Abstract
AlkB is an Escherichia coli protein that catalyses the oxidative demethylation of 1-methyladenine and 3-methylcytosine in DNA and RNA. The enzyme activity of AlkB is dependent on a 2-oxoglutarate- and Fe(II)-dependent (2OG-Fe[II]) oxygenase domain. Human AlkB homologues (hABH), hABH1, hABH2 and hABH3, which also possess the 2OG-Fe(II) oxygenase domain, have previously been identified. Recent bioinformatics analysis suggests the existence of an additional five ABH genes in humans. In this study, we identified the hABH4–hABH7 mRNAs and determined their expression in human tissues. Moreover, an hABH2 splice variant lacking the 2OG-Fe(II) oxygenase domain and a new gene, hABH8, were cloned from testis cDNA. hABH8 possesses not only the 2OG-Fe(II) oxygenase domain but both an RNA-binding motif and a methyl-transferase domain. mRNA of the eight hABH molecules was detected in the 16 normal human tissues examined. The sub-cellular localization of EmGFP-hABH8 was restricted to the cytoplasm. EmGFP-hABH1, 3, 4, 6 and 7 were localized in both the cytoplasm and nuclei. Interestingly, the EmGFP-hABH2 splice variant localized in nucleoplasm with a dot-like pattern. In some HeLa cells transfected with EmGFP-hABH5, dot-like fluorescence was also detected in the cytoplasm. These observations provide important information for the future annotation of the hABH family of molecules.
Keywords: AlkB, DNA repair, demethylation, 2-oxoglutarate-Fe(II) oxygenase domain
Introduction
DNA is frequently methylated by methylating substances present in the external environment in addition to those within cells themselves. In mammalian cells, the methylation of DNA causes mutations, cytotoxicity and carcinogenecity. However, living cells have acquired three mechanisms for the repair of methylation-damaged DNA. First, the base excision repair pathway, which is initiated by methylpurine-DNA glycosylase, excises methylated purines, such as 3-methyladenine and 7-methylguanine, in DNA [1]. The resulting apurinic site is then sequentially processed by an apurinic endonuclease, a DNA polymerase, and a DNA ligase, leading to complete repair of the methylated DNA. Secondly, O-methylat-ed bases, such as O6-methylguanine, are directly reversed by O6-methylguanine methyltransferase [2]. In the enzymatic reaction, the methyltransferase transfers a methyl group from the O6-methylguanine of DNA to its own cysteine residue, thereby repairing the methylated DNA in a single-step reaction. Finally, a recently identified repair mechanism is oxidative demethylation that is catalysed by the E. coli AlkB protein. Treatment of E. coli with methylating agents induces four types of methylation repair proteins–AlkA, Ada, AidB and AlkB–as an adaptive response [3]. AlkA and Tag are methylbase glycosydases, and Ada and Ogt are methyltransferases. For some time, the function of AlkB remained undetermined. Recently, however, an interesting function of AlkB was discovered. AlkB possesses a 2-oxoglutarate-and iron(II)-dependent (2OG-Fe[II]) oxygenase domain [4]. In the presence of 2-oxoglutarate and Fe2+, this domain and a nucleotide recognition lid mediate the demethylation of 1-methyladenine (1-meA) and 3-methylcytosine (3-meC) previously methylated by SN2 methylating agents [5–7].
A human AlkB homologue was first identified from a human synovial sarcoma cDNA library and designated ABH (AlkB homologue) [8]. The amino acid sequences of ABH and AlkB are conserved in the 2OG-Fe(II) oxygenase domain with 34% identity. Subsequently, two research groups determined that hABH2 and hABH3 function as human homologues of AlkB [9, 10]. hABH2 and hABH3 catalyse the demethylation of 1-meA and 3-meC damages in DNA by a mechanism involving oxidative demethylation. Interestingly, it was demonstrated that both AlkB and hABH3 catalyse the demethylation of RNA methylated by methylating agents [10, 11]. Mice lacking functional mABH2 and mABH3 genes have recently been generated [12]. In the absence of exposure to exogenous methylating agents, ABH2-deficient mice accumulate significant levels of 1-meA in the genome, suggesting a physiologically important role for ABH2 in demethylation.
Bioinformatics analyses suggest the existence of five new AlkB family members in humans that possess a highly similar region to the 2OG-Fe(II) oxygenase domain of AlkB. These molecules have been designated hABH4 through hABH8 [13]. The nucleotide sequences of hABH4 to hABH7, each of which contains an open reading frame (ORF), have been registered in the database. The hABH8 nucleotide sequence, however, was indicated to be incomplete as a consequence of a split-domain conformation and alternative splicing [13, 14]. Osada et al. proposed the existence of a human gene that is highly similar to the ABH8 gene of Macaca fascicularis[15]. However, the nucleotide sequence and domain structure of hABH8 remain to be clarified. Moreover, hABH4–hABH7, as well as hABH1, are experimentally un-characterized and poorly annotated [16].
