, and Ki67 in the partially reprogrammed cell line SC4. Oct4/Klf4 expressing cell clusters that precede the highly SC4-like population are indicated by magenta dashed lines, and the progression of time is indicated by black arrows.
(B) Time-resolved FLOW-MAP plots for the three reprogramming systems (Figures 3D–3F) colored by Oct4 and Klf4 protein expression.
(C) 2D cluster analysis of day 16 subpopulations in combination with differentiated, pluripotent, and partially reprogrammed samples. Day 16 samples for the three reprogramming systems were hierarchically clustered into five populations each and then cluster analysis was performed on these populations to determine their similarity to day 0 MEFs, ESCs, and the partially reprogrammed iPSC line SC4. Heatmap plots of the markers used for clustering are shown, along with dendrograms for the 2D cluster analysis performed on each reprogramming system.
(D) Oct4× Klf4 biaxial plots of the Oct4-GFP primary MEF reprogramming time course, gated to show the CD104highCD54low and CD104lowCD54high populations.
(E) Time-resolved FLOW-MAP plots (Figures 3D–3F) colored by EpCAM protein expression level. The SC4-like population is indicated by a magenta dashed line in each plot, and the progression of time is indicated by black arrows.
(F) Intracellular sort gating strategy for mRNA extraction from the day 8 Nanog-Neo Secondary MEF sample. The Oct4lowKlf4low gate (O−K−) is colored blue, the Oct4highKlf4low gate (O+K−) is colored purple, the Oct4highKlf4lhighEpCAMlow gate (O+K+E−) is colored green, and the Oct4highKlf4highEpCAMhigh gate (O+K+E+) is colored red.
(G) Hierarchical clustering of mRNA microarray data from the four intracellular-sorted populations, which are labeled and colored as in (F).
(H) Selected mRNA expression level measurements for the four intracellular-sorted Nanog-Neo Secondary MEF day 8 populations, which are labeled and colored as in (F).
(I) Ki67 × CD73 biaxial plots for day 16 reprogramming samples. CD73highKi67low population is indicated by a magenta dashed line.
(J) Time-resolved FLOW-MAP plots for Oct4-GFP primary MEFs, colored by CD73 and Ki67 protein expression level. The CD73highKi67low population is indicated by a magenta dashed line in each plot.
See also Figure S5.
To further characterize this Oct4highKlf4highEpCAMlow transitional SC4-like population, a sample of methanol-permeabilized day 8 Nanog-Neo MEFs was FACS-sorted by Oct4, Klf4, and EpCAM protein expression (Figures 5F and S5A) and then mRNA was extracted from these sorted populations for microarray analysis (Figures 5G, S5B, and S5C). Oct4, Klf4, and EpCAM mRNA expression levels follow protein expression levels as measured by the FACS antibodies (Figure 5H), although an increase in mRNA expression of the epithelial marker EpCAM precedes protein expression in the EpCAMlow SC4-like population. Other epithelial markers such as E-cadherin and Crb3 show elevated mRNA expression to varying degrees in this Oct4highKlf4highEpCAMlow transitional population, but mesenchymal markers such as Vimentin and Slug remain elevated as well, indicating in the SC4-like population has both mesenchymal and epithelial characteristics.
During late reprogramming, high levels of CD73 become associated with low Ki67 expression (Figure 5I) indicating that a subset of the CD73high population begins to exit the cell cycle. In the Oct4-GFP primary system, a well-defined trajectory from the CD73highKi67low population toward MEF reversion is present in the FLOW-MAP layout (Figure 5J), suggesting that elevated CD73 expression marks cells that will not successfully reprogram. This CD73highKi67low population is almost completely absent from the Nanog-GFP secondary reprogramming MEFs (Figure 5I), and there is very little MEF reversion in this system as well (Figure 3F). This suggests that CD73highKi67low represents an unstable population distinct from the SC4-like state that is prone to revert to a fibroblast-like population.
Cellular Reprogramming End States Are Controlled by Cell Signaling
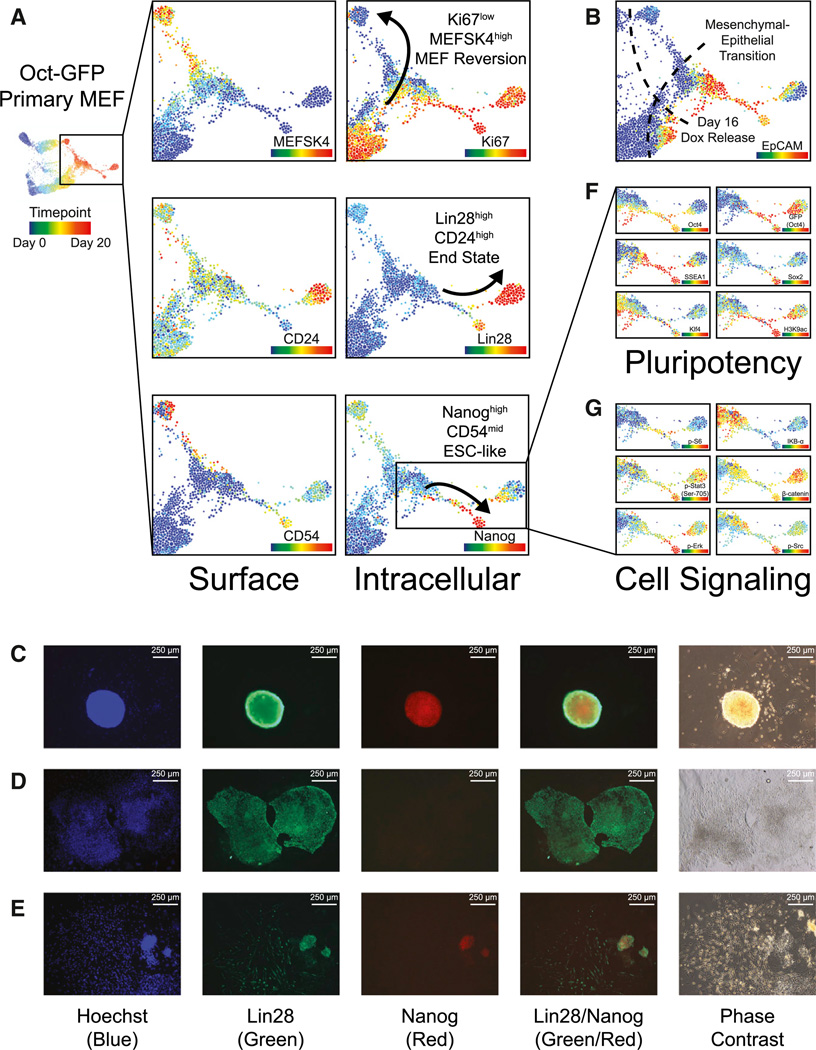
After Dox release on day 16, reprogramming MEFs progress to one of three primary cell populations: (1) MEFSK4high, (2) Lin28high, or (3) Nanoghigh (Figure 6A). The MEFSK4high population appears to be a reversion to a MEF-like state and is preceded by a CD73highKi67low population and correlated with EpCAMlow MET failure (Figure 6B). The MEF-like MEFSK4high population also arises from OSKM non-expressing cells in two of the three reprogramming system FLOW-MAP plots, which gradually change over the time course with decreased Thy1 and increased PDGFR-α and CD54 expression. The shift in proteomic signature of these MEF-like cells is likely due to changes in their local environment, because the MEFs are plated very sparsely, but become highly confluent by the end of the reprogramming time course.
