Abstract
Dendroecological research uses information stored in tree rings to understand how single trees and even entire forest ecosystems responded to environmental changes and to finally reconstruct such changes. This is done by analyzing growth variations back in time and correlating various plant-specific parameters to (for example) temperature records. Integrating wood anatomical parameters in these analyses would strengthen reconstructions, even down to intra-annual resolution. We therefore present a protocol on how to sample, prepare, and analyze wooden specimen for common macroscopic analyses, but also for subsequent microscopic analyses. Furthermore we introduce a potential solution for analyzing digital images generated from common small and large specimens to support time-series analyses. The protocol presents the basic steps as they currently can be used. Beyond this, there is an ongoing need for the improvement of existing techniques, and development of new techniques, to record and quantify past and ongoing environmental processes. Traditional wood anatomical research needs to be expanded to include ecological information to this field of research. This would support dendro-scientists who intend to analyze new parameters and develop new methodologies to understand the short and long term effects of specific environmental factors on the anatomy of woody plants.
Keywords: Environmental Sciences, Issue 97, Cell parameters, dendroecology, image analysis, micro sectioning, microtomes, sample preparation, wood anatomy
Introduction
Trees, as well as shrubs, dwarf shrubs, and even herbs, show manifold response patterns related to changes in their environment. These patterns have been subject to botany and plant physiology since the mid-19th century. Back then, research on woody plants focused mostly on trees and a descriptive analysis of the structure and variability of annual rings in an ecological context1. When Andrew Ellicott Douglass invented the cross-dating technique for tree-ring research2, this ecological context was more or less suppressed by the new ability to accurately date wooden findings in archaeology. Cross-dating for the first time enabled the accurate dating of tree rings to the calendar year and is until now regarded as the backbone of tree-ring research in all fields of its application1.
In parallel, since the end of the 19th century, wood anatomy evolved into an important research discipline related to many other fields of natural and applied sciences3. Two main domains are established: the systematic wood anatomy, which is the basis for identifying wood in archaeology4, and the applied wood anatomy, related to wood technology, physiology, pathology, and ecology3,5.
In tree-ring research, dendroecology nowadays is defined as a topic encompassing tree-ring related studies focusing on environmental studies such as geomorphic processes (dendrogeomorphology), temperature and precipitation reconstructions (dendroclimatology), water level changes (dendrohydrology) or even glacier fluctuations (dendroglaciology)6. As this definition indicates, tree-ring analyses have become increasingly important in the field of dating and reconstructing environmental processes such as (i) past climate conditions by analyzing annual variations in ring-width7,8, wood density9 or isotopes10, or (ii) the recurrence intervals of geomorphic processes11. These very detailed studies about ring-width variations and their isotopic content demonstrate the need to analyze rings in more detail, i.e., to study the anatomical structure of the rings. However, detailed studies of wood anatomical features within the annual rings related to environmental changes are rare12,13. Although these microscopic features are known14, they have rarely been applied on a microscopic level to dendroecological research. Furthermore, the accurate timing of these growth reactions in naturally grown trees, essential for exact dating purposes, has rarely been documented recently15.
Regarding the effects of the global warming16, the improvement of existing and development of new techniques to record and quantify past and ongoing environmental processes is required, especially in terms of climate impact research11. By expanding traditional wood anatomical research to an ecologically based wood anatomy17, dendro-scientists can analyze new parameters and develop new methodologies to understand the short- and long-term effects of specific environmental factors on the anatomy of woody plants18. Detailed knowledge about variations in different cell parameters within individual rings related to specific drivers (e.g., mechanical forces, climate variations) is the basic requirement for understanding the variability in tree ring formation. Compared to common ring-width measurements, identifying wood anatomical variations requires more complex and expansive preparation techniques that require a lot of labor and time. Detailed procedures of sample cutting, staining, and embedding are manifold and are always dependent on the aim of the study19.
For macroscopic analysis of ring width in conifers or even structures for number, size or distribution of vessels in hardwoods, the surface of a sample is commonly polished using fine abrasive paper or special grinding machines20. A disadvantage of this procedure is the filling of the individual cells with dust that prevents further semiautomatic microscopic analysis21. The best results for macroscopic sample preparation are achieved when sample surface are cut using a razor blade or another sharp knife.
While for small samples, razor blades are a perfect tool; bigger samples as cores require the cutting of plane surfaces over the whole extent of cores. In contrast to sanding, the cells are not filled with dust, which enables further preparation for the successive image analysis. Furthermore, the open cell lumen, the properly cut cell walls, and the plane surface of the entire sample enable the application of high frequency densitometry22 to the whole extent of the core. For image analyses, the surface of samples (cell walls) can be stained using dark ink and the open cell lumen can subsequently be filled with white chalk to enhance the contrast between the cell wall and the lumen area19,23. This rather simple technique enables a basic macroscopic assessment of larger cell structures for vessel size measurements.