In this study, we identified the mRNA expression of the proposed hABH family members, including hABH2 and hABH3, and compared their expression and sub-cellular localization in order to obtain fundamental information for analysing their physiological function. The expression of hABH1–hABH8 mRNAs was detected in testis. Sequence analysis of these molecules revealed not only the hABH1–hABH7 ORFs contained in the database, but also the existence of an alternatively spliced form of hABH2 that lacks the 2OG-Fe(II) oxygenase domain. Moreover, a full-length sequence of the hABH8 ORF was determined. Real-time PCR analysis indicated the expression of hABH mRNAs in all the tissues examined. Sub-cellular localization was examined by expressing EmGFP-hABHs fusion proteins. hABH1, hABH4, hABH5, hABH6 and hABH7 localized in the nuclei and cytoplasm, whereas hABH8 localized only in the cytoplasm. Interestingly, a dot-like localization was detected in some cells expressing hABH5. Moreover, localization of the alternative splice form of hABH2 also exhibited a dot-like pattern in the nucleoplasm that was clearly different from that of hABH2 in nuclei.
Materials and methods
Reverse transcription (RT)-PCR of hABH1 to hABH8
The expression of the mRNA for hABH family molecules was evaluated in testis cDNA (Clontech Takara). The cDNA was subjected to PCR for 35 cycles using LA Taq DNA polymerase (Takara) for hABH1, hABH3 and hABH8, using KOD DNA polymerase (Toyobo) for for hABH2, hABH4, hABH5 and hABH7, and using PrimeStar HS DNA poly-merase (Takara) for hABH6, in 25-μl reactions. The GenBank accession numbers and primer sequences used for the amplification of each hABH are listed in Table 1. The reaction products were resolved by electrophoresis on a 2% agarose gel for the verification of the product sizes. The resolved products were excised, purified and then cloned into a pT7 blue vector (Promega) for sequencing (Applied Biosystems).
1.
GenBank accession number, primer sequences, and predicted sizes of the amplified products for hABH family molecules
| Gene | Accession no. | Primers | Predicted size (bp) |
|---|---|---|---|
| hABH1 | NM_006020 | F:CGCGAGATGGGGAAGATGGCA | 1184 |
| R:TCCAAGTCTCAGCTGTGAGG | |||
| hABH2 | NM_001001655 | F:GAATTCGCGAGGATGGACAGATTCC | 811 |
| R:GTTAAAAATGTTTTTATTTTTTAGTAAGC | |||
| hABH3 | NM_139178 | F:GGAATTCCTGAAAGCTCGGAGCAGAAGC | 1094 |
| R:CTGTTCCCACAGTAGACC | |||
| hABH4 | NM_017621 | F:GAATTCGCGATGGCGGCGGCTGCCGC | 929 |
| R:CTCGAGGCGGTTCACACGGGTCTTCC | |||
| hABH5 | NM_017758 | F:GGAGGACCCTAGAGCAGC | 1410 |
| R:CCCCTATTGATGCCAACAGC | |||
| hABH6 | NM_032878 | F:CACCATGGCTGGGAGGGGGATG | 844 |
| R:GCAGGAACCTGGGAATCCGAGG | |||
| hABH7 | NM_32306 | F:GAATTCGGGATTATGGCCGGGACTGG | 699 |
| R:CTCGAGGTAGAAAGCTGGGGGTCAGC | |||
| hABH8 | AB218768 | F:CACCATGGACAGCAACCATCAAAG | 717 |
| R:ATCAGGCCTTTTGAAGAATCAC | |||
Real-time PCR
The relative quantities of hABH mRNAs in a panel of normalized cDNAs derived from 16 human tissues (Human MTC Panels I and II; Clontech Takara) were analysed by real-time PCR (Roche) using a SYBR ExScript reverse transcriptase-polymerase chain reaction (RT-PCR) kit (Takara). Details of the primer sequences for each hABH are provided in Table 2. In order to avoid the amplification of the entire genomic DNA, each primer was located in a different exon. Serial dilutions of hABH and glyceraldehyde-3-phophate dehydrogenase (GAPDH) cDNAs were used to construct standard curves. Each determination was performed three times for each cDNA sample as independent PCR runs. The data from each PCR were normalized by dividing the quantity of each hABH by the quantity of GAPDH to correct for the differences in each sample.
2.
Primer sequences used for real-time RT-PCR analysis of hABH family molecules
| Gene | Forward primer | Reverse primer | Size (bp) |
|---|---|---|---|
| hABH1 | AGATCTGTGGGAACAGAGCAAAG | ATAATGGTAGCCTACGGTCACCC | 109 |
| hABH3 | TTGCTCAGCCAGCTACCACTGC | GACAGGCTGATTTCATACACACC | 181 |
| hABH4 | CCCCCAGCGAAAACATAC | AAAGTCCTCGATCAGCATCACTC | 123 |
| hABH5 | CAGTTCAAGCCTATTCGGG | CTTGTCTTCCTGAGGATG | 161 |
| hABH6 | TCATCTCCAAAGAAGAGGAGGAG | CCCCAGTTCTGTAACTTTCTCCC | 100 |
| hABH7 | TCTCCTGTCTCCCAGCGTTATG | TGAGCCCCTAAGGATGTAGAGG | 103 |
| hABH8 | TGCTGAGACATGAAGGCATTGA | ACGGCTTGTTAGGTGGCATTAAAGA | 159 |
| GAPDH | CCATCACCATCTTCCAGGAG | AATGAGCCCCAGCCTTCTCC | 117 |
Construction of pcDNA6.2/N-EmGFP-hABHs vectors
The cloned hABH1–hABH8 cDNAs were amplified by PCR using LATaq DNA polymerase (Takara). The amplified products were then sub-cloned into Gateway entry vectors, either pENTR1A, pENTR3C or pENTR/D-TOPO (Invitrogen). The entire coding sequence of the hABH1–hABH8 cDNAs was confirmed by sequencing. The hABH cDNAs in the pENTR-hABHs entry vectors were transferred into the Gateway destination vector pcDNA6.2/N-EmGFP-DEST using the LR clonase enzyme mix (Invitrogen) according to the manufacturer's instructions. pcDNA6.2/N-EmGFP-DEST was designed to produce an N-terminal fusion protein with an EmGFP tag for detection using fluorescence microscopy. The alternatively spliced form of hABH2 cDNA was also cloned into pcDNA6.2/N-EmGFP-DEST in the same manner as that described for the hABH2 cDNA.
