Abstract
This protocol discusses the fabrication and usage of a disposable low cost microfluidic chip as sample introduction system for inductively coupled plasma mass spectrometry (ICPMS). The chip produces monodisperse aqueous sample droplets in perfluorohexane (PFH). Size and frequency of the aqueous droplets can be varied in the range of 40 to 60 µm and from 90 to 7,000 Hz, respectively. The droplets are ejected from the chip with a second flow of PFH and remain intact during the ejection. A custom-built desolvation system removes the PFH and transports the droplets into the ICPMS. Here, very stable signals with a narrow intensity distribution can be measured, showing the monodispersity of the droplets. We show that the introduction system can be used to quantitatively determine iron in single bovine red blood cells. In the future, the capabilities of the introduction device can easily be extended by the integration of additional microfluidic modules.
Keywords: Bioengineering, Issue 97, mass spectrometry, ICPMS, microfluidics, droplet microfluidics, monodisperse, sample introduction, chip, red blood cells, erythrocytes, single cell analysis
Introduction
Elemental analysis of liquid samples by inductively coupled plasma mass spectrometry (ICPMS) is commonly carried out using nebulizers in combination with spray chambers as introduction system1. In this sample introduction system the sample is sprayed by a nebulizer to generate a polydisperse aerosol. A downstream spray chamber is used to filter out large droplets. This method is associated with high sample consumption (>0.3 ml min-1)2 and an incomplete sample transport. Thus, it becomes impractical for applications where only microliter sample volumes are available, as in biological, forensic, toxicological and clinical studies3. To reduce the sample consumption, nebulizers with smaller nozzle dimensions were developed3. However, the reduced nozzle size increases the risk of clogging when samples of undigested biological fluids or concentrated salt solutions have to be analyzed3.
A different approach for sample introduction was proposed by Olesik et al.4. The authors injected a liquid into ICPMS in the form of monodisperse discrete microdroplets, which were produced by a piezo-electrically driven micropump. Even though this very system did not find wide application, it initiated the further development of the concept of discrete droplet introduction in ICPMS. Today, piezo-electrically driven dispensing systems, which can generate droplets in size of 30, 50, 70 and 100 µm and at frequencies of 100-2,000 Hz, can be purchased. The droplets can be transported into ICPMS with close to 100% efficiency5. These microdroplet dispensers have been applied for quantitatively measuring single nanoparticles5,6 as well as characterizing individual biological cells7. A similar system based on thermal inkjet technology8 was tested for analysis of biological samples9. Although the available single droplet introduction systems are very efficient, can be used for small sample volumes and are promising for the analysis of nanoparticles and cells, they have several limitations. For a fixed nozzle size, the droplet size can be varied only slightly (unless custom settings are used10). Changes of the physical properties of the liquid (pH, salt content) can alter the droplet characteristics (size, injection speed). Also, these devices are rather expensive, prone to clogging and are difficult to clean.
Another method to generate droplets is known in the field of droplet microfluidics11. In recent years droplet microfluidics has gained interest for (bio-)chemical reactions12-15 and for single cell studies16,17. Additionally, this technique was applied for introducing samples in electrospray ionization mass spectrometry18,19 and for preparing samples in matrix-assisted laser desorption/ionization mass spectrometry20,21.
Recently, we introduced a microfluidic based system for sample introduction in ICPMS22. The key component of our introduction system is the liquid assisted droplet ejection (LADE) chip. This chip consists completely of poly(dimethylsiloxane) (PDMS). In the first channel junction flow focusing is used to generate monodisperse droplets of an aqueous sample solution (Figure 1). For this purpose the highly volatile (boiling point of 58-60 °C23) and immiscible carrier phase perfluorohexane (PFH) is used (Figure 1). These PFH properties enable a stable droplet generation and fast removal of the carrier phase. Changes in the properties of the sample liquid influence this generation method less, compared to other droplet generators. The droplet size is adjustable over a wide range by changing the flow rates of the aqueous phase and the PFH. In a downstream secondary junction, more PFH is added to increase the flow speed to at least 1 m sec-1. At this speed the liquid can be ejected from the chip in stable and straight jet (Figure 1) without droplet destruction (Figure 1 inset). This double-junction design allows controlling the jet stability independent of droplet generation. The droplets are transported to the ICPMS with a customized transport system. This system comprises a falling tube and a membrane desolvator to remove the PFH. The dried residues of the aqueous droplets are subsequently ionized in the plasma of the ICPMS and a mass detector measures the ions. The front part of the chip is barrel-shaped to ensure a tight connection with the droplet transport system. The ejection of the aqueous sample as droplets in PFH is beneficial, because contact with the nozzle is avoided. This considerably lowers the risk of nozzle clogging, which can be a problem when working with cell suspensions or concentrated salt solutions. The LADE chips, fabricated by PDMS soft lithography, are cheap (material cost approximately $2 per chip), disposable and easy to modify. In combination with the fabrication that requires only a small amount of manual work each experiment can be performed with a new chip. Therefore, a laborious cleaning is not needed and cross contamination is minimized.
