Abstract
The deleted in liver cancer 1 (DLC-1) gene encodes a GTPase activating protein that acts as a negative regulator of the Rho family of small GTPases. Rho proteins transduce signals that influence cell morphology and physiology, and their aberrant up-regulation is a key factor in the neoplastic process, including metastasis. Since its discovery, compelling evidence has accumulated that demonstrates a role for DLC-1 as a bona fide tumour suppressor gene in different types of human cancer. Loss of DLC-1 expression mediated by genetic and epigenetic mechanisms has been associated with the development of many human cancers, and restoration of DLC-1 expression inhibited the growth of tumour cells in vivo and in vitro. Two closely related genes, DLC-2 and DLC-3, may also be tumour suppressors. This review presents the current status of progress in understanding the biological functions of DLC-1 and its relatives and their roles in neoplasia.
Keywords: deleted in liver cancer, GTPase activating protein, tumour suppressor, metastasis suppressor, cytoskeletal organization, tensin, focal adhesion, cancer detection, cancer therapy
Introduction
Small GTPases of the Ras superfamily are critical components of the signalling pathways that control many aspects of cell behaviour. The members of the Rho (ras homology) branch of the Ras family were originally characterized as regulators of the actin cytoskeleton, but have now been implicated in a wide range of cellular processes, such as proliferation, motility, morphogenesis, vesiscular trafficking and gene expression [1–3]. Rho-GTPase activity is frequently dysregulated in human cancers and may contribute to the growth and invasiveness of tumour cells [4, 5]. The human genome encodes 20–22 Rho proteins, of which the best characterized are RhoA, Rac1 and Cdc42 [6, 7].
In common with other Ras-like GTPases, Rho proteins act as molecular switches that undergo a nucleotide-regulated conformational change. In the GTP-bound state they transmit signals by activating downstream effector molecules, such as protein and lipid kinases, phospholipases and regulators of cytoskeletal organization [1, 3]. The Rho GTP-GDP cycle is controlled by two classes of regulatory proteins; guanine nucleotide exchange factors (GEFs) promote the exchange of bound GDP for GTP to generate the active state, and GTPase activating proteins (GAPs) stimulate GTP hydrolysis to reconstitute the inactive state [8].
In human beings, the ≍70 RhoGEFs and ≍80 RhoGAPs are structurally diverse multi-domain proteins, and their activities are regulated and targeted by interactions with other molecules [9–12]. A number of RhoGEFs are activated in response to stimulation of cell surface receptors, and several GEFs have been localized to complexes organized by scaffolding proteins that include downstream Rho effectors, which would increase the efficiency and specificity of Rho signalling [13, 14]. While fewer details are known about RhoGAP signalling pathways, their importance in normal cellular homeostasis is evidenced by the association of several human disorders with mutations in RhoGAP genes and by the abnormal phenotypes generated by targeted inactivation of mouse RhoGAP genes [12, 15]. One RhoGAP that is rapidly gaining recognition for its role as a tumour suppressor and a regulator of cell proliferation and cytoskeletal organization is DLC-1 (deleted in liver cancer 1). In this review we summarize the current state of knowledge of DLC-1 and its relatives.
The deleted in liver cancer family of RhoGAP domain proteins
The DLC-1 gene was identified by Yuan and colleagues as a genomic DNA segment under-represented in a human hepatocellular carcinoma (HCC) specimen, using representational difference analysis, a PCR-based subtractive hybridization technique [16]. The DLC-1 locus was mapped to the human chromosome 8p22 region frequently lost in HCC and other cancers (Fig. 1A), and it was named ‘deleted in liver cancer’ when found to be deleted in primary HCC and HCC cell lines [16]. Sequencing of the DLC-1 cDNA showed that it was the human orthologue of the rat p122RhoGAP protein, which had been cloned by Homma and Emori when screening a >11 expression library with anti-serum against phospholipase δ 1 (PLCδ 1) [17]. The predicted amino acid sequence of p122RhoGAP/DLC-1 contains a RhoGAP domain, a conserved region of around 200 amino acids responsible for the catalytic activity of RhoGAPs [10]. Further analysis of the DLC-1 polypeptide sequence revealed the presence of two additional conserved elements, an N-terminal sterile alpha motif (SAM) domain and a C-terminal steroidogenic acute regulatory protein (StAR)-related lipid-transfer (START) domain [18].