Figure 6. End-Stage Trajectories of Cellular Reprogramming: MEF-like, ESC-like, and Lin28high.
(A) Time-resolved FLOW-MAP plots for Oct4-GFP primary MEFs, colored by protein expression level of the indicated markers. The end-stage trajectories are indicated by black arrows.
(B) Overview FLOW-MAP plot of the late stage reprogramming Oct4-GFP primary MEFs. Day 16 Dox release and the MET are indicated by dashed lines.
(C–E) Immunofluorescence microscopy of day 24 primary reprogramming MEFs stained with anti-Lin28 antibody (Alexafluor-488 secondary antibody), anti-Nanog antibody (Alexafluor-594 secondary antibody), and Hoechst dye. Representative images of a Lin28highNanoghigh colony with mESC-like morphology (C), a Lin28high Nanoglow colony with epithelial cobblestone morphology (D), and a Lin28highNanoglow colony with fibroblast-like morphology adjacent to a Lin28high Nanoghigh colony with mESC-like morphology (E). (F and G) Lin28high and Nanoghigh branches of the Oct4-GFP primary MEF time-resolved FLOW-MAP, colored by markers of pluripotency (F) and cell signaling (G).
See also Figure S6.
EpCAM-expressing cells proceed to either the Nanoghigh or Lin28high end stage populations in the FLOW-MAP plots of all three reprogramming MEF systems. Immunofluorescence imaging identified separate colonies each containing one of these three populations (Figures 6C–6E), indicating that they arise in parallel as suggested by the FLOW-MAP trajectories. The Nanoghigh population is ESC-like with elevated expression of the pluripotency markers Oct4, Sox2, SSEA1, Klf4, and acetyl-Histone H3 (Lys9), as well as mid-level CD54 expression. The Lin28high population forms a separate diverging branch in the three reprogramming systems and is marked by elevated CD24 and PDGFR-α expression, as well as lower levels of the pluripotency markers Oct4, Sox2, SSEA1, Klf4, and acetyl-Histone H3 (Lys9) (Figure 6F). The Oct4-driven GFP reporter persists in this population longer than Oct4 protein itself, which could be due to GFP having a longer half-life than Oct4 in this cell type.
The Nanoghigh and Lin28high populations both diverge from a common branch of the FLOW-MAP plot, suggesting an unstable intermediate state that resolves into an ESC-like Nanoghigh state of pluripotency or a mesendoderm-like Lin28high state (Tada et al., 2005; Wang et al., 2012). In the Oct4-GFP primary MEFs, this common branch point is characterized by elevated levels of phospho-S6 (Ser235/236) and decreased levels of IκBα (Figure 6G). As cells progress from the branch point to the Lin28high and Nanoghigh populations, phospho-Stat3 (Tyr705) increases in both populations, while total β-catenin is higher in the mesendoderm-like Lin28high population and phospho-Erk and phospho-Src are higher in the ESC-like Nanoghigh population. Cell signaling is highly sensitive to changes in culture conditions such as cell density, colony size, and the relative depletion of the culture medium, especially for upstream and membrane proximal signaling proteins such as phospho-Erk, which was not elevated in the ESC-like Nanoghigh populations of the secondary reprogramming MEFs (Figures S6 and S7). The concerted changes observed in cell signaling in all three reprogramming systems, while somewhat variable between systems, still suggest a critical role for cell signaling in the final determination phase of iPSC reprogramming.
