Abstract
BACKGROUND
Alzheimer’s disease (AD) is the most common cause of dementia with no curative therapy currently available. Establishment of sensitive and non-invasive biomarkers that promote an early diagnosis of AD is crucial for the effective administration of disease-modifying drugs. MicroRNAs (miRNAs) mediate posttranscriptional repression of numerous target genes. Aberrant regulation of miRNA expression is implicated in AD pathogenesis, and circulating miRNAs serve as potential biomarkers for AD. However, data analysis of numerous AD-specific miRNAs derived from small RNA-sequencing (RNA-Seq) is most often laborious.
METHODS
To identify circulating miRNA biomarkers for AD, we reanalyzed a publicly available small RNA-Seq dataset, composed of blood samples derived from 48 AD patients and 22 normal control (NC) subjects, by a simple web-based miRNA data analysis pipeline that combines omiRas and DIANA miRPath.
RESULTS
By using omiRas, we identified 27 miRNAs expressed differentially between both groups, including upregulation in AD of miR-26b-3p, miR-28–3p, miR-30c-5p, miR-30d-5p, miR-148b-5p, miR-151a-3p, miR-186–5p, miR-425–5p, miR-550a-5p, miR-1468, miR-4781–3p, miR-5001–3p, and miR-6513–3p and downregulation in AD of let-7a-5p, let-7e-5p, let-7f-5p, let-7g-5p, miR-15a-5p, miR-17–3p, miR-29b-3p, miR-98–5p, miR-144–5p, miR-148a-3p, miR-502–3p, miR-660–5p, miR-1294, and miR-3200–3p. DIANA miRPath indicated that miRNA-regulated pathways potentially downregulated in AD are linked with neuronal synaptic functions, while those upregulated in AD are implicated in cell survival and cellular communication.
CONCLUSIONS
The simple web-based miRNA data analysis pipeline helps us to effortlessly identify candidates for miRNA biomarkers and pathways of AD from the complex small RNA-Seq data.
Keywords: Alzheimer’s disease, biomarkers, data analysis pipeline, DIANA miRPath, microRNA, omiRas, small RNA-Seq
Introduction
Alzheimer’s disease (AD) is the most common cause of dementia affecting 36 million people worldwide, showing the hallmark pathology of amyloid-β (Aβ) deposition, neurofibrillary tangle (NFT) formation, extensive synaptic loss, and neurodegeneration in the brain.1 The complex interaction between multiple genetic and environmental factors affecting various molecular pathways plays a key role in AD, although the precise molecular mechanism remains largely unknown.2 Currently, only four drugs with limited efficacies are available for the treatment of AD, including donepezil, rivastigmine, galantamine, and memantine. Development of disease-modifying drugs for AD, mainly targeting Aβ, has been unsuccessful, suggesting that both early diagnosis and early treatment are highly important to achieve maximal response to disease-modifying treatments.3 However, an accurate diagnosis of AD is often difficult, solely based on neuropsychological examinations, because the possibility of numerous dementing diseases, such as frontotemporal dementia (FTD), dementia with Lewy bodies (DLB), vascular dementia, HIV-associated dementia (HAD), and prion diseases, should be carefully excluded.4 Although decreased Aβ42 and increased pTau levels in the cerebrospinal fluid (CSF) serve as a biomarker for the diagnosis of AD, the collection of CSF is too invasive to apply to routine clinical works.5 Therefore, establishment of most reliable, non-invasive, ultrasensitive, high-throughput, and inexpensive biomarkers that promote the early diagnosis of AD would meet a growing demand from the public.6
MicroRNAs (miRNAs), a class of endogenous small non-coding RNAs (ncRNAs), mediate posttranscriptional regulation of protein-coding genes by binding to the 3′ untranslated region (UTR) of target mRNAs, leading to translational inhibition or mRNA destabilization or degradation.7 Currently, 1,881 precursor and 2,588 mature human miRNAs are registered in miRBase release 21 in June 2014. A single miRNA, capable of downregulating hundreds of target mRNAs concurrently, fine tunes diverse cellular functions involved in development, differentiation, proliferation, apoptosis, and metabolism.8 Overall, the whole human miRNome regulates greater than 60% of all protein-coding genes.9 Importantly, aberrant regulation of the miRNome plays a central role in pathological events underlying cancers and neurodegenerative diseases.10–12 Recently, we identified 852 experimentally validated target genes for more than 100 miRNAs downregulated in AD brains.13 The molecular network analysis of 852 genes revealed that aberrant expression of cell cycle regulators contributes to neurodegeneration in AD.
Increasing evidence indicated that various miRNAs, released extracellularly to biological fluids, serve as novel biomarkers for diagnosis and prediction of prognosis of AD, Parkinson’s disease (PD), and cancers.10,14,15 They circulate in the serum, plasma, urine and the CSF in a free form binding to RNA-binding proteins or in an encapsulated form within exosomes protected from endogenous RNase activity. Approximately 70% of presently identified miRNAs are expressed in the brain in a spatially and temporally controlled manner.16 Certain brain-enriched miRNAs are transported through the blood–brain barrier (BBB) into the circulatory system via unknown mechanisms.14
The recent breakthrough in deep sequencing technology has made a great impact on the field of genome research. Whole RNA-sequencing (RNA-Seq) represents an innovative tool for the genome-wide transcriptome profiling in a high-throughput and quantitative manner with excellent reproducibility.17 It identifies the unbiased expression of the complex transcriptome, composed of both mRNAs and ncRNAs, even in a single cell at a single base resolution. Small RNA-Seq, one of RNA-Seq applications, analyzes the differential expression of all kinds of small ncRNAs, without prior assumptions inevitable in quantitative PCR (qPCR) assays. Furthermore, this technology overcomes several drawbacks intrinsic to the microarray-based approach, such as the difficulty in detection of novel miRNAs and high backgrounds because of cross-hybridization. Actually, it provides a powerful approach to the comprehensive profiling of miRNAs circulating in biological fluids of AD.14 However, at present, it is most often laborious to clarify biological implications from billions of sequencing read data.
To identify circulating miRNA biomarkers for AD, we studied a publicly available small RNA-Seq dataset, composed of blood miRNA profiles derived from AD patients and normal control (NC) subjects.18 For this purpose, we established a simple web-based miRNA data analysis pipeline that combines omiRas19 and DIANA miRPath.20 We found that this pipeline helps us to effortlessly identify candidates for miRNA biomarkers and pathways of AD from the complex small RNA-Seq data.