Transient transfection
HeLa cells maintained in Dulbecco's modified Eagle's medium containing 10% foetal calf serum were plated at 1.4 x 105 in a 35- mm dish on the day before transfection. pcDNA6.2/N-EmGFP-hABHs plasmids (2 μg) were trans-fected into the cells using TransIT HeLa Monster (Mirus) according to the manufacturer's protocol.
Immunoblot analysis
Immunoblotting was performed as previously described [17]. Briefly, hABH transfectants were lysed in 1 ml of ice cold lysis buffer (1% Nonidet P-40, 150 mM NaCl, and 50 mM Tris, pH 7.4) with a protease inhibitor cocktail (Nakarai). The cell lysates were centrifuged, resolved by 12% sodium dodecyl sulfate-polyacrylamide gel elec-trophoresis (SDS-PAGE) and then transferred to a polyvinylidene fluoride (PVDF) membrane (BioRad). The membrane was blocked with 3% bovine serum albumin (BSA) in TBS-T (20 mM Tris-HCl, pH 8.0, 137 mM NaCl, and 0.1% Tween 20), incubated with rat anti-GFP antibody at room temperature for 1 hr, and then with horseradish peroxidase (HRP)-conjugated antirat IgG (Santa Cruz Biotechnology) diluted in TBS-T and incubated at room temperature for 1 hr. After three washings with TBS-T, the bound HRP conjugates were visualized with an ECL Plus Western blotting detection system (GE Healthcare) and detected with a cooled CCD camera system (Light-Capture, Atto).
Sub-cellular localization
Transiently transfected HeLa cells were fixed with 4% paraformaldehyde in PBS for 30 min. The DNA was stained with 0.1 μM 4,6-diamino-2-phenylindole (DAPI) fluorescent dye (Boehringer Mannheim) and washed three times with phosphate-buffered saline (PBS). Following the washes, a drop of mounting solution (90% glycerol in PBS) was placed directly onto the culture dish and a cover slip applied. Fluorescence was visualized with an Olympus BH2-RFCA fluorescence microscope.
Accession number
Accession numbers of hABH2 transcript variant 1 and hABH8 are AB277859 and AB218768, respectively.
Results
Detection of the hABH family members in testis
In our previous paper, we described the cloning of hABH3 as a highly expressed gene in human prostate cancer (Fig. 1A) [18]. Recently, based on in silico analysis, the existence of five members of the hABH family (hABH4 through hABH8) was predicted. Therefore, we first examined the existence of these experimentally un-characterized genes. Since we have observed the highest expression of hABH3 mRNA in human testis, the mRNA expression of the other hABH family members was also examined using human testis cDNA. The RT-PCR primer pairs used were designed to amplify the full-length of the ORF of each hABH mRNA (Table 1). As shown in Figure 2, with the exception of hABH8, the products amplified using each of the hABH primer pairs corresponded closely to the expected sizes. The nucleotide sequences of the amplified products of hABH1 through hABH7 were almost completely identical to those in the National Center for Biotechnology Information (NCBI) database, indicating that these six hABH family members, including hABH3, are at least expressed in the testis.
1.

Schematic diagram of the hABH family molecules. (A) The conserved 2OG-Fe(II) oxygenase domain, the methyltransferase domain and the RNA-binding motif are drawn as a solid box, a hatched box and a shaded box, respectively. In hABH7, the AlkB homologous region is drawn as a solid box. (B) Relationship of the exon structure and protein domains of hABH2 and ▵hABH2. The conserved 2OG-Fe(II) oxygenase domain is drawn as a solid box. A dotted box indicates a completely different amino acid sequence from hABH2 due to a frameshift.
2.
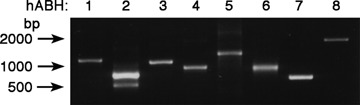
Reverse transcriptase-polymerase chain reaction (RT-PCR) analysis of the hABH family molecules in human testis. Expression of the mRNA of the hABH family molecules was evaluated in human testis cDNA by RT-PCR. A list of the primer pairs used for each hABH amplification is presented in Table 1.
Only in the hABH2-amplified product were two discrete bands detected:an upper band of approximately 800 bp, and a lower band of approximately 600 bp. The sequence analysis indicated that the amplified upper band was completely identical to the ORF of the hABH2 gene (accession no. NM_001001655). The hABH2 gene maps to chromosome region 12q24.1 and comprises at least four exons (Fig. 1B). The nucleotide sequence of the lower band in the hABH2-amplified products lacks the exon 3 of the hABH2 gene and encodes a product of 852 nucleotides in length, including an ORF of 157 amino acids. The sequence was submitted to the DNA Data Bank of Japan (DDBJ) data base as hABH2 transcript variant 1 (accession no. AB277859). The alternative splicing of the hABH2 gene results in a frameshift deletion of the C-terminal 2OG-Fe(II) oxy-genase domain; this is described as hABH2 in Figure 1B. The RT-PCR analysis indicated that there was no tissue-specific expression of hABH2 mRNAs in the normal tissues (data not shown).