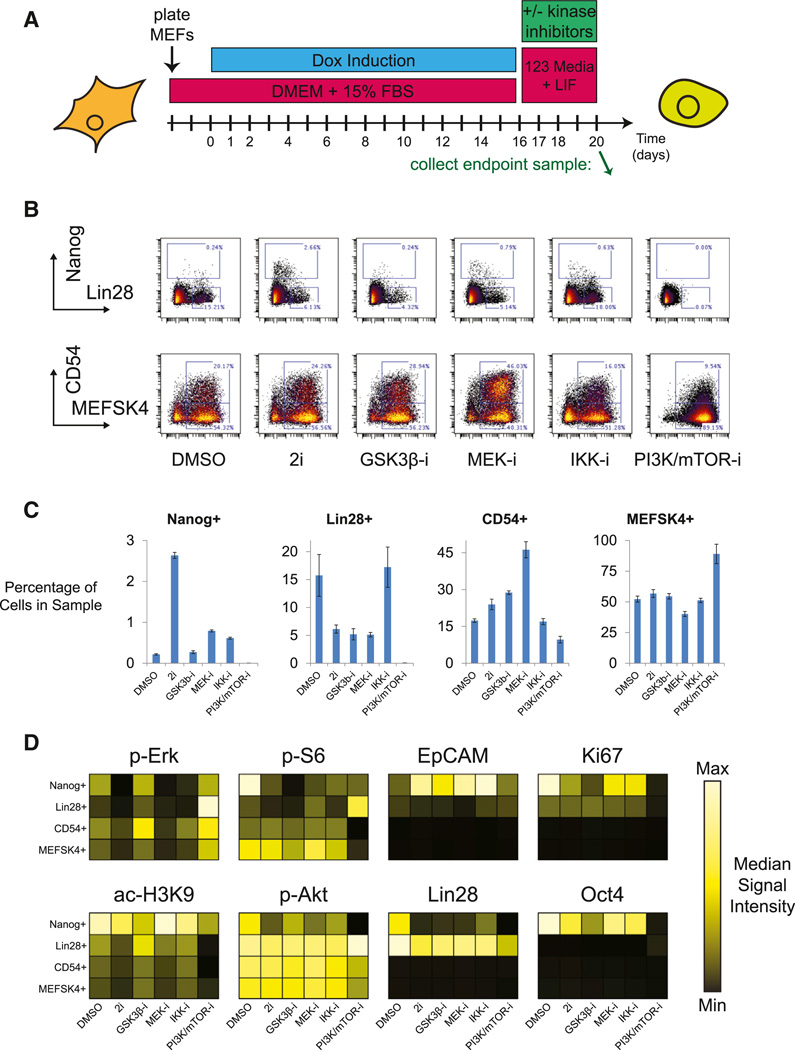
To investigate the functional relevance of the implied signaling pathways in the end stage reprogramming populations, Oct4-GFP primary reprogramming MEFs were treated with a panel of small molecule kinase inhibitors from the point of Dox removal on day 16 until sample collection on day 20 (Figure 7A). IKK Inhibitor X, an IκB kinase inhibitor, and BEZ-235, a dual PI3K/mTOR inhibitor, were chosen due to the changes observed in total IκB and phospho-S6 observed during progression through the FLOW-MAP branch leading to Lin28high and Nanoghigh cell populations. CHIR-99021, a GSK3 inhibitor, and PD-0325901, a MEK inhibitor, were chosen due to their known role in pluripotency maintenance and cellular reprogramming, in particular when used in combination as the “2i” condition (Silva et al., 2008; Ying et al., 2008).
Figure 7. Kinase Inhibitor Modulation of iPS Cell Reprogramming End-Stage Trajectories.
(A) Tissue culture schematic for kinase inhibitor modulation of Oct4-GFP primary MEF reprogramming, indicating kinase inhibitor treatment upon Dox removal from day 16 until day 20. Green arrow indicates the sample collection time point at day 20.
(B) Nanog × Lin28 and CD54 × MEFSK4 biaxial plots for day 20 reprogramming MEFs treated with the indicated small molecule inhibitors from days 16 to 20. 2D gates correspond to the MEFSK4high, CD54high, Lin28high, and Nanoghigh cell populations identified in (B).
(C) Number of cells falling into each gate from (B), as a percentage of the total number of cells in the sample. Experiment was performed in triplicate, and error bars indicate SEM.
(D) Kinase inhibitor modulation of selected signaling molecules in the MEFSK4high, CD54high, Lin28high, and Nanoghigh cell populations.
See also Figure S7.
Biaxial gating of the four mutually exclusive Nanoghigh, Lin28high (Nanoglow), CD54high, and MEFSK4high (CD54low) populations (Figure 7B) allowed quantification of the changes in population distribution between the inhibitor-treated Oct4-GFP primary MEF day 20 endpoint samples (Figure 7C). The most dramatic change in end-stage population distribution occurred after treatment with the PI3K/mTOR inhibitor, which resulted in almost complete loss of the Lin28high and Nanoghigh populations. This result demonstrates an essential role for the PI3K/Akt/mTOR pathway in late stage cellular reprogramming or pluripotency maintenance and indicates an important functional role for the burst of S6 phosphorylation observed in the EpCAMhigh branch of the Oct4-GFP primary MEF FLOW-MAP plot (Figure 6G). Previous studies have found roles for PI3K and mTOR signaling in pluripotency maintenance (Murakami et al., 2004; Paling et al., 2004), but the role of PI3K/Akt/mTOR signaling during cellular reprogramming is less clear, with some studies reporting increased reprogramming efficiency upon mTOR inhibition (Chen et al., 2011; He et al., 2012). 2i treatment from days 16–20 increased the percentage of Nanoghigh cells as expected, but MEK and IKK inhibition also increased this ESC-like population relative to the DMSO only control and the previous time course experiments. 2i, GSK3-, and MEK-inhibition dramatically reduced the percentage of Lin28high cells, but this decrease was not observed when IKK was inhibited. Along with an increase in Nanoghigh cells, the percentage of CD54high cells was also increased by 2i, MEK-, and GSK3-, but not IKK-inhibition, indicating a different reprogramming mode in the presence of an IKK inhibitor.
Heatmap plots of the gated end-stage populations were used to explore the molecular and cell signaling impact of the kinase inhibitor treatments (Figure 7D). As expected, MEK-inhibition, either alone or in the 2i combination, depressed phospho-Erk levels in all cell types. Decreased phospho-Erk correlated with decreased Histone H3 Lys9 acetylation in the MEFSK4high, CD54high, and Lin28high populations, indicating that MEK inhibition represses transcription via global chromatin compaction. However, this relationship was not observed in the ESC-like Nanoghigh population, suggesting that these cells have alternative cellular signaling pathways responsible for histone acetylation. PI3K/mTOR inhibition reduced Akt and S6 phosphorylation as expected and increased compensatory Erk phosphorylation in a manner similar to previous reports (Paling et al., 2004). The exception to this trend was the Lin28high population, which saw increased phosphorylation of Akt, S6, and Erk, although these cells experienced Oct4, Ki67, and H3K9ac loss just as the other cell populations upon PI3K/mTOR inhibition. The BEZ-235-induced increase in S6 phosphorylation observed in Lin28high cells may be due to an alternative pathway through p90 Ribosomal S6 Kinase (RSK)-dependent phosphorylation.
