Abstract
The Anopheles gambiae species complex includes the major malaria transmitting mosquitoes in Africa. Because these species are of such medical importance, several traits are typically characterized using molecular assays to aid in epidemiological studies. These traits include species identification, insecticide resistance, parasite infection status, and host preference. Since populations of the Anopheles gambiae complex are morphologically indistinguishable, a polymerase chain reaction (PCR) is traditionally used to identify species. Once the species is known, several downstream assays are routinely performed to elucidate further characteristics. For instance, mutations known as KDR in a para gene confer resistance against DDT and pyrethroid insecticides. Additionally, enzyme-linked immunosorbent assays (ELISAs) or Plasmodium parasite DNA detection PCR assays are used to detect parasites present in mosquito tissues. Lastly, a combination of PCR and restriction enzyme digests can be used to elucidate host preference (e.g., human vs. animal blood) by screening the mosquito bloodmeal for host-specific DNA. We have developed a multi-detection assay (MDA) that combines all of the aforementioned assays into a single multiplex reaction genotyping 33SNPs for 96 or 384 samples at a time. Because the MDA includes multiple markers for species, Plasmodium detection, and host blood identification, the likelihood of generating false positives or negatives is greatly reduced from previous assays that include only one marker per trait. This robust and simple assay can detect these key mosquito traits cost-effectively and in a fraction of the time of existing assays.
Keywords: Infectious Diseases, Issue 96, Mosquito, SNP genotyping, multiplex assay, iPLEX, MALDI-TOF, insecticide resistance, speciation islands, species diagnosis, parasite detection, blood source detection, host preference, infection status
Introduction
Anopheles arabiensis, Anopheles coluzzii and Anopheles gambiae are the major vectors responsible for malaria transmission in Africa1. These three species are morphologically indistinguishable2 and can only be distinguished by molecular assays3-9. In addition, there are many downstream assays routinely conducted to aid epidemiological and population genetics studies. These include (1) a genotyping assay for speciation islands10-12, (2) a genotyping assay recognizing non-synonymous SNPs in the 1014th amino acid codon position of para gene (the knock-down resistance, or kdr, SNP)13-18, (3) parasite detection PCR19-23, and (4) screening for host-specific DNA in mosquito midguts24,25.
We developed a multi-detection assay (MDA) that combines all these assays into a single multiplex reaction with the goal of analyzing epidemiologically important characteristics of malaria vectors in Africa. The MDA assay includes multiple markers for detecting (1) species (A. arabiensis, A. gambiae, A. coluzzii, or other (none of the three)), (2) insecticide resistance represented in kdr SNPs (both L1014Fand L1014S), (3) presence of two major malaria parasites, Plasmodium falciparum and P. vivax, and (4) blood source from one avian and six mammalian hosts.
Traditionally, these assays were conducted in separate polymerase chain reactions. When all these assays are done using a conventional PCR platform, it requires conducting 8-10 PCR reaction assays and accompanying gel electrophoresis steps. Each individual PCR reaction from preparation to documenting results takes 4-5 hr, while the MDA method presented here takes about 5 hr in totality. This is equivalent to a 90% savings in labor costs alone. The MDA presented here costs $5 per sample to genotype all 33 SNPs. This is considerably less expensive than the single agarose gel-based assays, which costs about $1.50 per sample. Assays detecting all the characteristics covered by the MDA would require a minimum of 8-10 separate agarose gel-based assays at a cost of $12-15 per sample. Moreover, the MDA greatly reduces the chance of generating a false positive or negative by utilizing at least three markers for each parasite or host source detection.
The platform we utilized is not limited to malaria vectors but can be used in a wide variety of applications such as medicine, veterinary medicine, and basic biology26-28. In-depth association studies or population genetics studies involving a large (in order of 100s) number of samples require cost-effective assays screening for multiple markers simultaneously. Most studies that utilize two or more separate PCR assays could implement the MDA for quicker results at a lower cost.
Protocol
1. PCR Amplification
Mix all the PCR primers (See supplemental Table S1) in a 1.5ml microtube. Each SNP has two primers (forward and reverse). Ensure that the stock concentration of each PCR primer is kept at 100 μM. Make enough primer mix for 500-1,000 reactions to reduce pipetting error and time at the bench (See supplemental Table S2). If desired, prepare aliquots for 100-200 reactions per tube and store at -20°C.
Prepare PCR cocktail as described in the manufacturer’s protocol (Table 1). The volume for 96 reactions includes an overhang of 20%. Vortex the tube containing the PCR cocktail lightly and centrifuge for 30 sec at 2,000 x g.
Dispense 3 μl of the PCR cocktail into each well of a non-skirted 96 well plate. A repeater pipette can save time for this process.