Molecular cloning of hABH8 cDNA amplified by RT-PCR
The hABH8 PCR product was approximately 2000 bp in length (Fig. 2), which is considerably longer than the nucleotide number of the predicted hABH8 ORF (717 bp) in the database (accession no. NM_138775). The nucleotide sequence of the amplified RT-PCR product suggested that the hABH8 registered in the NCBI database would result from the alternative splicing of our identified product. Therefore, we cloned the hABH8 cDNA encoding a product of 2144 nucleotides in length–including an ORF of 664 amino acids–based on a combination of M. mulatta (XM_001102947) cDNA and information obtained from the NCBI human genome resources database (Fig. 3). We submitted the sequence to the DDBJ database as hABH8 (accession no. AB218768). The NCBI human genome resources database indicated that the cloned hABH8 gene is located in chromosome region 11q23.1 and comprises at least 12 exons. An EMBL-EBI InterProScan sequence search (http://www. ebi. ac. uk/interpro/) indicated that the cloned hABH8 is a member of the 2OG-Fe(II) oxygenase superfamily. Moreover, the existence of an RNA recognition motif and a methyltransferase domain are recognized in hABH8 (Fig. 1A), suggesting that hABH8 might be a unique molecule that uses RNA as a substrate and expresses both methylation and demethylation enzymatic activities.
3.

Nucleotide and predicted amino acid sequences of hABH8. The conserved 2OG-Fe(II) oxy-genase domain is boxed, the RNA-binding motif is underlined and the methyltrans-ferase domain is shaded. Numbering of the nucleotide and amino acid positions is indicated on the left.
Real-time PCR analysis of hABHs in normal human tissues
The expression levels of hABH1 to hABH8 mRNAs in normal human tissues were determined in a normalized panel of 16 human tissues by quantitative realtime PCR. Table 2 lists the specific primer pairs for each hABH amplification and the number of nucleotide in the amplified fragments within the hABH ORFs. The hABH2 primers were purchased from Qiagen (Cat. No. QT00219093) and their nucleotide sequence are not disclosed. A 117-bp fragment within GAPDH was amplified as a housekeeping gene and the results were expressed as a relative fold against GAPDH. As shown in Figure 4, relatively high expression levels of hABH1 to hABH8 transcripts were detected in the pancreas and testis, although their expression was ubiquitously observed in all the human tissues examined.
4.
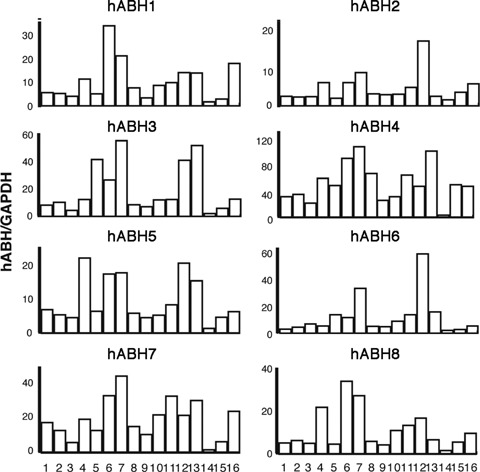
Real-time PCR analysis of the hABH family molecules in normal human tissues. The tissue-specific expression of hABH1–8 mRNAs was determined by a SYBR Green I-based quantitative PCR assay in normalized cDNA samples derived from 16 different human tissues. The expression levels of the hABH transcripts in the human tissues were normalized for the expression levels of the glyceraldehyde-3-phophate dehydrogenase (GAPDH) transcript in the same cDNAs. 1: brain, 2: thy-mus, 3:heart, 4:lung, 5: liver, 6: spleen, 7: pancreas, 8: small intestine, 9: colon, 10: kidney, 11: prostate, 12: testis, 13: ovary, 14: skeletal muscle, 15: peripheral blood leukocytes and 16: placenta. The results shown are the averages of three independent measurements per tissue.
Immunoblot and sub-cellular localization of EmGFP-hABHs
In order to examine the sub-cellular localization of the hABH family molecules, we generated hABH expression vectors in which EmGFP was fused at the N terminus of each hABH1-hABH8 coding region. HeLa cells were transiently transfected with the constructs encoding the EmGFP-hABH fusion proteins as well as EmGFP alone as a control. The cell lysates were prepared 24 hrs after transfection, resolved on an SDS-PAGE gel, and then transferred to a PVDF membrane. The membrane was immunoblotted with anti-GFP antiserum. As shown in Figure 5, discrete bands with the expected molecular masses were detected in each lysate of the EmGFP-hABH-transfected cells, indicating successful construction and transfection of each EmGFP-hABH expression vector. The sub-cellular localization of the EmGFP-hABH fusion proteins was examined with fluorescence microscopy 24 hrs after transfection. As shown in Figure 6A, the control EmGFP was diffusely distributed in the cell and did not exhibit any particular localization pattern. As previously reported, hABH2 accumulates in the nuclei with particularly bright nucleolar staining and hABH3 localizes in both the nuclei and cytoplasm. hABH1, hABH4, hABH5, hABH6 and hABH7 were observed throughout the cells but hABH8 was detected only in the cytoplasm. Interestingly, many bright EmGFP foci were detected in the cytoplasm of some EmGFP-hABH5-expressing HeLa cells after approximately 24 hrs of transfection (Fig. 6B). However, we could not detect the specific localization of EmGFP-hABH5 in the cell organelles.
