Abstract
Human leukocyte antigen-G (HLA-G) molecule exerts multiple immunoregulatory functions that have been suggested to contribute to the immune evasion of tumour cells. Studies on HLA-G expression in malignant haematopoietic diseases are controversial, and the functions of HLA-G on this context are limited. In the current study, HLA-G expression was analysed in different types of patients: de novo acute myeloid leukaemia (AML, n = 54), B cell acute lymphoblastic leukaemia (B-ALL, n= 13), chronic myeloid leukaemia (CML, n= 9) and myelodysplastic syndrome (MDS, n= 11). HLA-G expression was observed in 18.5% cases of AML, 22.2% in CML and 18.2% in MDS, but not in B-ALL patients. In AML, HLA-G-positive patients had a significant higher bone marrow leukaemic blast cell percentage when compared with that of HLA-G-negative patients (P < 0.01). Total T-cell percentage was dramatically decreased in HLA-G-positive patients (P < 0.05). Cytogenetic karyotyping results showed that all HLA-G-positive AML patients (n= 5) were cytogenetically abnormal, which was markedly different from that of HLA-G-negative patients (P < 0.01). Ex vivo cytotoxicity analysis revealed that HLA-G expression in AML leukaemic cells could directly inhibit NK cell cytolysis (P < 0.01). These findings indicated that HLA-G expression in AML is of unfavourable clinical implications, and that HLA-G could be a potential target for therapy.
Keywords: HLA-G, acute myeloid leukaemia, NK cell, cytogenetics
Introduction
The expression of non-classical HLA class I molecule HLA-G was initially reported to be restricted to extravillous cytotrophoblast and confers protection to foetus from maternal semia llograft immune responses [1, 2]. HLA-G features low level of allelic polymorphisms and encodes seven protein isoforms generated by alternative splicing of its primary mRNA [3]. Extensive studies have been carried out on its functions in foetal-maternal immune maintenance. Studies suggested that HLA-G serves as a protection factor for foetus from maternal semiallorecognition by the ability of HLA-G to modulate the functions of immune-competent cells, such as natural killer (NK) cells, T lymphocytes and antigen-presenting cells [4]. HLA-G functioned as an immunotolerant that is involved in its interaction with diverse receptors, such as the immunoglobulin-like transcript 2 (ILT2), ILT4 and killer immunoglobulin-like receptor 2DL4 (KIR2DL4) expressed on these immune cells [5–8].
Other than extra villous cytotrophoblasts, distribution of HLA-G expression recently has been found to be much broader than originally reported. An increasing number of studies addressed that HLA-G expression was observed in various malignancies, such as colorectal cancer, retinoblastoma, ovarian carcinoma, breast carcinoma, haematopoietic tumours, renal cell carcinoma, melanoma and lung cancer [9–17]. Few studies had been carried out for HLA-G antigen expression in malignant haematopoietic cells. However, data were controversial, and the functional roles of HLA-G in these diseases were limited [18–20].
In an effort to gain further insight into the role of HLA-G in malignant haematologic diseases, HLA-G cell surface expression was measured in bone marrow leukaemic blast cells from 87 malignant haematopoietic diseases and a variety of clinical and laboratory variables were analysed. Meanwhile, NK cell cytotoxicity against HLA-G-positive or-negative leukaemic cells isolated from AML patients was performed ex vivo.
Materials and methods
Patients
Eighty-seven patients with malignant haematopoietic diseases treated at the Department of Hematology, Wenzhou Medical College-affiliated Taizhou Hospital of Zhejiang Province were included in this retrospective study (between March 2006 and June 2007). Bone marrow samples were taken for diagnostic purposes, and surplus material was used in this study with informed consent of each patient according to institutional guidelines. Patient diagnosis was subclassified by morphological and by immunophenotyping. Patients with AML, CML, B-ALL or MDS at the time of diagnosis were characterized in Table 1.
1.
Clinical characteristics of patients
| Diagnosis | No. | Sex (M/F) | Age (y) * | No. of HLA- Gpos patients (%) |
|---|---|---|---|---|
| All patients | 87 | 46/41 | 50.3 ± 19.3 | 14 (16.1) |
| AML | 54 | 28/26 | 50 ± 18.2 | 10 (18.5) |
| AML-M1 | 1 | 0/1 | 70 | 1(100) |
| AML-M2 | 20 | 12/8 | 50.7 ± 20 | 4(20.0) |
| AML-M3 | 14 | 6/8 | 44.3 ± 16.7 | 2(14.3) |
| AML-M4 | 9 | 4/5 | 57.3 ± 17.7 | 0(0) |
| AML-M5 | 11 | 6/5 | 48.9 ± 19.6 | 3(27.3) |
| B-ALL | 13 | 5/8 | 43.7 ± 24.5 | 0(0) |
| CML | 9 | 5/4 | 52.0 ± 19.7 | 2(22.2) |
| MDS | 11 | 8/3 | 58.4 ± 18.4 | 2(18.2) |
| MDS-RA | 6 | 3/3 | 53.2 ± 20.2 | 0(0) |
| MDS-RAEB | 2 | 3/0 | 66 ± 8.5 | 0(0) |
| MDS-RAEBT | 3 | 3/0 | 63 ± 6.6 | 2(66.7) |
HLA-Gpos indicates HLA-G-positive patients.
