Abstract
The different genes that encode mammalian spectrins give rise to proteins differing in their apparent stiffness. To explore this, we have compared the thermal stabilities of the structural repeats of brain spectrin subunits (αII- and βII) with those of erythrocyte spectrin (αI- and βI). The unfolding transition mid-points (Tm) of the 36 αII- and βII-spectrin repeats extend between 24 and 82°C, with an average higher by some 10°C than that of the αI- and βI-spectrin repeats. This difference is reflected in the Tm –s of the intact brain and erythrocyte spectrins. Two of three tandem-repeat constructs from brain spectrin showed strong cooperative coupling, with elevation of the Tm of the less stable partner corresponding to coupling free energies of about −4.4 and −3.5 kcal mol−1. The third tandem-repeat construct, by contrast, showed negligible cooperativity. Tandem-repeat mutants, in which a part of the ‘linker’ helix that connects the two domains was replaced by a corresponding helical segment from erythroid spectrin, showed only minor perturbation of the thermal melting profiles, without breakdown of cooperativity. Thus the linker regions, which tolerate few point-mutations without loss of cooperative function, have evidently evolved to permit conformational coupling in specified regions. The greater structural stability of the repeats in αII- and βII-spectrin may account, at least in part, for the higher rigidity of brain compared to erythrocyte spectrin.
Spectrin arose in evolution with the metazoa to meet the need for structures that strengthen cell adhesions and stabilize the plasma membrane against the forces of animal movement (1). The protein also plays a part in organizing plasma membrane signalling complexes (1, 2). Spectrin occurs as an (αβ)2 tetramer, specialized for cross-linking actin filaments to membrane constituents. Both the α and β chains are largely made up of consecutive triple-helical repeating units of about 106 amino acids (3, 4); these act in ensemble as spacers between actin-binding domains in the β-subunits at opposite ends of the tetramers, and some contain binding sites for proteins such as ankyrin or for aminophospholipids (5, 6).
The number of spectrin genes increased during evolution with the advent of vertebrates. Invertebrates have one α-spectrin and, one β-spectrin with 16 complete triple-helical repeats and a βHeavy subunit with 30 complete triple helices. Vertebrates have four genes encoding ‘conventional’ β subunits (βI-IV) that have 16 complete triple helical modules, and one βHeavy subunit that has 30 triple helices (βV-spectrin) (1). Mammals have gained an additional α-spectrin by duplication of the pre-existing α-spectrin gene (7). There is now clear evidence of functional specialization of the two mammalian α-spectrins (αI and αII). The αII-spectrins, which are closest in sequence to invertebrate α-spectrins, are abundant in the cells of complex tissues (8). They are enriched in such locations as post-synaptic contacts (9) (where complexes of signalling molecules reside) and intercalated discs (10, 11) (where forces of muscle contraction are transmitted). The importance of spectrin in the resistance of tissues to the forces generated by muscle contraction is evident in all animals: thus for example, in the worm Caenorhabditis elegans spectrin is required to strengthen adhesion between the body wall and the muscles beneath (12, 13). By contrast, αI-spectrin is abundant in enucleate red blood cells, where it imparts to the membrane the elasticity needed to survive the rigors of circulation (7, 14-15).
The functional distinction between the spectrins of mammalian tissue and erythrocyte is reflected in their physical properties. Spectrin purified from brain (comprising mainly αII and βII polypeptides, also known as fodrin) has a stiffer and straighter appearance in the electron microscope than that of the erythrocyte (αIβI) spectrin (16-19), and this difference is also reflected in their hydrodynamic properties (16, 19). It might be conjectured that the “floppier” appearance of erythrocyte spectrin reflects its adaptation to the requirement for membrane elasticity.
Some 80-90% of each spectrin polypeptide is folded into the sequential triple helical modules, so it might be supposed that structural and functional differences between the proteins have a counterpart in the properties of their constituent domains. Having previously found that the repeating units of αI- and βI-spectrin vary widely in their thermostabilities (20), we were prompted to examine those of αII- and βII-spectrins to investigate the basis of the differences in the gross characteristics of the intact proteins. We have accordingly determined the thermal unfolding properties of each repeat of αII- and βII-spectrin, as well as some constructs comprising selected tandem pairs of repeats, and some with mutations in the single α-helix uniting them.
EXPERIMENTAL PROCEDURES
Materials
Pig brain was purchased from Pel-Freez Biologicals (Rogers, AR), human brain whole marathon-ready cDNA from Clotech (Mountain View, CA), Sephacryl S-500HR, pGEX-4T-2 vector and glutathione-Sepharose 4B from Amersham Biosciences (Piscataway, NJ), protease inhibitor cocktail sets II and III from Calbiochem (San Diego, CA), pET-31b(+) vector and nickel resin from Novagen (Madison, WI), QuickChange site-directed mutagenesis kit and BL21 (DE3) bacteria from Stratagene (La Jolla, CA). Restriction enzymes were from New England BioLabs (Beverly, MA), reduced-form glutathione, thrombin, IPTG, DFP and PMSF from Sigma (St. Louis, MO), SDS-PAGE and electrophoresis reagents from Bio-Rad (Hercules, CA), and GelCode staining reagent from Pierce Biotechnology, Inc (Rockford, IL). Human erythrocyte and pig brain spectrins were purified by the methods of Tyler et al. (21) and Davis and Bennett (19) respectively.