1.
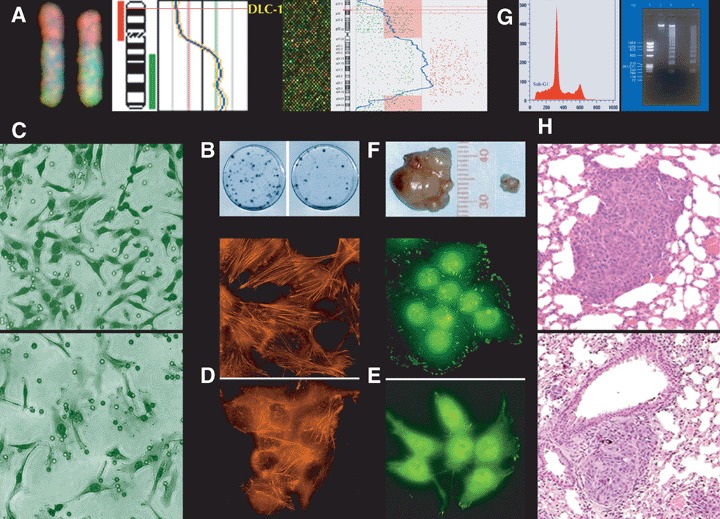
Tumour suppressor activity of DLC1. (A) The human DLC-1 gene is located in band 8p22, a chromosomal region of DNA copy-number losses in a number of cancers, as demonstrated by conventional (left) and array-based (right) comparative genom-ic hybridization. Re-expression of the DLC-1 cDNA in human tumour cells that lack expression of the endogenous gene results in suppression of colony formation (B), suppression of cell invasion (C), reduction of actin stress fibres (D), diminution of vin-culin-containing focal adhesions (E), reduction of tumour size in nude mice (F), induction of apoptosis (as demonstrated by cell accumulation of sub-G1 phase and chromatin fragmentation) (G), and suppression of formation of lung metastases in nude mice (H). In each pair of photographs, DLC-1-expressing cells are below (C, D, E, H) or to the right (B, F) of cells transfected with control vectors. Images in H are reproduced with permission from Cancer Research.
DLC-1 was later recognized as the founding member of a family of proteins that shares the SAM-RhoGAP-START domain organization, which now includes DLC-2 (also known as STARD13, for START domain-containing protein 13) [19, 20] and DLC-3 (also known as KIAA0189 and STARD8) [21, 22] (Fig. 2). The genes encoding the three human DLC proteins appear to be paralogues that arose through duplication of chromoso-mal segments [22]. Orthologues of each of the three DLC family proteins have been found in other vertebrates, including the mouse, rat, dog, chicken, frog and puffer fish (Ensembl Project; http://www.ensembl.org). The urochordate Ciona intestinalis and invertebrates such as Drosophila and C. elegans appear to possess a single gene encoding a DLC-1-like protein (Ensembl Project). The SAM-RhoGAP-START domain proteins apparently arose in multi-cellular organisms, as none of the 11 predicted RhoGAP domain proteins in S. cerevisiae is homologous to DLC-1 [23].
2.
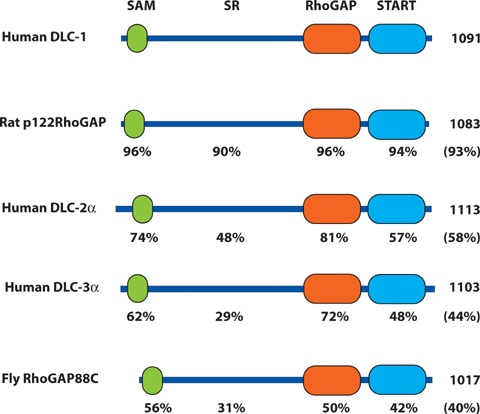
Comparison of the mammalian and Drosophila DLC family proteins. Schematic representation of the structure of human DLC-1, DLC-2α and DLC-3α, the rat DLC-1 orthologue p122RhoGAP and Drosophila RhoGAP88 C (cv-c). The SAM, serine-rich (SR), RhoGAP and START domains are indicated. The amino acid length of the polypeptide is given at the right. Beneath each domain is the percent identity to the corresponding domain of human DLC-1, and in parenthesis is the percent identity of the full-length polypeptide.