Age (year) was presented as mean ±standard deviation.
The diagnosis was based on standard cytological criteria according to the French–American–British (FAB) classification. Fifty-four patients with AML, 9 patients with CML, 13 patients with B-ALL and 11 patients with MDS were enrolled in the study, respectively. Mononuclear cell isolation from bone marrow was performed with standard Ficoll-Paque (Sigma, Steinheim, Germany) density-gradient centrifugation method. Mononuclear cells from HLA-G negative (n= 11) and HLA-G strong positive samples (n= 3) were isolated and enriched by CD34 microbeads (Miltenyi Biotec, Auburn, CA, USA) according to the manufacturer's instructions for NK cell cytotoxicity analysis.
Cytogenetic analysis
Cytogenetic analysis of 21 cases from AML was performed using standard G-band karyotyping techniques. After bone marrow cells were processed using a short-term culture (24 hrs), 10 banded metaphases were analysed. Results were described according to the International System for Human Cytogenetic Nomenclature [21].
Flow cytometry
Bone marrow mononuclear cells were separated from freshly drawn anti-coagulated by Ficoll-Paque density-gradient centrifugation. Blasts were identified by CD45 dim/low side scatter characteristics according to Lacombe et al.[22]. A peridinin chlorophyll protein (PerCP)-labelled anti-CD45 antibody (BD, San Jose, CA, USA) was used to differentiate leukaemic cells from normal lymphocytes in each case. HLA-G expression on leukaemic blast cells, which were differentiated from normal lymphocytes by CD45 dim/low side scatter fluorescence, was evaluated. 1 × 106 mononuclear cells were incubated in 200 μL PBS at room temperature for 30 min. in the dark with 10 μL (1 mg/ml) fluorescein isothiocyanate (FITC)-conjugated antihuman HLA-G-specific monoclonal antibody MEM/G9 (IgG1, Serotec, Oxford, UK), and an isotype IgG was added as negative control (BD). After two washing steps with PBS, HLA-G expression was quantified by flow cytometry (FACSCalibur, BD). Ten thousand cells were studied for each analysis. Data were acquired and analysed using CellQuest software (BD).
Cytotoxicity analysis and HLA-G-specific mAb 87G blocking assay
Cytotoxicity analysis was performed using CytoTox96® NonRadioactive Cytotoxicity Assay Kit (Promega, Madison, WI, USA). Effector NK-92 cells (ATCC, MD, USA) were cultured in alpha minimum essential (α-MEM) media without ribonucleosides and deoxyribonucleosides (Gibco BRL, Grand Island, NY, USA) supplemented with 12.5% heat-inactivated foetal calf serum (FCS) (Gibco BRL), 12.5% horse serum, 0.2 mM inositol, 0.1 mM 2-mercaptoethanol, 0.02 mM folic acid and 200 U/ml of rIL-2 (Bioscience, NY, USA) on the day before assay. Effector/target (E:T) ratio was optimized. During cytolysis assay, effector NK-92 cells were mixed with 1 × 104 CD34-enriched blast target cells at a 20:1 E:T ratio in V-bottom 96-well plates (Costar, Cambridge, MA, USA) as protocol instructed. For the cytotoxicity experiments with an HLA-G-specific mAb 87G (gifted by Dr. Geraghty, Fred Hutchinson Cancer Research Center, Seattle, WA, USA) blockade, targets were pre-incubated with 5 μg/ml and 10 μg/ml 87G, respectively, for 30 min. before the effector NK-92 cells were added. Isotype IgG was added as an internal control. Target cell spontaneous release and maximal release of lactic dehydrogenase (LDH) and the effector cell spontaneous release of LDH were determined by incubating these cells in medium alone. Each assay was performed in quadruplicate, and the results are expressed as percentages of lysis ± S.D. The percentage of specific lysis was determined as follows:
Statistical analysis
Comparison of clinical or laboratory parameters was performed using the nonparametric Mann–Whitney U test for continuous variables and the χ2 test for categoric data.
Cytotoxicity differences between groups were analysed for significance by a two-sided Student's t-test. Differences were regarded as significant at the P value less than 0.05.
Results
Cell surface HLA-G expression
HLA-G was expressed in 14 out of 87 (16.1%) patients. Among them, 18.5% in AML patients, 22.2% in CML patients, 18.2% in MDS patients, while no HLA-G-positive patients with B-ALL was observed. The proportion of malignant haematopoietic cells expressing HLA-G molecules varied from 3.47% to 99.69%. For each subgroup of the leukaemic malignancy, the proportion of HLA-G expression on leukaemic cells varied from 3.47% to 99.69%, from 3.8% to 70.0%, from 8.8% to 13.8% for the AML, CML and MDS, respectively (Figs 1 and 2).
1.