These techniques for cutting plane surfaces are sufficient for macroscopic analyses. For a detailed wood anatomical (i.e., microscopic) analysis, transmitted light microscopy is the most common method applied in dendro sciences. Xylem cells differentiate through complex processes encompassing cell-type determination, cell division, cell differentiation, and programmed cell death24. Since the timing and rate at which these processes occur determine cell anatomical characteristics, environmental conditions affecting these processes can generate anatomical deviations in the ring structure. As an important precondition for these analyses, micro sections need to be prepared with a microtome19. When preparing samples for sectioning, the visibility of the tracheid or fiber direction is crucial. The use of hand driven sliding microtomes is recommended to cut micro sections because this technique facilitates high-quality sections as needed for image analyses19. Depending on the specific aim of a certain study, micro sections are cut perpendicular or parallel to the longitudinal extent of the cells. These sections are then photographed below a microscope and cell dimensions measured using specialized image analyses software.
Until recently, the ability to prepare micro sections was restricted to small sample sizes only (approximately 1 cm x 1 cm). This is acceptable to analyze single events as disturbances in specific years, but this technique does not allow the extended time series analysis needed for environmental reconstructions. This effort can only be realized through the development of new, efficient and economic preparation procedures and analytical techniques. In recent years, the members of the tree-ring lab at the Swiss Federal Research Institute WSL in Switzerland have started intensive work on this topic. As a result, new devices and analyzing techniques have been developed to support the idea of integrating wood anatomical features to a broad range of environmental research topics.
Protocol
1. Sampling Techniques
- Core sampling
- For sampling tree stems, extract at least two cores using an increment corer from each stem to analyze its growth development. Vary the position of sampling on the research task, e.g., for common climate reconstructions take the cores parallel to the slope. When working with tropical species, take at least three or more cores.
- Use a sharpened increment corer (5, 10 or 12 mm in diameter) and place the corer perpendicular to the growing axis of the stem. NOTE: To have a controlled and stable position of the corer, use a pusher to avoid movements of the corer other than its rotation when drilling it into the tree. Bore into the stem all the way into and through the pith. To core very dense wood (i.e., tropical species), use special reconfigured chainsaw bodies with an adaptor of increment cores which allow to core even dense woods as Diospyrus ebenum (Ebony).
- Use an extractor and remove the resulting core from the tree. Turn out the corer and remove it from the stem.
- Add a specific ID to the extracted core using a soft pencil by writing on a tape, which is then placed directly on the core. Repeat the procedure on the opposite side of the stem.
- Store the two samples of the tree in plastic or paper tubes or special boxes to prevent any damage. NOTE: Paper straws are beneficial especially in tropical environments because they prevent the cores from molding.
- Finally, mount the cores (fiber direction upright) on wooden support beams. Use cold water resistant wood glue and let them dry. NOTE: Never mount the cores on wooden supports if the aim of the study includes further chemical, isotope or density analysis. For these purposes, fix the cores in open cable conduits or a single faced corrugated board.
- Micro-core sampling
- Use a special punching device or needle with an opening of 2 mm. Depending on the bark thickness use a chisel to remove a necessary portion of the bark.
- Place the device perpendicular to the growing axis on the stem as described above, and use a hammer to penetrate the xylem of the stem to about 2 cm in depth.
- Turn the needle to break the resulting micro core inside the tool and remove it from the stem. Store the micro cores in microcentrifuge vials and label them.
- Disc sampling
- In special cases (and especially in the tropics), e.g., when sampling stumps and fallen trees or whenever possible, take discs (cross sections perpendicular to the growing axis) with a chain saw. Simply label the discs and store them until further analysis. NOTE: This sampling procedure is most commonly applied for all dry-dead, historical, archaeological and sub-fossil material as well as very dense hardwoods that do not allow the use of increment corers.
2. Sample Preparation
- Sanding disks
- Place the discs on a sanding machine and grind the surface using a sequence of increasingly fine abrasives, starting with sandpaper of 80-grit, followed by 120, 220, 300-grit. A final polishing with 400-grit is sufficient for most conifers. For hardwoods and especially tropical species, use sanding grits up to 1,200 to distinguish the growth-ring boundaries.
- Cutting plane surfaces on cores
- Fix the increment cores in the core holder of a newly developed core microtome enabling to cut plane surfaces on cores over their entire extent. NOTE: Correct the orientation of the core within the holder to have the fiber direction of the sample upright, which is in a right angle to the knife of the microtome.
- Lift the core until it slightly touches the blade. Pull the blade, which is fixed on ball bearing guidance over the extent of the core to cut off a first part of the top.
- Push back the knife behind the core, lift the sample holder with the core about 10 µm and repeat the cutting procedure. Do this until approximately one third of the core’s diameter is cut down resulting in a continuous plane surface.
- Preparing micro sections by adding corn starch solution.
- For detailed image analysis of the anatomical structure, fix cores or other samples split off from discs in the holder of a microtome. Position the microtome blade, guided by a ball bearing guidance of a newly developed sledge microtome on the upper edge of the sample and pull over it.