Design and Subcloning of Recombinant Brain Spectrin Polypeptides
The boundaries of all repeats were defined by the SMART database (22). The design of all αII- and βII-spectrin single repeats and tandem-repeat fragments followed that for αI- and βI-spectrin fragments (20). The residue numbers of all sequences are given in Table 1. αIIR14, αIIR18, βIIR9 and βIIR11 were subcloned into the pET31b(+) vector, using NSiI and XhoI. All other brain spectrin single repeats and two-repeat fragments were subcloned into the pGEX-4T-2 vector, using EcoRI and SalI restriction enzymes upstream and downstream respectively. Three different αII-spectrin clones (23) and human brain cDNA were used to amplify the required α-II repeats. For αIIR1, αIIR2, αIIR3 and αIIR4, the template was clone JS1. For αIIR5, αIIR6, αIIR7 and αIIR8, the template was Marathon cDNA. Clone 2.7A was used as template to amplify αIIR9-10, αIIR11, αIIR12 and αIIR13. The template used to amplify the remaining αII-spectrin repeats was clone 3′DA. Similarly, three βII-spectrin clones (24) were used as templates to amplify βII-spectrin single repeats and tandem-repeat fragments. For βIIR1, βIIR2, βIIR3 and βIIR4, βIIR5, βIIR6, βIIR7 and βIIR8, the template was β5.1, while for βIIR9 and βIIR10 it was clone β2.1. Clone β1.20 was the template for amplification of the remaining βII repeats. The fidelity of all the constructs was confirmed by DNA sequencing.
Table 1. Information of αII and βII Spectrin Single Repeats.
| Name | Amino acids | Nucleotides | Name | Amino acids | Nucleotides |
|---|---|---|---|---|---|
| αIIR1 | 39-154 | 115-462 | βIIR1 | 284 - 422 | 850 - 1266 |
| αIIR2 | 145-260 | 433-780 | βIIR2 | 418 - 533 | 1252 -1599 |
| αIIR3 | 251-366 | 751-1098 | βIIR3 | 524 - 643 | 1570 -1929 |
| αIIR4 | 357-472 | 1069-1416 | βIIR4 | 634 - 749 | 1900 - 2247 |
| αIIR5 | 463-578 | 1387-1734 | βIIR5 | 740 - 854 | 2218 - 2562 |
| αIIR6 | 569-683 | 1705-2049 | βIIR6 | 845 - 960 | 2533 - 2880 |
| αIIR7 | 674-789 | 2020-2367 | βIIR7 | 951 - 1067 | 2851 - 3201 |
| αIIR8 | 780-895 | 2338-2685 | βIIR8 | 1058 -1174 | 3172 - 3522 |
| αIIR9-10 | 886-1096 | 2656-3283 | βIIR9 | 1165 -1280 | 3493 - 3840 |
| αIIR11 | 1087-1238 | 3265-3705 | βIIR10 | 1271 -1385 | 3811 - 4155 |
| αIIR12 | 1229-1344 | 3685-4032 | βIIR11 | 1376 -1490 | 4126 - 4470 |
| αIIR13 | 1335-1450 | 4003-4350 | βIIR12 | 1481 -1597 | 4441 - 4791 |
| αIIR14 | 1441-1556 | 4321-4668 | βIIR13 | 1588 -1703 | 4762 - 5109 |
| αIIR15 | 1547-1663 | 4639-4989 | βIIR14 | 1694 -1810 | 5080 - 5430 |
| αIIR16 | 1654-1769 | 4960-5307 | βIIR15 | 1801 -1916 | 5401 - 5748 |
| αIIR17 | 1760-1875 | 5278-5625 | βIIR16 | 1907 -2022 | 5719 - 6066 |
| αIIR18 | 1866-1981 | 5596-5943 | |||
| αIIR19 | 1972-2088 | 5914-6264 | |||
| αIIR20 | 2079-2202 | 6235-6606 | |||
| αIIR21 | 2193-2317 | 6577-6951 |
The boundaries of all repeats were defined by SMART database (http://smart.emblheidelberg.de/) with the exception that five amino acids are extended at both N-terminus and C-terminus to ensure proper folding. The N-terminal six amino acids are from pGEX-4T-2 vector.
Generation of Linker Exchange Constructs
To define the five-amino-acid linker region, we used ClustalW (25) to align the sequences of tandem-repeat fragments αIIR12-13, αIR12-13, αI3-4, αIIR12-13, βI12-13 and βI9-10 with the sequence of PDB entry 1S35. PDB: 1S35 is the tandem-repeat βIR8-9, for which the heptad repeats and linker region have been defined (26). The replacement of αIIR12-13 linker GDSHD by the corresponding αIR12-13 linker NEAQK or αIR3-4 linker QATYW was accomplished by site-directed mutagenesis using the Stratagene QuickChange kit. The same method was used to replace βIIR12-13 linker EEAHR by βIR12-13 linker RDANE or βIR9-10 linker RDNLE. All the sequences were confirmed by DNA sequencing.