DISCUSSION
Here, we have applied mass cytometry to three MEF reprogramming systems, providing the most comprehensive reference available for protein expression measured at the single cell level during cellular reprogramming. Using a newly developed algorithm for time-resolved progression analysis on the three time course data sets, we provide a global continuous map of reprogramming that includes several well defined stages that appear in all three reprogramming systems, including an early Oct4highKlf4high stage, an intermediate partially reprogrammed stage, and a divergent Lin28high end stage (Figures 3D–3F). The novel combination of high-dimensional mass cytometry and time-resolved progression analysis should serve as a useful tool for the study of additional reprogramming systems and for investigation into the dynamic changes that occur in other complex cell systems such as directed differentiation and oncogenic transformation.
In the three reprogramming systems examined here, we observed an initial period of mixed reprogramming factor stoichiometry, followed by the emergence of anOct4highKlf4high cell population. Previous studies have highlighted the importance of Oct4 and Klf4 expression for optimal reprogramming (Carey et al., 2011; Papapetrou et al., 2009). Here, we propose that the functional consequence of this specific reprogramming stoichiometry is the induction of a partially reprogrammed state that is similar to previously reported partially reprogrammed cell lines (Meissner et al., 2007; Theunissen et al., 2011; Wernig et al., 2008). This Oct4highKlf4high partially reprogrammed state preceded the MET and showed elevated expression of both mesenchymal and epithelial markers (Figure 5H). Previous observations of early stochastic and late hierarchic phases of reprogramming (Buganim et al., 2012), as well as the two transcriptional waves of reprogramming (Polo et al., 2012) may be explained by early mixed reprogramming factor stoichiometries, followed by the emergence of an OcthighKlf4high population that transitions through a partially reprogrammed state to the MET and then pluripotency.
Recently, dramatic progress has been made in improving the speed and efficiency of cellular reprogramming by genetic depletion of Mbd3, perhaps even rendering the reprogramming process deterministic (Rais et al., 2013). It is possible that Mbd3/NuRD complex acts as a “brake” that holds reprogramming MEFs in the SC4-like partially reprogrammed state. Analysis of published mRNA microarray data revealed that Mbd3fl/− reprogramming MEFs express elevated CD73 and CD104 and decreased CD54 at the day 4 time point, but return to baseline levels by day 11, suggesting that this culture system passes through an accelerated SC4-like partially reprogrammed state rather than bypassing it completely.
Efforts to identify reprograming conditions though small molecule inhibitors alone have also made great progress recently (Hou et al., 2013). Small molecule-based reprogramming has important advantages for translational applications, including ease of standardization as well as generating iPSCs without genetic modification. Single cell analysis could play an important role as these protocols are further optimized for mice and adapted to human reprogramming, because the speed and efficiency of small molecule reprogramming is much lower than transcription factor-based induction of pluripotency. The importance of rare population analysis is therefore increased in these systems, and mass cytometry is an attractive option for analysis because its antibody-based readout allows direct access to post-translational modifications and cell signaling—the mode of action for small molecule reprogramming. The methods and data sets reported here will prove a useful resource for reprogramming optimization and other mechanistic studies of cellular reprogramming, as well as for the study of additional complex cell populations that change dynamically over time.
EXPERIMENTAL PROCEDURES
Viral Preparation
HEK293T cells were transfected with lentiviral vectors containing Oct4, Sox2, Klf4, and c-Myc under the control of the tetracycline operator and a minimal CMV promoter, to produce replication-incompetent lentiviral particles with a VSV-G envelope.
Tissue Culture
MEFs were prepared as described previously (Wernig et al., 2008) and cultured in MEF medium (DMEM, 10% CCS, Pen/Strep). Cryogenically preserved passage 0 MEFs were thawed and expanded for three passages and then seeded onto gelatin-coated 10-cm plates at a density of 250,000 cells per plate. For primary MEF infection, freshly prepared lentiviral supernatant was added to the culture medium 1 day after seeding. One day after infection, reprogramming factor expression was induced (day 0) by the addition of 2 µg/ml Dox (Sigma-Aldrich). MEF culture medium with Dox was refreshed every 2 days until day 16, at which point the medium was replaced with 123 Medium (Knockout DMEM, 12% KSR, 3% CCS, beta-mercaptoethanol, Sodium Pyruvate, Non-essential Amino Acids, Pen/Strep) supplemented with Leukemia Inhibitory Factor and refreshed every 2 days. Neomycin selection was performed on the Nanog-Neo secondary MEF reprogramming culture from days 24 to 30 with 400 µg/ml G418. IdU incorporation was performed as previously described (Behbehani et al., 2012) at 1 µM for 15 min before dissociation. For kinase inhibitor treatment, DMSO stocks were added to the medium at 1:1,000, and the medium was refreshed daily. GSK3 inhibitor CHIR-99021 (Stemgent) was used at 3 µM, MEK inhibitor PD-0325901 (LCLabs) was used at 1 µM, IKK inhibitor × (Millipore) was used at 10 µM, and PI3K/mTOR inhibitor BEZ-235 (LCLabs) was used at 1 µM.
Cell Dissociation, Fixation, and Permeabilization
Reprogramming MEF cultures were washed once with 37°C HBSS and then incubated with 10× TrypLE (Life Technologies) for 3 min at 37°C. The cells were then triturated into a single-cell suspension with a 10-ml transfer pipet and filtered through a 40-µm filter. Filtered cells were fixed at room temperature with 1.6% paraformaldehyde and then permeabilized with ice-cold methanol, as previously described (Krutzik et al., 2005).
Mass-Tag Cell Barcoding
Cell samples were individually Mass-tag Cell Barcoded (MCB) as previously described (Bodenmiller et al., 2012; Zunder et al., 2015). Methanol-permeabilized cells were washed once with Cell Staining Medium (CSM, PBS with 0.5% BSA, 0.02% NaN3) and then once with PBS. Different combinatorial mixtures of Palladium-containing MCB reagents in DMSO were then added to the individual samples at 1:100 DMSO with vortexing and then incubated at room temperature for 30 min, followed by three washes with CSM. The individual samples were then pooled for antibody staining and mass cytometry analysis. These MCB modifications are discussed in greater detail in Supplemental Information.