Add 2 μl of the appropriate genomic DNA of mosquitoes in each well. NOTE: Process of obtaining genomic DNA from mosquito tissues (head/thorax, abdomen or whole body) is described in other literatures29-32. Choose an appropriate DNA extraction protocol to differentiate those with Plasmodium in the blood (abdomen) from infective mosquitoes (Plasmodium in the head/thorax). We typically use DNA extracted from body parts (head/thorax or abdomen) of mosquitoes, so DNA concentration varies from 0.05-2ng/μl. Although the total DNA input is lower than the manufacturer’s recommendation (5-10ng/rxn), we typically get robust SNP calls from 98% of DNA samples without whole genome amplification.
Seal the plate, gently mix or vortex, and centrifuge for 1 min at 370 x g.
Use the following thermocycler protocol to carry out PCR amplification: (1) 94°C for 2 min, (2) 94°C for 30 sec, (3) 56°C for 30 sec, (4) 72°C for 1 min, (5) repeat step (2)-(4) for 45 cycles, (6) 72°C for 1 min, and 4°C hold.
2. SAP Treatment
Prepare the SAP enzyme solution in a 96 well plate (See Table 2). Process 96 samples in a plate. The volume for 96 reactions includes an overhang of 20%.
Vortex the tube of SAP enzyme solution lightly and centrifuge for 30 sec at 2,000 x g. Dispense 2 μl of the SAP enzyme solution into each well. Again, a repeater pipette can save time for this process.
Seal the plate, gently mix or vortex, and centrifuge for 1 min at 370 x g. Incubate the plate as follows: (1) 37°C for 40 min, (2) 85°C for 5 min to inactivate the enzyme, (3) 4°C hold. At completion of this step, store the plate at -20°C before continuing. Process the stored plate within 2 weeks.
3. SNP Extension
If the sample plate is frozen after SAP treatment, allow it to thaw to RT before proceeding.
Prepare the extension primer mix (Supplemental Table S2). Each SNP has one extension primer. Keep the stock concentration of each extension primer at 500 μM. Optionally, aliquot for 100-200 reactions per tube and store at -20°C.
Prepare extension mix as described in Table 3. The 96 reaction volume includes an overhang of 20%.
Vortex the tube of extension cocktail lightly and centrifuge for 30 sec at 2,000 x g. Dispense 2 μl of the extension cocktail into each well. Again, a repeater pipette can save time for this process.
Seal the plate, gently mix or vortex, and centrifuge for 1 min at 370 x g. Thermocycle the plate as shown in Figure 1. At this point, proceed to mass spectrometry or store the plate at -20°C up to 2 weeks.
4. Conditioning the SNP Extension Product to Optimize Mass Spectrometry Analysis
If the sample plate is frozen after SNP extension, allow it to thaw to RT before proceeding.
Transfer 15mg of resin from its container onto the dimple plate using an elongated spoon. Use the scraper to spread resin into the wells of the dimple plate. Sweep the scraper from side to side across the dimple plate to spread the resin. Make sure there is resin in each well.
Scrape excess resin off the dimple plate using the scraper. Let the resin stand in the dimple plate for at least 20 min. While letting the resin stand in the dimple plate, add 41 μl of HPLC grade water to each well of the sample plate.
Gently place the sample plate upside-down, onto the dimple plate. Make sure the sample plate resets against the small rounded post of the dimple plate, which aligns the wells in the sample plate with the wells in the dimple plate.
Holding the sample plate and the dimple plate together, gently flip them over so the resin falls out of the dimple plate into the wells of the sample plate. Tap the dimple plate so the resin falls out into the sample plate. Make sure all the resin in the dimple plate falls out into the sample plate wells. Centrifuge the plate at 3,200 x g for 30 sec.
Rotate the sample plate on a rotator for 5 min at RT. Rotator must rotate the sample plate 360° about the long axis.
Centrifuge the sample plate at 3,200 x g for 5 min. After this step, store the sample plate at -20°C up to 2 weeks before proceeding to the next step.
5. MALDI-TOF Mass Spectrometry
Transfer the conditioned SNP extension product onto a bioarray such as SpectroCHIP as per manufacturer’s instructions. We use MassArray Nanodispenser for this step.
Once the transfer is complete, define how assays and plates are to be set up in the assay database. Name the assay and plates such that each plate has unique identifier that links to specific assay design (Supplemental Table S3).
After assay and plate have been defined, acquire spectra using a mass spectrometer.
6. Data Analysis
Perform data analysis using a real-time PCR analysis data interpretation software such as Typer Analyzer or TyperViewer (Sequenom). Obtain data in .xml format which contains the sample layout, mass spectrogram for each well, the marker information including name, expected mass for each variant of the corresponding SNP, and any error or warning messages associated with each reaction. See Figure 2 for the resulting mass spectrogram.
Carry out SNP genotype clustering analysis by setting the detection threshold (magnitude or signal to noise ratio) and tolerance/variance allowed for each genotype. Figure 3 shows an example of a 04707-KDR#2 SNP genotype clusters.
Export the resulting SNP genotype calls into a spreadsheet program.