Preparation of Recombinant Polypeptides
The cDNA encoding the desired polypeptide was transformed into E. coli BL21. The expression, thrombin-cleavage to remove GST and purification of the polypeptides were performed as described previously (20). The products were dialyzed against phosphate-buffered isotonic saline (buffer A: 10 mM sodium phosphate, pH 7.4, 150 mM sodium chloride) and clarified before use by ultracentrifugation at 230, 000g for 30 min at 4°C. Protein concentrations of the polypeptides were determined spectrophotometrically, using calculated specific absorptivities (27).
Mass Spectrometric Analysis and Sedimentation Equilibrium
Mass spectrometric analyses were performed using an Applied Biosystems (ABI, Foster City, CA) Voyager DE MALDI mass spectrometer. Spectrin polypeptide (2-10 mM) in low-salt solution was mixed with equal volume of a 10mg/mL 3,5-dimethoxy-4-hydroxy-cinnamic acid solution (Sigma). 1 μL of each mixture was spotted onto the MALDI plate. Spectra were calibrated against bovine serum albumin or myoglobin as external standards. Sedimentation equilibrium of all products in isotonic buffer A was performed in a Beckman Coulter ProteomeLab™ XL-A Analytical Ultracentrifuge, as described before. The rotor speed was set at 20,000 rpm and the equilibrium distributions were scanned at 280 nm. A good fit to a monodisperse ideal model was obtained in all cases.
Circular Dichroism Spectra and Thermal Unfolding
Far-UV CD spectra and temperature-induced unfolding were performed as described before (20). To determine the melting profiles, the ellipticity at 222 nm in buffer A was recorded at temperature intervals of 2°C. The data were corrected for the linear changes above and below the sigmoid melting transition, and expressed in terms of fraction unfolded. Coupling free energies, representing the conformational stabilization of one repeat in the tandem-repeat fragments by the other, were derived from van’t Hoff plots of the corrected melting profiles of the isolated repeats and the lower-melting phases of those of the tandem repeats (20). Approximate free energy of unfolding of various single repeats at 37°C was also calculated as described previously (20).
RESULTS
Recombinant αII- and βII-spectrin Polypeptides
Table I lists the boundaries of the αII- and βII-spectrin repeats. The fragments, expressed as GST-fusion constructs, contained six additional amino acids from the vector at their N-termini. Mass spectrometry confirmed that all had the expected molecular mass, and sedimentation equilibrium showed them to be monodisperse and monomeric in all cases. Circular dichroism spectra were characteristic of polypeptide chains of high α-helicity.
Temperature-induced Unfolding of αII- and βII-Spectrin Single Repeats
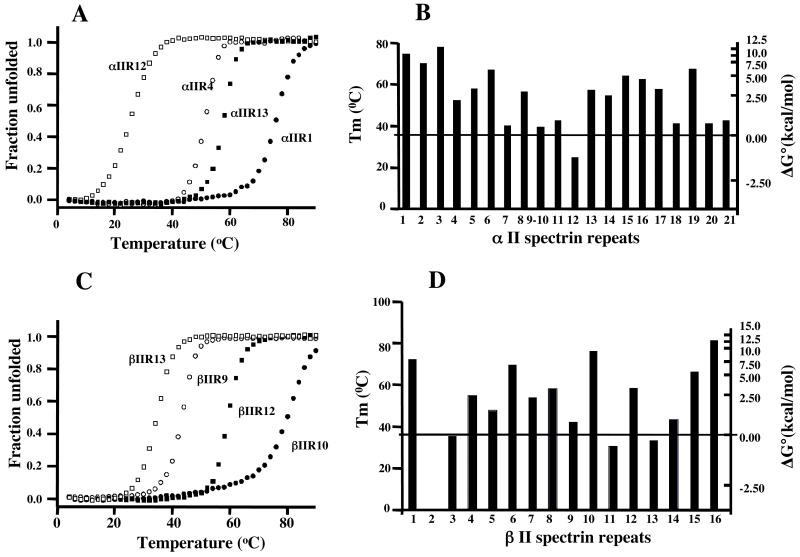
We expressed and purified all αII- and βII-spectrin repeats and measured their thermal stabilities. All the repeats gave rise to typical sigmoidal unfolding equilibrium profiles, measured by temperature-dependence of circular dichroism at 222 nm. Figs 1A and 1C show representative corrected unfolding profiles of αII- and βII-spectrin repeats respectively. Figs 1B and 1D display the stability distribution of the repeats, in terms of the denaturation midpoints (Tm), along the αII- and βII-spectrin chains. These reveal appreciable differences from the corresponding results for erythrocyte spectrin (20). Thus, (1) whereas eight of the 36 isolated repeats of the latter are more than 50% unfolded at physiological temperature, there are only four repeats of such low stability in αII/βII-spectrin. (2) While the Tm values of erythrocyte αI/βI-spectrin repeats lie between 21 and 72 °C, those of αII/βII-spectrin range from 25 to 81 °C. (3) The Tm averaged over all isolated repeats is higher by some 10 °C in αII/βII- than in αI/βI-spectrin (55.5, as against 46°C). The relationship between Tm and approximate free energy of unfolding of the repeats at 37°C is also shown in Figs 1B and 1D. It is approximate only because the relation between Tm and ΔG° at the reference temperature (37°C ) is not fixed inasmuch as it varies with the enthalpy of the unfolding transition.
Fig. 1.