Methods
miRNA-Seq dataset of blood samples of AD patients and NCs
First, we retrieved FASTQ-formatted files of a small RNA-Seq dataset from the DDBJ Sequence Read Archive (DRA) (trace.ddbj.nig.ac.jp/DRASearch) under the accession number of SRP022043. The researchers in Saarland University, Homburg, Germany carried out a study of original experiments.18 It included 48 samples of AD patients, composed of 23 males and 25 females with average age 70.3 ± 7.9 years and Mini-Mental State Examination (MMSE) score 18.7 ± 3.5, along with 22 samples of NC subjects, composed of 11 males and 11 females with average age 67.1 ± 7.5 years and MMSE score 29.3 ± 1.2. They obtained the samples mostly from the Biorepository and Tissue Bank PrecisionMed (www.precisionmed.com). Total RNA including miRNA was isolated from the blood by the PAXgene Blood miRNA Kit (Qiagen, Valencia, CA, USA). A total of 200 ng of RNA was processed to generate multiplexed sequencing libraries by using the TruSeq Small RNA Sample Kit (Illumina, San Diego, CA, USA). Then, DNA library products with the size of ∼50 bp were processed for single-end sequencing on HiSeq 2000 (Illumina).
miRNA-Seq data analysis by omiRas
After removing adapters, poly-A tails, and low-quality reads from the original sequencing data with cutadapt (code.google.com/p/cutadapt) and Qcleaner (Amelieff, Tokyo, Japan), we imported GZIP-compressed FASTQ files of cleaned data into the analysis tool named omiRas (tools.genxpro.net/omiras), a free web server established for differential expression analysis of miRNA-Seq data between two groups.19 On omiRas, bowtie processed mapping of short reads of each library, which are summarized to tags in a quantified FASTA format, on the human genome reference sequence hg19, allowing at most two mismatches. Mapping locus annotations were given for each tag, based on information derived from miRbase v19 (www.mirbase.org), snoRNABase (www-snorna.biotoul.fr), Repbase (www.girinst.org/repbase), Genomic tRNA database (gtrnadb.ucsc.edu), and Rfam (www.sanger.ac.uk/resources/databases/rfam.html). Following normalization of the number of reads for each tag with the number of mapping loci, differential expression analysis was carried out between two groups for each miRNA according to the DESeq algorithm (bioconductor.org/packages/release/bioc/html/DESeq.html) in the setting of false discovery rate (FDR)-corrected P-value <0.05.21 We excluded tRNAs, rRNAs, scRNAs, snoRNAs, and precursor miRNAs because of lack of the precise annotation of their targets. We also removed mature miRNAs showing normalized counts less than 10 in both groups because of their lower expression levels.
Receiver operating characteristic (ROC) analysis was performed by using SPSS Version 19 (IBM Japan). We imported normalized counts of individual miRNAs in the setting of the group discrimination number 1 for AD and 0 for NC. For ROC analysis of the entire panel of 27 miRNAs, comprising a mixture of upregulated and downregulated classes, we converted normalized counts into applicable values as follows: for miRNAs upregulated in AD, the summation of individual counts for each miRNA was adjusted to 1. For miRNAs downregulated in AD, the summation of individual counts subtracted from total counts was adjusted to 1. Finally, all adjusted values for each miRNA in individual samples were integrated for ROC analysis.
Pathway analysis of miRNA targets by DIANA miR-Path
We utilized DIANA miRPath v2.0 (diana.imis.athena-innovation.gr/DianaTools/index.php?r=mirpath/index), a web server established for identification of Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways corresponding to the networks of miRNA targets by superimposing numerous miRNA-target relationships on the merging and meta-analysis algorithm.20 This program predicts miRNA targets with high accuracy based on the DIANA-microT-CDS algorithm that considers the evolutional conservation of miRNA-binding sites. KEGG (www.genome.jp/kegg/kegg_ja.html) is a publicly accessible knowledgebase, composed of manually curated 364,925 pathways that cover a wide range of metabolic, genetic, environmental, and cellular processes and human diseases.22
We also utilized the interaction network tool of omiRas for identification of miRNA targets. It extracted overlapping targets for miRNAs predicted by five distinct algorithms, such as miRanda (www.microrna.org/microrna/home.do), TargetScan (www.targetscan.org), PicTar (pictar.mdc-berlin.de), miRDB (mirdb.org/miRDB), and PITA (genie.weizmann.ac.il/pubs/mir07/mir07_prediction.html). We imported them into the functional annotation tool of Database for Annotation, Visualization and Integrated Discovery (DAVID) v6.7 (david.abcc.ncifcrf.gov).23 DAVID identified gene ontology (GO) terms enriched in the set of imported genes and extracted relevant pathways constructed by KEGG, followed by statistical evaluation with the modified Fisher’s exact test and the correction by multiple comparison tests. We considered P-value <0.05 after Bonferroni correction as significant.
We also imported the set of miRNA targets into the core analysis tool of ingenuity pathway analysis (IPA) (Ingenuity Systems) (www.ingenuity.com). IPA is a commercial knowledgebase, comprising approximately 3,000,000 biological and chemical interactions with definite scientific evidence. By uploading the list of Gene IDs, the network-generation algorithm identified focused genes integrated in global molecular pathways and networks. IPA calculated the score P-value that reflects the statistical significance of association between the genes and the pathways and networks by Fisher’s exact test. We considered P-value <0.05 as significant. The present study complied with the principles of the Declaration of Helsinki.
Results
miRNA-Seq data analysis by omiRas identified miRNAs differentially expressed between AD and NC
By analyzing miRNA-Seq dataset of blood samples derived from 48 AD patients and 22 NC subjects with omiRas, we identified totally 27 differentially expressed mature miRNAs that satisfied FDR-corrected P-value <0.05 (Table 1). The omiRas quality control (QC) tool validated the acceptable quality of length and mapping of sequencing reads, presenting with a discernible correlation in the miRNA expression levels between both groups (Supplementary Fig. 1). The set of 27 miRNAs included 13 miRNAs upregulated in AD, such as miR-26b-3p, miR-28–3p, miR-30c-5p, miR-30d-5p, miR-148b-5p, miR-151a-3p, miR-186–5p, miR-425–5p, miR-550a-5p, miR-1468, miR-4781–3p, miR-5001–3p, and miR-6513–3p, and 14 miRNAs downregulated in AD, such as let-7a-5p, let-7e-5p, let-7f-5p, let-7g-5p, miR-15a-5p, miR-17–3p, miR-29b-3p, miR-98–5p, miR-144–5p, miR-148a-3p, miR-502–3p, miR-660–5p, miR-1294, and miR-3200–3p (Table 1).
Table 1.