5.
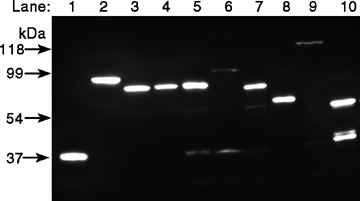
Immunoblot analysis of the EmGFP-hABH family of molecules in transfected HeLa cells. HeLa cells were transfected with pcDNA6.2/N-EmGFP (Mock), pcDNA6.2/N -EmGFP-hABH1–hABH8, and pcDNA6.2/N-EmGFP-hABH2 variant 1 (▵hABH2) family expression constructs. After incubation for 24 hrs, the transfected cells were lysed, electrophoresed on a 12% sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) gel, and transferred to a polyvinylidene fluoride (PVDF) membrane. The membrane was immunoblotted with rat anti-GFP antibody followed by HRP-conjugated antirat IgG. lane 1, EmGFP; lane 2, EmGFP-hABH1; lane 3, EmGFP-hABH2; lane 4, EmGFP-hABH3; lane 5, EmGFP-hABH4; lane 6, EmGFP-hABH5; lane 7, EmGFP-hABH6; lane 8, EmGFP-hABH7; lane 9, EmGFP-hABH8;and lane 10, EmGFP- hABH2.
6.
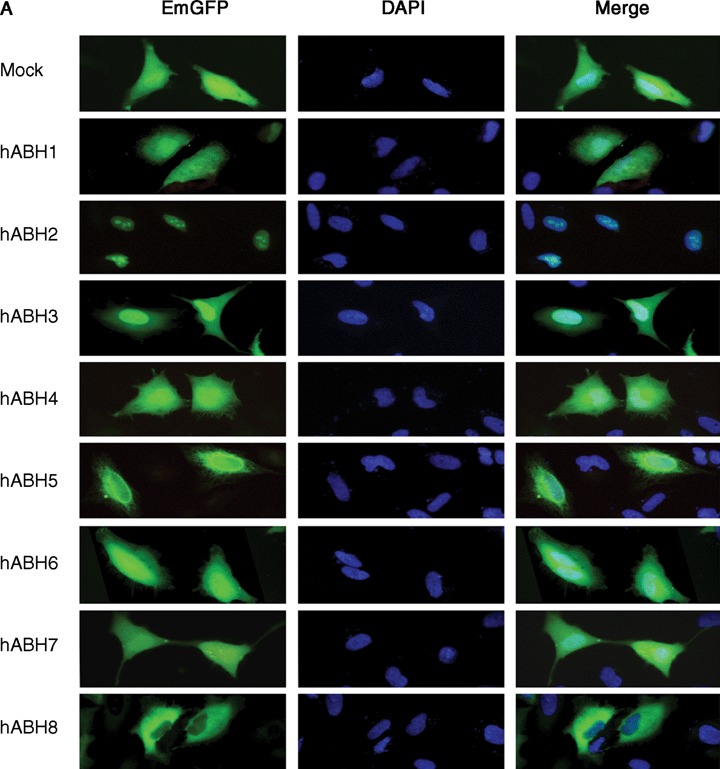
Sub-cellular localization of the hABH family molecules in transfected HeLa cells. HeLa cells were transfected with pcDNA6.2/N-EmGFP (Mock) and pcDNA6.2/N-EmGFP-hABH1–hABH8 expression constructs and incubated for 24 hrs (A), and with pcDNA6.2/N-EmGFP-hABH5 and pcDNA6.2/N-EmGFP-hABH2 variant 1 (▵hABH2) expression constructs and incubated for over 24 hrs (B). The cells were fixed in 4% paraformalde-hyde/PBS and processed for staining with DAPI to visualize the DNA. The expressed EmGFP-hABHs fusion proteins are false-coloured with green (EmGFP) and the DNA is false-coloured with blue (DAPI). The merged images are also shown on the right.
The sub-cellular localization of hABH2 variant 1 was compared to hABH2. The fluorescence of EmGFP-hABH2 transcript variant 1 (▵hABH2) was detected diffusely in the nuclei as well as in the cytoplasm. This localization pattern of hABH2 variant 1 is markedly different from that of ABH2 (Fig. 6A). Moreover, bright, dot-like fluorescence structures were detected in the nuclei but not in the nucleoli. There was no difference in the sub-cellular localization of EmGFP-tagged hABH proteins between the fixed and un-fixed cells. The expression of EmGFP-hABH2 variant 1 was also confirmed by western blotting using an anti-GFP antibody (Fig. 5).
