Abstract
Atomic force spectroscopy is an ideal tool to study molecules at surfaces and interfaces. An experimental protocol to couple a large variety of single molecules covalently onto an AFM tip is presented. At the same time the AFM tip is passivated to prevent unspecific interactions between the tip and the substrate, which is a prerequisite to study single molecules attached to the AFM tip. Analyses to determine the adhesion force, the adhesion length, and the free energy of these molecules on solid surfaces and bio-interfaces are shortly presented and external references for further reading are provided. Example molecules are the poly(amino acid) polytyrosine, the graft polymer PI-g-PS and the phospholipid POPE (1-palmitoyl-2-oleoyl-sn-glycero-3-phosphoethanolamine). These molecules are desorbed from different surfaces like CH3-SAMs, hydrogen terminated diamond and supported lipid bilayers under various solvent conditions. Finally, the advantages of force spectroscopic single molecule experiments are discussed including means to decide if truly a single molecule has been studied in the experiment.
Keywords: Physics, Issue 96, AFM, functionalization, single molecule, polymer, lipid, adhesion, atomic force microscopy, force spectroscopy
Introduction
Over the past 30 years, atomic force microscopy (AFM) has turned out to be a valuable imaging technique to study biological 1,2 and synthetic 3 materials and surfaces since it provides molecular spatial resolution in all three dimensions and can be operated in various solvent environments. In addition, AFM-single molecule force spectroscopy (SMFS) enables to measure forces ranging from the pN to µN regime and has given unprecedented insight for example into protein folding 4,5, polymer physics 6–8 , and single molecule-surface interaction9–12.The rationale behind studying single molecules rather than an ensemble of molecules is to avoid averaging effects which often mask rare events or hidden molecular states. Furthermore, a multitude of molecular parameters such as the contour length, the Kuhn length, the adhesion free energy, etc. can be obtained. This is detailed in the examples below. In a typical AFM-SMFS experiment, the probe molecule is coupled to a very sharp tip via a linker molecule. The tip itself is located at the end of a bendable cantilever. If the tip is brought into contact with the surface the probe molecule will interact with this surface. By observing the deflection of the cantilever upon retraction of the tip, the force, and hence the free energy, to detach the molecule from the surface can be determined. To obtain meaningful statistics, a large number of so called force-distance curves have to be acquired. Furthermore, to have true single molecule experiments (i.e., using one and the same probe molecule over the duration of the whole experiment) the probe molecule should be coupled covalently to the AFM tip. Here, an experimental protocol for cantilever functionalization with a single molecule via a covalent bond is presented. The single molecule can either be coupled via an amino or a thiol group to the AFM tip. The conjugation process can be performed in a broad variety of solvents (organic and aqueous) to account for the solvation properties of the polymers used.
In the first part, a general protocol to covalently attach a single molecule (“probe molecule”) via a linker molecule to an AFM tip is described. To this end, organic NHS- or maleimide-chemistry is used13. Along with the protocol for three example molecules, the data acquisition and data analysis processes are described and references for further reading are provided. The example molecules are: the (linear) polymer tyrosine, the graft polymer PI-g-PS and the lipid POPE. This includes slight variations of the protocol, for example to covalently attach cysteines. In addition, a section is dedicated to the preparation of different surfaces such as a diamond surface, a CH3-self-assembled monolayer and lipid bilayers. These interfaces have proven to be good references and examples.
Protocol
NOTE: See Figure 2 for an overview of the process flow comprising the preparation, the data acquisition and data analysis steps.
1. Reagent Setup
NOTE: All chemicals must be handled with care, and thus a lab coat, gloves and eye protection should be used. All operations must be performed in a laboratory hood. In particular, special gloves should be worn in case of chloroform use.
Use chemicals with low water content such as dry chloroform rapidly and store dry but not longer than a week. Store both chemicals at -20 °C and under nitrogen or argon gas because APTES ((3-aminopropyl)triethoxysilan) (see Table 1) and PEG (polyethylenglycol) are hygroscopic and PEG is subject to oxidation in air.
To avoid frequent exposure of the stock to atmospheric oxygen and moisture, prepare smaller aliquots, ideally within a glovebox system with a nitrogen atmosphere.
2. Equipment Setup
NOTE: To avoid possible cross-contamination, use fresh and clean vessels for each step.
Clean glassware and tweezers in detergent solution for 30 min in an ultrasonic bath at 60 °C.
Rinse and sonicate equipment from step 2.1 two times thoroughly with ultrapure water.
Heat equipment from step 2.1 in RCA solution (ultrapure water, hydrogen peroxide and ammonia (5:1:1)) to 75 °C in an oven for 45 min and subsequently rinse them with ultrapure water.
Finally, dry the glassware and tweezers under a stream of dry nitrogen or in an oven (100 °C, 3 hr).