The set of 27 differentially expressed microRNAs in blood samples of AD patients and NC subjects.
| TWENTY SEVEN DIFFERENTIALLY EXPRESSED microRNAs IN THE PRESENT STUDY | miRBase ACCESSION NUMBER | NORMALIZED COUNT_AD | NORMALIZED COUNT_NC | LOG2FC (AD/NC) | FC (AD/NC) | FDR-CORRECTED P-VALUE |
|---|---|---|---|---|---|---|
| hsa-miR-1468 | MIMAT0006789 | 75.5198 | 32.1045 | 1.234079414 | 2.352311981 | 0.0055675 |
| hsa-miR-4781–3p | MIMAT0019943 | 13.4487 | 6.19139 | 1.11913148 | 2.172161663 | 0.000231431 |
| hsa-miR-26b-3p | MIMAT0004500 | 65.9038 | 32.4001 | 1.024363387 | 2.034061623 | 0.008447968 |
| hsa-miR-5001–3p | MIMAT0021022 | 13.1559 | 7.02103 | 0.90595535 | 1.873784901 | 0.0055675 |
| hsa-miR-6513–3p | MIMAT0025483 | 15.1891 | 8.70471 | 0.803168248 | 1.744928895 | 0.002112453 |
| hsa-miR-148b-5p | MIMAT0004699 | 20.5116 | 12.0991 | 0.761540298 | 1.69529965 | 0.011220933 |
| hsa-miR-28–3p | MIMAT0004502 | 638.594 | 382.199 | 0.740574994 | 1.67084163 | 0.03701297 |
| hsa-miR-151a-3p | MIMAT0000757 | 2466.05 | 1495.83 | 0.721255828 | 1.648616487 | 0.002112453 |
| hsa-miR-30c-5p | MIMAT0000244 | 3411.48 | 2070.43 | 0.720467332 | 1.647715692 | 0.006802852 |
| hsa-miR-186–5p | MIMAT0000456 | 4234.72 | 2615.92 | 0.694948161 | 1.618826264 | 0.007140543 |
| hsa-miR-550a-5p | MIMAT0004800 | 47.0717 | 32.4523 | 0.536539227 | 1.450488871 | 0.007140543 |
| hsa-miR-30d-5p | MIMAT0000245 | 10108.7 | 6995.13 | 0.531174701 | 1.44510538 | 0.008842896 |
| hsa-miR-425–5p | MIMAT0003393 | 5195.81 | 3696.46 | 0.491204374 | 1.405617807 | 0.011388068 |
| hsa-let-7a-5p | MIMAT0000062 | 6253.08 | 10508.6 | −0.7489316 | 0.595044059 | 0.020304816 |
| hsa-miR-502–3p | MIMAT0004775 | 17.066 | 32.9147 | −0.947607095 | 0.518491738 | 0.033463494 |
| hsa-miR-29b-3p | MIMAT0000100 | 6.90953 | 13.6193 | −0.97899307 | 0.50733371 | 0.020304816 |
| hsa-let-7e-5p | MIMAT0000066 | 7.00419 | 15.2301 | −1.12063529 | 0.459891268 | 0.007140543 |
| hsa-miR-98–5p | MIMAT0000096 | 93.3802 | 207.101 | −1.149145936 | 0.450892077 | 0.002112453 |
| hsa-let-7g-5p | MIMAT0000414 | 1383.36 | 3092.06 | −1.160391668 | 0.44739106 | 0.011973772 |
| hsa-let-7f-5p | MIMAT0000067 | 3020.48 | 6927.54 | −1.197565302 | 0.436010474 | 0.000176286 |
| hsa-miR-1294 | MIMAT0005884 | 7.66566 | 20.889 | −1.446261513 | 0.366971133 | 0.011327892 |
| hsa-miR-15a-5p | MIMAT0000068 | 365.776 | 1044.86 | −1.514277327 | 0.35007178 | 0.007140543 |
| hsa-miR-17–3p | MIMAT0000071 | 18.3476 | 52.4116 | −1.514294791 | 0.350067542 | 0.011973772 |
| hsa-miR-660–5p | MIMAT0003338 | 32.2007 | 99.6604 | −1.629928314 | 0.323104262 | 0.008841428 |
| hsa-miR-148a-3p | MIMAT0000243 | 428.942 | 1345.96 | −1.649781046 | 0.31868852 | 0.007140543 |
| hsa-miR-144–5p | MIMAT0004600 | 328.701 | 1132.86 | −1.785121832 | 0.290151475 | 0.001282761 |
| hsa-miR-3200–3p | MIMAT0015085 | 14.9001 | 59.0867 | −1.987511413 | 0.252173501 | 0.008841428 |
| SEVEN DIFFERENTIALLY EXPRESSED microRNAs IN THE PREVIOUS STUDY | miRBASE ACCESSION NUMBER | NORMALIZED COUNT_AD | NORMALIZED COUNT_NC | LOG2FC (AD/NC) | FC (AD/NC) | FDR-CORRECTED P-VALUE |
| hsa-let-7d-3p | MIMAT0000065 | 354.327 | 198.231 | 0.837898811 | 1.787444951 | 0.496016391 |
| hsa-miR-5010–3p | MIMAT0021044 | 132.557 | 84.4256 | 0.650860425 | 1.570104329 | 0.085089651 |
| hsa-miR-26a-5p | MIMAT0000082 | 5663.92 | 5272.27 | 0.103376634 | 1.074284891 | 1 |
| hsa-miR-103a-3p | MIMAT0000101 | 1288.68 | 1981.45 | −0.620662499 | 0.650372202 | 0.43522577 |
| hsa-miR-532–5p | MIMAT0002888 | 368.938 | 622.771 | −0.755323373 | 0.592413584 | 0.10385145 |
| hsa-miR-107 | MIMAT0000104 | 124.417 | 249.563 | −1.004220432 | 0.498539447 | 0.102581668 |
| hsa-miR-26b-5p | MIMAT0000083 | 341.938 | 696.265 | −1.025901743 | 0.491103244 | 0.172704544 |
Notes: MicroRNA-Seq dataset SRP022043 composed of 48 AD and 22 NC blood samples was analyzed by omiRas. It identified 27 differentially expressed mature miRNAs between two groups. They are shown with miRNA name, miRBase accession number, normalized count, log2(fold change), fold change, and FDR-corrected P-value. From the identical dataset, a previous study (Ref. 18) identified the set of 12 differentially expressed miRNAs. Among them, the expression levels of seven miRNAs reanalyzed by omiRas are listed in the bottom.