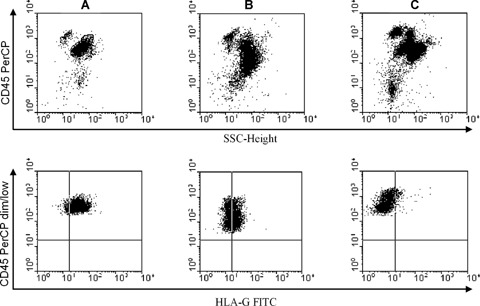
Representative cases of cell surface HLA-G expression in leukaemic blast cells. A PerCP-labelled anti-CD45 antibody was used to differentiate leukaemic cells from normal lymphocytes. Gating on leukaemic blasts characterized by CD45 dim/low side scatter (SSC) (upper panel). HLA-G-specific mAb FITC-labelled MEM-G/09 was used to detect HLA-G expression on the gated CD45 dim/low SSC cells (low panel).(A) High HLA-G expression.(B) Media HLA-G expression.(C) Low HLA-G expression.
2.
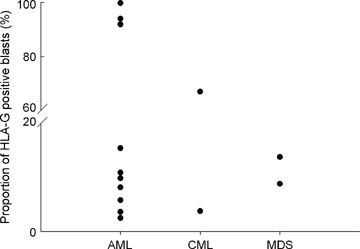
Distribution of HLA-G-positive cases from all patients. HLA-G cell surface expression was analysed by flow cytometry. The dot in vertical represents proportion of HLA-G-positive leukaemic blasts from AML, CML and MDS, respectively.
The relationship between the HLA-G expression and the percentage of blasts and immune cells
The percentage of malignant blasts or immune cells including total T lymphocytes, B lymphocyte and NK cells in HLA-G-positive and HLA-G-negative patients was analysed. Given that a wide variation of the proportion of HLA-G expression on leukaemic cells, which varied from 3.47% to 99.69% in AML patients, the percentage of the blasts and immune cells in these HLA-G-positive patients was analysed with different cut-off level of HLA-G expression. No dramatic difference was observed. In AML patients, the percentage of leukaemic blasts was significantly higher in HLA-G-positive patients (83.1%± 14.7%) than that in HLA-G-negative patients (66.3%± 17.9%) (P < 0.01). Furthermore, the percentage of total T lymphocytes was dramatically decreased in HLA-G-positive AML patients when compared with that in HLA-G-negative AML patients (47.1%± 22.5%versus 65.0%± 18.1%) (P < 0.05). However, no such difference was found between the HLA-G-positive and HLA-G-negative groups in both CML and MDS. Furthermore, no significant difference between HLA-G-positive and HLA-G-negative group was found for the percentage of B lymphocytes and NK cells in all patients studied (Table 2).
2.
Immunologic parameters (Part 1)
| Variable | AML | MDS | |||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| All patients n= 54 | HLA-Gposn= 10 | HLA-Gnegn= 44 | P* | All patients n= 11 | HLA-Gposn= 2 | HLA-Gnegn= 9 | P* | ||||||||||||||||||||||||||||||||||||
| Blast (%) | 69.3 ± 18.4 | 83.1 ± 14.7 | 66.3 ± 17.9 | <0.01 | 18.8 ± 7.7 | 26.9 ± 2.6 | 17.1 ± 7.4 | NS | |||||||||||||||||||||||||||||||||||
| T cell (%) | 61.9 ± 19.9 | 47.1 ± 22.5 | 65.0 ± 18.1 | <0.05 | 56.4 ± 11.7 | 65.0 ± 8.5 | 56.4 ± 12.1 | NS | |||||||||||||||||||||||||||||||||||
| B cell (%) | 13.0 ± 8.5 | 15.0 ± 8.3 | 12.2 ± 8.7 | NS | 10.6 ± 6.2 | 8.5 ± 0.7 | 10.6 ± 7.0 | NS | |||||||||||||||||||||||||||||||||||
| NK cell (%) | 23.0 ± 18.1 | 32.9.0 ± 24.2 | 19.7 ± 14.1 | NS | 30.9 ± 8.8 | 28.0 ± 5.7 | 30.9 ± 9.7 | NS | |||||||||||||||||||||||||||||||||||
2.
Immunologic parameters (Part 2)
| Variable | AML | MDS | |||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| All patients n= 9 | HLA-Gposn= 2 | HLA-Gnegn= 7 | P* | All patients n= 13 | HLA-Gposn= 0 | HLA-Gnegn= 13 | P* | ||||||||||||||||||||||||||||||||||||
| Blast (%) | 28.4 ± 32.0 | 28.43 ± 31.9 | 29.6 ± 34.6 | NS | 73.9 ± 17.9 | 73.9 ± 17.9 | |||||||||||||||||||||||||||||||||||||
| T cell (%) | 60.7 ± 31.0 | 89.0 ± 1.4 | 52.7 ± 30.5 | NS | 72.5 ± 21.6 | 72.5 ± 21.6 | |||||||||||||||||||||||||||||||||||||
| B cell (%) | 22.0 ± 28.1 | 6.0 ± 5.7 | 26.6 ± 30.6 | NS | 22.8 ± 31.1 | 22.8 ± 31.1 | |||||||||||||||||||||||||||||||||||||
| NK cell (%) | 17.2 ± 18.2 | 7.5 ± 5.0 | 20.0 ± 19.9 | NS | 11.8 ± 8.7 | 11.8 ± 8.7 | |||||||||||||||||||||||||||||||||||||
Data are presented as Mean ± standard deviation (S.D.).
NS indicates not significant.