Discussion
In the present paper, we have clarified the expression of five members of the human ABH family (hABH4–hABH8) and have compared their tissue distribution and sub-cellular localization with those of hABH1–hABH3. Wei et al. were the first to clone the human AlkB homologue (hABH) [8], the sequence of which was subsequently corrected and referred to as hABH1 [9, 10]. Although hABH was reported to partially rescue an E. coli AlkB mutant from methyl methanesulfonate-induced cell death, this function has not been confirmed for hABH1. Subsequently, hABH2 and hABH3 have also been identified as human homologues of AlkB and are confirmed to complement the AlkB mutant phenotype. hABH2 demethylates methylated double-stranded DNA, while hABH3 demethylates both methylated single-stranded DNA and methylated RNA. Recent in silico analysis suggested that more than five genes (hABH4–hABH8) that are experimentally un-characterized and poorly annotated comprise the 2OG-Fe(II) oxygenase superfamily together with hABH1–hABH3 [13]. We have detected the expression of the hABH4 to hABH7 mRNAs as well as that of hABH1–hABH3 in testis cDNA and verified the sequences of their complete ORFs. On the other hand, the hABH8 sequence that is presented in the database is assumed to be derived from an alternatively spliced form of hABH8 and has no in-frame 2OG-Fe(II) oxygenase domain. We have determined the mRNA sequence, including a complete ORF of hABH8. The cloned hABH8 cDNA comprises at least 12 exons in the 11q22.3 region and encodes 664 amino acids. The database hABH8 cDNA (accession no. NM_138775.1) is lacking exons 7, 10 and 11, resulting in an hABH8 isoform without the 2OG-Fe(II) oxygenase domain due to frameshift errors. The cloned hABH8 displays a similarity to M. mulatta CG17807-PA, transcript variant 3 (XM_001102947) and Mus musculus alkylation repair homologue 8 (NM_026303) with approximately 97% and 43% identity, respectively. Interestingly, not only the 2OG-Fe(II) oxygenase domain but also the RNA binding motif and the methyltransferase domain were detected in cloned hABH8 by motif and domain searches. Moreover, hABH8 possesses sequences that are similar to those of AdoMet binding motifs and to the C-terminal region of trm9 [18], suggesting that hABH8 might play an important role as a tRNA methylase. The EmGFP-hABH8 expressed in HeLa cells was localized exclusively in the cytoplasm. These results suggest that hABH8 might possess both demethylation and methylation activities in a single molecule and its target molecule as a substrate might be RNA. Studies on the interesting enzymatic activity of hABH8, its substrate specificity and the regulatory mechanisms of the two different enzymatic activities are now in progress.
The demethylation activities of hABH2 and hABH3 have been previously examined in vitro as well as in E. coli and in a bacteriophage system [9, 10]. These two hABHs are capable of demethylating the methylated residue at the N1 position of purines and the N3 position of pyrimidines [19–21]. hABH2 preferentially repairs alkylated double-stranded DNA, whereas hABH3 targets alkylated single-stranded DNA and alkylated RNA as substrates. Immunohistochemical analysis using an anti-hABH3 antibody indicated the localization of hABH3 in the nuclei of human prostate cancer specimens [22]. Moreover, EYFP-hABH2 and EYFP-hABH3 transfectants indicate that hABH2 accumulates in the nuclei, and particularly in the nucleoli, whereas hABH3 is present in both the nuclei and in the cytoplasm [10]; observations that were confirmed in the present study. Interestingly, the EmGFP- ▵hABH2 localized in the nucleoplasm with a dot-like pattern; however, no localization was observed in the nucleoli during any cell cycle. Due to the absence of a 2OG-Fe(II) oxygenase domain, the hABH2 splice variant is expected to be deficient in demethylation activity. Recently, mice lacking a functional mABH2 gene have been generated [12]. These mice are viable and no obvious phenotype was observed. However, the accumulation of significant levels of 1-meA was detected in the genome of mice lacking mABH2. These results indicate that ABH2 plays a major role in removing endogenously generated 1-meA in genomic DNA. Therefore, the dot-like fluorescence of EmGFP- ▵hABH2 might indicate the position of methylated or un-detected, damaged DNA. hABH1–8 mRNAs are ubiquitously expressed in normal human tissues, suggesting that the hABH family molecules play fundamental and crucial roles in these tissues. All organisms are continually exposed to alkylating substances generated endogenously and in the environment. The alkylating substances induce the alkylation of DNA, RNA and proteins, causing developmental abnormalities, immunological diseases, premature aging and cancer by disrupting the normal functions of these mole-cules. If the hABH family molecules function as repair molecules for alkylation damage to DNA and RNA, the broad and abundant expression in these organs is explicable. In contrast to demethylation, the methy-lation of genomic DNA is known to be catalysed by DNA methyltransferases (DNMTs). The eukaryotic DNMT family has five members: DNMT1, DNMT2, DNMT3a, DNMT3b and DNMT3L [23–25]. The DNMT family members are classified into two classes based on their preferred DNA substrate: The de novo methyltransferases DNMT3a and DNMT3b are mainly responsible for introducing cytosine methylation at previously unmethylated CpG sites, whereas the maintenance methyltransferase DNMT1 copies pre-existing methylation patterns onto the new DNA strand during DNA replication. In contrast, DNMT2 appears to function as a tRNA methyltransferase [26–29]. DNMT3L does not exhibit intrinsic DNA methyltransferase activity;however, it does modulate the catalytic activities of DNMT3a and DNMT3b [30]. DNMT1, DNMT3a and DNMT3b exhibit widespread expression in most normal tissues [31]. These characteristics and expression patterns of the DNMT family members are very similar to those of the ABH family members:at least eight ABH family molecules with the conserved AlkB homologous domain distribute in many tissues, the sub-cellular localization of the hABHs overlap, and there is a splice variant lacking the catalytic domain. Therefore, the findings of this study together with a comparison with the DNMTs may provide us with useful information for the functional analyses of the hABH family members [32].