3. Tip Functionalization
NOTE: Use tweezers, vessels, etc. made from stainless steel, PTFE, glass or any other material which is chemically stable in organic solutions if applicable. Unless specified otherwise, carry out all steps at RT. The amount of incubation solution needed depends on the number of cantilever chips. Make sure that the cantilevers are immersed in the respective solutions at all time.
NOTE: Use a hood to avoid inhalation of organic vapors.
- Formation of OH-groups on cantilever surface (‘activation’) (approximately 0.5 hr):
- Use tweezers to place fresh cantilever chips (material: SiN, spring constant: 10-100 pN/nm) on a clean glass slide and put them in a plasma chamber (100 W).
- Evacuate chamber (~0.1 mbar).
- Flood chamber with oxygen gas and evacuate again.
- Activate the plasma process (power: 20%, duration: 15 min, process pressure: 0.25 mbar).
- Amino-silanization of cantilevers (approximately 1 hr):
- Prepare the 2.5 ml of APTES solution (see Table 1) in a glass Petri dish – do this ideally during the plasma process.
- Immediately after the plasma process, dip each cantilever for 1 sec in acetone and place them immediately afterwards in the APTES solution.
- Incubate for 15 min at RT.
- Carefully rinse the cantilever chips twice in 10 ml acetone and once in 10 ml chloroform.
- Optional final step: Place cantilever chips from previous step on a clean glass slide and bake them for 30 min at 70 °C. Note that there are also alternative strategies for surface activation, e.g., UV treatment 12.
- PEGylation (approximately 2 hr): NOTE: Carry out steps 3.3.1-3.3.4 during amino-silanization. NHS and maleimide groups are subject to hydrolysis in aqueous environments and PEG itself is subject to oxidation in air. Therefore timing (especially in between the steps) is a critical parameter. Refer to 13 for further information.
- Prepare the chloroform solution (see Table 1).
- To avoid condensation, warm PEG powders up to RT before opening the aliquot and weighing the appropriate amount.
- For the coupling of polymers or lipids with amino groups, separately solve NHS-PEG-NHS (6 kDa) and methyl-PEG-NHS (5 kDa) in the chloroform solution by vortexing them until they are completely solved. Refer to Table 1 for concentrations.
- Alternatively. for the coupling of polymers with thiol groups separately solve Mal-NHS- PEG and methyl-PEG-NHS in the chloroform solution.
- Mix the solutions as required to adjust a certain number ratio between the NHS- or the maleimide- and the methyl-terminated PEG molecules (typically 1:500). Note that the ideal ratio has to be determined iteratively in a series of preparation-experiment cycles.
- Incubate cantilever chips in the PEG solution for 1 hr within a chloroform saturated atmosphere to prevent evaporation of the chloroform.
- Probe molecule conjugation (> 1 hr): NOTE: In the following 3.4.1-3.4.3, three examples for the covalent coupling of different probe molecules to the AFM tip are described. For each molecule the protocol has to be adjusted. Further note, that in example 1 and 3 NHS-chemistry is used whereas in example 2 the thiol functionalized polymer PI-g-PS is coupled to the PEG via maleimide-chemistry. For details see 14.
- For poly(amino acid) poly-D-tyrosine (40-100 kDa)
- Dissolve the polytyrosine conjugated probe molecule in 1 M NaOH. Adjust the concentration to 1 mg/ml.
- Exchange the NaOH for sodium borate buffer (pH 8.1) using spin desalting columns (7 kDa MWCO) immediately before the functionalization
- Rinse the cantilevers first with 5 ml of chloroform, then with 5 ml of ethanol and finally with 5 ml of borate buffer.
- Incubate the cantilever chips for 1 hr in the polytyrosine borate buffer solution and then rinse with borate buffer and ultrapure water. Store in ultrapure water until the measurement.
- For polymers with a linear backbone (polyisoprene, 119 kDa) having grafted side chains (polystyrene, 88 kDa) 14
- Dissolve the PI-g-PS in dry chloroform. Adjust the concentration of the conjugation solution to 4 mg/ml.
- Rinse the cantilevers with 5 ml of chloroform and incubate for at least 1 hr in the conjugation solution.
- Rinse again in 5 ml of chloroform and store in chloroform in a chloroform-saturated environment (to prevent chloroform evaporation) until the measurement.
- For the lipid POPE
- Dissolve POPE in dry chloroform. Adjust the concentration to 20 mM.
- Rinse the cantilevers with 5 ml of ethanol and 5 ml of ultrapure water before incubation of the cantilevers for 1 hr in the lipid solution.
- After incubation, rinse cantilevers with 5 ml of chloroform, 5 ml of ethanol and 5 ml of warm, ultrapure water (in this order) to remove unbound probe molecules and to get rid of chloroform residues. Use functionalized tips immediately. Alternatively, store them in chloroform until needed.