Here, the fabrication of the LADE chip by soft lithography and its application for ICPMS are described. Examples of measurements with an aqueous solution and a cell suspension are presented.
Protocol
1. SU-8 Master Fabrication (Figure 2)
NOTE: Perform the fabrication of the SU-8 master molds in a clean room to prevent defects caused by dust particles. Two wafers are needed for the fabrication, one wafer with microfluidic features and one without.
- Prepare the master molds for the microfluidic chip. First apply an adhesion layer to the silicon wafer.
- Dehydrate a silicon wafer for 10 min at 200 °C. Cool the wafer down to RT and load it on to a spin coater and spin coat it with SU-8 2002 with the following protocol.
- Dispense about 3 ml resist onto the wafer.
- Spin the wafer at 500 rpm for 10 sec to spread the resist over the whole wafer.
- Spin the wafer at 2,000 rpm for 30 sec to achieve a resist height of approximately 2 µm.
Remove excess resist from the edge of the wafer with an acetone soaked swab, to prevent sticking of the wafer to the hot plate in the next step. Bake the wafer for 60 sec at 95 °C on a hot plate.
Expose the whole wafer with ultraviolet light (80 mJ/cm2 at 365 nm). Post-bake the wafer for 120 sec to 95 °C.
- Cool the wafer down and immediately spin coat the wafer again using the following protocol for SU-8 2050:
- Spin the wafer at 100 rpm for 20 sec (dispense about 3 ml SU-8 resist during this step).
- Spin the wafer at 500 rpm for 10 sec to spread the resist over the whole wafer.
- Spin the wafer at 3,250 rpm for 30 sec resulting in a resist thickness of approximately 40 µm.
Again, remove excess resist from the edge of the wafer with an acetone soaked swab and soft bake the wafer on a hot plate for 180 sec at 65 °C and for 360 sec at 95 °C.
Prepare the photomask by sticking it to a soda-lime glass. See Figure 3 for the mask design. Use a mask aligner to expose the resist with ultraviolet light (160 mJ/cm2, measured at 365 nm) through the prepared mask. Bake the exposed wafer again on a hot plate for 60 sec at 65 °C and for 360 sec at 95 °C.
After cooling down the wafer to RT, immerse it in a glass Petri dish filled with developer for 5 min to develop the resist. Gently agitate the petri dish to remove unexposed SU-8. Rinse the wafer with isopropanol and blow it dry with a nitrogen gun.
Examine the wafer under a microscope. In case undeveloped resist remains on the features, develop the wafer again for a few minutes, as described in step 1.7.
Remove any residual solvent by baking the wafers for 2 hr at 200 °C. Check the height of the features with a step profiler. In case the measured height differs from the desired height begin with this protocol again and adapt the spin speed in step 1.1.4.
- To prevent sticking of the PDMS to the wafer silanize it by placing it in desiccator together with 50 µl of 1H,1H,2H,2H-perfluorodecyltrichlorosilane in a small porcelain dish. Reduce the pressure in the desiccator to 100 mbar and incubate the wafer for 12 hr.
- For the blank PDMS parts silanize another silicon wafer using the method of step 1.10. To save time silanize both wafers at the same time in a single desiccator.
2. LADE Chip Fabrication
NOTE: The LADE chip is made out of two PDMS pieces that are bonded together by adhesive bonding24. The first part contains the microfluidic features. The other part is flat and used to seal the channels. Bonded together, they form the round shape necessary to interface the chip with the droplet transport system. Here, the fabrication of the two parts and their bonding is described. All process steps are shown in Figure 4.
Prepare 44 g of PDMS by mixing 40 g of prepolymer with 4 g of the PDMS curing agent (this will result in up to 6 chips). Degas the PDMS in a desiccator until it is bubble free (this will take about 20 min).
- Replica molding of the structured halves.
- Place the casting form on top of the wafer and snap it into place using the guiding structures around the design (see Figure 5). Skip the snapping into place for the flat PDMS halve.
- Pour approximately 3 to 4 g of the degased PDMS in the casting form and place it for 6 min on a hot plate at 150 °C. Cool down the cured PDMS in the casting form and carefully lift the casting form wafer using a spatula.
- In order to prevent any contamination of the microfluidic channels cover the side of the chip that was previously in contact with the wafer with tape. Carefully cut the tape along the edge of the PDMS part to remove excess PDMS.