- Push back the blade to lift up the sample by about 20 to 30 µm and pull the blade over the sample again. Repeat this procedure until a continuous surface on top of the sample is created.
- Cover the resulting surface with a cornstarch solution (10 g of cornstarch, 8 ml of water, and 7 g of 100% glycerol) with a common brush. NOTE: The starch grains fill up the open cells of the sample to stabilize the structure for the subsequent cutting of the thin sections.
- Lift the sample by 15 µm, place a brush on the surface of the sample and pull the blade slowly towards the sample starting to cut off a thin section (below the brush). While cutting, the resulting section slides on the blade guided by the brush.
- When the entire section is cut, remove the micro section from the blade with a brush and water (to reduce friction) and place the section on a glass slide for further preparation. To prevent them from drying, cover them using a small amount of glycerol (33% = one part of 100% glycerol and two parts of water).
3. Microslide Preparation
For the following preparation of permanent slides, first wash the glycerol away from the micro section with a pipette and water.
Cover the wet section with a few drops of Safranin (1 g of Safranin powder + 100 ml of water) and Astrablue (0.5 g of Astra blue powder + 2 ml of 100% acetic acid + 100 ml of water) to distinguish between lignified and non-lignified structures, but also to enhance the contrast for the subsequent image analysis.
Let the section rest for 5 min covered by the dye and then wash it away with a pipette and water again.
As soon as the excessive dye is removed, dehydrate the section with pipettes by washing them with a sequence of 75% diluted, 96% and finally 100% ethanol.
Use pipettes to rinse the samples on the glass slide instead of placing the samples in a bath to reduce the processing time to about two to 3 min for the entire dehydration process. NOTE: Dehydration with ethanol prevents fragile samples from breaking.
Thereafter rinse the section with 100% xylol. NOTE: Xylene is carcinogenic and ventilation is obligatory in the lab when using it. Other products exist replacing xylol, but if there is intent to store the samples over longer time periods (years, decades) for a potential reanalysis, Canada balsam (step 3.5.2) is the only embedding medium that stays clear without changing color over time. Unfortunately Canada balsam needs to be used in combination with xylol.
As long as the xylol turns milky (whitish), repeat the dehydration process as it is not sufficiently dehydrated.
As soon as the xylol stays clear, cover the section with a drop of 100% Canada balsam and cover it with a cover glass of at least the size of the section. NOTE: Be aware of removing air bubbles by gently pressing the cover glass down.
Place the resulting micro slide between two strips of heat resistant plastics and place it on a metal plate. Place a weight (e.g., a magnet) on top of the slide to press it down to keep the section flat during the following drying process.
Place the slides in an oven at 60 °C for about 12 hr. After 12 hr, take the slides out of the oven and place them on a shelf to let them cool down to RT (about 20 °C). When cool, remove the weight and the plastic strip and clean the slide using razor blades to remove the surplus Canada balsam.
4. Visualizing Cell Contents
- Prepare Nawashin solution25.
- Prepare a solution (A) with 5 g of chromic acid, 50 ml of 96% acetic acid, and 320 ml of deionized water. Prepare another solution (B) with 200 ml of formalin with 175 ml of deionized water.
- Mix solution A and solution B in a 1:1 ratio to create the Nawashin solution.
To visualize the cell contents, fix the sample with Nawashin solution for 10 min to stop all degradation processes within the cell. NOTE: The fixation preserves cell nuclei allowing for an estimation of cell longevity (cell longevity = nuclei present / nuclei absent; the presence or absence of the nucleus indicates whether the cell is living or dead).
After fixation, wash the sections with water to fully remove the Nawashin solution before staining them with Safranin-Astra blue and before additionally staining them with picric-aniline blue (1 part saturated water-soluble anilin blue + 4 parts of 10% picric acid).
Carefully heat the sample to about 80 °C (in any case below boiling point) to accelerate the staining process. For dehydration and embedding of the samples follow the protocol steps 3.4 to 3.8.
5. Preparing Digital Images of Anatomical Features
- Create digital images of core surfaces for vessel analysis.
- Cut the core surface plane using the core microtome and stain the resulting surface black by simply using a felt marker.
- As soon as the dye is dried, rub the surface of the core with white chalk. Press the chalk in the cells by simply rubbing the chalk over the surface using a finger. Also remove the surplus chalk by this procedure. NOTE: As a result the cell walls are black and the lumen parts of the vessels are white. This contrast is a precondition for an automated image analysis.
- Place the prepared core below a binocular microscope equipped with a digital camera. Take a sequence of slightly overlapping (approx. 10%) images starting on one side of the core until the entire surface is captured. Stitch the single images to create a complete image of the core surface.
- Create digital images from micro slides
- Place the cleaned micro slides under a microscope and take overlapping images from the entire section.
- Define a specific magnification depending on the structures to be analyzed, ranging between 40X and 1,000X magnification.
- Stitch the single images to create one complete image of the section. To avoid possible distortions during the stitching process, use “plan”-type objective lenses with the microscope.