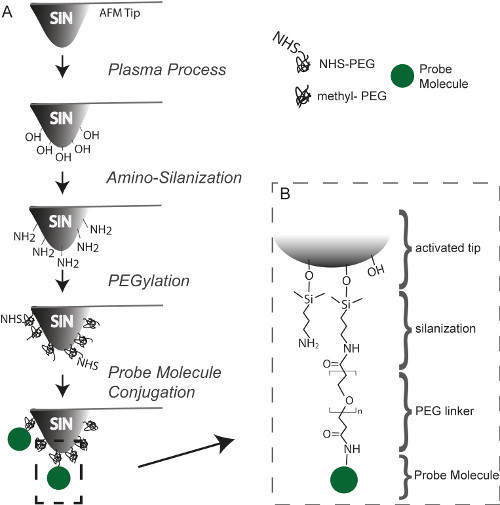 Figure 1. (A) Schematic showing the tip functionalization process using the example of NHS chemistry. (B) Chemical bonding employed to attach a probe molecule to the tip via an amino group. Please click here to view a larger version of this figure.
Figure 1. (A) Schematic showing the tip functionalization process using the example of NHS chemistry. (B) Chemical bonding employed to attach a probe molecule to the tip via an amino group. Please click here to view a larger version of this figure.
4. Surface Preparation
- Self-assembled monolayer (SAM) NOTE: As surface for the first example, a self-assembled monolayer was chosen. Refer to literature 15 for details.
- Clean glass slides with detergent solution and then twice in ultrapure water in an ultrasonic bath for 30 min each. Then, as an additional cleaning step, place them in the RCA solution (see step 2.3) at 75 °C for 15 min.
- Coat the slides with 10 nm chromium nickel and 100 nm gold in a vacuum coater. Then store in the fridge and clean again in the RCA solution directly prior to the next step.
- Incubate the slides of the previous step in 2 mM 1-dodecanthiol/ethanol solution for 12 hr. The thiol groups bind to the gold surface and a hydrophobic monolayer self assembles. Rinse the slides with ethanol and ultrapure water and then dry by a stream of nitrogen gas. Confirm the hydrophobicity of the prepared surface with static contact angle measurements in a goniometer with a CCD camera10.
Hydrogen terminated diamond NOTE: As a surface for the second example, a hydrogen terminated diamond was chosen. The surface preparation was performed as described previously16.
Supported lipid bilayer NOTE: In the last example, a surface supported lipid bilayer (SLB) was used as a surface. To obtain such a surface, a solution (0.1 mg/ml) of large unilamellar vesicles (LUVs) consisting of the phospholipid POPC were formed by the extrusion method. Then 50 µl of the LUV solution were put on a freshly cleaved mica sheet (A = 1 cm²) and incubated for 30 min. Finally the solution was rinsed with 20 ml of ultrapure water. See 17,18 for a detailed description of the preparation of SLBs.
5. Data Acquisition
NOTE: For the experiments, use an AFM, which provides the capability to measure in liquids. The data acquisition and data analysis procedures are applicable regardless of the AFM model used. Furthermore, in some experiments it is advantageous to have the possibility to control the temperature within the liquid cell. Cantilever deflection was detected via the laser beam deflection method 19. Spring constants were determined with the thermal noise method 20.
Insert the functionalized cantilever into an AFM cantilever holder which is suitable for measuring in fluids.
Put the surface (preparation see above) to be sampled into the AFM and cover it with liquid. Both, cantilever and surface should now be immersed in the liquid with the cantilever tip pointing towards the surface. Note that in many cases a drop of liquid is enough. In any case evaporation of the fluid should be avoided, e.g., by using a closed fluid cell.
If applicable, adjust desired temperature on the controller.
Let the system equilibrate for at least half an hour.
Record a thermal noise spectrum of the cantilever with the cantilever far away from the surface in order to exclude any surface damping effects. Note that usually 10 or more spectra have to be accumulated to obtain a sufficient signal-to-noise ratio.
Approach the surface. Note that in order to conserve tip geometry and tip functionalization the approach process has to be carried out carefully. This can be accomplished by, for example, approaching in intermittent contact mode.
- Determine the inverse optical lever sensitivity (InvOLS) measured in nm/volt. Do this by pushing the cantilever tip against a hard surface.
- Determine the InvOLS by measuring the slope of the piezo travel distance vs. the change in photodiode voltage. Fit a line to the part of the force-distance curve where the cantilever tip is in contact with the surface. In experiments involving a soft surface like lipid bilayers, carry out this step on areas which are not covered with a lipid bilayer (‘defect spots’).
Determine the spring constant (pN/nm) by fitting a harmonic oscillator to the thermal noise spectrum. Use automated software function for spring constant determination; otherwise consult literature 20.
- Start the experiment.