HLA-Gpos indicates HLA-G-positive patients; HLA-Gneg indicates HLA-G-negative patients.
Comparison between the HLA-G-positive and HLA-G-negative patients using the nonparametric Mann–Whitney U test.
HLA-G expression and the bone marrow cytogenetic karyotype
Twenty-one AML samples (5 for HLA-G-positive and 16 for HLA-G-negative) were available for both the HLA-G expression measurement and the cytogenetic karyotyping. All five patients with HLA-G-positive showed cytogenetic karyotype abnormality, including two cases with t(15;17); two cases t(8;21) and 1 case with t(9;11), respectively, while 6 out of 16 HLA-G-negative patients are with cytogenetic karyotype abnormality, including two cases with t(7;11); two cases t(15;17), one case with t(6;9) and one case with +8, +8, +15, respectively. When compared, a significant difference was obtained (P < 0.01) (Table 3).
3.
HLA-G expression status and cytogenetic karyotype
| Patient no. | Diagnosis | Sex | Age | HLA-G status | Karyotype | P* |
|---|---|---|---|---|---|---|
| 1 | AML-M2 | F | 75 | Positive | 46XX, t(15;17)(q22;q21) | <0.01 |
| 2 | AML-M5 | M | 39 | Positive | 46XY, t(9;11)(p11;p13) | |
| 3 | AML-M2 | F | 40 | Positive | 46XX, t(8;21)(q22;q22) | |
| 4 | AML-M3 | M | 19 | Positive | 46XY, t(15;17)(q22;q21) | |
| 5 | AML-M2 | M | 75 | Positive | 46XY, t(8;21)(q22;q22) | |
| 6 | AML-M2a | M | 59 | Negative | 46XY, t(7;11)(p15;p15) | |
| 7 | AML-M2a | M | 52 | Negative | 46XY, t(7;11)(p11;p13) | |
| 8 | AML-M4 | M | 27 | Negative | 46XY, t(6;9)(p23;q34) | |
| 9 | AML-M3 | F | 53 | Negative | 46XX, t(15;17)(q22;q11) | |
| 10 | AML-M3 | M | 62 | Negative | 46XY, t(15;17)(q22;q11) | |
| 11 | AML-M5b | M | 83 | Negative | 49XY, +8, +8, +15 | |
| 12 | AML-M5 | M | 36 | Negative | 46XY | |
| 13 | AML-M3 | M | 28 | Negative | 46XY | |
| 14 | AML-M3 | M | 52 | Negative | 46XY | |
| 15 | AML-M2 | M | 15 | Negative | 46XY | |
| 16 | AML-M2 | M | 59 | Negative | 46XY | |
| 17 | AML-M2 | M | 37 | Negative | 46XY | |
| 18 | AML-M5 | F | 24 | Negative | 46XX | |
| 19 | AML-M3 | F | 16 | Negative | 46XX | |
| 20 | AML-M2 | F | 27 | Negative | 46XX | |
| 21 | AML-M2 | F | 39 | Negative | 46XX |
Comparison between the HLA-G-positive and HLA-G-negative patients using the chi-square test.
HLA-G expression and NK cytolysis against AML leukaemic blasts
HLA-G expression on tumour cells has been supposed to be an important way for malignant cells escaping from host immunosurveillance. We then investigated whether HLA-G protein expression in AML leukaemic cells could be associated with a decreased susceptibility to NK cytolysis. For this purpose, the IL-2-dependent leukaemic cell line NK-92 was used as a model of the NK effector cells. The cell line NK-92 was proved to be an excellent model system to study NK cell biology and KIR functions for its strong target cell cytolysis and well-defined cell surface markers, such as receptors ILT2 and KIR2DL4, which were involved in HLA-G-specific recognition [23, 24]. To test whether HLA-G expression on leukaemic cells from AML could directly exert an inhibition effect on NK cell cytolysis, blockade experiments with the HLA-G-specific mAb 87G were performed, which could exclude the involvement of other HLA molecules present on cells in the inhibition of NK-92 cytolysis. In this ex vivo study, no markedly different cytolysis was observed for the HLA-G-negative leukaemic cells when HLA-G was blocked. However, for the HLA-G-positive leukaemic cells, data showed that the NK cell lysis could be dramatically enhanced in a antibody dose-dependent manner when the cell surface HLA-G was blocked (Fig. 3), suggesting that HLA-G expression could directly protect AML leukaemic cells from NK-92 cell cytolysis.
3.