From the identical dataset, a previous study of original experiments identified the set of 12 miRNAs differentially expressed between AD and NC groups.18 The panel included seven miRNAs upregulated in AD, such as let-7d-3p, miR-26a-5p, brain-miR-112, miR-151a-3p, brain-miR-161, miR-1285–5p, and miR-5010–3p, and five miRNAs downregulated in AD, such as let-7f-5p, miR-26b-5p, miR-103a-3p, miR-107, and miR-532–5p. Unexpectedly, only upregulation of miR-151a-3p and downregulation of let-7f-5p in AD were shared between the present and previous observations. Therefore, we reevaluated by omiRas the expression levels of previously reported seven miRNAs, such as let-7d-3p, miR-26a-5p, miR-26b-5p, miR-103a-3p, miR-107, miR-532–5p, and miR-5010–3p. Surprisingly, we found that the differential expression of none of them reached the levels of statistical significance (Table 1).
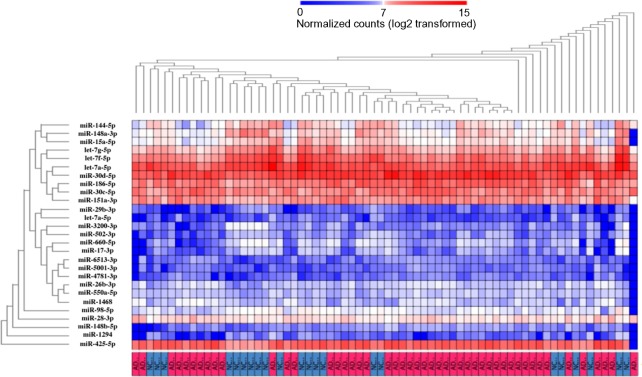
By hierarchical clustering analysis (HCA) with omiRas, the set of 27 miRNAs separated several AD subgroups from NC clusters, where the largest AD cluster contained 23 patients (47.9%) (Fig. 1). Thus, the 27 miRNA panel did not perfectly discriminate AD from NC on HCA. The expression profiles of individual 27 miRNAs were different between both groups, although the expression levels were highly variable among each sample (Fig. 2A–F and Supplementary Figs. 2 and 3).
Figure 1.
HCA of 27 miRNAs differentially expressed in blood of AD and NC. miRNA-Seq dataset of blood samples derived from 48 AD patients and 22 NC subjects was analyzed on omiRas. It identified 27 differentially expressed mature miRNAs between two groups. They separated several AD subgroups (red) from NC clusters (blue), where the largest AD cluster included 23 patients (47.9%).
Figure 2.
The expression profile of miRNAs differentially expressed in blood of AD and NC. By omiRas, we identified the set of 27 miRNAs differentially expressed in blood samples of AD (blue) and NC (red). The representative profiles of (A) miR-26b-3p, (B) miR-148b-5p, (C) miR-186–5p, (D) miR-148–3p, (E) miR-17–3p, and (F) let-7g-5p are shown. All 27 profiles are shown in Supplementary Figures 2 and 3.
By ROC analysis, the set of differentially expressed miRNAs, except for let-7e-5p and miR-29b-3p, showed statistically significant levels of the sensitivity and the specificity for discrimination between AD and NC (Fig. 3A–C and Supplementary Figs. 4 and 5). ROC analysis of the entire 27 miRNA panel revealed the area under the curve (AUC) = 0.801 (P = 0.00006) with the sensitivity = 70.8% and the specificity = 81.8% for discrimination of both groups (Fig. 3D).
Figure 3.
ROC analysis of miRNAs differentially expressed in blood of AD and NC. By omiRas, we identified the set of 27 miRNAs differentially expressed between AD and NC blood samples. ROC analysis was performed by importing normalized counts of individual miRNAs into SPSS in the setting of the group discrimination number 1 for AD and 0 for NC, where 1 is defined as a state variable for miRNAs upregulated in AD. The representative profiles of (A) miR-26b-3p, (B) miR-148b-5p, (C) miR-186–5p, and (D) the entire 27 miRNA panel are shown. All 27 profiles are shown in Supplementary Figures 4 and 5.
Pathway analysis by DIANA miRPath characterized biological pathways of miRNA targets differentially expressed between AD and NC
By DIANA miRPath, we identified KEGG biological pathways constructed by predicted targets for differentially expressed miRNAs based on the DIANA-microT-CDS algorithm. For the set of 13 miRNAs upregulated in AD, without the inclusion of miR-6513–3p whose information is absent in DIANA-microT, it identified top three most significant pathways potentially downregulated in AD, such as dopaminergic synapse (hsa04728) (P = 2.100E−16), long-term potentiation (hsa04720) (P = 7.706E−15) (Fig. 4), and ubiquitin-mediated proteolysis (hsa04120) (P = 6.233E−12) (Table 2). For the set of 14 miRNAs downregulated in AD, it identified top three most significant pathways potentially upregulated in AD, such as PI3K-Akt signaling pathway (hsa04151) (P = 4.359E−33) (Fig. 5), ECM–receptor interaction (hsa04512) (P = 1.404E−21), and focal adhesion (hsa04510) (P = 1.404E−21).
Figure 4.
DIANA miRPath analysis of target genes for miRNAs upregulated in AD blood. The set of 13 miRNAs upregulated in AD, except for hsa-miR-6513–3p whose information is absent in DIANA-microT, was imported into DIANA miRPath. It identified the second-rank significant KEGG pathway potentially downregulated in AD, termed long-term potentiation (hsa04720), relevant to target gene network for imported miRNAs. The genes targeted by more than one miRNA are colored by orange, while the genes targeted by a single miRNA are colored by yellow.
Table 2.
Top 10 KEGG pathways of target genes for microRNAs differentially expressed in blood samples of AD patients and NC subjects.