A. Typical thermal unfolding profiles, measured by circular dichroism at 222 nm, corrected for linear changes above and below the sigmoidal unfolding transition, of expressed single repeats of αII spectrin. The curves, from left to right, show unfolding of repeats 12, 4, 13 and 1. B. The Tm of all repeats along the αII spectrin chain. The horizontal line corresponds to physiological temperature. The scale on the right shows the approximate free energy of unfolding of the repeats at 37°C, and applies also to the β-chain repeats in panel D. C. Typical thermal unfolding profiles for single repeats of βII spectrin, corrected as above. The curves, from left to right, are show unfolding of repeats 13, 9, 12 and 10. D. The Tm of all repeats along the βII spectrin chain.
Temperature-induced Unfolding of Tandem-Repeat Fragments
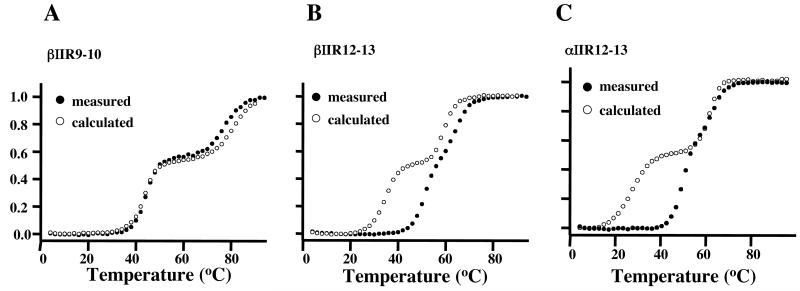
The thermal unfolding profiles of tandem-repeat fragments of erythrocyte spectrin demonstrated varying coupling, with stabilization of the less stable by the more stable unit (20); in all the cases that we examined, however, the biphasic character persisted, showing that cooperativity was by no means complete. Here we have examined the unfolding of three tandem-repeat fragments of brain spectrin, αIIR12-13, βIIR9-10 and βIIR12-13. (Two additional such fragments, βIIR10-11 and βIIR11-12, were also prepared, but problems of aggregation vitiated the quality of the data, which we did not attempt to analyze.) The fragments were chosen to embody two folding units differing most widely in stability, as seen in Fig. 1, and affording the best reflection therefore of cooperativity. As Fig 2 shows, the cooperativity is minimal in one of the fragments but large in the other two. Thus in both βIIR12-13 (Fig 2B) and αIIR12-13 (Fig 2C) the conformation of the repeat of lower stability is substantially stabilized by interaction with its more stable neighbor (ΔTm ca.16 and 19° respectively). The corresponding coupling free energies could be readily calculated as before (20), most usefully at the Tm of the less stable repeat in its isolated state (where its unfolding free energy is zero), and were, found to be ca. −4.4 and −3.5 kcal mol−1 for αIIR12-13 and αIIR12-13 respectively. In the case of βII12-13 (Fig 3B) there is evidence (besides the large stabilization of the conformation of the less stable repeat) of a small but distinct stabilization of the more stable repeat by the attached unfolded repeat. This is not in itself remarkable, and indeed has been reported before for a similar fragment (αII16-17) of chicken brain spectrin (28).
Fig. 2.
Thermal unfolding profiles of tandem repeat constructs from αII and βII spectrin chains. A. Fragment comprising repeats 9 and 10 of βII-spectrin; B. Fragment of repeats 12 and 13 of βII-spectrin; C. Fragment of repeats 12 and 13 of αII-spectrin. All curves are corrected for linear slopes above and below the transition. Closed circles are observed data; open circles are calculated for independent unfolding of the two repeats. Note strong cooperativity of melting of the constituent repeats in B and C, while there is no perceptible coupling of the repeats in A.
Fig. 3.
Thermal unfolding profiles of two-repeat fragments of brain spectrin with mutated linker regions. A. Fragment comprising repeats 12 and 13 of the αII chain: wild-type (solid line); substituent linker of αI-spectrin repeats 12 and 13 (●); substituent linker of αI-spectrin repeats 3 and 4 (○). B. Fragment comprising repeats 12 and 13 of the βII chain: wild-type (solid line); substituent linker of βI-spectrin repeats 12 and 13 (●); substituent linker of βI-spectrin repeats 9 and 10 (○). Note that in no case do the substitutions cause any perceptible loss of cooperativity.