Mass Cytometry Antibodies
The mass cytometry antibodies used in this study are summarized in Table S1, including antibody clone, vendor, metal isotope, and staining concentration. All antibodies were conjugated to isotopically pure lanthanide metal using the MaxPAR antibody conjugation kit (DVS Sciences), according to the manufacturer’s recommended protocol. Labeled antibodies were stored at 4°C in PBS-based Antibody Stabilizer (Candor Bioscience). A single antibody staining cocktail was prepared for the three reprogramming time courses and then split into one-third and two-thirds volumes for addition of anti-Sox2 antibody, which needed to be used at a lower concentration of 200 ng/ml on the Nanog-GFP secondary MEF samples. This was required because higher-induced Sox2 expression resulted in CyTOF detector saturation at 2000 ng/ml as was used for the Oct4-GFP and Nanog-Neo MEFs. A separate staining cocktail using the same antibody concentrations was prepared for the MCB-multiplexed kinase inhibitor-treated samples.
Mass Cytometry Measurement and Data Processing
MCB-multiplexed, antibody-stained cell samples were analyzed on a CyTOF mass cytometer (DVS Sciences) in several runs at a rate of 500 cells per second or less. Normalization for detector sensitivity was performed as previously described (Finck et al., 2013), using normalization beads containing Lanthanum-139, Praseodymium-141, Terbium-159, Thulium-169, and Lutetium-175. After measurement and normalization, the individual FCS files for each barcoded sample were concatenated, and each barcoded sample was de-barcoded to the individual samples as previously described (Bodenmiller et al., 2012). Cell events with zero signal on the Histone H3 acetyl-Lys9 and Rb phospho-Ser807/11 channels were gated out as non-nucleated cell fragments.
FLOW-MAP Analysis
Density-dependent downsampling, hierarchical clustering, cluster upsampling, and extraction of parameter medians was performed by the SPADE package (http://www.cytospade.org) as described in the main text and as previously described (Linderman et al., 2012; Qiu et al., 2011). Cell clusters and their attendant median values were used to build FLOW-MAP graphs by scripts written with the R language and environment for statistical computing (http://www.R-project.org/) and the igraph package (http://igraph.sf.net) (E.R.Z., P.F.G., and G.P.N., unpublished data) and are described in the Supplemental Information. Force-directed layout of the FLOW-MAP graphs was performed using the ForceAtlas2 algorithm (Jacomy et al., 2014) from the Gephi software package (http://www.gephi.org) (Bastian et al., 2009).
Intracellular FACS Sort for RNA Extraction and Microarray Analysis
Isolation of RNA from intracellularly stained, FACS sorted samples is described in the Supplemental Experimental Procedures. Amplified cDNA was prepared from the extracted RNA using the Ovation Pico WTA System V2 (Nugen) according to the manufacturer’s instructions. cDNA samples were analyzed by Mouse Genome 430 2.0 Microarray (Affymetrix) at the Stanford PAN facility, deposited in NCBI’s Gene Expression Omnibus (Edgar et al., 2002), and are accessible through GEO Series accession number GSE56764 (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?token=abehwqeihnsrzoh&acc=GSE56764). Cluster analysis was performed on the microarray data sets using Cluster (http://rana.lbl.gov/EisenSoftware.htm) and Java Treeview (http://jtreeview.sourceforge.net/) as previously described (Eisen et al., 1998; Saldanha, 2004).
Immunofluorescence Imaging
MEFs were seeded onto 6-well plates and subjected to reprogramming conditions via lentiviral infection and doxycycline induction. At the indicated time points, the cultures were washed once with PBS and then fixed by incubation with 4% paraformaldehyde in PBS at room temperature for 15 min. After fixation, the wells were washed three times with PBS and then stored at 4°C. Before antibody staining, the wells were incubated with blocking solution (5% FBS, 0.1% Triton X-100 in PBS) for 30 min at room temperature. The wells were then incubated with the indicated primary antibodies in blocking solution for 30 min at room temperature, washed three times with PBS, then incubated with the indicated fluorophore-conjugated secondary antibodies in blocking solution for 30 min at room temperature shielded from light and washed three times with PBS. Before imaging, the wells were incubated with 1 µg/ml Hoechst 33342 dye in PBS for 30 min at room temperature shielded from light and then washed once with PBS. Images were acquired using an Axio Observer. A1 microscope, an AxioCam MRC camera, and AxioVision software 4.8.1 (Zeiss).
Supplementary Material
Highlights.
Single-cell analysis of three fibroblast reprogramming systems by mass cytometry
Time-resolved high-dimensional progression analysis with FLOW-MAP algorithm
Oct4highKlf4high transition to CD73highCD104highCD54low state preceding the M-E-T
Alternative NanoglowLin28highCD24highPDGFR-αhigh reprogramming trajectory
ACKNOWLEDGMENTS
We thank Angelica Trejo and Astraea Jager for mass cytometry quality control and instrument maintenance. We thank the Stanford Shared FACS Facility for assistance with cell sorting. We thank the Stanford PAN Facility for assistance with microarray hybridization and imaging. We thank Rob Bruggner for patiently answering our questions about the R programming language. We thank Sean Bendall and David Burns for helpful discussions. We thank Alex Chang for assistance with immunofluorescence microscopy. E.R.Z. was supported by NIH NRSA F32 GM093508-01. E.L. was supported by the California Institute for Regenerative Medicine (CIRM) Predoctoral Fellowship TG2-01159 and the National Science Foundation (NSF) Graduate Research Fellowship DGE-114747. M.W. is a New York Stem Cell Foundation-Robertson Investigator and a Tashia and John Morgridge Faculty Scholar at the Child Health Research Institute at Stanford. This work was supported by NIH (U19 AI057229, U54CA149145, N01-HV-00242, 1U19AI100627, 5R01AI07372405, R01CA184968, 1 R33 CA183654, and R33 CA183692), NIH-Baylor Research Institute (41000411217), NIH-Northrop Grumman (7500108142), CIRM (DR1-01477 and RB2–01592), Department of Defense (OC110674 and 11491122), European Commission (Health.2010.1.2-1), Food and Drug Administration (HHSF223201210194C), Bill and Melinda Gates Foundation (OPP 1017093), the Alliance for Lupus Research, the Entertainment Industry Foundation (NWCRA grant), and the Rachford and Carlota A. Harris Endowed Professorship to G.P.N. G.P.N. has personal financial interest in the company Fluidigm, the manufacturer of the mass cytometer used in this manuscript.