6. Quantifying Anatomical Features
To detect cells and ring borders using the image analysis tool ROXAS18 or similar software, load a complete image of a micro section and the associated spatial calibration to obtain quantitative results in metric units. Calculate the spatial calibration by measuring the number of pixels between the scales of a micrometer stage images taken at the same magnification and dividing the obtained value by the scale spacing in metric units (e.g. 1,000 µm).
- Select from the provided list (provided in the software) a suitable configuration before starting the automatic part of the analysis. NOTE: A configuration is a previously optimized set of program settings that, for example, takes into account the staining color of the sample and the size and shape range of the cells to be detected. Tailored configurations thus allow to produce optimal recognition results for different species and image qualities.
- Select further options such as the usage of regions in the image that are to be included or excluded to avoid, e.g., blank image margins or cracks in the sample, if required.
Start the analysis by pushing the Analyze button. If opted, define regions in the image that are to be included or excluded using the polygon, rectangle or circle tool. The following automatic analysis uses flexible algorithms to correct some image deficiencies (e.g., poor contrast) and improve contrast based on the quality of the image.
Representative Results
All dendroecological analyses depend on accurate samples, no matter if discs, cores, or micro cores are taken. For this, the devices need to be in perfect shape (accurately sharpened) to avoid micro cracks within the wooden sample. When preparing surfaces on increment cores, the use of a core microtome is essential. The ability to have open cells, which can be further treated to enhance contrast for image analyses and vessel size measurements (Figure 1), is a first important step towards the adaption of anatomical structures into time series analyses. Sometimes the density of a hardwood sample prevents the use of a microtome. In that case a proper polishing and subsequent removal of excessive sawdust from the vessels with a compressor or vacuum is the best option.
For more detailed analyses of smaller cell structures as earlywood and latewood tracheids in conifers, high quality micro sections are needed. Here, potential artifacts such as secondary cell walls being stripped off the primary wall need to be avoided (Figure 2). If these artifacts occur in the digital images, an automated analysis of the cell dimensions is no longer possible. The artifacts then need to be manually corrected, which is time consuming and in most cases results in incorrect measurements of cell dimensions. The simple application of a Non-Newtonian fluid, i.e., a corn starch solution, to the top of the sample supports the stability of the structure while reducing the occurrence of artifacts to a minimum (Figure 2)26. This application of corn starch, suitable for all wooden samples including tropical species, makes the application of the embedding procedure to the samples before cutting redundant.
Micro sections enable a more secure determination of annual rings. This is especially the case for conifers growing on their natural limits, i.e., at the tree line in high alpine areas. Extremely narrow rings are frequent and hard to detect macroscopically (Figure 3). In extreme cases, the rings consist of one or two rows of earlywood cells and one row of flattened latewood cells, which lack (in contrast to common latewood cells) thickened cell walls. For this they are better or even only visible when using micro sections. Furthermore, density fluctuations can be differentiated from ring boundaries more clearly, which simplifies the detection of annual rings especially in the Mediterranean and the tropics (Figure 3).
When analyzing images at a magnification of 40X or higher, single cells are visible and the thickness of their cell walls is also detectable. Semi-automatic analysis software enables the measurement of specific parameters along defined paths following the direction of their temporal and spatial development (Figure 4). With this, changes of single parameters such as the cell lumen or cell wall thickness can be determined over the whole extent of an annual ring (Figure 4). This can be done for all rings visible within the image and this fully supports the need for an extended time series analysis.
Image analyses can also be used to determine the developmental phases of annual rings within the vegetation period (Figure 5). When analyzing the image of a section stained with Safranin and Astra-blue using polarized light, even the different stages of lignification, beginning in the outermost corners of the cell walls until the full lignification of the secondary cell wall, become visible. This is because the lignified (mature) cell walls shine up in polarized light (Figure 5). Detailed information can be related to environmental data documented for the respective vegetation period to determine for example a more detailed climate-growth relationship.
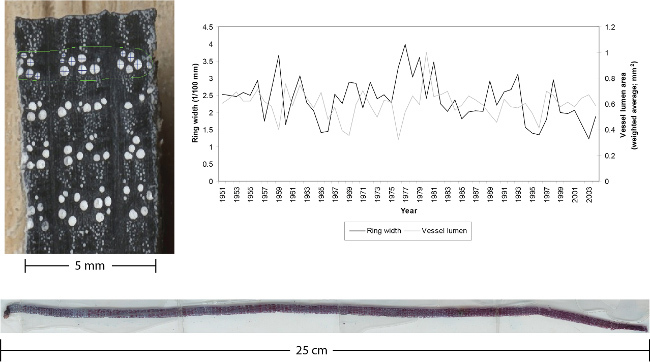 Figure 1.Examples of a micro section and a prepared plane surface of an oak including ring-width and vessel size measurements. Left: outermost part of an oak increment core. The 5 mm diameter core was cut using a core microtome. The surface was then stained black using a felt marker and cells were filled with white chalk after the stain was dry. Right: the graph is indicating ring-width and vessel size measurements done on the surface of the core shown on the left. No micro sections were needed to do these measurements due to the clear surface created by the core microtome (modified after21). Bottom: Micro section cut off an increment core, stained, dehydrated and fixed in Canada balsam. Thickness: 20 µm, length: 25 cm. Please click here to view a larger version of this figure.