- Record numerous force-distance curves at each experimental condition. Typically, use the following values for the experimental parameters: Sampling rate of 5 kHz, tip speed of 1 µm/sec, retract distance of 1 µm. NOTE: Sometimes, depending on the dynamics of the studied experiment, it might be necessary to wait a certain amount of time with the tip in contact with the surface (‘dwell time’). NOTE: The experiment specific parameters can deviate substantially from the values given above.
- During long term measurements, zero the laser position on the photodiode from time to time by repositioning the photodiode.
- Optional: After every force curve, relocate the AFM-tip to another x-y position.
- At the end of the experiment, determine the sensitivity and the spring constant again to check for consistency and stability of the system. NOTE: Comparing the spring constant and sensitivity before and after the experiment is also a means to ensure that the properties of the system and the properties of the tip remained unchanged over the course of the experiment. If the difference of the spring constants is not too large (<=15%), they can be averaged, since the intrinsic uncertainty of spring constant determination is also around 15%. For larger differences, it is advisable to first find and eliminate the cause for the changing apparent spring constant before continuing with further experiments.
6. Data Preparation
NOTE: In this section general data preparation steps which are typically carried out independent of the specific type of experiment to convert units to newton and nanometer as well as to correct the data are described. The experiment specific data analyses are briefly described further below in the respective representative example section.
Typically, carry out all data preparation and analysis steps automatically by a home-written algorithm, e.g., based on the software IGOR Pro 6.
Convert the raw deflection signal (Defl) [V] into force by multiplying it with the InvOLS [nm/V] and the spring constant [nN/nm].
Offset the force in such a way that the force on the cantilever in sufficient distance from the surface (unloaded cantilever) becomes 0 nN.
Calculate the actual tip position (also called separation when measured relative to the surface) z*, which takes the bending of the cantilever into account:

Offset z* in such a way that z* = 0 corresponds to the z position of the surface. NOTE: Here, F is the force which acts on the tip and hence bends the cantilever (determined in step 6.2.), z is the measured z-sensor position (directly given by the instrument) and k represents the spring constant (determined in step 5.8).
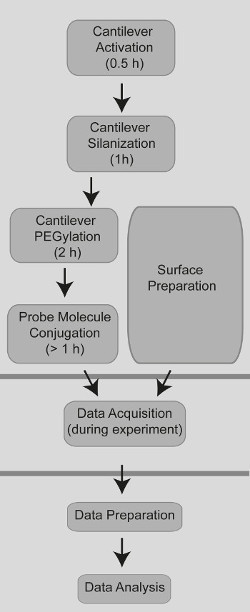 Figure 2. Process flow diagram showing the sample preparation, the data acquisition and the data analysis.
Please click here to view a larger version of this figure.
Figure 2. Process flow diagram showing the sample preparation, the data acquisition and the data analysis.
Please click here to view a larger version of this figure.
Representative Results
In the following, the results for the above described example molecules, namely the polymers poly(amino acid) polytyrosine, the graft polymer PI-g-PS and the phospholipid POPE, are presented. First for each example, experiment specific details for the data acquisition and data preparation are provided. Then, the exemplary results for experiments where these molecules were desorbed from different surfaces (CH3-SAMs, hydrogen terminated diamond and lipid bilayers) are shown. Determination of the adhesion force, the adhesion length and the free energy are introduced.
Example 1: Desorption of polytyrosine from self-assembled monolayer
The desorption of polytyrosine from a hydrophobic SAM in different solutions with a varying content of ethanol was measured. In each solution the cantilever InvOLS and spring constants were determined as described above and at least 300 force distance traces were recorded for each solution. All experiments of one experimental set were done with a single cantilever on a single day to ensure that one and the same single poly(amino acid) was measured. The extension/retraction velocity of the cantilever was 0.5 µm/sec and the dwell time on the surface was 1 sec. The force distance traces showed plateaus of constant force. Each plateau was fitted with a sigmoidal curve and the plateau force as well as the plateau length were determined and plotted in a histogram. The histograms were fitted with a Gaussian and the peak value as well as the standard variation (error) of the distribution was extracted. Until the very end of the force plateau, the system is in equilibrium and the area underneath the plateau therefore equals the free energy per polymer length. Only the detachment process itself at the end of the plateau is in non-equilibrium. Because of this, for typical experiments the plateau length is longer than the equilibrium length (about 30%) and the area underneath the sigmoidal fits an upper estimate for the adhesion free energy 21.