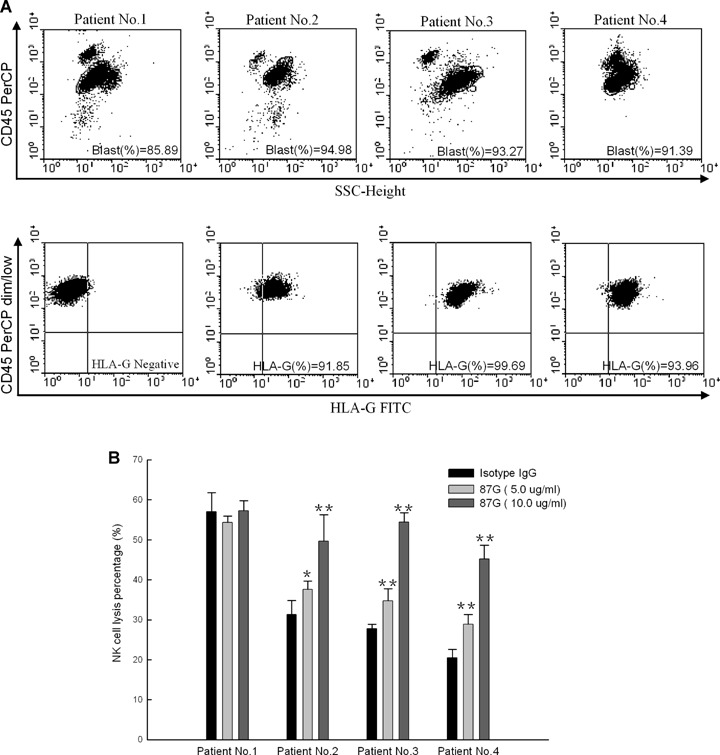
Cytotoxicity of NK cell against leukaemic cells isolated from AML patients and restored with an HLA-G-specific mAb 87G. Results were shown for 5-hr cytotoxicity assay using CD34-enriched leukaemic blasts isolated from different AML patients as the targets with an E:T ratio at 20:1. Experiments were performed in quadruplicate and the results were expressed as percentage of specific lysis ± S.D. (A) A PerCP-labelled anti-CD45 antibody was used to gate blasts that were characterized by CD45 dim/low side scatter (SSC) (upper panel). HLA-G-specific mAb FITC-labelled MEM-G/09 was used to measure HLA-G expression on the gated CD45 dim/low cells (low panel).(B) Cytotoxicity of NK cell against leukaemic cells isolated from AML patients and restored with an HLA-G-specific mAb 87G. For the blockade experiments, target cells were pre-incubated with 5 and 10 μg/ml 87G or an isotype-matched IgG as an internal control, respectively. For HLA-G-negative patients no.1 (n= 11).*P < 0.05, **P < 0.01.
Discussion
HLA-G could inhibit function(s) of immune effector cells through binding to diverse types of receptors, such as ILT2, ILT4 and KIR2DL4 [25]. Although its initial significance had been established in foetal–maternal immunotolerance, HLA-G has been suggested to provide tumour cells with an effective pathway to escape from anti-tumour immune responses [26–28]. In this context, Paul et al.[29] have described for the first time that expression of HLA-G was found in solid tumours, and recently HLA-G expression has been observed in various tumours, including ovarian cancer, lung cancer, colon cancer, melanoma, breast cancer and renal cancer [30].
Few studies had been performed on HLA-G expression in different types of leukaemia patients, however, and data are limited and conclusions remain controversial. A group of previous studies addressed that no cell surface HLA-G was expressed in various types of haematopoietic diseases, such as AML, ALL, CLL and CML [19, 20, 31]. In contrast to these previous studies, a recent report by Nuckel et al.[18] concluded that all 47 cases of B-CLL samples express cell surface HLA-G antigen in a variable proportion of leukaemic tumour cells. Methodology difference such as indirect immunofluorescence binding with the 87G antibody or direct immunofluorescence binding with the MEM/G9 antibody may partially address this controversy.
In the current study, cell surface HLA-G expression varies from 18.5% in AML, 22.2% in CML and 18.2% in MDS patients, respectively. However, no HLA-G expression was detected in B-ALL patients. A recent study by Gros et al.[32] showed that soluble HLA-G antigen were increased during acute leukaemia, especially in subtypes of AML-M4 and AML-M5, as well as in both B- and T-ALL patients. In our study, 3 out of 11 cases of AML-M5 patients were HLA-G-positive, but not detected in both AML-M4 and B-ALL patients. These data are consistent with the point that soluble HLA-G protein seems to be more frequently expressed than membrane-bound HLA-G molecules in haematologic malignancies, such as lymphoproliferative disorders [33].
HLA-G molecules possess potent immune-suppressive properties by inhibition of NK cell functions and proliferation of CD4+ T lymphocytes, and by the induction of CD8+ T lymphocytes apoptosis and CD4+ T lymphocytes anergy [34–36], and by trogo-cytosis, HLA-G could lead to instantly generate T lymphocytes from effectors to regulatory cells [37]. HLA-G expression in CLL patients was found to be associated with both humoural and cellular immnuosuppression [18]. In this study, the total T-cell percentage was dramatically lower in HLA-G-positive AML patients when compared with that in the HLA-G-negative group, while no such significance was found for both B lymphocytes and NK cells. Ex vivo cytotoxicity analysis in our study showed that HLA-G expressing in AML blasts could directly inhibit NK cell cytolysis, which could be restored by blockade of HLA-G with the HLA-G-conformational specific mAb 87G. However, whether functionality of NK cells in AML patients is altered remains unknown.A previous study indicated that NK cytolytic function was impaired in MDS patients partly due to the reduced NK-activating receptors [38]. Apart from this, HLA-G expression could also up-regulate NK cell inhibitory receptors, such as ILT2, ILT3, ILT4 and KIR2DL4, which may take part in immune evasion mechanisms [39]. These data indicated that HLA-G expression in malignant cells may result in host immune suppression, thus favouring tumour cells escaping from anti-tumour immunosurveillance and immune responses.