In conclusion, we have experimentally verified the existence of hABH4, hABH5, hABH6 and hABH7 mRNAs containing the entire length of ORFs. Moreover, we have cloned the hABH2 splice variant and hABH8 containing an RNA recognition motif, a methyltransferase domain, and a 2OG-Fe(II) oxygenase domain. The hABH2-splicing variant has no 2OG-Fe(II) oxygenase domain and exhibits unique sub-cellular localization. The eight ABH family molecules containing the conserved AlkB homologous regions are expressed ubiquitously in human tissues, suggesting either the existence of specific substrates and/or redundant substrate specificity as backup enzymes. An examination of the biological functions of the ABH family of molecules is necessary for the clarification of the DNA and RNA repair systems.
Acknowledgments
We thank Dr Shinobu Fujita (Mitsubishi Chemical) for providing rat anti-GFP antibody. This work is supported in part by a Grant-in-Aid from the Ministry of Education, Culture, Sports, Science and Technology of Japan (17013058). Additional support was also obtained through Long-range Research Initiative (LRI) by Japan Chemical Industry Association (JCIA), LinkGenomics, Inc., and PCA InterMed, Inc.
References
- 1.Lindahl T. New class of enzymes acting on damaged DNA. Nature. 1976;259:64–6. doi: 10.1038/259064a0. [DOI] [PubMed] [Google Scholar]
- 2.Olsson M, Lindahl T. Repair of alkylated DNA in Escherichia coli. Methyl group transfer from O6-methylguanine to a protein cysteine residue. J Biol Chem. 1980;255:10569–71. [PubMed] [Google Scholar]
- 3.Lindahl T, Sedgwick B, Sekiguchi M, Nakabeppu Y. Regulation and expression of the adaptive response to alkylating agents. Annu Rev Biochem. 1988;57:133–57. doi: 10.1146/annurev.bi.57.070188.001025. [DOI] [PubMed] [Google Scholar]
- 4.Aravind L, Koonin EV. The DNA-repair protein AlkB, EGL-9, and leprecan define new families of 2-oxoglu-tarate- and iron-dependent dioxygenases. Genome Biol. 2001;2:RESEARCH0007. doi: 10.1186/gb-2001-2-3-research0007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Trewick SC, Henshaw TF, Hausinger RP, Lindahl T, Sedgwick B. Oxidative demethylation by Escherichia coli AlkB directly reverts DNA base damage. Nature. 2002;419:174–8. doi: 10.1038/nature00908. [DOI] [PubMed] [Google Scholar]
- 6.Falnes PO, Johansen RF, Seeberg E. AlkB-mediated oxidative demethylation reverses DNA damage in Escherichia coli. Nature. 2002;419:178–82. doi: 10.1038/nature01048. [DOI] [PubMed] [Google Scholar]
- 7.Koivisto P, Duncan T, Lindahl T, Sedgwick B. Minimal methylated substrate and extended substrate range of Escherichia coli AlkB protein, a 1-methyladenine-DNA dioxygenase. J Biol Chem. 2003;278:44348–54. doi: 10.1074/jbc.M307361200. [DOI] [PubMed] [Google Scholar]
- 8.Wei YF, Carter KC, Wang RP, Shell BK. Molecular cloning and functional analysis of a human cDNA encoding an Escherichia coli AlkB homolog, a protein involved in DNA alkylation damage repair. Nucleic Acids Res. 1996;24:931–37. doi: 10.1093/nar/24.5.931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Duncan T, Trewick SC, Koivisto P, Bates PA, Lindahl T, Sedgwick B. Reversal of DNA alkylation damage by two human dioxygenases. Proc Natl Acad Sci USA. 2002;99:16660–5. doi: 10.1073/pnas.262589799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Aas PA, Otterlei M, Falnes PO, Vagbo CB, Skorpen F, Akbari M, Sundheim O, Bjoras M, Slupphaug G, Seeberg E, Krokan HE. Human and bacterial oxidative demethylases repair alkylation damage in both RNA and DNA. Nature. 2003;421:859–63. doi: 10.1038/nature01363. [DOI] [PubMed] [Google Scholar]
- 10.Ougland R, Zhang CM, Liiv A, Johansen RF, Seeberg E, Hou YM, Remme J, Falnes PO. AlkB restores the biological function of mRNA and tRNA inactivated by chemical methylation. Mol Cell. 2004;16:107–16. doi: 10.1016/j.molcel.2004.09.002. [DOI] [PubMed] [Google Scholar]
- 11.Ringvoll J, Nordstrand LM, Vagbo CB, Talstad V, Reite K, Aas PA, Lauritzen KH, Liabakk NB, Bjork A, Doughty RW, Falnes PO, Krokan HE, Klungland A. Repair deficient mice reveal mABH2 as the primary oxidative demethylase for repairing 1meA and 3meC lesions in DNA. EMBO J. 2006;25:2189–98. doi: 10.1038/sj.emboj.7601109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kurowski MA, Bhagwat AS, Papaj G, Bujnicki JM. Phylogenomic identification of five new human homologs of the DNA repair enzyme AlkB. BMC Genomics. 2003;4:48. doi: 10.1186/1471-2164-4-48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Drablos F, Feyzi E, Aas PA, Vaagbo CB, Kavli B, Bratlie MS, Pena-Diaz J, Otterlei M, Slupphaug G, Krokan HE. Alkylation damage in DNA and RNA–repair mechanisms and medical significance. DNA Repair. 2004;3:1389–407. doi: 10.1016/j.dnarep.2004.05.004. [DOI] [PubMed] [Google Scholar]