Representative Results
Species identification:
The following 5 SNPs together identify three species (A. arabiensis, A. coluzzii and A. gambiae) (Table 4). If a sample is not one of the three species, the three SNPs (01073-213, 04679-157 and 10313-052) fail to amplify.
kdr genotype for inferring insecticide resistance:
The 1014th codon of the para voltage-gated sodium channel corresponds to kdr. Two SNPs on the 2nd and 3rd base of the 1014th codon determine the three possible amino acids. The possible combinations of the genotypes are listed in Table 5. This SNP works for three species of African malaria vectors: A. arabiensis, A. coluzzii and A. gambiae. These SNPs fail to amplify in A. darlingi, a Brazilian malaria vector. This is not surprising given that mitochondrial genome sequence similarity between A. darlingi and A. gambiae is only 88.9% while mitochondrial genome sequence similarity among three African malaria vectors are over 98%. Other species have not been tested. We were successful in amplification from samples collected in Mali, Cameroon, Tanzania and Zambia.
Plasmodium detection:
If intact Plasmodium DNA is present in the mosquito tissue, we expect to detect P. falciparum or P. vivax specific DNA among the DNA sample extracted from the mosquito. The species specific SNPs were identified from sequence alignment of the cytochrome b sequence of 123 P. falciparum, 97 P. vivax, 11 P. malariae and 18 P. ovale isolate sequences from Africa available on Genbank. We designed 6 SNPs which produce unique SNP genotypes to distinguish P. falciparum or P. vivax from other Plasmodium species (Table 6). We have tested this assay on mosquito samples collected in Mali, Cameroon, Tanzania, Zambia and Brazil and confirmed that this particular set of markers works regardless of mosquito species.
Bloodmeal detection PCR:
Cytochrome b sequences from 7 host species (human, chicken, cow, dog, goat, pig and sheep) were collected from Genbank and consensus sequences were generated. SNPs informative for each host were then selected for genotyping. We tested this with blood sources from 7 different animals (Table 7).
Because PCR fragments are short (80-120 bp), this works on somewhat degraded DNA or on partially digested bloodmeals from mosquito tissue. The probability of false positive calls is greatly reduced by having multiple markers per host type.
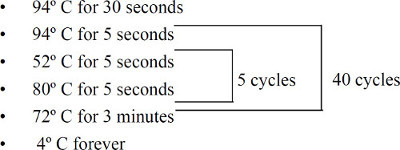 Figure 1. Thermocycler program for SNP extension step. The amplification cycle is total of 200 (5x40).
Figure 1. Thermocycler program for SNP extension step. The amplification cycle is total of 200 (5x40).
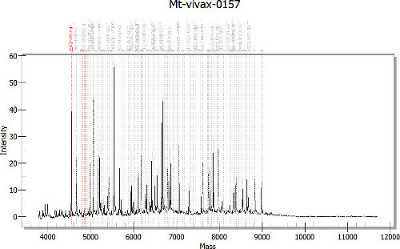 Figure 2. Mass-spectrogram for the MDA. The X axis is mass (Da) of SNP extension products and the Y axis is signal intensity. The Red lines indicate the expected mass of Unincorporated Extension primer, SNP allele 1 and allele 2 for the selected marker. Gray lines indicate expected mass of other markers in the MDA. This plot is generated from data analysis software (Typer Analyzer or Typer Viewer) from the output data file after Step 5. Please click here to view a larger version of this figure.
Figure 2. Mass-spectrogram for the MDA. The X axis is mass (Da) of SNP extension products and the Y axis is signal intensity. The Red lines indicate the expected mass of Unincorporated Extension primer, SNP allele 1 and allele 2 for the selected marker. Gray lines indicate expected mass of other markers in the MDA. This plot is generated from data analysis software (Typer Analyzer or Typer Viewer) from the output data file after Step 5. Please click here to view a larger version of this figure.
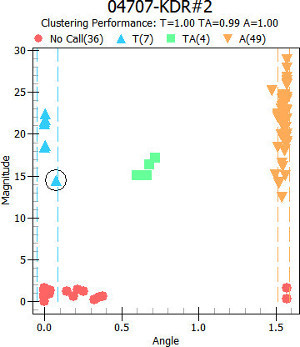 Figure 3. Individual SNP genotype cluster view The X axis is the arctangent value (labeled as “Angle”) of the signal intensities of SNP allele 1 and SNP allele 2. The Y axis is the magnitude of genotype call signal. This plot is provided in the data analysis software (Typer Analyzer or Typer Viewer). Any particular data can be selected with a click of a mouse button and selected sample data is indicated by circle. Clustering analysis provided in the software clusters genotypes with a user-defined variance threshold and automatically colors three genotypes of a biallelic SNP. Example shown here indicates homozygous T (TT) individuals as blue triangles, heterozygote (TA) as green squares and homozygous A (AA) individuals as orange inverted triangles. Red circles represent No call (no amplification). Please click here to view a larger version of this figure.