In addition, HLA-G expression association with a significantly higher percentage of bone marrow blast in AML patients was observed. In the current study, 21 cases of AML patients were available with both HLA-G expression status and cytogenetic karyotyping performed. Unexpectedly, all the HLA-G-positive AML patients were cytogenetic abnormal. The frequency of cytogenetic abnormality was markedly different between the HLA-G-positive and HLA-G-negative AML patients. Be noted, four out of five HLA-G-positive patients were with ‘favourable’ karyotypes, such as t(15;17) and t(8;21) [40, 41]. However, clinical implications of these associations remain unknown, and a larger cohort trial is necessary to confirm this.
In summary, for the first time, we demonstrated that various proportion of leukaemic cells express cell surface HLA-G. HLA-G expression is correlated to a significantly lower total T lymphocyte percentage, rather higher percentage of leukaemic blast cells and cytogenetic abnormalities in AML patients. Furthermore, HLA-G expression could directly inhibit the NK cell cytolysis against the malignant cells. These findings indicated that HLA-G exerts unfavourable clinical roles in AML.
Acknowledgments
This work was supported by the grants from Natural Science Foundation of Zhejiang Province, China (Y205531, Y205575), from the Health Bereau of Zhejiang Province (2007A195) and a grant from Ministry of Personnel, China. We are grateful to Dr. Geraghty (Fred Hutchinson Cancer Research Center, Seattle, WA, USA) for generously providing us antibodies.
References
- 1.Kovats S, Main EK, Librach C, Stubblebine M, Fisher SJ, DeMars R. A class I antigen, HLA-G, expressed in human trophoblasts. Science. 1990;248:220–3. doi: 10.1126/science.2326636. [DOI] [PubMed] [Google Scholar]
- 2.Carosella ED, Moreau P, Le Maoult J, Le Discorde M, Dausset J, Rouas-Freiss N. HLA-G molecules: from maternal-fetal tolerance to tissue acceptance. Adv Immunol. 2003;81:199–252. doi: 10.1016/s0065-2776(03)81006-4. [DOI] [PubMed] [Google Scholar]
- 3.Paul P, Cabestre FA, Ibrahim EC, Lefebvre S, Khalil-Daher I, Vazeux G, Quiles RM, Bermond F, Dausset J, Carosella ED. Identification of HLA-G7 as a new splice variant of the HLA-G mRNA and expression of soluble HLA-G5, -G6, and -G7 transcripts in human transfected cells. Hum Immunol. 2000;61:1138–49. doi: 10.1016/s0198-8859(00)00197-x. [DOI] [PubMed] [Google Scholar]
- 4.Hunt JS, Petroff MG, McIntire RH, Ober C. HLA-G and immune tolerance in pregnancy. FASEB J. 2005;19:681–93. doi: 10.1096/fj.04-2078rev. [DOI] [PubMed] [Google Scholar]
- 5.Shiroishi M, Kuroki K, Rasubala L, Tsumoto K, Kumagai I, Kurimoto E, Kato K, Kohda D, Maenaka K. Structural basis for recognition of the nonclassical MHC molecule HLA-G by the leukocyte Ig-like receptor B2 (LILRB2/LIR2/ILT4/CD85d) Proc Natl Acad Sci USA. 2006;103:16412–7. doi: 10.1073/pnas.0605228103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rajagopalan S, Long EO. A human histocompatibil-ity leukocyte antigen (HLA)-G-specific receptor expressed on all natural killer cells. J Exp Med. 1999;189:1093–100. doi: 10.1084/jem.189.7.1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yan WH, Fan LA. Residues Met76 and Gln79 in HLA-G alpha1 domain involve in KIR2DL4 recognition. Cell Res. 2005;15:176–82. doi: 10.1038/sj.cr.7290283. [DOI] [PubMed] [Google Scholar]
- 8.Shiroishi M, Tsumoto K, Amano K, Shirakihara Y, Colonna M, Braud VM, Allan DS, Makadzange A, Rowland-Jones S, Willcox B, Jones EY, Van Der Merwe PA, Kumagai I, Maenaka K. Human inhibitory receptors Ig-like transcript 2 (ILT2) and ILT4 compete with CD8 for MHC class I binding and bind preferentially to HLA-G. Proc Natl Acad Sci USA. 2003;100:8856–61. doi: 10.1073/pnas.1431057100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ye SR, Yang H, Li K, Dong DD, Lin XM, Yie SM. Human leukocyte antigen G expression: as a significant prognostic indicator for patients with colorectal cancer. Mod Pathol. 2007;20:375–83. doi: 10.1038/modpathol.3800751. [DOI] [PubMed] [Google Scholar]
- 10.Adithi M, Kandalam M, Ramkumar HL, Subramanian A, Venkatesan N, Krishnakumar S. Retinoblastoma: expression of HLA-G. Ocul Immunol Inflamm. 2006;14:207–13. doi: 10.1080/09273940600826497. [DOI] [PubMed] [Google Scholar]