To fabricate the flat PDMS halves repeat the above-mentioned steps 2.2.1 to 2.2.3 with the blank silanized wafer.
Peel of the tape and punch fluid connection holes into the structured halves with a biopsy puncher. Protect the structures with tape during storage.
- Bond the PDMS parts together by adhesive bonding using the PDMS curing agent24.
- Take an untreated silicon wafer and spin coat it with PDMS curing agent for 30 sec at 6,000 rpm. Take the wafer out of the spin coater.
- Remove the tape from the structured halves and place them with the structures facing downwards onto the wafer. Gently push on top of the PDMS to remove air bubbles.
- Remove the tape from the blank PDMS halves. Take a structured halve from the wafer and manually align it on top of the flat PDMS halve. Gently squeeze the part together to remove air bubbles and let the assembled chip cure for 24 hr at RT. Do not push the parts together with force as this can cause the channels to collapse.
Cut the tip of the chip along the indicator line orthogonal to the nozzle channel with a utility knife to open the outlet nozzle. Use an alignment device to ensure a straight cut, which is necessary for a straight liquid ejection. Inspect the chip under a microscope for defects in the microfluidic channels and dust particles. Put a tape over the inlet holes to protect the chips during storage.
Connect a Woulff bottle with tubing to a dry nitrogen source and to all inlets of the LADE chip. Deposit 50 µl of 1H,1H,2H,2H-perfluorodecyltrichlorosilane at the bottom of the Woulff bottle and close it.
Silanize the microfluidic channels by flushing all channels for 20 min with the nitrogen stream carrying 1H,1H,2H,2H-perfluorodecyltrichlorosilane at a flow rate of approximately 1 ml/sec. The chips are ready for experiments and can be stored for at least several weeks at RT.
3. Preparations for Measurement / Droplet Transport System
NOTE: Build the whole droplet transport system on top of an optical table, since it is necessary to construct a stable supporting structure for the setup. A scheme of the whole droplet transportation system is depicted in Figure 6.
Install a custom cyclonic poly(methyl methacrylate) (PMMA) adapter with an 50 cm attached stainless steel tube vertically. Attach the adapter to a helium source with a mass flow controller. Attach a (high-speed) camera and a lamp to the adapter on the opposite sites for droplet visualization.
Place a cartridge heater in the middle of the steel tube and use poly(vinyl chloride) (PVC) tubing and a Legris tube connector to connect the end of the steel tube with the inlet of the membrane desolvator.
Connect the outlet of the desolvator with another PVC tubing to a laminar flow adapter, which is directly connected to the ICPMS inlet. Connect the laminar flow adapter to an argon source with a mass flow controller and later on use it to admix Argon to achieve a stable operation condition.
Align the adapter as well as the steel tube vertical with a spirit level. If the alignment is not accurate, it may lead to significant losses of droplets. Insert a plug into the adapter to prevent gases leaking out during the system warm up time.
Start all above-mentioned gas flows and devices using the settings from Table 1. Allow the system to warm up for 15 min. The cartridge heater needs 2 hr to stabilize the temperature, switch it on in advance.
Place the syringe pumps on a rack at the height of cyclonic helium adapter. Keep the distance between the syringe pumps and the adapter as short as possible.
4. Measurements
NOTE: The following protocol is written in general terms because of the variety of solutions and suspensions that can be used. However, cell suspensions should be diluted to a concentration of < 1 x 107 cells/ml, when single cell analysis is performed, to ensure that the majority of the droplets carry only one cell. For measurements with cells place the syringe pumps at an angle so that the outlet of the syringes point downwards and install the tubing in such a way that they point downwards.
Attach tubing to the syringes. Load two 5 ml syringes with perfluorohexane and one 1 ml syringe with a sample solution or suspension. Remove all bubbles trapped in the syringes and tubing.
- Install the syringes in a syringe pump and connect them to the inlets of the chip. Start the syringe pumps using the start settings from Table 1 (or higher flow rates). Give the flows 3 to 5 min to stabilize.
- Remove excess liquid from the tip of the chip with a tissue. The liquids should now eject from the chip in a straight jet. If a straight ejection cannot be achieved by wiping with a tissue replace the chip and start over with this step.
Remove the plug from the adapter and carefully insert the chip into the adapter. Lubricate the chip with FC-40 if necessary. A chip can be used for at least 2 hr of experiments.
Change the flow rate to be within the recommended measurement settings from Table 1. Lower flow rate of the PFH not only saves PFH but also reduces signal background, caused by isobaric interferences.