Figure 1.Examples of a micro section and a prepared plane surface of an oak including ring-width and vessel size measurements. Left: outermost part of an oak increment core. The 5 mm diameter core was cut using a core microtome. The surface was then stained black using a felt marker and cells were filled with white chalk after the stain was dry. Right: the graph is indicating ring-width and vessel size measurements done on the surface of the core shown on the left. No micro sections were needed to do these measurements due to the clear surface created by the core microtome (modified after21). Bottom: Micro section cut off an increment core, stained, dehydrated and fixed in Canada balsam. Thickness: 20 µm, length: 25 cm. Please click here to view a larger version of this figure.
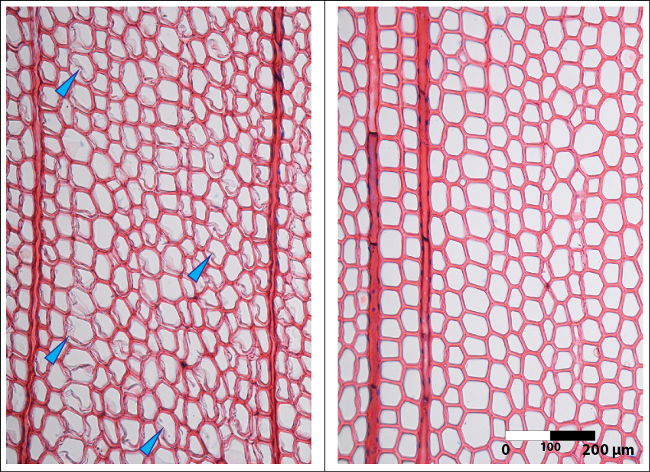 Figure 2.Images of micro sections cut without stabilization versus a section cut using the corn starch solution. Left: Micro section showing cutting artifacts in the earlywood tracheids of a conifer. The sample was cut without embedding and as a result the rather thin secondary walls of the earlywood cells were split off the primary wall (blue arrows). Right: Micro section without any artifacts in the earlywood tracheids of a conifer. This section (same sample as shown on the left) was cut after applying the cornstarch solution with a brush on top of the sample surface. Please click here to view a larger version of this figure.
Figure 2.Images of micro sections cut without stabilization versus a section cut using the corn starch solution. Left: Micro section showing cutting artifacts in the earlywood tracheids of a conifer. The sample was cut without embedding and as a result the rather thin secondary walls of the earlywood cells were split off the primary wall (blue arrows). Right: Micro section without any artifacts in the earlywood tracheids of a conifer. This section (same sample as shown on the left) was cut after applying the cornstarch solution with a brush on top of the sample surface. Please click here to view a larger version of this figure.
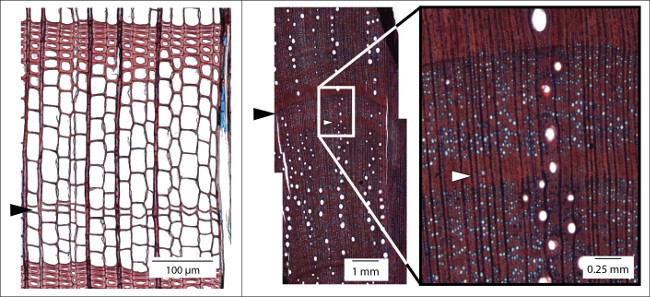 Figure 3.Examples of hardly detectable annual ring boundaries. Left: Micro section of an increment core (here: Larix decidua) showing a ring boundary (black arrow) indicated by a single row of flattened latewood cells without any thickening of the cell walls. This ring would not be visible macroscopically. Right: Intra-annual density fluctuations (white arrow) are common in Mediterranean species (micro section: Quercus ilex). The gradual change of the cell structure towards latewood and back to earlywood structure (white arrow) allows for differentiating density fluctuations from real ring boundaries (black arrow). Please click here to view a larger version of this figure.
Figure 3.Examples of hardly detectable annual ring boundaries. Left: Micro section of an increment core (here: Larix decidua) showing a ring boundary (black arrow) indicated by a single row of flattened latewood cells without any thickening of the cell walls. This ring would not be visible macroscopically. Right: Intra-annual density fluctuations (white arrow) are common in Mediterranean species (micro section: Quercus ilex). The gradual change of the cell structure towards latewood and back to earlywood structure (white arrow) allows for differentiating density fluctuations from real ring boundaries (black arrow). Please click here to view a larger version of this figure.