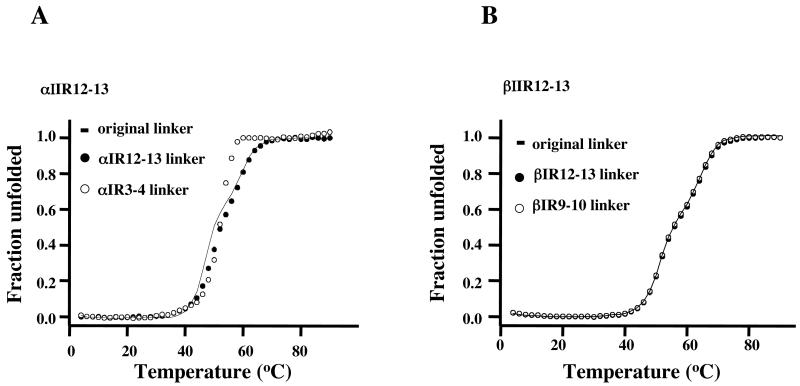
Contribution of “Linker” Region to Cooperativity
MacDonald and Cummings (29) have inferred from secondary structure predictions by two methods that the α-helical propensities of sequences (‘linkers’) of 5 residues at the center of the α-helix uniting adjacent repeats of erythrocyte spectrin govern the conformational stability of the tandem-repeat fragment. To determine whether the properties of the two tandem repeats (αIIR12-13 and βIIR12-13) that we have found to display strong cooperativity are affected by the nature of the linker, we prepared two mutants of each. In αIIR12-13 the sequence in question was replaced by the corresponding residues of αIR12-13 and of αIR3-4; in βIIR12-13 the linker residues were replaced by the analogous residues from βIR12-13 and βIR9-10. The substituents were chosen because all the erythrocyte spectrin tandem-repeat fragments from which they derive display only weak to moderate cooperativity (20). As Fig 3A shows, the substitutions have relatively small effects on the unfolding profiles, although that of the erythroid αIR12-13 linker causes a discernible increase in stability, with no loss of cooperativity. The erythroid αIR3-4 linker engenders in addition a perceptible increase in cooperativity. Again (Fig 3B) the substitutions caused no major perturbation of the unfolding profiles of βII12-13.
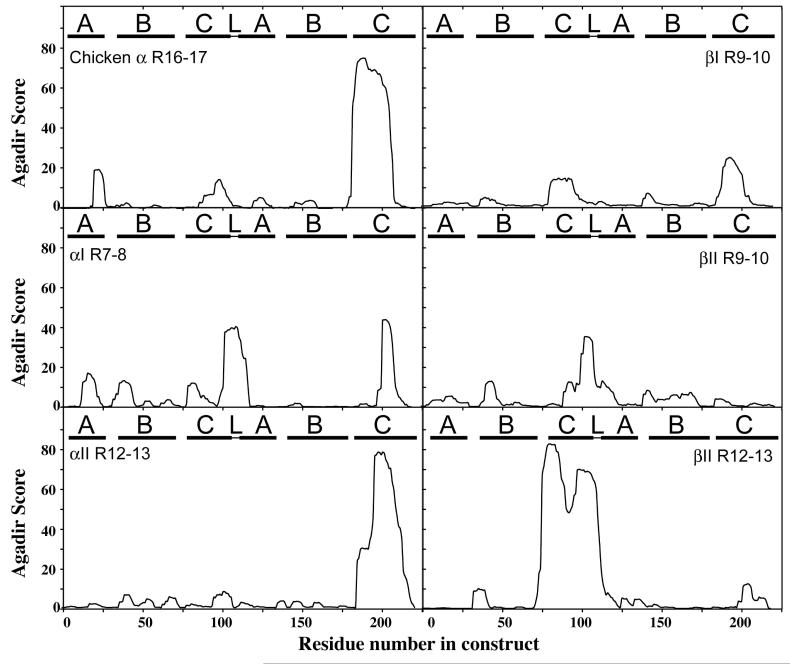
MacDonald and Cummings (29) suggested that the secondary structure of the linker regions dictates the stability of folding in pairs of adjacent repeats. They also noted a partial breakdown of cooperativity when the linker region was predicted not to be helical, but this was not a large effect, because in no cases, whether the denaturing agency was heat or urea, separate melting phases were observed. MacDonald and Cummings used for predictions first the DSC routine (30), which failed to predict correctly the structure of one linker that had been determined by crystallography, and secondly the PSIPRED routine (31), which delivered the correct answer. Since our data indicate that the relationship between cooperative coupling and linker sequence is more complex than the predictions of MacDonald and Cummings might suggest, we sought alternative means of investigating it. We made sequence alignments of highly cooperative and weakly cooperative pairs from multiple organisms, and we also ran predictions of the propensity of the sequences in the tandem pairs to form α-helices, using the program Agadir (32). Inspection of the multiple sequence alignments disclosed no motifs or characteristic arrangement of types of amino acids that distinguished the different classes of pairs (Fig 4). Likewise, Agadir gave no consistent indication of the contribution of helical propensity either in the linker or in the helices that it joins.
Fig. 4.
Agadir analysis of helical propensity of triple helical pairs. The figure shows predictions of helical propensity for the indicated paired spectrin repeats: chicken α-spectrin repeats 16-17 (from sequence given in PDB:1U4Q), human αI R7-8, human αII R12-13, human βI R9-10, human βII R9-10, human βII R12-13. Sequence corresponding to the helices A, B and C of the repeats is indicated as well as the linker (L) sequence.
At present, we cannot envisage any mechanism that might allow the linker to engender cooperative folding between repeats. Batey et al. (28) show that mutations in the linker sequence can eliminate coupling. The only conclusion that seems compatible with all available data is that linkers between triple helical repeats have evolved to allow or exclude cooperative coupling as required.