- 14.Osada N, Hida M, Kusuda J, Tanuma R, Hirata M, Hirai M, Terao K, Suzuki Y, Sugano S, Hashimoto K. Prediction of unidentified human genes on the basis of sequence similarity to novel cDNAs from cynomolgus monkey brain. Genome Biol. 2002;3:RESEARCH0006. doi: 10.1186/gb-2001-3-1-research0006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lee DH, Jin SG, Cai S, Chen Y, Pfeifer GP, O'Connor TR. Repair of methylation damage in DNA and RNA by mammalian AlkB homologues. J Biol Chem. 2005;280:39448–59. doi: 10.1074/jbc.M509881200. [DOI] [PubMed] [Google Scholar]
- 16.Tsujikawa K, Kawakami N, Uchino Y, Ichijo T, Furukawa T, Saito H, Yamamoto H. Distinct functions of the two protein tyrosine phosphatase domains of LAR (leukocyte common antigen-related) on tyrosine dephosphorylation of insulin receptor. Mol Endocrinol. 2001;15:271–80. doi: 10.1210/mend.15.2.0592. [DOI] [PubMed] [Google Scholar]
- 17.Kalhor HR, Clarke S. Novel methyltransferase for modified uridine residues at the wobble position of tRNA. Mol Cell Biol. 2003;23:9283–92. doi: 10.1128/MCB.23.24.9283-9292.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Delaney JC, Essigmann JM. Mutagenesis, genotoxicity, and repair of 1-methyladenine, 3-alkylcytosines, 1-methylguanine, and 3-methylthymine in alkB Escherichia coli. Proc Natl Acad Sci USA. 2004;101:14051–6. doi: 10.1073/pnas.0403489101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Falnes PO. Repair of 3-methylthymine and 1-methyl-guanine lesions by bacterial and human AlkB proteins. Nucleic Acids Res. 2004;32:6260–7. doi: 10.1093/nar/gkh964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Koivisto P, Robins P, Lindahl T, Sedgwick B. Demethylation of 3-methylthymine in DNA by bacterial and human DNA dioxygenases. J Biol Chem. 2004;279:40470–4. doi: 10.1074/jbc.M407960200. [DOI] [PubMed] [Google Scholar]
- 21.Konishi N, Nakamura M, Ishida E, Shimada K, Mitsui E, Yoshikawa R, Yamamoto H, Tsujikawa K. High expression of a new marker PCA-1 in human prostate carcinoma. Clin Cancer Res. 2005;11:5090–7. doi: 10.1158/1078-0432.CCR-05-0195. [DOI] [PubMed] [Google Scholar]
- 22.Bestor TH. The DNA methyltransferases of mammals. Hum Mol Genet. 2000;9:2395–402. doi: 10.1093/hmg/9.16.2395. [DOI] [PubMed] [Google Scholar]
- 23.Margot JB, Ehrenhofer-Murray AE, Leonhardt H. Interactions within the mammalian DNA methyltrans-ferase family. BMC Mol Biol. 2003;4:7. doi: 10.1186/1471-2199-4-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Turek-Plewa J, Jagodzinski PP. The role of mammalian DNA methyltransferases in the regulation of gene expression. Cell Mol Biol Lett. 2005;10:631–47. [PubMed] [Google Scholar]
- 25.Okano M, Xie S, Li E. Cloning and characterization of a family of novel mammalian DNA (cytosine-5) methyltransferases. Nat Genet. 1998;19:219–20. doi: 10.1038/890. [DOI] [PubMed] [Google Scholar]
- 26.Pradhan S, Esteve PO. Mammalian DNA (cytosine-5) methyltransferases and their expression. Clin Immunol. 2003;109:6–16. doi: 10.1016/s1521-6616(03)00204-3. [DOI] [PubMed] [Google Scholar]
- 27.Goll MG, Kirpekar F, Maggert KA, Yoder JA, Hsieh CL, Zhang X, Golic KG, Jacobsen SE, Bestor TH. Methylation of tRNAAsp by the DNA methyltransferase homolog Dnmt2. Science. 2006;311:395–8. doi: 10.1126/science.1120976. [DOI] [PubMed] [Google Scholar]
- 28.Rai K, Chidester S, Zavala CV, Manos EJ, James SR, Karpf AR, Jones DA, Cairns BR. Dnmt2 functions in the cytoplasm to promote liver, brain, and retina development in zebrafish. Genes Dev. 2007;21:261–6. doi: 10.1101/gad.1472907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Suetake I, Morimoto Y, Fuchikami T, Abe K, Tajima S. Stimulation Effect of Dnmt3L on the DNA Methylation Activity of Dnmt3a2. J Biochem. 2006;140:553–9. doi: 10.1093/jb/mvj185. [DOI] [PubMed] [Google Scholar]
- 30.Robertson KD, Uzvolgyi E, Liang G, Talmadge C, Sumegi J, Gonzales FA, Jones PA. The human DNA methyltransferases (DNMTs) 1, 3a and 3b: coordinate mRNA expression in normal tissues and overexpression in tumors. Nucleic Acids Res. 1999;27:2291–8. doi: 10.1093/nar/27.11.2291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sedgwick B, Bates PA, Paik J, Jacobs SC, Lindahl T. Repair of alkylated DNA: recent advances. DNA Repair. 2007;6:429–42. doi: 10.1016/j.dnarep.2006.10.005. [DOI] [PubMed] [Google Scholar]