| RANK | KEGG PATHWAY OF TARGET GENES FOR UPREGULATED microRNAs IN AD | NO. OF TARGET GENES | NO. OF microRNAs | FDR-CORRECTED P-VALUE |
|---|---|---|---|---|
| 1 | Dopaminergic synapse (hsa04728) | 51 | 11 | 2.09951E-16 |
| 2 | Long-term potentiation (hsa04720) | 30 | 9 | 7.70571E-15 |
| 3 | Ubiquitin mediated proteolysis (hsa04120) | 48 | 8 | 6.23343E-12 |
| 4 | Endometrial cancer (hsa05213) | 22 | 10 | 6.70195E-11 |
| 5 | Aldosterone-regulated sodium reabsorption (hsa04960) | 18 | 10 | 1.24983E-10 |
| 6 | ErbB signaling pathway (hsa04012) | 34 | 11 | 1.24983E-10 |
| 7 | Axon guidance (hsa04360) | 46 | 10 | 2.66356E-10 |
| 8 | mRNA surveillance pathway (hsa03015) | 32 | 10 | 1.41697E-09 |
| 9 | Wnt signaling pathway (hsa04310) | 50 | 10 | 2.08198E-09 |
| 10 | Colorectal cancer (hsa05210) | 25 | 11 | 3.23945E-09 |
| RANK | KEGG PATHWAY OF TARGET GENES FOR DOWNREGULATED microRNAs IN AD | NO. OF TARGET GENES | NO. OF microRNAs | FDR-CORRECTED P-VALUE |
| 1 | PI3K-Akt signaling pathway (hsa04151) | 108 | 14 | 4.36E-33 |
| 2 | ECM-receptor interaction (hsa04512) | 29 | 12 | 1.40E-21 |
| 3 | Focal adhesion (hsa04510) | 67 | 13 | 1.40E-21 |
| 4 | p53 signaling pathway (hsa04115) | 31 | 12 | 4.3261E-20 |
| 5 | mTOR signaling pathway (hsa04150) | 29 | 14 | 6.97691E-19 |
| 6 | Pathways in cancer (hsa05200) | 94 | 14 | 6.97912E-19 |
| 7 | Prostate cancer (hsa05215) | 34 | 12 | 2.78621E-17 |
| 8 | Small cell lung cancer (hsa05222) | 32 | 12 | 5.05832E-15 |
| 9 | Melanoma (has05218) | 28 | 12 | 6.45591E-15 |
| 10 | MAPK signaling pathway (hsa04010) | 71 | 13 | 1.2189E-12 |
Notes: DIANA miRpath identified KEGG pathways constructed by predicted targets for differentially expressed miRNAs between AD and NC based on the DIANA-microT-CDS algorithm. Top ten pathways are shown with rank, pathway name, number of target genes and microRNAs, and FDR-corrected P-value. The upper half represents the pathways potentially downregulated in AD, while the lower half indicates the pathways potentially upregulated in AD.
Figure 5.
DIANA miRPath analysis of target genes for miRNAs downregulated in AD blood. The set of 14 miRNAs downregulated in AD was imported into DIANA miRPath. It identified the most significant KEGG pathway potentially upregulated in AD, termed PI3K-Akt signaling pathway (hsa04151), relevant to target gene network for imported miRNAs. The genes targeted by more than one miRNA are colored by orange, while the genes targeted by a single miRNA are colored by yellow.
To verify these results, we studied miRNA targets overlapping in the prediction by miRanda, TargetScan, PicTar, miRDB, and PITA on the interaction network tool of omiRas. We identified the set of 749 target genes for miRNAs upregulated in AD, including those for miR-30c-5p, miR-30d-5p, miR-186–5p, and miR-425–5p, and the set of 829 target genes for miRNAs downregulated in AD, including those for let-7a-5p, let-7g-5p, let-7e-5p, let-7f-5p, miR-15a-5p, miR-29b-3p, miR-98a-5p, and miR-148a-3p (Supplementary Table 1). Overlaps were not found for other miRNAs by the prediction on five distinct algorithms described above.
For the set of 749 targets potentially downregulated in AD, DAVID indicated their relevance to the KEGG pathway termed long-term potentiation (hsa042720) (P = 0.00774 corrected by Bonferroni), while IPA showed the most significant relationship with the canonical pathways defined as axonal guidance signaling (P = 6.37E−06) and synaptic long-term potentiation (P = 4.53E−05). Thus, they are well consistent with the pathway-finding results of DIANA miRPath. For the set of 829 targets potentially upregulated in AD, DAVID revealed their relevance to the KEGG pathways termed focal adhesion (hsa04510) (P = 7.793E−06 corrected by Bonferroni) and ECM–receptor interaction (hsa04512) (P = 9.082E−05 corrected by Bonferroni), again well consistent with the results of DIANA miRPath. IPA showed the most significant relationship with the canonical pathways defined as hepatic fibrosis and hepatic stellate cell activation (P = 3.84E−14) and PTEN signaling (1.56E−07). The latter is equivalent to the KEGG pathway termed as PI3K-Akt signaling pathway (hsa04151), which was identified by DIANA miRPath.
Discussion
Utility of the miRNA-Seq data analysis pipeline for identification of AD blood biomarkers
Increasing evidence indicates that miRNAs circulating in biological fluids serve as non-invasive biomarkers for diagnosis of AD.14 Recently, small RNA-Seq has been utilized as a powerful approach to the comprehensive profiling of disease-relevant circulating miRNAs. However, it is most often laborious to clarify biological implications from billions of sequencing read data. Here, we established a simple web-based miRNA data analysis pipeline by combining omiRas and DIANA miRPath that effortlessly identifies candidates for miRNA biomarkers and pathways of AD from the complex small RNA-Seq data. We applied this pipeline to a publicly available small RNA-Seq dataset, comprising blood samples derived from 48 AD patients and 22 NC subjects.
First, we identified 27 differentially expressed miRNAs between both groups. They included upregulation in AD of miR-26b-3p, miR-28–3p, miR-30c-5p, miR-30d-5p, miR-148b-5p, miR-151a-3p, miR-186–5p, miR-425–5p, miR-550a-5p, miR-1468, miR-4781–3p, miR-5001–3p, and miR-6513–3p and downregulation in AD of let-7a-5p, let-7e-5p, let-7f-5p, let-7g-5p, miR-15a-5p, miR-17–3p, miR-29b-3p, miR-98–5p, miR-144–5p, miR-148a-3p, miR-502–3p, miR-660–5p, miR-1294, and miR-3200–3p. Among them, previous studies reported downregulation of miR-29b targeting BACE1 in the anterior temporal cortex and the serum of AD,24,25 downregulation of miR-148a-3p in the parietal lobe of AD,26 and upregulation of miR-186–5p in the prefrontal cortex of AD.27 ROC analysis indicated that discrimination between AD and NC on the entire 27 miRNA panel satisfies the accuracy over 80%.
Next, we characterized miRNA-regulated pathways deregulated in AD blood. We found that the pathways potentially downregulated in AD are linked with neuronal synaptic functions, such as dopaminergic synapse and long-term potentiation, while those upregulated in AD are implicated in cell survival and cellular communication, such as PI3K-Akt signaling pathway, ECM–receptor interaction, and focal adhesion. Notably, it takes only a couple of days to accomplish all analyses online on a PC with standard specifications, suggesting that the pipeline process seems less time consuming. Even though the gold standard for small RNA-Seq data analysis has not been presently established, our pipeline potentially applicable to large-scale deep sequencing data, including those of familial AD patients, would serve as a general standard for AD biomarker mining.
Possible explanations for the discrepancy between the present and previous studies
The previous study of original small RNA-Seq experiments identified the set of 12 miRNAs, containing two unannotated ones, differentially expressed between AD and NC blood samples.18 They also verified the results by qPCR on a larger cohort composed of 202 samples, containing those derived from 94 AD patients, 21 NC subjects, and 87 patients with other neuropsychiatric diseases. They found that the discrimination of AD from other neuropsychiatric diseases is possible with 74–78% accuracy.