- Give the system 2-5 min to stabilize (depending on the chosen flow rates). Optimize the ICPMS for the highest signal intensity of the analytes of interest.
- Successively adjust the flows of all the gases on the mass flow controllers (see the recommended ranges in Table 1) until the maximum signal intensity of the analytes of interest is achieved. Tune the plasma power and the focusing lens voltages (according to the manufacturer's recommended ranges) on the ICPMS in the same way.
Set the ICPMS to a dwell time of 10 msec (applied for the very ICPMS used but can be adjusted with other instruments to ensure a time-resolved acquisition). Start recording the signal of a particular m/z using manufacturer’s protocol.
After the measurement, transfer the raw data to data analysis program for evaluation. Bin the data, given in counts per 10 msec, with a built-in-function, and plot resulting bin center values against counts. Fit each peak in the plotted frequency distribution histogram with a Gauss function. The mean and sigma of the fit represent the mean signal intensity and its standard deviation, respectively.
5. Calibration Concept
Measure a single or multi-element standard solution containing the element or elements of interest at the same flow rates as the sample.
Place a LADE chip in a petri dish on a microscope. For a better image quality use a non-round chip. Fabricate this chip as described in step 2, but using a simple rectangular casting form instead of the partially round shaped one.
Follow the steps 4.1 to 4.2.1 to start the droplet generation. Set the flow rates to the flow rates used in step 5.1.
Record images of the aqueous droplets with a high-speed camera attached to a microscope (20X objective). Use an image analysis software like the droplet morphometry and velocimetry software by Basu25 to obtain the average droplet diameter from the recordings.
- Use average droplet diameter to calculate the droplet volume, assuming the droplet is a spherical object.
- From this volume and the known concentration of an analyte in the droplet calculate the number of corresponding atoms. Divide the number of measured ions per droplet by the number of atoms to obtain the detection efficiency. Use this detection efficiency to calculate the number of atoms in an unknown sample. NOTE: Since the variations between individual chips are small22, it is not necessary to repeat the measurement of the droplet size for every chip or solution if the flow rates remain the same. A list of the droplet sizes and frequencies according to the specific flow rates is published by Verboket et al.22.
Representative Results
The presented system can be employed to measure small volumes of solutions or suspensions containing cells or nanoparticles. Examples of a measurement of a standard solution and characterization of single cells are shown here. More examples can be found in Verboket et al.22.
Typically the signal of a single droplet of a solution is a very short event. It usually lasts for a few 100 µsec26. With the ICPMS used here (dwell time 10 msec) short signals like these cannot be temporally resolved. Figure 7a and Figure 7b show the signals and frequency distribution histogram of a Na standard solution. The droplets arrive at the plasma with a temporal jitter > 10 msec. The detection is unsynchronized. Signals of one (201 ± 24), two (381 ± 34) or three (560 ± 45) droplets are detected within one dwell time. Low variation in signal intensity suggests high droplet monodispersity. The first tailing peak is likely the result of droplet fragments; the cause of this fragmentation is still under investigation.
The calibration approach described in 5 (using Fe standard solution) was tested for the determination of Fe content of single bovine/calf red blood cells (6-7 µm in diameter) suspended in phosphate buffered saline (PBS). The suspension of 1 x 107 cells ml-1 was used to ensure that the majority of droplets carry only one cell. Figure 8 shows the signals from cells as frequency distribution histogram. On average every cell contained 5.3 ± 1.2 x 108 Fe atoms (see Verboket et al.22).
 Figure 1. Micrograph of droplet generation, acceleration and ejection. In a flow focusing junction, monodisperse aqueous droplets are generated in the stream of PFH. Additional PFH is added at a second junction. Subsequently, the liquid stream is ejected from the LADE chip through a nozzle. Arrows indicate the direction of the flow. Scale bar is 500 µm. Inset: Micrograph of an aqueous droplet in a PFH shell after ejection from the LADE chip. Scale bar 100 µm. Adapted with permission from22. Copyright 2014 American Chemical Society. This figure has been modified with data from research conducted since publication in the laboratory of P. S. Dittrich. Please click here to view a larger version of this figure.
Figure 1. Micrograph of droplet generation, acceleration and ejection. In a flow focusing junction, monodisperse aqueous droplets are generated in the stream of PFH. Additional PFH is added at a second junction. Subsequently, the liquid stream is ejected from the LADE chip through a nozzle. Arrows indicate the direction of the flow. Scale bar is 500 µm. Inset: Micrograph of an aqueous droplet in a PFH shell after ejection from the LADE chip. Scale bar 100 µm. Adapted with permission from22. Copyright 2014 American Chemical Society. This figure has been modified with data from research conducted since publication in the laboratory of P. S. Dittrich. Please click here to view a larger version of this figure.