Intact Brain Spectrin
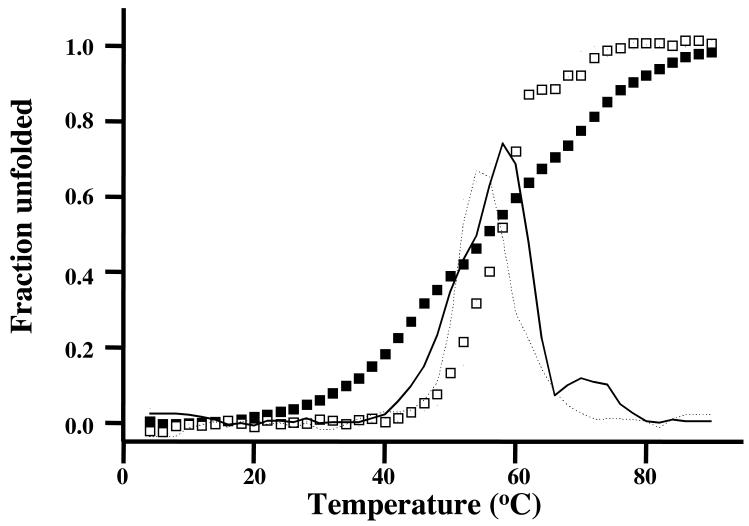
The denaturation profile of pig brain spectrin tetramer is shown in Fig 4. This demonstrates first that there is extensive cooperative coupling between many of the repeats along the brain spectrin chains, since unfolding occurs much more sharply than calculated by merely summing the profiles of all individual repeats; secondly that the thermal stability of brain spectrin is greater than that of erythroid spectrin (Tm respectively 49.5 and 58 °C). We have assumed that the result for the brain spectrin tetramer will hold good also for the human protein, in view of the high degree of sequence conservation in mammalian species (7).
DISCUSSION
We have shown that the individual brain spectrin repeats, like those of erythroid spectrin, exhibit a wide range of thermal stabilities. The extent to which the low stability of certain of the repeats, such as αIIR12 and βIIRll, which are substantially unfolded at 37°C (Fig 1), impacts on the hydrodynamic flexibility of the intact protein remains uncertain in consequence of the extensive (though varying) stabilizing interactions between successive repeats along the chain. Thus, the tandem-repeat construct, βIIR9-10 shows little interaction between its constituent parts, whereas in βIIR12-13 the free energy of coupling at the Tm of the isolated βIIR13 is some 4.4 kcal mol−1. Even here, however, the shape of the unfolding profile indicates that cooperativity is incomplete, for the perceptible shoulder in the region of the mid-point reflects a significant weight of the half-unfolded form. This contrasts with the tandem repeat αIIR16-17 of chicken spectrin, which by this criterion exhibits completely cooperative melting (28). We cannot of course, on the basis of a circular dichroism profile alone, exclude small deviations from two-state behavior, such as, for instance, comparison with a fluorescence-based profile can reveal (28).
Law et al (33, 34) have demonstrated that some triple helices can be unfolded in physiological conditions by an applied pulling force, well within the range of the shearing forces encountered by the red cell in the circulation. Law et al. (34) have also shown that the magnitude of conformational coupling in forced unfolding diminishes with increasing temperature. As an extreme example of this, the 5-repeat fragment of erythrocyte spectrin βIR5-9 loses resistance to pulling in the atomic force microscope in two of its constituent repeats (R8-9) below 37°C (20). The resulting local extension of a repeat to 4-5 times its folded length must be supposed to occur reversibly in red cells in the circulation.
Our data reveal major evolutionary adaptations between αI- and αII-spectrins. The αI-spectrin confers on the erythrocyte membrane the elasticity required for survival in the circulation. It has 6 repeats with Tm at or below 37°C (20). These repeats show substantially incomplete conformational coupling with their neighbors. By contrast αII-spectrin has only one repeat with Tm below 37°C, and this is conformationally coupled to R13, so that the αIIR12-13 pair has Tm substantially higher than 37°C.
It remains to consider the structural basis for the much greater stiffness of brain spectrin than erythroid spectrin, apparent from its hydrodynamic properties (16, 19) and its appearance in the electron microscope (16-19). Fig 4 shows that brain spectrin also has the greater thermal stability, as previously observed by Di Stasi et al. (35) for bovine brain and erythrocyte spectrins, and also high conformational coupling throughout. A high degree of stiffness could perhaps be related to the close lateral association between the αII- and βII-chains, apparent in electron microscope images of brain spectrin. Law et al. (36) have shown that lateral association (which in erythroid spectrin is confined to a run of four repeats in each chain at one end of the dimer (37)) promotes cooperative unfolding. Constructs of αII- and βII-spectrin, other than those containing the corresponding four repeats, show, like those of the αI- and βI-proteins (38), little interaction in vitro (unpublished data in this laboratory). At the same time, the greater average structural stability along the array of repeats in both brain spectrin chains, even if a few partly unfolded in the low-temperature range at the foot of the melting profile, must be expected to reduce the incidence of conformationally weak regions. The asymmetry apparent in the derivative curve in Fig. 4 suggests that there is some clustering of stable and less stable repeats in the chains, and/or of regions in which cooperativity is greater or less. The data we have presented for the entire range of repeats should assist in the selection of chain elements for further study.
Fig. 5.
Thermal unfolding profile of intact αIIβII-spectrin (open squares) and the summed melting profile of all single repeats (closed squares). The first order derivative for αIIβII-spectr in (solid line) and αI βI-spectrin (dashed line) are also shown.