Footnotes
ACCESSION NUMBERS
The NCBI Gene Expression Omnibus accession number for the microarray data sets reported in this paper is GSE56764.
SUPPLEMENTAL INFORMATION
Supplemental Information includes Supplemental Experimental Procedures, three figures, one table, and three Supplemental Files and can be found with this article online at http://dx.doi.org/10.1016/j.stem.2015.01.015.
AUTHOR CONTRIBUTIONS
E.R.Z., E.L., M.W., and G.P.N. designed research. E.R.Z. and E.L. conducted reprogramming culture experiments. E.R.Z. conducted CyTOF analysis. E.R.Z. developed and implemented the FLOW-MAP algorithm. E.R.Z. and Y.G. performed cell sorting, RNA extraction, and microarray analysis. E.R.Z., E.L., Y.G., M.W., and G.P.N. analyzed data. E.R.Z., E.L., M.W., and G.P.N. wrote the manuscript.
REFERENCES
- Bandura DR, Baranov VI, Ornatsky OI, Antonov A, Kinach R, Lou X, Pavlov S, Vorobiev S, Dick JE, Tanner SD. Mass cytometry: technique for real time single cell multitarget immunoassay based on inductively coupled plasma time-of-flight mass spectrometry. Anal. Chem. 2009;81:6813–6822. doi: 10.1021/ac901049w. [DOI] [PubMed] [Google Scholar]
- Bastian M, Heymann S, Jacomy M. Gephi: an open source software for exploring and manipulating networks; Third International AAAI Conference on Weblogs and Social Media; 2009. [Google Scholar]
- Behbehani GK, Bendall SC, Clutter MR, Fantl WJ, Nolan GP. Single-cell mass cytometry adapted to measurements of the cell cycle. Cytometry A. 2012;81:552–566. doi: 10.1002/cyto.a.22075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bendall SC, Simonds EF, Qiu P, Amir AD, Krutzik PO, Finck R, Bruggner RV, Melamed R, Trejo A, Ornatsky OI, et al. Single-cell mass cytometry of differential immune and drug responses across a human hematopoietic continuum. Science. 2011;332:687–696. doi: 10.1126/science.1198704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bodenmiller B, Zunder ER, Finck R, Chen TJ, Savig ES, Bruggner RV, Simonds EF, Bendall SC, Sachs K, Krutzik PO, Nolan GP. Multiplexed mass cytometry profiling of cellular states perturbed by small-molecule regulators. Nat. Biotechnol. 2012;30:858–867. doi: 10.1038/nbt.2317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buganim Y, Faddah DA, Cheng AW, Itskovich E, Markoulaki S, Ganz K, Klemm SL, van Oudenaarden A, Jaenisch R. Single-cell expression analyses during cellular reprogramming reveal an early stochastic and a late hierarchic phase. Cell. 2012;150:1209–1222. doi: 10.1016/j.cell.2012.08.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carey BW, Markoulaki S, Hanna J, Saha K, Gao Q, Mitalipova M, Jaenisch R. Reprogramming of murine and human somatic cells using a single polycistronic vector. Proc. Natl. Acad. Sci. USA. 2009;106:157–162. doi: 10.1073/pnas.0811426106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carey BW, Markoulaki S, Hanna JH, Faddah DA, Buganim Y, Kim J, Ganz K, Steine EJ, Cassady JP, Creyghton MP, et al. Reprogramming factor stoichiometry influences the epigenetic state and biological properties of induced pluripotent stem cells. Cell Stem Cell. 2011;9:588–598. doi: 10.1016/j.stem.2011.11.003. [DOI] [PubMed] [Google Scholar]
- Chen T, Shen L, Yu J, Wan H, Guo A, Chen J, Long Y, Zhao J, Pei G. Rapamycin and other longevity-promoting compounds enhance the generation of mouse induced pluripotent stem cells. Aging Cell. 2011;10:908–911. doi: 10.1111/j.1474-9726.2011.00722.x. [DOI] [PubMed] [Google Scholar]
- Chen J, Liu H, Liu J, Qi J, Wei B, Yang J, Liang H, Chen Y, Chen J, Wu Y, et al. H3K9 methylation is a barrier during somatic cell reprogramming into iPSCs. Nat. Genet. 2013;45:34–42. doi: 10.1038/ng.2491. [DOI] [PubMed] [Google Scholar]
- Dufner A, Thomas G. Ribosomal S6 kinase signaling and the control of translation. Exp. Cell Res. 1999;253:100–109. doi: 10.1006/excr.1999.4683. [DOI] [PubMed] [Google Scholar]
- Edgar R, Domrachev M, Lash AE. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 2002;30:207–210. doi: 10.1093/nar/30.1.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisen MB, Spellman PT, Brown PO, Botstein D. Cluster analysis and display of genome-wide expression patterns. Proc. Natl. Acad. Sci. USA. 1998;95:14863–14868. doi: 10.1073/pnas.95.25.14863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng Z, Zhang H, Levine AJ, Jin S. The coordinate regulation of the p53 and mTOR pathways in cells. Proc. Natl. Acad. Sci. USA. 2005;102:8204–8209. doi: 10.1073/pnas.0502857102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finck R, Simonds EF, Jager A, Krishnaswamy S, Sachs K, Fantl W, Pe’er D, Nolan GP, Bendall SC. Normalization of mass cytometry data with bead standards. Cytometry A. 2013;83:483–494. doi: 10.1002/cyto.a.22271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanna J, Saha K, Pando B, van Zon J, Lengner CJ, Creyghton MP, van Oudenaarden A, Jaenisch R. Direct cell reprogramming is a stochastic process amenable to acceleration. Nature. 2009;462:595–601. doi: 10.1038/nature08592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi K, Lopes SMC, de S, Tang F, Surani MA. Dynamic equilibrium and heterogeneity of mouse pluripotent stem cells with distinct functional and epigenetic states. Cell Stem Cell. 2008;3:391–401. doi: 10.1016/j.stem.2008.07.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He J, Kang L, Wu T, Zhang J, Wang H, Gao H, Zhang Y, Huang B, Liu W, Kou Z, et al. An elaborate regulation of Mammalian target of rapamycin activity is required for somatic cell reprogramming induced by defined transcription factors. Stem Cells Dev. 2012;21:2630–2641. doi: 10.1089/scd.2012.0015. [DOI] [PubMed] [Google Scholar]