 Figure 2. Schematic of the SU-8 processing. First an adhesion layer is coated. A silicon wafer is spin coated with SU-8 2002, soft baked, flood exposed with ultraviolet light and post baked. On top of this layer the microfluidic structures are produced. The wafer is spin coated with SU-8 2050 and soft baked. The design of the microfluidic structures is transferred to the wafer by exposing it with ultraviolet light through a photomask. After a postexposure bake, the photoresist is developed and a hardbake is performed. Please click here to view a larger version of this figure.
Figure 2. Schematic of the SU-8 processing. First an adhesion layer is coated. A silicon wafer is spin coated with SU-8 2002, soft baked, flood exposed with ultraviolet light and post baked. On top of this layer the microfluidic structures are produced. The wafer is spin coated with SU-8 2050 and soft baked. The design of the microfluidic structures is transferred to the wafer by exposing it with ultraviolet light through a photomask. After a postexposure bake, the photoresist is developed and a hardbake is performed. Please click here to view a larger version of this figure.
 Figure 3. Design of the photomask for the LADE chip containing the following features: a) guiding structures for the casting form, b) an inlet for the PFH for droplet acceleration, c) an inlet for PFH for droplet generation and d) an inlet for the aqueous sample. e) Indicator line for cutting off the tip of the chip. Channel widths f) = 40 µm, g) = 20 µm and h) = 25 µm. Please click here to view a larger version of this figure.
Figure 3. Design of the photomask for the LADE chip containing the following features: a) guiding structures for the casting form, b) an inlet for the PFH for droplet acceleration, c) an inlet for PFH for droplet generation and d) an inlet for the aqueous sample. e) Indicator line for cutting off the tip of the chip. Channel widths f) = 40 µm, g) = 20 µm and h) = 25 µm. Please click here to view a larger version of this figure.
 Figure 4.Process chart of the LADE chip fabrication. First the structured and the flat PDMS parts are fabricated by replica molding. The two pieces are bonded together by adhesive bonding. Finally, the tip of the chip is cut off and the microfluidic channels are silanized. Adapted with permission from22. Copyright 2014 American Chemical Society. This figure has been modified since publication. Please click here to view a larger version of this figure.
Figure 4.Process chart of the LADE chip fabrication. First the structured and the flat PDMS parts are fabricated by replica molding. The two pieces are bonded together by adhesive bonding. Finally, the tip of the chip is cut off and the microfluidic channels are silanized. Adapted with permission from22. Copyright 2014 American Chemical Society. This figure has been modified since publication. Please click here to view a larger version of this figure.
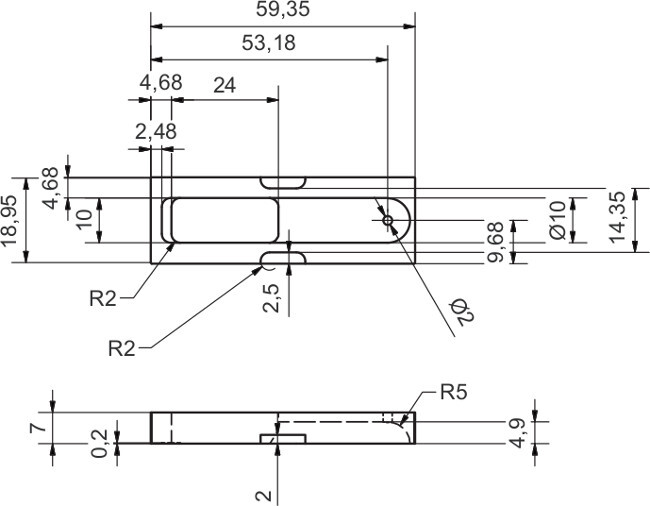 Figure 5. Technical drawing of the aluminum casting form for the LADE chip.
Please click here to view a larger version of this figure.
Figure 5. Technical drawing of the aluminum casting form for the LADE chip.
Please click here to view a larger version of this figure.
 Figure 6. Schematic drawing of the setup (not to scale). The system consists of the LADE chip, a cyclonic adapter, a heated steel tube, a membrane desolvator, and an ICPMS. Please click here to view a larger version of this figure.
Figure 6. Schematic drawing of the setup (not to scale). The system consists of the LADE chip, a cyclonic adapter, a heated steel tube, a membrane desolvator, and an ICPMS. Please click here to view a larger version of this figure.