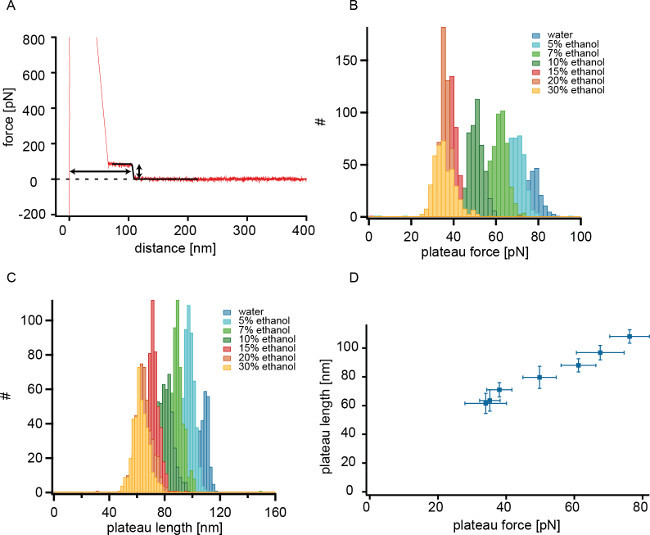 Figure 3. (A) Force distance trace (retraction) of polytyrosine on a methyl-SAM in pure water (shown in red). Only a single molecule desorbs as can be seen in the single step drop of the force to the baseline (zero force). The sigmoidal fit is shown in black and the extracted plateau length and plateau force are indicated by arrows. (B) Histograms of the extracted plateau force for measurements in pure water and in water/ethanol mixtures with different molar ethanol content. (C) Plateau length histograms of the same experimental set. (D) Peak plateau length vs. peak plateau force.” Please click here to view a larger version of this figure.
Figure 3. (A) Force distance trace (retraction) of polytyrosine on a methyl-SAM in pure water (shown in red). Only a single molecule desorbs as can be seen in the single step drop of the force to the baseline (zero force). The sigmoidal fit is shown in black and the extracted plateau length and plateau force are indicated by arrows. (B) Histograms of the extracted plateau force for measurements in pure water and in water/ethanol mixtures with different molar ethanol content. (C) Plateau length histograms of the same experimental set. (D) Peak plateau length vs. peak plateau force.” Please click here to view a larger version of this figure.
Figure 3A shows a single force-distance trace of polytyrosine measured in pure water. Plateau force and plateau length are extracted. If there is only one molecule on the tip that is desorbed from the SAM, the plateau length histogram shows a narrow and monomodal distribution (typically in experiments the standard deviation in a gauss fit of the length histogram is around 5–20 nm). Vice versa, if there’s only one narrow peak in the plateau length histogram, one can be relatively sure that the whole experimental set was done with one and the same polymer. The reason for this is that the polytyrosines are highly polydispersive in size (50 to 100 kDa). Therefore, two polymers of exactly the same length are highly improbable. Adding ethanol weakens the hydrophobic interaction between polytyrosine and the surface 10,22 and leads to a lower adsorption free energy and thus to a lower plateau force as can be seen in the plateau force histograms for the different solvent conditions shown in Figure 3B. Now it can be investigated how the plateau length of a single polytyrosine changes for different adsorption free energies. As can be seen in Figure 3C the plateau length decreases with the addition of ethanol. At the same time the plateau force decreases (Figure 3D).
In order to obtain all relevant parameters of the desorption process namely the contour length, the Kuhn length, the adsorption free energy and the intrinsic monomeric desorption rate, one has to combine the above described measurements with constant distance experiments. For details see 21.
Example 2: Desorption of the graft polymer PI-g-PS from a hydrophobic diamond
In Figure 4A and 4B typical force-distance curves obtained with PI-g-PS on hydrogenated diamond in water at RT are shown. The dwell time on the surface was 1 sec and the velocity of the cantilever was 1 µm/sec. Either plateaus of constant force (Figure 4A) or rupture events were observed (Figure 4B). For the plateaus of constant force, the height of the plateaus was determined with a sigmoidal fit and the values were plotted in a histogram (Figure 4C). To exclude the unspecific adhesion peak caused by direct interaction of the tip with the surface (first peak in the force-distance curve in Figure 4B), the maximal force in some distance from the surface was determined. The obtained values for these rupture events were then plotted in a histogram (Figure 4D). For further analysis the histograms were fitted with a Gaussian and the maximal height was chosen as an average value for the data point. With this kind of experiment the influence of the presence of side chains and their architecture was investigated 14.
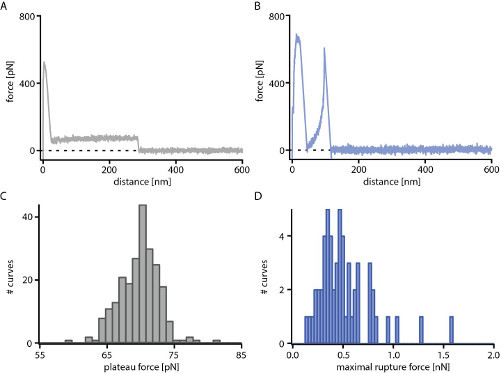 Figure 4. Force-distance retraction curves performed with a linear polyisoprene backbone grafted with polystyrene side chains (PI-g-PS). Either plateaus of constant force (A) or rupture events (B) are observed. For plateaus of constant force, the height of the plateaus is determined with a sigmoidal fit and plotted in a histogram (C) whereas for rupture events, the maximal rupture force is determined (D). Please click here to view a larger version of this figure.