However, only 2 of the 12 miRNAs, such as upregulation of miR-151a-3p and downregulation of let-7f-5p in AD, were reproduced between the present and previous observations, regardless of the identical dataset employed. Importantly, omiRas indicated that differential expression of let-7d-3p, miR-26a-5p, miR-26b-5p, miR-103a-3p, miR-107, miR-532–5p, and miR-5010–3p reported previously does not reach the levels of statistical significance (Table 1, the bottom). Since the corresponding qPCR dataset is publicly unavailable, we could not currently clarify the precise reason for the inconsistency between both by reanalyzing qPCR data.
We suggested the possibility that these discrepancies are attributable to differences in data processing, including the methods of cleaning raw data of Illumina short reads, ie, cutadapt and Qcleaner in the present study versus fastx clipper in the previous study and mapping loci annotations based on miRbase v19 in the present study versus miRbase v18 in the previous study or the methods of statistical analysis utilized to identify differentially expressed miRNAs, such as DESeq in the present study versus the Wilcoxon-Mann–Whitney test with Benjamini–Hochberg adjustment in the previous study. The study that includes independent cohorts of large numbers of AD patients and controls is required for validation of the 27 blood miRNA signatures in AD we identified. It is worthy of note that omiRas successfully identified previously overlooked 19 miRNAs differentially expressed in the prefrontal cortex between Huntington’s disease (HD) patients and controls.28
Previous studies identified variable profiles of circulating miRNAs in AD
Accumulating studies indicated that expression profiles of circulating miRNAs are highly variable among the patients with AD, patients with mild cognitive impairment (MCI), and NC subjects.29–33 The lack of reproducibility is mostly attributable to great variations in samples, ie, the plasma, serum, whole blood, and exosomes, affected by sampling time and confounding factors such as smoking and medications; study populations, ie, the stage of diseases, age, sex, ethnicity, the selection of adequate controls, and the size of study population to obtain sufficient statistical power; and analytical methods, ie, qPCR, microarray, and the most advanced technology of deep sequencing. Our observations suggested that data processing pipelines contribute to one of major factors greatly affecting the results.
A recent study by qPCR of the plasma miRNAs isolated from 50 AD and 50 NC subjects showed that both the miR-132 family composed of miR-128, miR-132, and miR-874 and the miR-134 family composed of miR-134, miR-323–3p, and miR-382, all of which are upregulated in MCI, differentiate MCI from NC with 87–96% accuracy.29 In contrast, another study by qPCR of the plasma miRNAs derived from 110 AD patients and 123 NC subjects indicated that the levels of miR-34c, which represses the genes involved in cell survival and anti-oxidative defense, are upregulated in AD.30 A previous study was performed by microarray analysis of miRNAs isolated from the commercially available plasma derived from 11 AD patients and 20 NC subjects on nCounter containing features of approximately 700 human and human-associated viral miRNAs.31 It showed that the 7-miRNA signature, composed of let-7d-5p, let-7g-5p, miR-15b-5p, miR-142–3p, miR-191–5p, miR-301a-3p, and miR-545–3p, all of which are downregulated in AD, discriminates AD from NC with greater than 95% accuracy, where the miRNA targets are involved in lipid metabolism. However, among them, we only verified downregulation of let-7g-5p in the miRNA-Seq dataset of AD blood.
A recent small RNA-Seq study performed on an Ion Personal Genome Machine using exosomal miRNAs isolated from 23 AD patients and 23 NC subjects showed that miR-15a-5p, miR-18b-5p, miR-20a-5p, miR-30e-5p, miR-93–5p, miR-101–3p, miR-106a-5p, miR-106b-5p, miR-143–3p, miR-335–5p, miR-361–5p, miR-424–5p, miR-582–5p, and miR-3065–5p are upregulated, while miR-15b-3p, miR-342–3p, and miR-1306–5p are downregulated in AD.32 Surprisingly, these observations partially contradicted our results that miR-15a-5p is downregulated in AD blood, suggesting that the source of miRNAs, either purified exosomes or unfractionated whole blood, serves as a critical factor responsible for differences in miRNA expression profiles. A different small RNA-Seq study performed on HiSeq 2000 using the serum miRNAs derived from 50 AD patients and 50 NC subjects revealed that the expression levels of let-7d-5p, miR-98–5p, miR-191–5p, miR-342–3p, miR-483–3p, and miR-885–5p are reduced in AD.33 They also found a trend for upregulation of miR-26b-3p and downregulation of let-7g-5p in AD. Among them, we verified upregulation of miR-26b-3p and downregulation of miR-98–5p and let-7g-5p in AD blood. Importantly, the expression levels of miR-26b-5p, the opposite strand of miR-26b-3p elevated in postmortem brains at the early stage of AD, represses retinoblastoma protein Rb1, resulting in acceleration of tau phosphorylation by activating Cdk5.34
Biological pathways regulated by circulating miRNAs aberrantly regulated in AD blood
By pathway analysis of target genes for differentially expressed miRNAs with DIANA miRPath, we found that biological pathways potentially downregulated in AD are related to neuronal synaptic functions. These findings might reflect profound disturbances of synapse integrity in AD brains. In contrast, the pathways potentially upregulated in AD exhibited a significant relationship with biological pathways of cell survival and cellular communication functions. These pathways are likely to be induced and activated by compensatory responses to extensive neurodegeneration in AD brains. Importantly, the pathway-finding results obtained by DIANA miRPath are well consistent with those from the prediction of overlapping targets by five distinct algorithms, followed by pathway analysis with DAVID and IPA. Importantly, various brain-enriched miRNAs are transported through BBB into the circulatory system.14 Therefore, our observations suggested that real-time profiles of AD blood miRNAs might reflect a disruption of the miRNA–mRNA interaction network in AD brains affected by ongoing neurodegeneration.
Conclusions
We reanalyzed a publicly available small RNA-Seq dataset, composed of blood samples derived from 48 AD patients and 22 NC subjects, by a newly established, simple web-based miRNA data analysis pipeline that combines omiRas and DIANA miRPath. We identified 27 differentially expressed miRNAs, including 13 upregulated miRNAs and 14 downregulated miRNAs in AD. The miRNA-regulated pathways potentially downregulated in AD were linked with neuronal synaptic functions, while those upregulated in AD were related to cell survival and cellular communication. This pipeline helps us to effortlessly identify candidates for miRNA biomarkers and pathways of AD from the complex small RNA-Seq data.