- 11.Lin A, Yan WH, Xu HH, Gan MF, Cai JF, Zhu M, Zhou MY. HLA-G expression in human ovarian carcinoma counteracts NK cell function. Ann Oncol. 2007;18:1804–9. doi: 10.1093/annonc/mdm356. [DOI] [PubMed] [Google Scholar]
- 12.Singer G, Rebmann V, Chen YC, Liu HT, Ali SZ, Reinsberg J, McMaster MT, Pfeiffer K, Chan DW, Wardelmann E, Grosse-Wilde H, Cheng CC, Kurman RJ, Shih IeM. HLA-G is a potential tumor marker in malignant ascites. Clin Cancer Res. 2003;9:4460–4. [PubMed] [Google Scholar]
- 13.Sebti Y, Le Friec G, Pangault C, Gros F, Drénou B, Guilloux V, Bernard M, Lamy T, Fauchet R, Amiot L. Soluble HLA-G molecules are increased in lymphoproliferative disorders. Hum Immunol. 2003;64:1093–101. doi: 10.1016/j.humimm.2003.08.345. [DOI] [PubMed] [Google Scholar]
- 14.Bukur J, Rebmann V, Grosse-Wilde H, Luboldt H, Ruebben H, Drexler I, Sutter G, Huber C, Seliger B. Functional role of human leukocyte antigen-G up-regulation in renal cell carcinoma. Cancer Res. 2003;63:4107–11. [PubMed] [Google Scholar]
- 15.Lefebvre S, Antoine M, Uzan S, McMaster M, Dausset J, Carosella ED, Paul P. Specific activation of the non-classical class I histocompatibility HLA-G antigen and expression of the ILT2 inhibitory receptor in human breast cancer. J Pathol. 2002;196:266–74. doi: 10.1002/path.1039. [DOI] [PubMed] [Google Scholar]
- 16.Urosevic M, Kurrer MO, Kamarashev J, Mueller B, Weder W, Burg G, Stahel RA, Dummer R, Trojan A. Human leukocyte antigen G up-regulation in lung cancer associates with high-grade histology, human leukocyte antigen class I loss and interleukin-10 production. Am J Pathol. 2001;159:817–24. doi: 10.1016/S0002-9440(10)61756-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Paul P, Cabestré FA, Le Gal FA, Khalil-Daher I, Le Danff C, Schmid M, Mercier S, Avril MF, Dausset J, Guillet JG, Carosella ED. Heterogeneity of HLA-G gene transcription and protein expression in malignant melanoma biopsies. Cancer Res. 1999;59:1954–60. [PubMed] [Google Scholar]
- 18.Nuckel H, Rebmann V, Durig J, Duhrsen U, Grosse-Wilde H. HLA-G expression is associated with an unfavorable outcome and immunodeficiency in chronic lymphocytic leukemia. Blood. 2005;105:1694–8. doi: 10.1182/blood-2004-08-3335. [DOI] [PubMed] [Google Scholar]
- 19.Polakova K, Krcova M, Kuba D, Russ G. Analysis of HLA-G expression in malignant hematopoetic cells from leukemia patients. Leuk Res. 2003;27:643–8. doi: 10.1016/s0145-2126(02)00228-x. [DOI] [PubMed] [Google Scholar]
- 20.Mizuno S, Emi N, Kasai M, Ishitani A, Saito H. Aberrant expression of HLA-G antigen in interferon gamma-stimulated acute myelogenous leukaemia. Br J Haematol. 2000;111:280–2. doi: 10.1046/j.1365-2141.2000.02345.x. [DOI] [PubMed] [Google Scholar]
- 21.Mitelman F. International system for human cytogenetic nomenclature. Basel: S. Karger; 1995. [Google Scholar]
- 22.Lacombe F, Durrieu F, Briais A, Dumain P, Belloc F, Bascans E, Reiffers J, Boisseau MR, Bernard P. Flow cytometry CD45 gating for immunophenotyping of acute myeloid leukemia. Leukemia. 1997;11:1878–86. doi: 10.1038/sj.leu.2400847. [DOI] [PubMed] [Google Scholar]
- 23.Maki G, Klingemann HG, Martinson JA, Tam YK. Factors regulating the cytotoxic activity of the human natural killer cell line, NK-92. J Hematother Stem Cell Res. 2001;10:369–83. doi: 10.1089/152581601750288975. [DOI] [PubMed] [Google Scholar]
- 24.Yan WH, Lin A, Chen BG, Zhou MY, Dai MZ, Chen XJ, Gan LH, Zhu M, Shi WW, Li BL. Possible roles of KIR2DL4 expression on uNK cells in human pregnancy. Am J Reprod Immunol. 2007;57:233–42. doi: 10.1111/j.1600-0897.2007.00469.x. [DOI] [PubMed] [Google Scholar]
- 25.Hofmeister V, Weiss EH. HLA-G modulates immune responses by diverse receptor interactions. Semin Cancer Biol. 2003;13:317–23. doi: 10.1016/s1044-579x(03)00022-1. [DOI] [PubMed] [Google Scholar]
- 26.Rouas-Freiss N, Moreau P, Ferrone S, Carosella ED. HLA-G proteins in cancer: do they provide tumor cells with an escape mechanism? Cancer Res. 2005;65:10139–44. doi: 10.1158/0008-5472.CAN-05-0097. [DOI] [PubMed] [Google Scholar]
- 27.Rouas-Freiss N, Moreau P, Menier C, Carosella ED. HLA-G in cancer: a way to turn off the immune system. Semin Cancer Biol. 2003;13:325–36. doi: 10.1016/s1044-579x(03)00023-3. [DOI] [PubMed] [Google Scholar]