Figure 4. Force-distance retraction curves performed with a linear polyisoprene backbone grafted with polystyrene side chains (PI-g-PS). Either plateaus of constant force (A) or rupture events (B) are observed. For plateaus of constant force, the height of the plateaus is determined with a sigmoidal fit and plotted in a histogram (C) whereas for rupture events, the maximal rupture force is determined (D). Please click here to view a larger version of this figure.
Example 3: Extraction of a single phospholipid (POPE) from a lipid bilayer
Figure 5A shows a schematic representation of the single molecule extraction process. Extraction measurements were performed by vertically approaching the POPE-functionalized AFM tip towards the supported lipid bilayer (not shown). Upon contact of the tip with the POPC bilayer, a small force (~100 pN) is kept constant by a feedback loop for about 4 sec. If the POPE molecule inserts into the bilayer during this dwell time, it is pulled out or ‘extracted’ from the bilayer upon retracting the AFM tip from the bilayer. This results in characteristic force-distance curves. Their shape is basically determined by the stretching elasticity of the PEG linker and can be fitted by a wormlike chain (WLC) model 23. A superposition of more than 50 curves is shown in Figure 5B. True single molecule events can be recognized by single peaked force histograms (Figure 5C). Note that in order to gain further insight into the size of certain molecular parameter (e.g., the mean life time of lipids in bilayers), it is necessary to carry out measurements with different force loading rates. For further analysis of single lipid extraction experiments see 24.
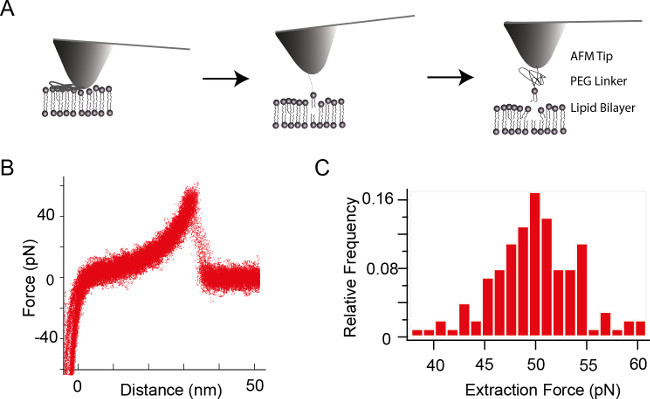 Figure 5. (A) Schematic for the extraction of a single molecule from a lipid bilayer. Note that the lipid molecule is covalently coupled to the tip. Therefore, many hundred or even thousands of force-extraction cycles can be carried out with one and the same molecule. (B) Superposition of more than 50 extraction curves 9. (C) Histogram of the extraction force distribution of the example shown in (B). Please click here to view a larger version of this figure.
Figure 5. (A) Schematic for the extraction of a single molecule from a lipid bilayer. Note that the lipid molecule is covalently coupled to the tip. Therefore, many hundred or even thousands of force-extraction cycles can be carried out with one and the same molecule. (B) Superposition of more than 50 extraction curves 9. (C) Histogram of the extraction force distribution of the example shown in (B). Please click here to view a larger version of this figure.
| Step | Purpose | Solute | Solvent | Typical ratio or concentration |
| Silanization | Create amino groups on cantilever surface | APTES (3-Aminopropyl) tri-ethoxysilane (e.g., Vectabond™) | dry acetone | 1:50 (v/v) – 1:100 (v/v) |
| PEGylation | Provide cantilever surface with amino or thiol reactive groups | NHS-PEG-NHS or Mal-PEG-NHS | dry chloroform | 0.25–25 mM |
| PEGylation | Passivate cantilever surface | methyl-PEG-NHS | dry chloroform | 25 mM |
| Conjugation of probe molecule | Couple probe molecule via an amino or thiol group to bi-functional PEG | probe molecule (polymer, lipid, etc.) | dry chloroform or borate buffer | 0.1–10 mM |
Table 1. Solutions needed for the tip functionalization.
Discussion
During the last decades, single molecule experiments have provided unprecedented insights into molecular mechanisms and turned out to be an invaluable approach in life science and beyond. To achieve good and meaningful statistics from SMFS experiments, ideally one and the same molecule is used over the whole course of the experiment. In contrast to experiments with ensembles of molecules, SMFS experiments are able to detect rare events and hidden molecular states. Another advantage of single molecule experiments is that they can be modeled by means of molecular dynamics simulations 24,25.