 Figure 7.(A) Signals of droplets made from of a 1 mg kg-1 Na standard solution. (B) Frequency distribution histogram of these signals. In the 10 msec dwell time signals of one (yellow), two (red) or three (blue) droplets were recorded. Mean and standard deviation of the signals were determined by fitting Gaussian functions. The flow rates used were 0.5, 50, and 60 µl min−1 of aqueous sample, PFH for droplet generation, and PFH for droplet acceleration, respectively. Please click here to view a larger version of this figure.
Figure 7.(A) Signals of droplets made from of a 1 mg kg-1 Na standard solution. (B) Frequency distribution histogram of these signals. In the 10 msec dwell time signals of one (yellow), two (red) or three (blue) droplets were recorded. Mean and standard deviation of the signals were determined by fitting Gaussian functions. The flow rates used were 0.5, 50, and 60 µl min−1 of aqueous sample, PFH for droplet generation, and PFH for droplet acceleration, respectively. Please click here to view a larger version of this figure.
 Figure 8. Frequency distribution histogram of 56Fe+ signals generated by red blood cells. Mean and standard deviation of the signals were determined by fitting a Gaussian function. The flow rates used were 2, 80, and 80 µl min−1 of cell suspension, PFH for droplet generation, and PFH for droplet acceleration, respectively.
Figure 8. Frequency distribution histogram of 56Fe+ signals generated by red blood cells. Mean and standard deviation of the signals were determined by fitting a Gaussian function. The flow rates used were 2, 80, and 80 µl min−1 of cell suspension, PFH for droplet generation, and PFH for droplet acceleration, respectively.
| He gas flow rate | 0.6-0.8 L min-1 | |
| Cartridge heater | 30 W | |
| Desolvator membrane temperature | 160 °C | |
| Desolvator sweep gas flow rate | 3-4 L min-1 | |
| Ar gas flow rate | 0.1 L min-1 | |
| ICP plasma power | 1,300 W | |
| Sample flow rate | start up | 1 µl min-1 |
| measurement | 0.3-15 µl min-1 | |
| Flow rate of PFH droplet generation | start up | 60 µl min-1 |
| measurement | 35-80 µl min-1 | |
| Flow rate of PFH droplet acceleration | start up | 60 µl min-1 |
| measurement | 35-80 µl min-1 |
Table 1. Start settings and recommended measurement settings for the ICPMS and the syringe pumps.
Discussion
Although the fabrication of the chips is very reliable there are some critical points during the fabrication that require special attention. First, cleanliness during the assembly is highly important to prevent contamination of the chip by dust. The dust can block the channels and prevent a stable droplet generation. Second, it is especially important that the tip is cut orthogonal to the nozzle channel. The angle of the cut strongly influences the ejection angle. If the liquid is ejected at an angle it can cause a loss of the ejected droplets.
When building the setup ensure that it is stable. The vertical alignment of the metal tube and the adapter are important. Also during the measurements there are some points that need special attention. The insertion of the chip into the adapter has to be performed carefully. It can happen that during the insertion the jet is disrupted and stops. For measurements with cells the position and orientation of the syringe pumps and tubing are important. Their proper placement can reduce settling of the cells in the syringe and the tubing.
The LADE chip presented here has several advantages over existing commercial droplet generators. The system is more robust, provides a wider droplet size range, which can be further extended by modification of the channel geometry, and is disposable. A single use device is of a particular interest for the analysis of samples with a high content of salts or solid residues, as for instance nanoparticles or cell suspensions, which can clog tiny channels and cannot always be easily washed out. The transport of single microdroplets generated by the LADE into the MS is still a limiting step in our system and has to be further optimized. The current droplet transport assembly, although removes the PFH vapor, which would otherwise create additional spectral and non-spectral interferences and cause plasma instabilities, but still results in a high temporal jitter of droplet arrival at the MS and an incomplete droplet transport. In comparison to the commercial available droplet introduction systems the transport system for this setup requires more equipment. The current chip is designed for sample introduction only. However, with slight changes of the design advanced introduction and sample preparation steps cloud be implemented on-chip, e.g., dilution27-29, ultra fast mixing30, chemical reactions31, separation32-34, or cell sorting35,36. Under advanced introduction devices we understand for example the introduction of sample and standard droplets sequentially or parallel with a single chip. This would increase the throughput and improve the accuracy of quantitative analysis.
Disclosures
The authors declare that they have no competing financial interests.
Acknowledgments
This work was supported by the European Research Council (ERC Starting Grant nμLIPIDS, No. 203428) and ETH Zurich (project number: ETH-49 12-2). The authors of this manuscript would like to thank Bodo Hattendorf for help with the ICP-MS and F. Kurth for cell counting. The authors also would like to thank Christoph Bärtschi and Roland Mäder for their support with building the mechanical setup. The clean room facility FIRST at ETH Zurich is acknowledged for support in microfabrication.