- 28.Shih IeM. Application of human leukocyte antigen-g expression in the diagnosis of human cancer. Hum Immunol. 2007;68:272–6. doi: 10.1016/j.humimm.2007.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Paul P, Rouas-Freiss N, Khalil-Daher I, Moreau P, Riteau B, Le Gal FA, Avril MF, Dausset J, Guillet JG, Carosella ED. HLA-G expression in melanoma: a way for tumor cells to escape from immunosurveillance. Proc Natl Acad Sci USA. 1998;95:4510–5. doi: 10.1073/pnas.95.8.4510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tripathi P, Agrawal S. Non-classical HLA-G antigen and its role in the cancer progression. Cancer Invest. 2006;24:178–86. doi: 10.1080/07357900500524579. [DOI] [PubMed] [Google Scholar]
- 31.Amiot L, Onno M, Drenou B, Monvoisin C, Fauchet R. HLA-G class I gene expression in normal and malignant hematopoietic cells. Hum Immunol. 1998;59:524–8. doi: 10.1016/s0198-8859(98)00041-x. [DOI] [PubMed] [Google Scholar]
- 32.Gros F, Sebti Y, De Guibert S, Branger B, Bernard M, Fauchet R, Amiot L. Soluble HLA-G molecules increase during acute leukemia, especially in subtypes affecting monocytic and lymphoid lineages. Neoplasia. 2006;8:223–30. doi: 10.1593/neo.05703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sebti Y, Le Friec G, Pangault C, Gros F, Drénou B, Guilloux V, Bernard M, Lamy T, Fauchet R, Amiot L. Soluble HLA-G molecules are increased in lymphoproliferative disorders. Hum Immunol. 2003;64:1093–101. doi: 10.1016/j.humimm.2003.08.345. [DOI] [PubMed] [Google Scholar]
- 34.LeMaoult J, Krawice-Radanne I, Dausset J, Carosella ED. HLA-G1-expressing antigen-presenting cells induce immunosuppressive CD4+ T cells. Proc Natl Acad Sci USA. 2004;101:7064–9. doi: 10.1073/pnas.0401922101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fournel S, Aguerre-Girr M, Huc X, Lenfant F, Alam A, Toubert A, Bensussan A, Le Bouteiller P. Cutting edge: soluble HLA-G1 triggers CD95/CD95 ligand-mediated apoptosis in activated CD8+ cells by interacting with CD8. J Immunol. 2000;164:6100–4. doi: 10.4049/jimmunol.164.12.6100. [DOI] [PubMed] [Google Scholar]
- 36.Rouas-Freiss N, Goncalves RM, Menier C, Dausset J, Carosella ED. Direct evidence to support the role of HLA-G in protecting the fetus from maternal uterine natural killer cytolysis. Proc Natl Acad Sci USA. 1997;94:11520–5. doi: 10.1073/pnas.94.21.11520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.LeMaoult J, Caumartin J, Daouya M, Favier B, Le Rond S, Gonzalez A, Carosella ED. Immune regulation by pretenders: cell-to-cell transfers of HLA-G make effector T cells act as regulatory cells. Blood. 2007;109:2040–8. doi: 10.1182/blood-2006-05-024547. [DOI] [PubMed] [Google Scholar]
- 38.Epling-Burnette PK, Bai F, Painter JS, Rollison DE, Salih HR, Krusch M, Zou J, Ku E, Zhong B, Boulware D, Moscinski L, Wei S, Djeu JY, List AF. Reduced natural killer (NK) function associated with high-risk myelodysplastic syndrome (MDS) and reduced expression of activating NK receptors. Blood. 2007;109:4816–24. doi: 10.1182/blood-2006-07-035519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.LeMaoult J, Zafaranloo K, Le Danff C, Carosella ED. HLA-G up-regulates ILT2, ILT3, ILT4, and KIR2DL4 in antigen presenting cells, NK cells, and T cells. FASEB J. 2005;19:662–4. doi: 10.1096/fj.04-1617fje. [DOI] [PubMed] [Google Scholar]
- 40.Grimwade D, Walker H, Harrison G, Oliver F, Chatters S, Harrison CJ, Wheatley K, Burnett AK, Goldstone AH. Medical Research Council Adult Leukemia Working Party. The predictive value of hierarchical cytogenetic classification in older adults with acute myeloid leukemia (AML): analysis of 1065 patients entered into the United Kingdom Medical Research Council AML11 trial. Blood. 2001;98:1312–20. doi: 10.1182/blood.v98.5.1312. [DOI] [PubMed] [Google Scholar]
- 41.Grimwade D, Walker H, Oliver F, Wheatley K, Harrison C, Harrison G, Rees J, Hann I, Stevens R, Burnett A, Goldstone A. The importance of diagnostic cytogenetics on outcome in AML: analysis of 1,612 patients entered into the MRC AML 10 trial. The Medical Research Council Adult and Children's Leukaemia Working Parties. Blood. 1998;92:2322–33. [PubMed] [Google Scholar]