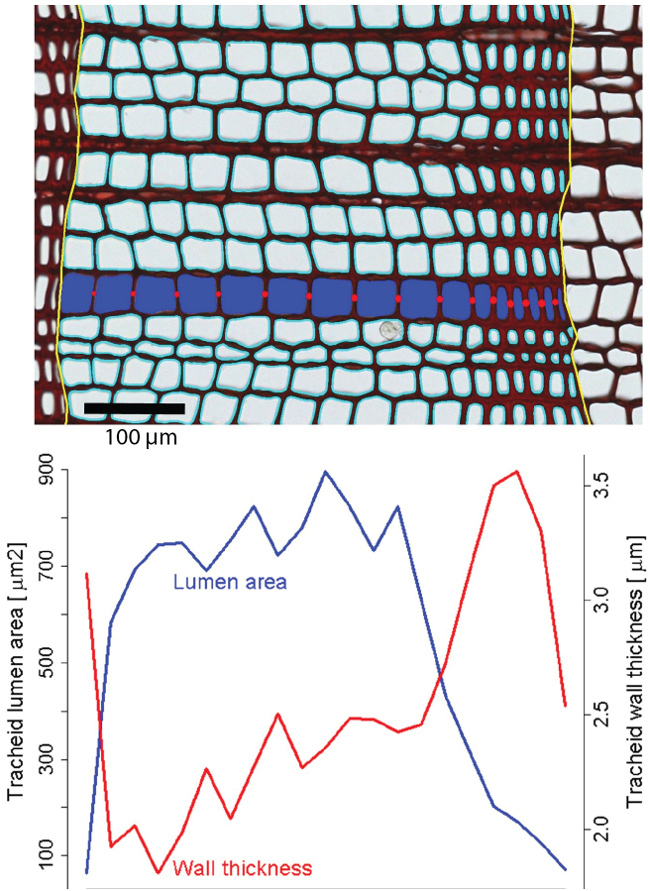 Figure 4. Illustration of lumen area and wall thickness measurements within an annual ring of a conifer. Top: Cut-out image showing an exemplary results of ROXAS analysis in a tree-ring of Pinus sylvestris (Scots pine). Ring borders are shown in yellow and outlines of tracheid lumina in cyan. For one radial file (blue tracheid lumina) the measured cell wall thicknesses is represented by red circles. Black scale bar = 100 µm. Bottom: The intra-annual changes in tracheid lumen area and tracheid cell-wall thickness for the entire annual ring. Please click here to view a larger version of this figure.
Figure 4. Illustration of lumen area and wall thickness measurements within an annual ring of a conifer. Top: Cut-out image showing an exemplary results of ROXAS analysis in a tree-ring of Pinus sylvestris (Scots pine). Ring borders are shown in yellow and outlines of tracheid lumina in cyan. For one radial file (blue tracheid lumina) the measured cell wall thicknesses is represented by red circles. Black scale bar = 100 µm. Bottom: The intra-annual changes in tracheid lumen area and tracheid cell-wall thickness for the entire annual ring. Please click here to view a larger version of this figure.
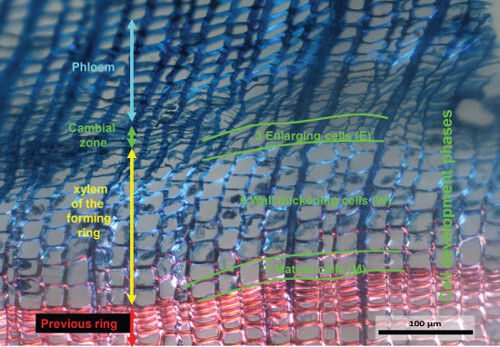 Figure 5.Example of a forming annual ring. The image has been captured under a light microscope with polarized light from a Safranin and Astra-blue stained micro-section obtained from a micro-core sampled on July 7th, 2007 from a Larix decidua growing in the Lötschental at 1,300 m above sea level. On this micro-section the cambial cells, the cells in the enlargement phase, the cells in the wall thickening phase and the mature cells are recognizable. The tangential width of the image covers ~1 mm of the xylem cross-section. Please click here to view a larger version of this figure.
Figure 5.Example of a forming annual ring. The image has been captured under a light microscope with polarized light from a Safranin and Astra-blue stained micro-section obtained from a micro-core sampled on July 7th, 2007 from a Larix decidua growing in the Lötschental at 1,300 m above sea level. On this micro-section the cambial cells, the cells in the enlargement phase, the cells in the wall thickening phase and the mature cells are recognizable. The tangential width of the image covers ~1 mm of the xylem cross-section. Please click here to view a larger version of this figure.
Discussion
The challenges of a successful and sustainable integration of wood anatomy into dendroecological research are, apart from manifold analytical problems, mostly due to technical aspects. These challenges range from principle sampling approaches to creating high quality micro sections and their subsequent analysis19.