Supplementary Files
Supplementary Figure 1. Quality control of microRNA-Seq data on omiRas. MicroRNA-Seq dataset of blood samples derived from 48 AD patients and 22 NC subjects was analyzed on omiRas. The mapping statistics of a sample of AD numbered SRR837437 are shown. The panels (A–D) indicate (A) general overview, (B) mapping region specificity, (C) sequence length distribution, and (D) the scatter plot of the expression levels between AD (x-axis) and NC (y-axis).
Supplementary Figure 2. The expression profile of 13 miRNAs upregulated in AD blood. By omiRas, we identified the set of 27 miRNAs differentially expressed in blood samples of AD (blue) and NC (red). The profiles of 13 miRNAs upregulated in AD are shown.
Supplementary Figure 3. The expression profile of 14 miRNAs downregulated in AD blood. By omiRas, we identified the set of 27 miRNAs differentially expressed in blood samples of AD (blue) and NC (red). The profiles of 14 miRNAs downregulated in AD are shown.
Supplementary Figure 4. ROC analysis of 13 miRNAs upregulated in AD blood. By omiRas, we identified the set of 27 miRNAs differentially expressed in blood samples of AD and NC. ROC analysis was performed by importing normalized counts of individual miRNAs into SPSS in the setting of the group discrimination number 1 for AD and 0 for NC, where 1 is defined as a state variable. The profiles of 13 miRNAs upregulated in AD are shown.
Supplementary Figure 5. ROC analysis of 14 miRNAs downregulated in AD blood. By omiRas, we identified the set of 27 miRNAs differentially expressed in blood samples of AD and NC. ROC analysis was performed by importing normalized counts of individual miRNAs into SPSS in the setting of the group discrimination number 1 for AD and 0 for NC, where 0 is defined as a state variable. The profiles of 14 miRNAs downregulated in AD are shown.
Supplementary Table 1. Predicted target genes for microRNAs differentially expressed in blood samples of AD patients and NC subjects.
Footnotes
ACADEMIC EDITOR: Karen Pulford, Editor in Chief
FUNDING: This work was supported by the Science Research Promotion Fund of the Promotion and Mutual Aid Corporation for Private Schools of Japan, the JSPS KAKENHI (C25430054), the Dementia Drug Development Research Center (DRC) project, the Ministry of Education, Culture, Sports, Science and Technology (MEXT), and the Longevity Sciences (26–20) from the National Center for Geriatrics and Gerontology (NCGG), Japan. The authors confirm that the funder had no influence over the study design, content of the article, or selection of this journal.
COMPETING INTERESTS: Authors disclose no potential conflicts of interest.
Paper subject to independent expert blind peer review by minimum of two reviewers. All editorial decisions made by independent academic editor. Upon submission manuscript was subject to anti-plagiarism scanning. Prior to publication all authors have given signed confirmation of agreement to article publication and compliance with all applicable ethical and legal requirements, including the accuracy of author and contributor information, disclosure of competing interests and funding sources, compliance with ethical requirements relating to human and animal study participants, and compliance with any copyright requirements of third parties. This journal is a member of the Committee on Publication Ethics (COPE).
Author Contributions
Designed the methods, analyzed the data, and drafted the manuscript: JS. Helped with the data analysis: YK, SN. All authors have read and approved the final manuscript.
REFERENCES
- 1.Serrano-Pozo A, Frosch MP, Masliah E, Hyman BT. Neuropathological alterations in Alzheimer disease. Cold Spring Harb Perspect Med. 2011;1(1):a006189. doi: 10.1101/cshperspect.a006189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gatz M, Reynolds CA, Fratiglioni L, et al. Role of genes and environments for explaining Alzheimer disease. Arch Gen Psychiatry. 2006;63(2):168–74. doi: 10.1001/archpsyc.63.2.168. [DOI] [PubMed] [Google Scholar]
- 3.Galimberti D, Scarpini E. Disease-modifying treatments for Alzheimer’s disease. Ther Adv Neurol Disord. 2011;4(4):203–16. doi: 10.1177/1756285611404470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Geldmacher DS, Whitehouse PJ., Jr Differential diagnosis of Alzheimer’s disease. Neurology. 1997;48(5 suppl 6):S2–9. doi: 10.1212/wnl.48.5_suppl_6.2s. [DOI] [PubMed] [Google Scholar]
- 5.McKhann GM, Knopman DS, Chertkow H, et al. The diagnosis of dementia due to Alzheimer’s disease: recommendations from the National Institute on Aging-Alzheimer’s Association workgroups on diagnostic guidelines for Alzheimer’s disease. Alzheimers Dement. 2011;7(3):263–9. doi: 10.1016/j.jalz.2011.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Antila K, Lötjönen J, Thurfjell L, et al. The PredictAD project: development of novel biomarkers and analysis software for early diagnosis of the Alzheimer’s disease. Interface Focus. 2013;3(2):20120072. doi: 10.1098/rsfs.2012.0072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Guo H, Ingolia NT, Weissman JS, Bartel DP. Mammalian microRNAs predominantly act to decrease target mRNA levels. Nature. 2010;466(7308):835–40. doi: 10.1038/nature09267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Selbach M, Schwanhäusser B, Thierfelder N, Fang Z, Khanin R, Rajewsky N. Widespread changes in protein synthesis induced by microRNAs. Nature. 2008;455(7209):58–63. doi: 10.1038/nature07228. [DOI] [PubMed] [Google Scholar]
- 9.Friedman RC, Farh KK, Burge CB, Bartel DP. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res. 2009;19(1):92–105. doi: 10.1101/gr.082701.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Keller A, Leidinger P, Bauer A, et al. Toward the blood-borne miRNome of human diseases. Nat Methods. 2011;8(10):841–3. doi: 10.1038/nmeth.1682. [DOI] [PubMed] [Google Scholar]
- 11.Satoh J. Molecular network analysis of human microRNA targetome: from cancers to Alzheimer’s disease. BioData Min. 2012;5(1):17. doi: 10.1186/1756-0381-5-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hayes J, Peruzzi PP, Lawler S. MicroRNAs in cancer: biomarkers, functions and therapy. Trends Mol Med. 2014;20(8):460–9. doi: 10.1016/j.molmed.2014.06.005. [DOI] [PubMed] [Google Scholar]
- 13.Satoh J. Molecular network of microRNA targets in Alzheimer’s disease brains. Exp Neurol. 2012;235(2):436–46. doi: 10.1016/j.expneurol.2011.09.003. [DOI] [PubMed] [Google Scholar]