Figure 3. Individual SNP genotype cluster view The X axis is the arctangent value (labeled as “Angle”) of the signal intensities of SNP allele 1 and SNP allele 2. The Y axis is the magnitude of genotype call signal. This plot is provided in the data analysis software (Typer Analyzer or Typer Viewer). Any particular data can be selected with a click of a mouse button and selected sample data is indicated by circle. Clustering analysis provided in the software clusters genotypes with a user-defined variance threshold and automatically colors three genotypes of a biallelic SNP. Example shown here indicates homozygous T (TT) individuals as blue triangles, heterozygote (TA) as green squares and homozygous A (AA) individuals as orange inverted triangles. Red circles represent No call (no amplification). Please click here to view a larger version of this figure.
| Reagent | Concentration in 5 μl | Volume | Volume |
| (1 rxn) | (96 rxns) | ||
| Water (HPLC grade) | NA | 0.8 μl | 92.2 μl |
| 10x PCR Buffer with 20 mM MgCl2 | 1x (2 mM MgCl2) | 0.5 μl | 57.6 μl |
| MgCl2 (25 mM) ** | 2 mM | 0.4 μl | 46.1 μl |
| dNTP mix (25 mM each) *** | 500 μM | 0.1 μl | 11.5 μl |
| Primer mix (500 nM each) | 100 nM | 1.0 μl | 115.2 μl |
| PCR Enzyme (5 U/μl) | 1.0 U/rxn | 0.2 μl | 23.0 μl |
| Total Volume: | 3.0 μl | 345.6 μl |
Table 1. Multiplex PCR cocktail recipe. The volume of each reagent for the PCR extension step for one reaction and 96 reactions with a 20% overhang.
| Reagent | Volume (1 rxn) | Volume (96 rxn) |
| Water (HPLC grade) | 1.53 μl | 176.3 μl |
| SAP Buffer (10x) | 0.17 μl | 19.6 μl |
| SAP enzyme (1.7 U/μl) | 0.30 μl | 34.6 μl |
| Total Volume | 2.00 μl | 230.4 μl |
Table 2. SAP enzyme solution. The volume of each reagent for the Shrimp Alkaline Phosphatase treatment for one reaction and 96 reactions with a 20% overhang.
| Reagent | Conc. In 9 μl | Volume (1rxn) | Volume (96 rxns) |
| Water (HPLC grade) | NA | 0.6190 μl | 71.3 μl |
| iPLEX Buffer Plus (10x) | 0.222x | 0.2000 μl | 23.0 μl |
| iPLEX Termination mix | 1x | 0.2000 μl | 23.0 μl |
| Extension Primer mix | 0.9400 μl | 108.3 μl | |
| iPLEX enzyme | 1x | 0.0410 μl | 4.7 μl |
| Total Volume | 2.0000 μl | 230.4 μl |
Table 3. SNP Extension cocktail The volume of each reagent for the SNP extension step for one reaction and 96 reactions with a 20% overhang.
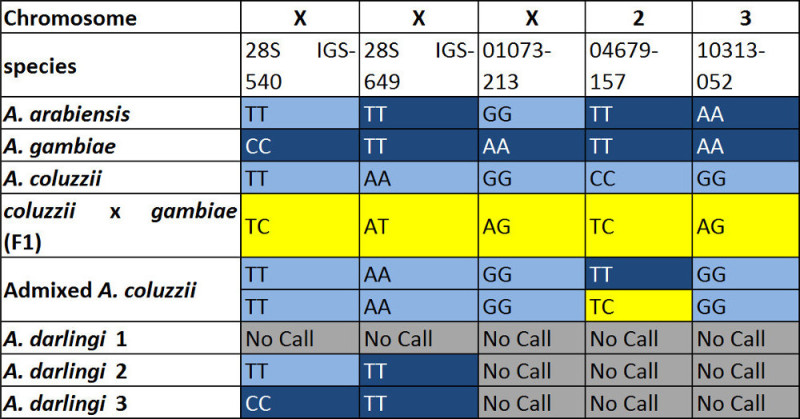 Table 4. SNP genotypes for each species. 28S IGS-540, 28S IGS-649 and 01073-213 are located on the X chromosome, 04679-157 is on chromosome 2 and 10313-052 is on chromosome 3. Hybrid genotype (heterozygote) is highlighted in yellow. Fixed variant in A. coluzzii is marked in light blue and fixed variant in A. gambiae is marked in dark blue.
Table 4. SNP genotypes for each species. 28S IGS-540, 28S IGS-649 and 01073-213 are located on the X chromosome, 04679-157 is on chromosome 2 and 10313-052 is on chromosome 3. Hybrid genotype (heterozygote) is highlighted in yellow. Fixed variant in A. coluzzii is marked in light blue and fixed variant in A. gambiae is marked in dark blue.