At first glance, the sampling of cores or even discs is a simple procedure that has been known for many years now. There are many things that can be done wrong and a small inaccuracy in sampling can result in severe problems during the subsequent preparation and analysis phases. Small inaccuracies such as coring that is not exactly perpendicular to the stem axis or using an imperfectly sharpened corer are not an issue if the aim of the study is restricted to ring-width measurements. However, when aiming for microscopic analysis of the samples, an incorrect sampling direction might result in optical distortions of cell walls, while the use of blunt corers results in micro cracks within the core. As a result, when trying to cut micro sections of these cores, the thin sections just fall apart and an efficient preparation is no longer guaranteed. The same is true for micro-core sampling. A blunt tip will result in high pressure when the puncher is hammered into the stem wood. Consequently the cambial layer will be compressed. The cambial cells (Figure 5) are consequently squeezed and cannot be analyzed.
Disc sampling is indeed the best strategy when analyzing growth variations to relate them to environmental changes. Unfortunately it is simply impossible to take discs from all trees intended to be sampled for further analyses. Nevertheless, especially in case of tropical dendrochronology, a certain amount of stem disks is needed in combination with increment cores. The disks are used as a base to define ring boundaries and for this to support the boundaries defined based on analyzing increment cores12,27,28.
The pros and cons of sanding versus cutting are frequently discussed1,11,21. As it is mentioned above, the best procedure always depends on the research question and the parameters to be analyzed (macroscopic or microscopic). If isotopic or chemical analyses are projected in a further working step, it is of utmost importance that the abrasive dust created by sanding that may fill into cell lumina over the entire sample, is carefully removed by vacuuming or pressure air.
Cutting micro sections is for all microscopic analyses the most appropriate way of preparing samples for further analyses. First of all, the section is cut off the sample, which then can be kept without any contamination for potential further analyses. Second these sections allow for high resolution measurements of single cell parameters. Furthermore, avoiding the time consuming embedding technique by using a cornstarch solution26 to stabilize the cells is a big advantage in micro sectioning.
A disadvantage of micro sectioning is still the limited sample size resulting in long preparation times. For real time series analyses going back in time over centuries or even millennia, there is a need to further develop existing cutting devices17,19, but also image processing and analysis18. A first step into this direction is the development of the core microtome21, initially manufactured to cut plane surfaces on cores (Figure 1). Recent tests revealed the ability to cut micro sections of entire cores using this device (Figure 1).
High quality micro sections provide the basic principle for an effective image analysis. Taking the images under a microscope is a common procedure19, but their effective analysis is still a task that needs to be further developed17. All existing image analysis systems are semi-automatic, i.e., they need to be more or less intensely controlled by the technician. In many cases, the images need to be corrected or even new images have to be done to enhance contrast for a better registration of the structures by the software without changing cell wall thickness within the image.
Specialized image analysis tools such as ROXAS18, WinCell or specific scripts for ImageJ29 are able to provide basic anatomical data such as cell number, cell dimension, cell wall thickness and cell position within the annual ring. Many additional anatomical metrics that are relevant in a dendroecological context can be calculated from these basic measurements such as size of the largest conduits, size distribution of conduits, size of earlywood or the first row of conduits, (optical) wood density, intra-annual profiles of conduit size and cell wall thickness, and grouping patterns of conduits (solitary, multiples, etc.).
Using the software ROXAS18, the outlines of conduit lumina (i.e., water conducting cell) and annual ring borders are automatically recognized and visually represented as overlays over the original image. Detection algorithms for conduits are based on color, size and shape information, detection algorithms for ring borders on the local context of each conduit. A toolbox allows us to manually improve these results by directly editing the overlay features, i.e., deleting, adding and modifying ring borders and conduit outlines. After editing, the final data output, including cell wall thickness (conifers), is automatically generated and saved into a spreadsheet. Fully automated systems are currently not available, not even for conifers showing a relatively simple structure, but this is a goal for future developments. This would strongly support the full integration of wood anatomical parameters in time series analyses.
Disclosures
The authors declare that they have no competing financial interests.
Acknowledgments
The authors would like to acknowledge the effort of Sandro Lucchinetti (Schenkung Dapples, Zürich) for constructing the devices needed to guarantee progress in sample preparation.