Acknowledgments
This work was supported in part by NIH grants DK 26263, DK 32094 and HL31579
The abbreviations used are
- CD
circular dichroism
- Tm (°C)
thermal denaturation midpoint
REFERENCES
- 1.Bennett V, Baines AJ. Spectrin and ankyrin-based pathways: metazoan inventions for integrating cells into tissues. Physiol Rev. 2001;81:1353–92. doi: 10.1152/physrev.2001.81.3.1353. [DOI] [PubMed] [Google Scholar]
- 2.Pinder JC, Baines AJ. A protein accumulator. Nature. 2000;406:253–4. doi: 10.1038/35018679. [DOI] [PubMed] [Google Scholar]
- 3.Yan Y, Winograd E, Viel A, Cronin T, Harrison SC, Branton D. Crystal structure of the repetitive segments of spectrin. Science. 1993;262:2027–30. doi: 10.1126/science.8266097. [DOI] [PubMed] [Google Scholar]
- 4.Speicher DW, Marchesi VT. Erythrocyte spectrin is comprised of many homologous triple helical segments. Nature. 1984;311:177–80. doi: 10.1038/311177a0. [DOI] [PubMed] [Google Scholar]
- 5.Kennedy SP, Warren SL, Forget BG, Morrow JS. Ankyrin binds to the 15th repetitive unit of erythroid and nonerythroid beta-spectrin. J Cell Biol. 1991;115:267–77. doi: 10.1083/jcb.115.1.267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.An X, Guo X, Sum H, Morrow J, Gratzer W, Mohandas N. Phosphatidylserine binding sites in erythroid spectrin: location and implications for membrane stability. Biochemistry. 2004;43:310–5. doi: 10.1021/bi035653h. [DOI] [PubMed] [Google Scholar]
- 7.Salomao M, An X, Guo X, Gratzer W, Mohandas N, Baines AJ. Mammalian αI-spectrin is a neofunctionalized polypeptide adapted to small, highly deformable erythrocytes. Proc Natl Acad Sci U S A. 2006;103:643–8. doi: 10.1073/pnas.0507661103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Prchal JT, Papayannopoulou T, Yoon SH. Patterns of spectrin transcripts in erythroid and non-erythroid cells. J Cell Physiol. 1990;144:287–94. doi: 10.1002/jcp.1041440215. [DOI] [PubMed] [Google Scholar]
- 9.Carlin RK, Bartelt DC, Siekevitz P. Identification of fodrin as a major calmodulin-binding protein in postsynaptic density preparations. J Cell Biol. 1983;96:443–8. doi: 10.1083/jcb.96.2.443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bennett PM, Baines AJ, Lecomte MC, Maggs AM, Pinder JC. Not just a plasma membrane protein: in cardiac muscle cells alpha-II spectrin also shows a close association with myofibrils. J Muscle Res Cell Motil. 2004;25:119–26. doi: 10.1023/b:jure.0000035892.77399.51. [DOI] [PubMed] [Google Scholar]
- 11.Baines AJ, Pinder JC. The spectrin-associated cytoskeleton in mammalian heart. Front Biosci. 2005;10:3020–33. doi: 10.2741/1759. [DOI] [PubMed] [Google Scholar]
- 12.Moorthy S, Chen L, Bennett V. Caenorhabditis elegans beta-G spectrin is dispensable for establishment of epithelial polarity, but essential for muscular and neuronal function. J Cell Biol. 2000;149:915–30. doi: 10.1083/jcb.149.4.915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hammarlund M, Davis WS, Jorgensen EM. Mutations in beta-spectrin disrupt axon outgrowth and sarcomere structure. J Cell Biol. 2000;149:931–42. doi: 10.1083/jcb.149.4.931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Knowles WJ, Morrow JS, Speicher DW, Zarkowsky HS, Mohandas N, Mentzer WC, Shohet SB, Marchesi VT. Molecular and functional changes in spectrin from patients with hereditary pyropoikilocytosis. J Clin Invest. 1983;71:1867–77. doi: 10.1172/JCI110942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.An X, Lecomte MC, Chasis JA, Mohandas N, Gratzer W. Shear-response of the spectrin dimer-tetramer equilibrium in the red blood cell membrane. J Biol Chem. 2002;277:31796–800. doi: 10.1074/jbc.M204567200. [DOI] [PubMed] [Google Scholar]
- 16.Burridge K, Kelly T, Mangeat P. Nonerythrocyte spectrins: actin-membrane attachment proteins occurring in many cell types. J Cell Biol. 1982;95:478–86. doi: 10.1083/jcb.95.2.478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bennett V, Davis J, Fowler WE. Brain spectrin, a membrane-associated protein related in structure and function to erythrocyte spectrin. Nature. 1982;299:126–31. doi: 10.1038/299126a0. [DOI] [PubMed] [Google Scholar]
- 18.Glenney JR, Jr., Glenney P, Osborn M, Weber K. An F-actin- and calmodulin-binding protein from isolated intestinal brush borders has a morphology related to spectrin. Cell. 1982;28:843–54. doi: 10.1016/0092-8674(82)90063-0. [DOI] [PubMed] [Google Scholar]
- 19.Davis J, Bennett V. Brain spectrin. Isolation of subunits and formation of hybrids with erythrocyte spectrin subunits. J Biol Chem. 1983;258:7757–66. [PubMed] [Google Scholar]