- 14.Cheng L, Quek CY, Sun X, Bellingham SA, Hill AF. The detection of microRNA associated with Alzheimer’s disease in biological fluids using next-generation sequencing technologies. Front Genet. 2013;4:150. doi: 10.3389/fgene.2013.00150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Vallelunga A, Ragusa M, Di Mauro S, et al. Identification of circulating microRNAs for the differential diagnosis of Parkinson’s disease and multiple system atrophy. Front Cell Neurosci. 2014;8:156. doi: 10.3389/fncel.2014.00156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fineberg SK, Kosik KS, Davidson BL. MicroRNAs potentiate neural development. Neuron. 2009;64(3):303–9. doi: 10.1016/j.neuron.2009.10.020. [DOI] [PubMed] [Google Scholar]
- 17.Ozsolak F, Milos PM. RNA sequencing: advances, challenges and opportunities. Nat Rev Genet. 2011;12(2):87–98. doi: 10.1038/nrg2934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Leidinger P, Backes C, Deutscher S, et al. A blood based 12-miRNA signature of Alzheimer disease patients. Genome Biol. 2013;14(7):R78. doi: 10.1186/gb-2013-14-7-r78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Müller S, Rycak L, Winter P, Kahl G, Koch I, Rotter B. omiRas: a Web server for differential expression analysis of miRNAs derived from small RNA-Seq data. Bioinformatics. 2013;29(20):2651–2. doi: 10.1093/bioinformatics/btt457. [DOI] [PubMed] [Google Scholar]
- 20.Vlachos IS, Kostoulas N, Vergoulis T, et al. DIANA miRPath v.2.0: investigating the combinatorial effect of microRNAs in pathways. Nucleic Acids Res. 2012;40(Web Server issue):W498–504. doi: 10.1093/nar/gks494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Anders S, Huber W. Differential expression analysis for sequence count data. Genome Biol. 2010;11(10):R106. doi: 10.1186/gb-2010-11-10-r106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kanehisa M, Goto S, Sato Y, Furumichi M, Tanabe M. KEGG for integration and interpretation of large-scale molecular data sets. Nucleic Acids Res. 2012;40(Database issue):D109–14. doi: 10.1093/nar/gkr988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4(1):44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 24.Hébert SS Horré K, L Nicolaï, et al. Loss of microRNA cluster miR-29a/b-1 in sporadic Alzheimer’s disease correlates with increased BACE1/β-secretase expression. Proc Natl Acad Sci U S A. 2008;105(17):6415–20. doi: 10.1073/pnas.0710263105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Geekiyanage H, Jicha GA, Nelson PT, Chan C. Blood serum miRNA: non-invasive biomarkers for Alzheimer’s disease. Exp Neurol. 2012;235(2):491–6. doi: 10.1016/j.expneurol.2011.11.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hu CW, Tseng CW, Chien CW, et al. Quantitative proteomics reveals diverse roles of miR-148a from gastric cancer progression to neurological development. J Proteome Res. 2013;12(9):3993–4004. doi: 10.1021/pr400302w. [DOI] [PubMed] [Google Scholar]
- 27.Lau P, Bossers K, Janky R, et al. Alteration of the microRNA network during the progression of Alzheimer’s disease. EMBO Mol Med. 2013;5(10):1613–34. doi: 10.1002/emmm.201201974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Müller S. In silico analysis of regulatory networks underlines the role of miR-10b-5p and its target BDNF in Huntington’s disease. Transl Neurodegener. 2014;3:17. doi: 10.1186/2047-9158-3-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sheinerman KS, Tsivinsky VG, Abdullah L, Crawford F, Umansky SR. Plasma microRNA biomarkers for detection of mild cognitive impairment: biomarker validation study. Aging (Albany NY) 2013;5(12):925–38. doi: 10.18632/aging.100624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bhatnagar S, Chertkow H, Schipper HM, et al. Increased microRNA-34c abundance in Alzheimer’s disease circulating blood plasma. Front Mol Neurosci. 2014;7:2. doi: 10.3389/fnmol.2014.00002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kumar P, Dezso Z, MacKenzie C, et al. Circulating miRNA biomarkers for Alzheimer’s disease. PLoS One. 2013;8(7):e69807. doi: 10.1371/journal.pone.0069807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cheng L, Doecke JD, Sharples RA, et al. Prognostic serum miRNA biomarkers associated with Alzheimer’s disease shows concordance with neuropsychological and neuroimaging assessment. Mol Psychiatry. 2014 doi: 10.1038/mp.2014.127. [DOI] [PubMed] [Google Scholar]
- 33.Tan L, Yu JT, Tan MS, et al. Genome-wide serum microRNA expression profiling identifies serum biomarkers for Alzheimer’s disease. J Alzheimers Dis. 2014;40(4):1017–27. doi: 10.3233/JAD-132144. [DOI] [PubMed] [Google Scholar]
- 34.Absalon S, Kochanek DM, Raghavan V, Krichevsky AM. MiR-26b, upregulated in Alzheimer’s disease, activates cell cycle entry, tau-phosphorylation, and apoptosis in postmitotic neurons. J Neurosci. 2013;33(37):14645–59. doi: 10.1523/JNEUROSCI.1327-13.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure 1. Quality control of microRNA-Seq data on omiRas. MicroRNA-Seq dataset of blood samples derived from 48 AD patients and 22 NC subjects was analyzed on omiRas. The mapping statistics of a sample of AD numbered SRR837437 are shown. The panels (A–D) indicate (A) general overview, (B) mapping region specificity, (C) sequence length distribution, and (D) the scatter plot of the expression levels between AD (x-axis) and NC (y-axis).
Supplementary Figure 2. The expression profile of 13 miRNAs upregulated in AD blood. By omiRas, we identified the set of 27 miRNAs differentially expressed in blood samples of AD (blue) and NC (red). The profiles of 13 miRNAs upregulated in AD are shown.
Supplementary Figure 3. The expression profile of 14 miRNAs downregulated in AD blood. By omiRas, we identified the set of 27 miRNAs differentially expressed in blood samples of AD (blue) and NC (red). The profiles of 14 miRNAs downregulated in AD are shown.
Supplementary Figure 4. ROC analysis of 13 miRNAs upregulated in AD blood. By omiRas, we identified the set of 27 miRNAs differentially expressed in blood samples of AD and NC. ROC analysis was performed by importing normalized counts of individual miRNAs into SPSS in the setting of the group discrimination number 1 for AD and 0 for NC, where 1 is defined as a state variable. The profiles of 13 miRNAs upregulated in AD are shown.
Supplementary Figure 5. ROC analysis of 14 miRNAs downregulated in AD blood. By omiRas, we identified the set of 27 miRNAs differentially expressed in blood samples of AD and NC. ROC analysis was performed by importing normalized counts of individual miRNAs into SPSS in the setting of the group discrimination number 1 for AD and 0 for NC, where 0 is defined as a state variable. The profiles of 14 miRNAs downregulated in AD are shown.
Supplementary Table 1. Predicted target genes for microRNAs differentially expressed in blood samples of AD patients and NC subjects.