References
- Schweingruber FH. Tree Rings and Environment: Dendroecology. Bern: Haupt; 1996. [Google Scholar]
- Sheppard PR. Dendroclimatology: extracting climate from trees. Wiley Interdisciplinary Reviews: Climate Change. 2010;1:343–352. [Google Scholar]
- Wagenführ R. Anatomie des Holzes: Strukturanalytik - Identifizierung - Nomenklatur - Mikrotechnologie. DRW-Verlag Weinbrenner; 1999. [Google Scholar]
- Tennessen D, Blanchette RA, Windes TC. Differentiating Aspen and Cottonwood in Prehistoric Wood from Chacoan Great House Ruins. Journal of Archaeological Science. 2002;29:521–527. [Google Scholar]
- Rowell RM. Handbook of Wood Chemistry and Wood Composites. Boca Raton, FL: CRC Press, Taylor & Francis Group; 2005. [Google Scholar]
- Kaennel M, Schweingruber FH. Multilingual Glossary of Dendrochronology. Terms and definitions in English, German, French, Spanish, Italian, Portuguese and Russian. Berne: Haupt; 1995. [Google Scholar]
- Esper J, Frank DC, Luterbacher J, et al. A changing world: challenges for landscape research. In: Kienast F, editor. On selected issues and challenges in dendroclimatology. Dordrecht: Springer; 2007. pp. 113–132. [Google Scholar]
- Esper J, Frank DC, Wilson RJS, Büntgen U, Treydte K. Uniform growth trends among central Asian low and high elevation juniper tree sites. Trees. 2007;21(2):141–150. [Google Scholar]
- Frank DC, Nievergelt D, Esper J. Summer temperature variations in the European Alps, AD 755-2004. Journal of Climate. 2006;19(2):5606–5623. [Google Scholar]
- Treydte K, et al. Millennium-long precipitation record from tree-ring oxygen isotopes in northern Pakistan. Nature. 2006;440:1179–1182. doi: 10.1038/nature04743. [DOI] [PubMed] [Google Scholar]
- Heinrich I. Dendrogeomorphology. In: Elias SA, editor. The Encyclopedia of Quaternary Science. Vol. 2. Amsterdam, Elsevier; 2013. pp. 91–103. [Google Scholar]
- Verheyden A, De Ridder F, Schmitz N, Beeckman H, Koedam N. High-resolution time series of vessel density in Kenyan mangrove trees reveal a link with climate) New Phytologist. 2005;167:425–435. doi: 10.1111/j.1469-8137.2005.01415.x. [DOI] [PubMed] [Google Scholar]
- Abrantes J, Campelo F, García-González I, Nabais C. Environmental control of vessel traits in Quercus ilex under Mediterranean climate: relating xylem anatomy to function. Trees. 2013;27:655–662. [Google Scholar]
- Fengel D, Wegener GWood. Wood: Chemistry, Ultrastructure, Reactions. Kessel Verlag, Remagen: 2003. [Google Scholar]
- Heinrich I, Gärtner H, Monbaron M. Tension wood formed in Fagus sylvatica and Alnus glutinosa after simulated mass movement events. IAWA-Journal. 2007;28(1):39–48. [Google Scholar]
- Stocker TF, et al., editors. Contribution of Working Group I to the Fifth Assessment Report of the Intergovernmental Panel on Climate Change. Cambridge, United Kingdom and New York, NY: Cambridge University Press; 2013. Climate Change 2013: The Physical Science Basis. [Google Scholar]
- Lucchinetti S, Schweingruber FH. New perspectives for wood anatomical analysis in Dendrosciences: The GSL1-microtome) Dendrochronologia. 2014;32(1):47–51. [Google Scholar]
- Arx G, Carrer M. ROXAS - a new tool to build centuries-long tracheid-lumen chronologies in conifers. Dendrochronologia. 2014;32(3):290–293. [Google Scholar]
- Schweingruber FH. Microscopic Preparation Techniques for Plant Stem Analysis. Verlag Dr Kessel, Remagen: 2013. [Google Scholar]
- Hoadley RB. Identifying wood: Accurate results with simple tools. Newtown, CT: The Taunton Press; 1990. [Google Scholar]
- Nievergelt D. The core-microtome. A new tool for surface preparation on cores and time series analysis of varying cell parameters. Dendrochronologia. 2010;28(2):85–92. [Google Scholar]
- Von Schnakenburg P, Bräuning A, Helle G. Detecting annual growth rhythms from high-frequency densitometry and carbon isotopes in tropical mountain rain forest trees in southern Ecuador. In: Elferts D, Brumelis G, Gärtner H, Helle G, Schleser G, editors. Tree Rings in Archaeology, Climatology and Ecology. Vol. 6. Potsdam: GFZ Potsdam; 2008. pp. 96–99. [Google Scholar]
- Garcia Gonzalez I, Fonti P. Ensuring a representative sample of earlywood vessels for dendroecological studies: an example from two ring-porous species. Trees. 2008;22(2):237–244. [Google Scholar]
- Fonti P, et al. Studying global change through plastic responses of xylem anatomy in tree rings. New Phytologist. 2010;185(1):42–53. doi: 10.1111/j.1469-8137.2009.03030.x. [DOI] [PubMed] [Google Scholar]
- Purvis MJ, Collier DC, Walls D. Laboratory techniques in Botany. Butterworth, London: 1966. [Google Scholar]
- Schneider L, Gärtner H. The advantage of using non-Newtonian fluids to prepare micro sections. Dendrochronologia. 2013;31(3):175–178. [Google Scholar]
- De Ridder M, Vanden Bulcke J, Van Ackera J, Beeckman H. Tree-ring analysis of an African long-lived pioneer species as a tool for sustainable forest management. Forest Ecology and Management. 2013;304:417–426. [Google Scholar]
- De Ridder M, et al. A tree-ring based comparison of Terminalia superba climate–growth relationships in West and Central. Trees. 2013;27(5):1225–1238. [Google Scholar]
- Redband WS. ImageJ. Bethesda, Maryland, USA: U.S. National Institutes of Health; 1997. Available from: http://imagej.nih.gov/ij. [Google Scholar]