References
- Todoli J-L, Mermet J-M. Liquid sample introduction in ICP spectrometry: A Practical Guide. Amsterdam: Elsevier; 2008. pp. 10–1016. [Google Scholar]
- Sutton KL, B'Hymer C, Caruso JA. Ultraviolet absorbance and inductively coupled plasma mass spectrometric detection for capillary electrophoresis - A comparison of detection modes and interface designs. J. Anal. At. Spectrom. 1998;13(9):885–891. [Google Scholar]
- Todoli J-L, Mermet J-M. Sample introduction systems for the analysis of liquid microsamples by ICP-AES and ICP-MS. Spectrochim. Acta, Part B. 2006;61(3):239–283. [Google Scholar]
- Olesik JW, Hobbs SE. Monodisperse dried microparticulate injector - A new tool for studying fundamental processes in inductively-coupled plasma. Anal. Chem. 1994;66(20):3371–3378. [Google Scholar]
- Gschwind S, Hagendorfer H, Frick DA, Günther D. Mass quantification of nanoparticles by single droplet calibration using inductively coupled plasma mass spectrometry. Anal. Chem. 2013;85(12):5875–5883. doi: 10.1021/ac400608c. [DOI] [PubMed] [Google Scholar]
- Garcia CC, Murtazin A, Groh S, Horvatic V, Niemax K. Characterization of single Au and SiO2 nano- and microparticles by ICP-OES using monodisperse droplets of standard solutions for calibration. J. Anal. At. Spectrom. 2010;25(5):645–653. [Google Scholar]
- Shigeta K, et al. Sample introduction of single selenized yeast cells (Saccharomyces cerevisiae) by micro droplet generation into an ICP-sector field mass spectrometer for label-free detection of trace elements. J. Anal. At. Spectrom. 2013;28(5):637–645. [Google Scholar]
- Orlandini v. Niessen JO, Schaper JN, Petersen JH, Bings NH. Development and characterization of a thermal inkjet-based aerosol generator for micro-volume sample introduction in analytical atomic spectrometry. J. Anal. At. Spectrom. 2011;26(9):1781–1789. [Google Scholar]
- Orlandini v. Niessen JO, Petersen JH, Schaper JN, Bings NH. Comparison of novel and conventional calibration techniques for the analysis of urine samples using plasma source mass spectrometry combined with a new dual-drop-on-demand aerosol generator. J. Anal. At. Spectrom. 2012;27(8):1234–1244. [Google Scholar]
- Shigeta K, et al. Application of a micro-droplet generator for an ICP-sector field mass spectrometer - optimization and analytical characterization. J. Anal. At. Spectrom. 2013;28(5):646–656. [Google Scholar]
- Teh S-Y, Lin R, Hung L-H, Lee AP. Droplet microfluidics. Lab on a Chip. 2008;8(2):198–220. doi: 10.1039/b715524g. [DOI] [PubMed] [Google Scholar]
- Zheng B, Tice JD, Ismagilov RF. Formation of arrayed droplets by soft lithography and two-phase fluid flow, and application in protein crystallization. Adv. Mater. 2004;16(15):1365–1368. doi: 10.1002/adma.200400590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theberge AB, et al. Microfluidic platform for combinatorial synthesis in picolitre droplets. Lab Chip. 2012;12(7):1320–1326. doi: 10.1039/c2lc21019c. [DOI] [PubMed] [Google Scholar]
- Li L, et al. Nanoliter microfluidic hybrid method for simultaneous screening and optimization validated with crystallization of membrane proteins. Proc. Natl. Acad. Sci. U. S. A. 2006;103(51):19243–19248. doi: 10.1073/pnas.0607502103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Q, Liu X, Liu D, Gai H. Ultra-small droplet generation via volatile component evaporation. Lab Chip. 2014;14(8):1395–1400. doi: 10.1039/c3lc51183a. [DOI] [PubMed] [Google Scholar]
- Baret JC, Beck Y, Billas-Massobrio I, Moras D, Griffiths AD. Quantitative cell-based reporter gene assays using droplet-based microfluidics. Chem. Biol. 2010;17(5):528–536. doi: 10.1016/j.chembiol.2010.04.010. [DOI] [PubMed] [Google Scholar]