The cantilever functionalization protocol described above is a modified version of procedures published previously 26–28. After activation and silanization, the AFM cantilever is functionalized with a mixture of two different types of polyethylene-glycol (PEG) molecules (linker molecules).
The first linker-PEG is terminated by an inert methyl group (“methyl-PEG”) and the second by an amino-reactive N-hydroxysuccinimide( NHS) group (or a thiol reactive maleimide group), where later on the probe molecule is conjugated (“NHS-PEG”, “Mal-PEG”). See Figure 1 for a schematic of the tip functionalization. The purpose of the PEG is threefold: First, it passivates the tip surface and therefore prevents unspecific adsorption. Second, using a certain methyl-PEG/NHS-PEG ratio allows for adjusting the NHS-PEG concentration on the tip surface. And third, it separates the tip from the probe molecule and therefore allows for quantification of the molecule-surface interaction without interference from the direct tip-surface interaction. Then the probe molecule is covalently coupled to the tip via the NHS-PEG (or Mal-PEG).
This protocol makes sure that all bonds (*) in the “coupling chain” tip*OH*silane*PEG*probe molecule are covalent bonds and therefore very strong. This in turn, allows that one and the same probe molecule is measured over the entire duration of the experiment. This has been exemplified above in the representative results section showing true single molecule force spectroscopy.
How can be verified that truly a single molecule is measured? For single traces with plateaus of constant force, a single drop to the baseline is sufficient. If more than one polymer is desorbed at the same time, the force drops to zero in several discrete steps, as each polymer detaches at a different point. More difficult is to decide if it is each time the same single molecule. Therefore the distribution of detachment lengths has to be evaluated. A narrow distribution with a single peak is a good indication that one and the same single molecule was probed in every force-extension trace. For single traces with rupture events, the superposition of all traces is the best method to decide if (one and the same) single molecule was measured. Alternatively, each trace can be fitted with a (WLC) model and then both the persistence length and the rupture lengths at a defined force can be plotted. The distribution of these two parameters has to be narrow and single peaked.
In conclusion, a preparation procedure for the attachment of a large variety of single molecules to AFM tips has been presented. The AFM’s small tip (radius ~ 10 nm), its lateral positioning capabilities and its force sensitivity make it the ideal tool to study the adhesion properties of single molecules on biological and non-biological surfaces in almost any liquid.
Disclosures
The authors declare that they have no competing financial interests.
Acknowledgments
The authors thank the DFG (Hu 997/2-2) for financial support. FS acknowledges the Hanns-Seidel-Stiftung (HSS). SKr was supported by the Elitenetzwerk Bayern in the framework of the doctorate program Material Science of Complex Interfaces. SKi thanks the SFB 863 for financial support.
References
- Scheuring S, Sapra K, Müller D. Probing Single Membrane Proteins by Atomic Force Microscopy. Handbook of Single-Molecule Biophysics. 2009. pp. 449–485.
- Kodera N, Yamamoto D, Ishikawa R, Ando T. Video imaging of walking myosin V by high-speed atomic force microscopy. Nature. 2010;468(7320):72–76. doi: 10.1038/nature09450. [DOI] [PubMed] [Google Scholar]
- Magonov SN. Atomic Force Microscopy in Analysis of Polymers. Encyclopedia of Analytical Chemistry. 2006.
- Rief M, Clausen-Schaumann H, Gaub HE. Sequence-dependent mechanics of single DNA molecules. Nature Structural Biology. 1999;6(4):346–349. doi: 10.1038/7582. [DOI] [PubMed] [Google Scholar]
- Li H, Linke Wa, et al. Reverse engineering of the giant muscle protein titin. Nature. 2002;418(6901):998–1002. doi: 10.1038/nature00938. [DOI] [PubMed] [Google Scholar]
- Bustamante C, Smith SB, Liphardt J, Smith D. Single-molecule studies of DNA mechanics. Current opinion in structural biology. 2000;10(3):279–285. doi: 10.1016/s0959-440x(00)00085-3. [DOI] [PubMed] [Google Scholar]
- Zhang W, Zhang X. Single molecule mechanochemistry of macromolecules. Progress in Polymer Science. 2003;28(8):1271–1295. [Google Scholar]
- Hugel T, Rief M, Seitz M, Gaub H, Netz R. Highly Stretched Single Polymers: Atomic-Force-Microscope Experiments Versus Ab-Initio Theory. Physical Review Letters. 2005;94(4):048301. doi: 10.1103/PhysRevLett.94.048301. [DOI] [PubMed] [Google Scholar]