- 20.An X, Guo X, Zhang X, Baines AJ, Debnath G, Moyo D, Salomao M, Bhasin N, Johnson C, Discher D, Gratzer WB, Mohandas N. Conformational stabilities of the structural repeats of erythroid spectrin and their functional implications. J Biol Chem. 2006;281:0527–32. doi: 10.1074/jbc.M513725200. [DOI] [PubMed] [Google Scholar]
- 21.Tyler JM, Hargreaves WR, Branton D. Purification of two spectrin-binding proteins: biochemical and electron microscopic evidence for site-specific reassociation between spectrin and bands 2.1 and 4.1. Proc Natl Acad Sci U S A. 1979;76:519296. doi: 10.1073/pnas.76.10.5192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schultz J, Milpetz F, Bork P, Ponting CP. SMART, a simple modular architecture research tool: identification of signaling domains. Proc Natl Acad Sci U S A. 1998;95:5857–64. doi: 10.1073/pnas.95.11.5857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Moon RT, McMahon AP. Generation of diversity in nonerythroid spectrins. Multiple polypeptides are predicted by sequence analysis of cDNAs encompassing the coding region of human nonerythroid alpha-spectrin. J Biol Chem. 1990;265:4427–33. [PubMed] [Google Scholar]
- 24.Hu RJ, Watanabe M, Bennett V. Characterization of human brain cDNA encoding the general isoform of beta-spectrin. J Biol Chem. 1992;267:18715–22. [PubMed] [Google Scholar]
- 25.Higgins DG, Sharp PM. CLUSTAL: a package for performing multiple sequence alignment on a microcomputer. Gene. 1988;73:237–44. doi: 10.1016/0378-1119(88)90330-7. [DOI] [PubMed] [Google Scholar]
- 26.Grum VL, Li D, MacDonald RI, Mondragon A. Structures of two repeats of spectrin suggest models of flexibility. Cell. 1999;98:523–35. doi: 10.1016/s0092-8674(00)81980-7. [DOI] [PubMed] [Google Scholar]
- 27.Perkins SJ. Protein volumes and hydration effects. The calculations of partial specific volumes, neutron scattering matchpoints and 280-nm absorption coefficients for proteins and glycoproteins from amino acid sequences. Eur J Biochem. 1986;157:169–80. doi: 10.1111/j.1432-1033.1986.tb09653.x. [DOI] [PubMed] [Google Scholar]
- 28.Batey S, Randles LG, Steward A, Clarke J. Cooperative folding in a multi-domain protein. J Mol Biol. 2005;349:1045–59. doi: 10.1016/j.jmb.2005.04.028. [DOI] [PubMed] [Google Scholar]
- 29.MacDonald RI, Cummings JA. Stabilities of folding of clustered, two-repeat fragments of spectrin reveal a potential hinge in the human erythroid spectrin tetramer. Proc Natl Acad Sci U S A. 2004;101:1502–7. doi: 10.1073/pnas.0308059100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.King RD, Sternberg MJ. Identification and application of the concepts important for accurate and reliable protein secondary structure prediction. Protein Sci. 1996;5:2298–310. doi: 10.1002/pro.5560051116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jones DT. Protein secondary structure prediction based on position-specific scoring matrices. J Mol Biol. 1999;292:195–202. doi: 10.1006/jmbi.1999.3091. [DOI] [PubMed] [Google Scholar]
- 32.Munoz V, Serrano L. Development of the multiple sequence approximation within the AGADIR model of alpha-helix formation: comparison with Zimm-Bragg and Lifson-Roig formalisms. Biopolymers. 1997;41:495–509. doi: 10.1002/(SICI)1097-0282(19970415)41:5<495::AID-BIP2>3.0.CO;2-H. [DOI] [PubMed] [Google Scholar]
- 33.Law R, Carl P, Harper S, Dalhaimer P, Speicher DW, Discher DE. Cooperativity in forced unfolding of tandem spectrin repeats. Biophys J. 2003;84:533–44. doi: 10.1016/S0006-3495(03)74872-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Law R, Liao G, Harper S, Yang G, Speicher DW, Discher DE. Pathway shifts and thermal softening in temperature-coupled forced unfolding of spectrin domains. Biophys J. 2003;85:3286–93. doi: 10.1016/S0006-3495(03)74747-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Di Stasi AM, Petrucci TC, Minetti M. Comparison of thermal properties of bovine spectrin and fodrin. Arch Biochem Biophys. 1987;256:144–9. doi: 10.1016/0003-9861(87)90432-2. [DOI] [PubMed] [Google Scholar]
- 36.Law R, Harper S, Speicher DW, Discher DE. Influence of lateral association on forced unfolding of antiparallel spectrin heterodimers. J Biol Chem. 2004;279:16410–6. doi: 10.1074/jbc.M313107200. [DOI] [PubMed] [Google Scholar]
- 37.Ursitti JA, Kotula L, DeSilva TM, Curtis PJ, Speicher DW. Mapping the human erythrocyte beta-spectrin dimer initiation site using recombinant peptides and correlation of its phasing with the alpha-actinin dimer site. J Biol Chem. 1996;271:6636–44. doi: 10.1074/jbc.271.12.6636. [DOI] [PubMed] [Google Scholar]
- 38.Speicher DW, Weglarz L, DeSilva TM. Properties of human red cell spectrin heterodimer (side-to-side) assembly and identification of an essential nucleation site. J Biol Chem. 1992;267:14775–82. [PubMed] [Google Scholar]