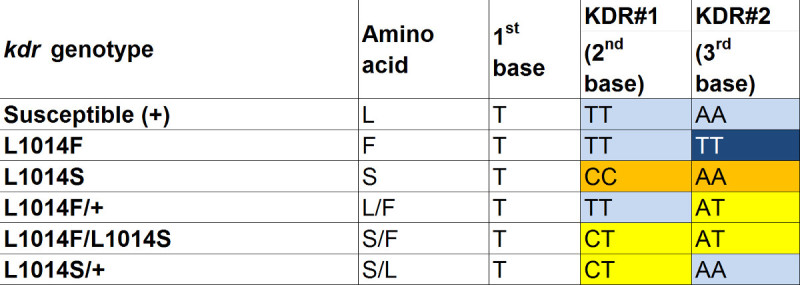 Table 5. SNP genotypes for kdr. Two SNPs KDR#1 and KDR#2 constitute a genotype for the 1014th codon of para gene. The first base is invariable so it is not included in the assay. Susceptible homozygous genotypes are marked in blue. The most resistant L1014F homozygote genotype is marked in dark blue. Yellow color indicates heterozygote. The L1014S, intermediately resistant genotype, is shown in orange.
Table 5. SNP genotypes for kdr. Two SNPs KDR#1 and KDR#2 constitute a genotype for the 1014th codon of para gene. The first base is invariable so it is not included in the assay. Susceptible homozygous genotypes are marked in blue. The most resistant L1014F homozygote genotype is marked in dark blue. Yellow color indicates heterozygote. The L1014S, intermediately resistant genotype, is shown in orange.
 Table 6. SNP genotypes for P. falciparum and P. vivax. Total 6 SNPs are used to determine the presence of malaria parasite. Pl-0041, Pl-1269 and Pl-1549 contain P. falciparum specific alleles (G, T and T respectively). Simultaneous amplification of the three alleles indicates presence of relatively intact P. falciparum DNA in sample DNA. Amplification of alternative alleles (A for Pl-0041 and Pl-1269) indicates presence of other Plasmodium species (e.g., P. vivax, P. ovale or P. malariae) in the sample. Pf-1549 is located in regions with many more P. falciparum specific flanking regions thus this SNP is only amplified when P. falciparum is present. Pl-0071, Pl-1245 and Pl-0157 contain P. vivax specific alleles (G, G and T respectively). Amplification of alternative alleles (A, A and C respectively) indicates presence of other Plasmodium DNA in the sample. Uninfected mosquitoes will have no amplification on all 6 markers.
Table 6. SNP genotypes for P. falciparum and P. vivax. Total 6 SNPs are used to determine the presence of malaria parasite. Pl-0041, Pl-1269 and Pl-1549 contain P. falciparum specific alleles (G, T and T respectively). Simultaneous amplification of the three alleles indicates presence of relatively intact P. falciparum DNA in sample DNA. Amplification of alternative alleles (A for Pl-0041 and Pl-1269) indicates presence of other Plasmodium species (e.g., P. vivax, P. ovale or P. malariae) in the sample. Pf-1549 is located in regions with many more P. falciparum specific flanking regions thus this SNP is only amplified when P. falciparum is present. Pl-0071, Pl-1245 and Pl-0157 contain P. vivax specific alleles (G, G and T respectively). Amplification of alternative alleles (A, A and C respectively) indicates presence of other Plasmodium DNA in the sample. Uninfected mosquitoes will have no amplification on all 6 markers.
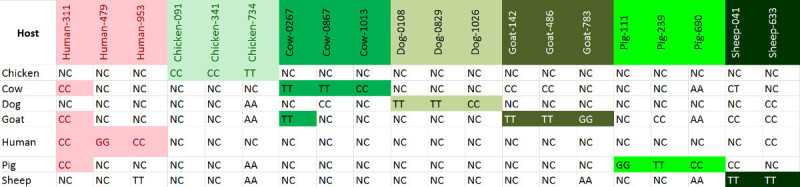 Table 7. SNP genotypes for 7 hosts: Human, Chicken, Cow, Dog, Goat, Pig and Sheep. “NC” stands for “No Call” (no amplification) in the following table. Note that some non-specific amplification may occur at some loci, but not for all the SNPs of a particular host. Please click here to view a larger version of this figure.
Table 7. SNP genotypes for 7 hosts: Human, Chicken, Cow, Dog, Goat, Pig and Sheep. “NC” stands for “No Call” (no amplification) in the following table. Note that some non-specific amplification may occur at some loci, but not for all the SNPs of a particular host. Please click here to view a larger version of this figure.
Discussion
The MDA is composed of five major steps: PCR amplification, shrimp alkaline phosphatase (SAP) reaction, SNP extension, extension product conditioning, and matrix-assisted laser desorption/ionization – time of flight (MALDI-TOF) mass spectrometry33-37. The first PCR amplification step amplifies DNA flanking each SNP so that enough template DNA will be available at the SNP extension step. The SAP reaction neutralizes unused dNTPs which can interfere with the following SNP extension step. SNP extension involves a single extension primer per SNP locus and mass-modified terminator nucleotides. The 3’-end modified nucleotides prevent subsequent nucleotides from being incorporated into the extension primer. Thus the mass of the single base indicates the genotype of the corresponding SNP.