- Brouzes E, et al. Droplet microfluidic technology for single-cell high-throughput screening. Proc. Natl. Acad. Sci. U. S. A. 2009;106(34):14195–14200. doi: 10.1073/pnas.0903542106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pei J, Li Q, Lee MS, Valaskovic GA, Kennedy RT. Analysis of samples stored as individual plugs in a capillary by electrospray ionization mass spectrometry. Anal. Chem. 2009;81(15):6558–6561. doi: 10.1021/ac901172a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly RT, Page JS, Marginean I, Tang K, Smith RD. Dilution-free analysis from picoliter droplets by nano-electrospray ionization mass spectrometry. Angew. Chem., Int. Ed. 2009;48(37):6832–6835. doi: 10.1002/anie.200902501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Küster SK, et al. Interfacing droplet microfluidics with matrix-assisted laser desorption/ionization mass spectrometry: label-free content analysis of single droplets. Anal. Chem. 2013;85(3):1285–1289. doi: 10.1021/ac3033189. [DOI] [PubMed] [Google Scholar]
- Pabst M, Jefimovs K, Zenobi R, Dittrich PS. High-Resolution Droplet-Based Fractionation of Nano-LC Separations onto Microarrays for MALDI-MS Analysis. Analytical Chemistry. 2014;86(10):4848–4855. doi: 10.1021/ac4041982. [DOI] [PubMed] [Google Scholar]
- Verboket PE, Borovinskaya O, Meyer N, Günther D, Dittrich PS. A New Microfluidics-Based Droplet Dispenser for ICPMS. Analytical Chemistry. 2014;86(12):6012–6018. doi: 10.1021/ac501149a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ammerman CN, You SM. Determination of the boiling enhancement mechanism caused by surfactant addition to water. J. Heat Transfer. 1996;118(2):429–435. [Google Scholar]
- Samel B, Chowdhury MK, Stemme G. The fabrication of microfluidic structures by means of full-wafer adhesive bonding using a poly(dimethylsiloxane) catalyst. J Micromech Microeng. 2007;17(8):1710–1714. [Google Scholar]
- Basu AS. Droplet morphometry and velocimetry (DMV): a video processing software for time-resolved, label-free tracking of droplet parameters. Lab Chip. 2013;13(10):1892–1901. doi: 10.1039/c3lc50074h. [DOI] [PubMed] [Google Scholar]
- Dziewatkoski MP, Daniels LB, Olesik JW. Time-resolved inductively coupled plasma mass spectrometry measurements with individual, monodisperse drop sample introduction. Anal. Chem. 1996;68(7):1101–1109. doi: 10.1021/ac951013b. [DOI] [PubMed] [Google Scholar]
- Abate AR, Hung T, Mary P, Agresti JJ, Weitz DA. High-throughput injection with microfluidics using picoinjectors. Proc. Natl. Acad. Sci. U. S. A. 2010;107(45):19163–19166. doi: 10.1073/pnas.1006888107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bremond N, Thiam AR, Bibette J. Decompressing emulsion droplets favors coalescence. Phys. Rev. Lett. 2008;100(2):024501. doi: 10.1103/PhysRevLett.100.024501. [DOI] [PubMed] [Google Scholar]
- Niu X, Gulati S, Edel JB, deMello AJ. Pillar-induced droplet merging in microfluidic circuits. Lab Chip. 2008;8(11):1837–1841. doi: 10.1039/b813325e. [DOI] [PubMed] [Google Scholar]
- Song H, Ismagilov RF. Millisecond kinetics on a microfluidic chip using nanoliters of reagents. J. Am. Chem. Soc. 2003;125(47):14613–14619. doi: 10.1021/ja0354566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song H, Chen DL, Ismagilov RF. Reactions in droplets in microflulidic channels. Angew. Chem., Int. Ed. 2006;45(44):7336–7356. doi: 10.1002/anie.200601554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lombardi D, Dittrich PS. Droplet microfluidics with magnetic beads: a new tool to investigate drug-protein interactions. Anal. Bioanal. Chem. 2011;399(1):347–352. doi: 10.1007/s00216-010-4302-7. [DOI] [PubMed] [Google Scholar]
- Edgar JS, et al. Compartmentalization of chemically separated components into droplets. Angew. Chem., Int. Ed. 2009;48(15):2719–2722. doi: 10.1002/anie.200805396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar JS, et al. Capillary electrophoresis separation in the presence of an immiscible boundary for droplet analysis. Anal. Chem. 2006;78(19):6948–6954. doi: 10.1021/ac0613131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baret JC, et al. Fluorescence-activated droplet sorting (FADS): efficient microfluidic cell sorting based on enzymatic activity. Lab Chip. 2009;9(13):1850–1858. doi: 10.1039/b902504a. [DOI] [PubMed] [Google Scholar]
- Agresti JJ, et al. Ultrahigh-throughput screening in drop-based microfluidics for directed evolution. Proc. Natl. Acad. Sci. U. S. A. 2010;107(9):4004–4009. doi: 10.1073/pnas.0910781107. [DOI] [PMC free article] [PubMed] [Google Scholar]