- Hong H, Takahashi K, Ichisaka T, Aoi T, Kanagawa O, Nakagawa M, Okita K, Yamanaka S. Suppression of induced pluripotent stem cell generation by the p53-p21 pathway. Nature. 2009;460:1132–1135. doi: 10.1038/nature08235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou P, Li Y, Zhang X, Liu C, Guan J, Li H, Zhao T, Ye J, Yang W, Liu K, et al. Pluripotent stem cells induced from mouse somatic cells by small-molecule compounds. Science. 2013;341:651–654. doi: 10.1126/science.1239278. [DOI] [PubMed] [Google Scholar]
- Ichida JK, Blanchard J, Lam K, Son EY, Chung JE, Egli D, Loh KM, Carter AC, Di Giorgio FP, Koszka K, et al. A small-molecule inhibitor of TGF-β signaling replaces sox2 in reprogramming by inducing nanog. Cell Stem Cell. 2009;5:491–503. doi: 10.1016/j.stem.2009.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacomy M, Venturini T, Heymann S, Bastian M. ForceAtlas2, a continuous graph layout algorithm for handy network visualization designed for the Gephi software. PLoS ONE. 2014;9:e98679. doi: 10.1371/journal.pone.0098679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawamura T, Suzuki J, Wang YV, Menendez S, Morera LB, Raya A, Wahl GM, Izpisúa Belmonte JC. Linking the p53 tumour suppressor pathway to somatic cell reprogramming. Nature. 2009;460:1140–1144. doi: 10.1038/nature08311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koche RP, Smith ZD, Adli M, Gu H, Ku M, Gnirke A, Bernstein BE, Meissner A. Reprogramming factor expression initiates wide-spread targeted chromatin remodeling. Cell Stem Cell. 2011;8:96–105. doi: 10.1016/j.stem.2010.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krutzik PO, Clutter MR, Nolan GP. Coordinate analysis of murine immune cell surface markers and intracellular phosphoproteins by flow cytometry. J. Immunol. 2005;175:2357–2365. doi: 10.4049/jimmunol.175.4.2357. [DOI] [PubMed] [Google Scholar]
- Lee J, Sayed N, Hunter A, Au KF, Wong WH, Mocarski ES, Pera RR, Yakubov E, Cooke JP. Activation of innate immunity is required for efficient nuclear reprogramming. Cell. 2012;151:547–558. doi: 10.1016/j.cell.2012.09.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lengner CJ, Camargo FD, Hochedlinger K, Welstead GG, Zaidi S, Gokhale S, Scholer HR, Tomilin A, Jaenisch R. Oct4 expression is not required for mouse somatic stem cell self-renewal. Cell Stem Cell. 2007;1:403–415. doi: 10.1016/j.stem.2007.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Collado M, Villasante A, Strati K, Ortega S, Cañamero M, Blasco MA, Serrano M. The Ink4/Arf locus is a barrier for iPS cell reprogramming. Nature. 2009;460:1136–1139. doi: 10.1038/nature08290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li R, Liang J, Ni S, Zhou T, Qing X, Li H, He W, Chen J, Li F, Zhuang Q, et al. A mesenchymal-to-epithelial transition initiates and is required for the nuclear reprogramming of mouse fibroblasts. Cell Stem Cell. 2010;7:51–63. doi: 10.1016/j.stem.2010.04.014. [DOI] [PubMed] [Google Scholar]
- Lin CY, Lovén J, Rahl PB, Paranal RM, Burge CB, Bradner JE, Lee TI, Young RA. Transcriptional amplification in tumor cells with elevated c-Myc. Cell. 2012;151:56–67. doi: 10.1016/j.cell.2012.08.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linderman MD, Bjornson Z, Simonds EF, Qiu P, Bruggner RV, Sheode K, Meng TH, Plevritis SK, Nolan GP. CytoSPADE: high-performance analysis and visualization of high-dimensional cytometry data. Bioinformatics. 2012;28:2400–2401. doi: 10.1093/bioinformatics/bts425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marión RM, Strati K, Li H, Murga M, Blanco R, Ortega S, Fernandez-Capetillo O, Serrano M, Blasco MA. A p53-mediated DNA damage response limits reprogramming to ensure iPS cell genomic integrity. Nature. 2009;460:1149–1153. doi: 10.1038/nature08287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meissner A, Wernig M, Jaenisch R. Direct reprogramming of genetically unmodified fibroblasts into pluripotent stem cells. Nat. Biotechnol. 2007;25:1177–1181. doi: 10.1038/nbt1335. [DOI] [PubMed] [Google Scholar]
- Mikkelsen TS, Hanna J, Zhang X, Ku M, Wernig M, Schorderet P, Bernstein BE, Jaenisch R, Lander ES, Meissner A. Dissecting direct reprogramming through integrative genomic analysis. Nature. 2008;454:49–55. doi: 10.1038/nature07056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murakami M, Ichisaka T, Maeda M, Oshiro N, Hara K, Edenhofer F, Kiyama H, Yonezawa K, Yamanaka S. mTOR is essential for growth and proliferation in early mouse embryos and embryonic stem cells. Mol. Cell. Biol. 2004;24:6710–6718. doi: 10.1128/MCB.24.15.6710-6718.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Malley J, Skylaki S, Iwabuchi KA, Chantzoura E, Ruetz T, Johnsson A, Tomlinson SR, Linnarsson S, Kaji K. High-resolution analysis with novel cell-surface markers identifies routes to iPS cells. Nature. 2013;499:88–91. doi: 10.1038/nature12243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paling NRD, Wheadon H, Bone HK, Welham MJ. Regulation of embryonic stem cell self-renewal by phosphoinositide 3-kinase-dependent signaling. J. Biol. Chem. 2004;279:48063–48070. doi: 10.1074/jbc.M406467200. [DOI] [PubMed] [Google Scholar]