The most critical step in ensuring the success of this approach is having a good dataset of existing polymorphisms in target organisms. We used well characterized SNPs for species identification (N>900) 10,12,38-40 and kdr (N>1,000)15,18. The species specific SNPs for Plasmodium were identified from sequence alignment of the cytochrome b sequences of 123 P. falciparum, 97 P. vivax, 11 P. malariae and 18 P. ovale isolate sequences from Africa which were available on Genbank. Cytochrome b sequences from 7 host species (human, chicken, cow, dog, goat, pig and sheep) were collected from Genbank for host-specific SNP identification. Based on this large body of sequence data, we were able to design a robust diagnostic assay using an assay designer software.
The number of markers each reaction can accommodate is determined by the biochemical compatibility of the primer mix given a tolerance to primer dimer potential (0.9 of 0-1 scale). Authors used the default value (1; most strict) for false priming potential. Relaxation of this parameter can potentially increase the number of SNPs that can be assayed in a single reaction in addition to the SNPs included in this study.
The PCR amplification steps are robust and typically do not require adjustment from the initial assay design. The SNP extension mix (Table 3), however, may need modification in relative quantity of each primer using mass spectrometry to ensure a sufficient signal to noise ratio is achieved for each primer. The provided SNP extension primer recipe is the result of these adjustments. Compatibility among extension primers may limit the total number of SNPs that can be multiplex in a single reaction. Since the reaction volume is small (5-8 µl), the sample plate needs to be properly sealed to prevent evaporation during amplification steps.
This platform can simultaneously score up to 35 SNPs per reaction, while other SNP genotyping platforms can accommodate over one thousand SNPs per reaction41. With the advances in genome sequencing technology, one can acquire millions of SNPs using next-generation sequencing 29,42,43. However, the technique used here provides an ideal application for studies that require genotyping a relatively small number of markers for many (100s-1,000s) individuals. Additional discussion of the design constraints of different SNP genotyping platform is covered in previous studies 41.
The MDA is aimed at characterizing epidemiologically important traits of malaria vectors and provides an ideal protocol for medium-throughput SNP genotyping assays analyzing thousands of mosquitoes collected from natural populations. The MDA presented here provides (1) over a 90% reduction in labor time, (2) a 60% reduction in reagent costs, and (3) reduction in false positives/negatives by utilizing multiple markers. The proposed assay can enhance epidemiological studies by greatly facilitating data collection. Also, it can improve genome-wide association studies by characterizing the mosquito samples with respect to population origin, insecticide resistance, parasite susceptibility and host choice. Finally, this MDA may provide inspiration and a template for designing other multiplex assays using a MALDI-TOF based SNP genotyping platform.
Disclosures
The authors have nothing to disclose.
Acknowledgments
We thank Drs. Anthony Cornel and Laura Norris at UC Davis and Dr. Katharina Kreppel at the University of Glasgow for providing mosquito specimens from Tanzania. We thank Ms. Smita Das and Dr. Douglas Norris from Johns Hopkins School of Public Health for sharing mosquito samples from Zambia. We also thank Mr. Lee V. Millon at the Veterinary Genetics Laboratory for training on assay design. This work was supported by the National Institute of Health grant R01AI 078183 and R21AI062929.
References
- Ayala FJ, Coluzzi M. Chromosome speciation: humans, Drosophila, and mosquitoes. Proc Natl Acad Sci U S A. 2005;102(1):6535–6542. doi: 10.1073/pnas.0501847102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coetzee M, Craig M, le Sueur D. Distribution of African malaria mosquitoes belonging to the Anopheles gambiae complex. Parasitol Today. 2000;16:74–77. doi: 10.1016/s0169-4758(99)01563-x. [DOI] [PubMed] [Google Scholar]
- Favia G, Lanfrancotti A, Spanos L, Siden-Kiamos I, Louis C. Molecular characterization of ribosomal DNA polymorphisms discriminating among chromosomal forms of Anopheles gambiae s.s. Insect Mol Biol. 2001;10:19–23. doi: 10.1046/j.1365-2583.2001.00236.x. [DOI] [PubMed] [Google Scholar]
- Fanello C, Santolamazza F, Torre della, A Simultaneous identification of species and molecular forms of the Anopheles gambiae complex by PCR-RFLP. Med Vet Entomol. 2002;16:461–464. doi: 10.1046/j.1365-2915.2002.00393.x. [DOI] [PubMed] [Google Scholar]