- Stetter FWS, Cwiklik L, Jungwirth P, Hugel T. Single Lipid Extraction - The Anchoring Strength of Cholesterol in Liquid Ordered and Liquid Disordered Phases. Biophysical journal. 2014;107(5) doi: 10.1016/j.bpj.2014.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pirzer T, Hugel T. Adsorption mechanism of polypeptides and their location at hydrophobic interfaces. Chemphyschem. 2009;10(16):2795–2799. doi: 10.1002/cphc.200900574. [DOI] [PubMed] [Google Scholar]
- Butt H-J, Cappella B, Kappl M. Force measurements with the atomic force microscope: Technique, interpretation and applications. Surface Science Reports. 2005;59(1-6):1–152. [Google Scholar]
- Nash MA, Gaub HE. Single-Molecule Adhesion of a Copolymer to Gold. ACS NANO. 2012;6(12):10735–10742. doi: 10.1021/nn303963m. [DOI] [PubMed] [Google Scholar]
- Hermanson G. Bioconjugate Techniques. New York: Academic Press; 1996. [Google Scholar]
- Kienle S, et al. Effect of molecular architecture on single polymer adhesion. Langmuir the ACS journal of surfaces and colloids. 2014;30(15):4351–4357. doi: 10.1021/la500783n. [DOI] [PubMed] [Google Scholar]
- Folkers JP, Laibinis PE, Whitesides GM. Self-assembled monolayers of alkanethiols on gold: comparisons of monolayers containing mixtures of short- and long-chain constituents with methyl and hydroxymethyl terminal groups. Langmuir. 1995;8(5):1330–1341. [Google Scholar]
- Dankerl M, et al. Diamond Transistor Array for Extracellular Recording From Electrogenic Cells. Advanced Functional Materials. 2009;19(18):2915–2923. [Google Scholar]
- Leonenko ZV, Carnini A, Cramb DT. Supported planar bilayer formation by vesicle fusion: the interaction of phospholipid vesicles with surfaces and the effect of gramicidin on bilayer properties using atomic force microscopy. Biochimica et biophysica acta. 2000;1509(1-2):131–147. doi: 10.1016/s0005-2736(00)00288-1. [DOI] [PubMed] [Google Scholar]
- Stetter FWS, Hugel T. The Nanomechanical Properties of Lipid Membranes are Significantly Influenced by the Presence of Ethanol. Biophysical Journal. 2013;104(5):1049–1055. doi: 10.1016/j.bpj.2013.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Putman CAJ, De Grooth BG, Hulst NF, Greve J. A detailed analysis of the optical beam deflection technique for use in atomic force microscopy. Journal of Applied Physics. 1992;72(1):6–12. [Google Scholar]
- Hutter JL, Bechhoefer J. Calibration of atomic-force microscope tips. Review of Scientific Instruments. 1993;64(7):1868. [Google Scholar]
- Krysiak S, Liese S, Netz RR, Hugel T. Peptide desorption kinetics from single molecule force spectroscopy studies. Journal of the American Chemical Society. 2014;136(2):688–697. doi: 10.1021/ja410278r. [DOI] [PubMed] [Google Scholar]
- Li ITS, Walker GC. Interfacial free energy governs single polystyrene chain collapse in water and aqueous solutions. Journal of the American Chemical Society. 2010;132(18):6530–6540. doi: 10.1021/ja101155h. [DOI] [PubMed] [Google Scholar]
- Dudko OK, Hummer G, Szabo A. Theory, analysis, and interpretation of single-molecule force spectroscopy experiments. Proceedings of the National Academy of Sciences of the United States of America. 2008;105(41):15755–15760. doi: 10.1073/pnas.0806085105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stetter FWS, Cwiklik L, Jungwirth P, Hugel T. Single Lipid Extraction: The Anchoring Strength of Cholesterol in Liquid-Ordered and Liquid-Disordered Phases. Biophysical Journal. 2014;107(5):1167–1175. doi: 10.1016/j.bpj.2014.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horinek D, et al. Peptide adsorption on a hydrophobic surface results from an interplay of solvation, surface, and intrapeptide forces. Proc Natl Acad Sci U S A. 2008;105(8):2842–2847. doi: 10.1073/pnas.0707879105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebner A, et al. Functionalization of probe tips and supports for single-molecule recognition force microscopy. Topics in current chemistry. 2008;285(April):29–76. doi: 10.1007/128_2007_24. [DOI] [PubMed] [Google Scholar]
- Geisler M, Balzer BN, Hugel T. Polymer Adhesion at the Solid-Liquid Interface Probed by a Single-Molecule Force Sensor. Small. 2009;5(24):2864–2869. doi: 10.1002/smll.200901237. [DOI] [PubMed] [Google Scholar]
- Morfill J, et al. Affinity-matured recombinant antibody fragments analyzed by single-molecule force spectroscopy. Biophysical journal. 2007;93(10):3583–3590. doi: 10.1529/biophysj.107.112532. [DOI] [PMC free article] [PubMed] [Google Scholar]