- Papapetrou EP, Tomishima MJ, Chambers SM, Mica Y, Reed E, Menon J, Tabar V, Mo Q, Studer L, Sadelain M. Stoichiometric and temporal requirements of Oct4, Sox2, Klf4, and c-Myc expression for efficient human iPSC induction and differentiation. Proc. Natl. Acad. Sci. USA. 2009;106:12759–12764. doi: 10.1073/pnas.0904825106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polo JM, Anderssen E, Walsh RM, Schwarz BA, Nefzger CM, Lim SM, Borkent M, Apostolou E, Alaei S, Cloutier J, et al. A molecular roadmap of reprogramming somatic cells into iPS cells. Cell. 2012;151:1617–1632. doi: 10.1016/j.cell.2012.11.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu P, Simonds EF, Bendall SC, Jr, Gibbs KD, Jr, Bruggner RV, Linderman MD, Sachs K, Nolan GP, Plevritis SK. Extracting a cellular hierarchy from high-dimensional cytometry data with SPADE. Nat. Biotechnol. 2011;29:886–891. doi: 10.1038/nbt.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rais Y, Zviran A, Geula S, Gafni O, Chomsky E, Viukov S, Mansour AA, Caspi I, Krupalnik V, Zerbib M, et al. Deterministic direct reprogramming of somatic cells to pluripotency. Nature. 2013;502:65–70. doi: 10.1038/nature12587. [DOI] [PubMed] [Google Scholar]
- Roy S, Gascard P, Dumont N, Zhao J, Pan D, Petrie S, Margeta M, Tlsty TD. Rare somatic cells from human breast tissue exhibit extensive lineage plasticity. Proc. Natl. Acad. Sci. USA. 2013;110:4598–4603. doi: 10.1073/pnas.1218682110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saldanha AJ. Java Treeview—extensible visualization of microarray data. Bioinformatics. 2004;20:3246–3248. doi: 10.1093/bioinformatics/bth349. [DOI] [PubMed] [Google Scholar]
- Samavarchi-Tehrani P, Golipour A, David L, Sung HK, Beyer TA, Datti A, Woltjen K, Nagy A, Wrana JL. Functional genomics reveals a BMP-driven mesenchymal-to-epithelial transition in the initiation of somatic cell reprogramming. Cell Stem Cell. 2010;7:64–77. doi: 10.1016/j.stem.2010.04.015. [DOI] [PubMed] [Google Scholar]
- Silva J, Barrandon O, Nichols J, Kawaguchi J, Theunissen TW, Smith A. Promotion of reprogramming to ground state pluripotency by signal inhibition. PLoS Biol. 2008;6:e253. doi: 10.1371/journal.pbio.0060253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith ZD, Nachman I, Regev A, Meissner A. Dynamic single-cell imaging of direct reprogramming reveals an early specifying event. Nat. Biotechnol. 2010;28:521–526. doi: 10.1038/nbt.1632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soufi A, Donahue G, Zaret KS. Facilitators and impediments of the pluripotency reprogramming factors’ initial engagement with the genome. Cell. 2012;151:994–1004. doi: 10.1016/j.cell.2012.09.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sridharan R, Tchieu J, Mason MJ, Yachechko R, Kuoy E, Horvath S, Zhou Q, Plath K. Role of the murine reprogramming factors in the induction of pluripotency. Cell. 2009;136:364–377. doi: 10.1016/j.cell.2009.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stadtfeld M, Maherali N, Breault DT, Hochedlinger K. Defining molecular cornerstones during fibroblast to iPS cell reprogramming in mouse. Cell Stem Cell. 2008;2:230–240. doi: 10.1016/j.stem.2008.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tada S, Era T, Furusawa C, Sakurai H, Nishikawa S, Kinoshita M, Nakao K, Chiba T, Nishikawa S. Characterization of mesendoderm: a diverging point of the definitive endoderm and mesoderm in embryonic stem cell differentiation culture. Development. 2005;132:4363–4374. doi: 10.1242/dev.02005. [DOI] [PubMed] [Google Scholar]
- Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- Theunissen TW, van Oosten AL, Castelo-Branco G, Hall J, Smith A, Silva JCR. Nanog overcomes reprogramming barriers and induces pluripotency in minimal conditions. Curr. Biol. 2011;21:65–71. doi: 10.1016/j.cub.2010.11.074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Utikal J, Polo JM, Stadtfeld M, Maherali N, Kulalert W, Walsh RM, Khalil A, Rheinwald JG, Hochedlinger K. Immortalization eliminates a roadblock during cellular reprogramming into iPS cells. Nature. 2009;460:1145–1148. doi: 10.1038/nature08285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Cao N, Yuan M, Cui H, Tang Y, Qin L, Huang X, Shen N, Yang H-T. MicroRNA-125b/Lin28 pathway contributes to the mesendodermal fate decision of embryonic stem cells. Stem Cells Dev. 2012;21:1524–1537. doi: 10.1089/scd.2011.0350. [DOI] [PubMed] [Google Scholar]
- Wernig M, Lengner CJ, Hanna J, Lodato MA, Steine E, Foreman R, Staerk J, Markoulaki S, Jaenisch R. A drug-inducible transgenic system for direct reprogramming of multiple somatic cell types. Nat. Biotechnol. 2008;26:916–924. doi: 10.1038/nbt1483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ying Q-L, Wray J, Nichols J, Batlle-Morera L, Doble B, Woodgett J, Cohen P, Smith A. The ground state of embryonic stem cell self-renewal. Nature. 2008;453:519–523. doi: 10.1038/nature06968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zunder ER, Finck R, Behbehani GK, Amir AD, Krishnaswamy S, Gonzalez VD, Lorang CG, Bjornson Z, Spitzer MH, Bodenmiller B, et al. Palladium-based mass tag cell barcoding with a doublet-filtering scheme and single-cell deconvolution algorithm. Nat. Protoc. 2015;10:316–333. doi: 10.1038/nprot.2015.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.