- Santolamazza F. Comparative analyses reveal discrepancies among results of commonly used methods for Anopheles gambiae molecular form identification. Malar J. 2011;10:215. doi: 10.1186/1475-2875-10-215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santolamazza F. Insertion polymorphisms of SINE200 retrotransposons within speciation islands of Anopheles gambiae molecular forms. Malar J. 2008;7:163. doi: 10.1186/1475-2875-7-163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santolamazza F, Della Torre A, Caccone A. Short report: A new polymerase chain reaction-restriction fragment length polymorphism method to identify Anopheles arabiensis from An. gambiae and its two molecular forms from degraded DNA templates or museum samples. Am J Trop Med Hyg. 2004;70:604–606. [PubMed] [Google Scholar]
- Lee Y, Marsden CD, Nieman C, Lanzaro GC. A new multiplex SNP genotyping assay for detecting hybridization and introgression between the M and S molecular forms of Anopheles gambiae. Mol Ecol Resour. 2013. [DOI] [PMC free article] [PubMed]
- Scott JA, Brogdon WG, Collins FH. Identification of single specimens of the Anopheles gambiae complex by the polymerase chain reaction. Am J Trop Med Hyg. 1993;49:520–529. doi: 10.4269/ajtmh.1993.49.520. [DOI] [PubMed] [Google Scholar]
- White BJ, Cheng C, Simard F, Costantini C, Besansky NJ. Genetic association of physically unlinked islands of genomic divergence in incipient species of Anopheles gambiae. Mol Ecol. 2010;19:925–939. doi: 10.1111/j.1365-294X.2010.04531.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahn MW, White BJ, Muir CD, Besansky NJ. No evidence for biased co-transmission of speciation islands in Anopheles gambiae. Philos Trans R Soc Lond B Biol Sci. 2012;367:374–384. doi: 10.1098/rstb.2011.0188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Y, Marsden CD, Nieman C, Lanzaro GC. A new multiplex SNP genotyping assay for detecting hybridization and introgression between the M and S molecular forms of Anopheles gambiae. Mol Ecol Resour. 2014;14:297–305. doi: 10.1111/1755-0998.12181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reimer L. Relationship between kdr mutation and resistance to pyrethroid and DDT insecticides in natural populations of Anopheles gambiae. J Med Entomol. 2008;45:260–266. doi: 10.1603/0022-2585(2008)45[260:rbkmar]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- Weill M. The kdr mutation occurs in the Mopti form of Anopheles gambiae s.s. through introgression. Insect Mol Biol. 2000;9:451–455. doi: 10.1046/j.1365-2583.2000.00206.x. [DOI] [PubMed] [Google Scholar]
- Tripet F. Longitudinal survey of knockdown resistance to pyrethroid (kdr) in Mali, West Africa, and evidence of its emergence in the Bamako form of Anopheles gambiae s.s. Am J Trop Med Hyg. 2007;76:81–87. [PubMed] [Google Scholar]
- Martinez-Torres D. Molecular characterization of pyrethroid knockdown resistance (kdr) in the major malaria vector Anopheles gambiae s.s. Insect Mol Biol. 1998;7:179–184. doi: 10.1046/j.1365-2583.1998.72062.x. [DOI] [PubMed] [Google Scholar]
- Diabate A. KDR mutation, a genetic marker to assess events of introgression between the molecular M and S forms of Anopheles gambiae (Diptera: Culicidae) in the tropical savannah area of West Africa. J Med Entomol. 2003;40:195–198. doi: 10.1603/0022-2585-40.2.195. [DOI] [PubMed] [Google Scholar]
- Chandre F. Current distribution of a pyrethroid resistance gene (kdr) in Anopheles gambiae complex from west Africa and further evidence for reproductive isolation of the Mopti form. Parassitologia. 1999;41:319–322. [PubMed] [Google Scholar]
- Fornadel CM, Norris LC, Franco V, Norris DE. Unexpected anthropophily in the potential secondary malaria vectors Anopheles coustani s.l. and Anopheles squamosus in Macha, Zambia. Vector Borne Zoonotic Dis. 2011;11:1173–1179. doi: 10.1089/vbz.2010.0082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snounou G. High sensitivity of detection of human malaria parasites by the use of nested polymerase chain reaction. Molecular and biochemical parasitology. 1993;61:315–320. doi: 10.1016/0166-6851(93)90077-b. [DOI] [PubMed] [Google Scholar]
- Singh B. A genus- and species-specific nested polymerase chain reaction malaria detection assay for epidemiologic studies. Am J Trop Med Hyg. 1999;60:687–692. doi: 10.4269/ajtmh.1999.60.687. [DOI] [PubMed] [Google Scholar]
- Oyedeji SI. Comparison of PCR-based detection of Plasmodium falciparum infections based on single and multicopy genes. Malar J. 2007;6:112. doi: 10.1186/1475-2875-6-112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gama BE. Real-time PCR versus conventional PCR for malaria parasite detection in low-grade parasitemia. Experimental parasitology. 2007;116:427–432. doi: 10.1016/j.exppara.2007.02.011. [DOI] [PubMed] [Google Scholar]
- Kent RJ, Norris DE. Identification of mammalian blood meals in mosquitoes by a multiplexed polymerase chain reaction targeting cytochrome. B. Am J Trop Med Hyg. 2005;73:336–342. [PMC free article] [PubMed] [Google Scholar]
- Fornadel CM, Norris DE. Increased endophily by the malaria vector Anopheles arabiensis in southern Zambia and identification of digested blood meals. Am J Trop Med Hyg. 2008;79:876–880. [PMC free article] [PubMed] [Google Scholar]
- Teeter KC. The variable genomic architecture of isolation between hybridizing species of house mice. Evolution. 2010;64:472–485. doi: 10.1111/j.1558-5646.2009.00846.x. [DOI] [PubMed] [Google Scholar]
- Han JY. A genome-wide association study of survival in small-cell lung cancer patients treated with irinotecan plus cisplatin chemotherapy. The pharmacogenomics journal. 2014;14:20–27. doi: 10.1038/tpj.2013.7. [DOI] [PubMed] [Google Scholar]
- Chakraborti S. Interaction of polyethyleneimine-functionalized ZnO nanoparticles with bovine serum albumin. Langmuir : the ACS journal of surfaces and colloids. 2012;28:11142–11152. doi: 10.1021/la3007603. [DOI] [PubMed] [Google Scholar]
- Marsden CD. Diversity, differentiation, and linkage disequilibrium: prospects for association mapping in the malaria vector Anopheles arabiensis. G3 (Bethesda) 2014;4:121–131. doi: 10.1534/g3.113.008326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Methods in Anopheles Research. Second edition. MR4; 2010. pp. 1725–1732. Available from: http://www.mr4.org/AnophelesProgram/TrainingMethods.aspx. [Google Scholar]
- Tripet F, et al. DNA analysis of transferred sperm reveals significant levels of gene flow between molecular forms of Anopheles gambiae. Mol Ecol. 2001;10:1725–1732. doi: 10.1046/j.0962-1083.2001.01301.x. [DOI] [PubMed] [Google Scholar]
- Slotman MA. Evidence for subdivision within the M molecular form of Anopheles gambiae. Mol Ecol. 2007;16:639–649. doi: 10.1111/j.1365-294X.2006.03172.x. [DOI] [PubMed] [Google Scholar]
- Basu P, et al. MassARRAY Spectrometry is More Sensitive Than PreTect HPV-Proofer and Consensus PCR for Type-Specific Detection of High-Risk Oncogenic HPV Genotypes in Cervical Cancer. J Clin Microbiol. 2011. [DOI] [PMC free article] [PubMed]
- Fu JF. MassARRAY assay: a more accurate method for JAK2V617F mutation detection in Chinese patients with myeloproliferative disorders. Leukemia. 2008;22:660–663. doi: 10.1038/sj.leu.2404931. [DOI] [PubMed] [Google Scholar]
- Gabriel S, Ziaugra L, Tabbaa D. SNP genotyping using the Sequenom MassARRAY iPLEX platform. Curr Protoc Hum Genet. 2009;2(Unit 2.12) doi: 10.1002/0471142905.hg0212s60. [DOI] [PubMed] [Google Scholar]
- Jurinke C, van den Boom D, Cantor CR, Koster H. The use of MassARRAY technology for high throughput genotyping. Adv Biochem Eng Biotechnol. 2002;77:57–74. doi: 10.1007/3-540-45713-5_4. [DOI] [PubMed] [Google Scholar]
- Wright WT. Multiplex MassARRAY spectrometry (iPLEX) produces a fast and economical test for 56 familial hypercholesterolaemia-causing mutations. Clin Genet. 2008;74:463–468. doi: 10.1111/j.1399-0004.2008.01071.x. [DOI] [PubMed] [Google Scholar]
- Turner TL, Hahn MW, Nuzhdin SV. Genomic islands of speciation in Anopheles gambiae. PLoS Biol. 2005;3:e285. doi: 10.1371/journal.pbio.0030285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner TL, Hahn MW. Locus-population-specific selection and differentiation between incipient species of Anopheles gambiae. Mol Biol Evol. 2007;24:2132–2138. doi: 10.1093/molbev/msm143. [DOI] [PubMed] [Google Scholar]
- Stump AD. Centromere-proximal differentiation and speciation in Anopheles gambiae. Proc Natl Acad Sci U S A. 2005;102:15930–15935. doi: 10.1073/pnas.0508161102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Y, Seifert SN, Fornadel CM, Norris DE, Lanzaro GC. Single-nucleotide polymorphisms for high-throughput genotyping of Anopheles arabiensis in East and southern Africa. J Med Entomol. 2012;49:307–315. doi: 10.1603/me11113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holt RA. The genome sequence of the malaria mosquito Anopheles gambiae. Science. 2002;298:129–149. doi: 10.1126/science.1076181. [DOI] [PubMed] [Google Scholar]
- Lawniczak MK. Widespread divergence between incipient Anopheles gambiae species revealed by whole genome sequences. Science. 2010;330:512–514. doi: 10.1126/science.1195755. [DOI] [PMC free article] [PubMed] [Google Scholar]