DLC-1
The human DLC1 gene is organized into 14 exons and yields a major transcript with a size of ≍6.3 kb [24]. The longest open reading frame of the cDNA (NM_006094) encodes a 1091-amino acid (aa) polypeptide with a predicted size of 123 kD [16]. However, translation initiation at an in-frame, downstream AUG codon in a better Kozak context would yield a protein of 1083 aa, and there is evidence that the second AUG is the predominant start site (A. Papageorge, X. Qian, D. Lowy, unpublished results). The human DLC-1 aa sequence is 93% identical to the rat p122RhoGAP [17] and 92% identical to the mouse protein [24, 25]. The rat Dlc1 gene on chromosome 16q12.2 (Rat Genome Database; http://rgd.mcwu.edu) and the mouse Dlc1 gene (formerly called Arhgap7) on chromosome 8B1 are located on regions syntenic with human 8p22 and have exon/intron structures nearly identical to that of the human gene, except for the presence of an extra codon in exon 5 of the mouse gene [24, 25].
Several variant transcripts are associated with the DLC1 locus (Table 1 and Fig. 3A). The 7.4 kb KIAA1723 cDNA clone was isolated from a human hippocampus library and has the potential to encode a larger DLC-1 isoform of 1528 aa [26]. The KIAA1723 cDNA contains exons 2–14 of DLC1, but exon 1 is replaced by a novel 1.7 kb sequence, distributed on 5 exons upstream of the start of the major transcript. The putative promoter region of KIAA1723 is approx.400 kb upstream of exon 2, and there are also shorter transcripts originating from this promoter that would not encode a DLC-1-related protein (NM_024767). The existence of a larger DLC-1 polypeptide has not yet been verified experimentally. The second alternative transcript contains a novel first exon located 160 kb upstream of exon 2 and was cloned from HepG2 hepatoblastoma cells, where it seems to be enriched, based on the number of promoter tags in the Fantom database isolated from these cells (http://fantom.gsc.riken.go.jp). The HepG2-enriched first exon would substitute 47 aa for the first 13 aa of DLC-1. As a transcript with a similar 5' end was identified in the mouse, this DLC-1 iso-form may be functionally conserved.
1.
Human and rodent DLC family members
| Isoform | cDNA sequence (GenBank #) | Protein Size (Amino acids) | Protein Sequence (UniProt #) |
|---|---|---|---|
| DLC-1 | |||
| Human | NM_006094 | 1091 | Q96QB1 |
| Mouse | NM_015802 | 1092 | Q9R0Z9 |
| Rat | D31962 | 1083 | Q63744† |
| DLC-1 long variant (KIAA1723) | |||
| Human | NM_182643 | 1528 | Q7Z5R8 |
| DLC-1 variant (HepG2) | |||
| Human | AK025544 | 1125‡ | na |
| Mouse | AK147539 | 1126 | Q3UH75 |
| DLC-2α | |||
| Human | NM_178006 | 1113 | Q9Y3M8–1 |
| Mouse | NM_146258 | 1113 | Q923Q2 |
| DLC-2β | |||
| Human | NM_178007 | 1105 | Q9Y3M8–2 |
| DLC-2γ | |||
| Human | NM_052851 | 995 | Q9Y3M8–3 |
| DLC-3α | |||
| Human | CR749411 | 1103 | Q68DG7 |
| Mouse | BB613875§ | 1099 | na |
| DLC-3β | |||
| Human | NM_014725 | 1023 | Q92502 |
| Mouse | NM_199018 | 1019 | Q8K031 |
na, not available.
The 1083-aa protein described in [17] originates from the downstream AUG codon, and the 1091-aa protein in Q63744 derives from the upstream AUG codon.
The AK025544nt sequence contains a sequencing error in the region shared with NM_006094 that would lead to premature translation termination. The predicted polypeptide length was obtained from the corrected sequence.
This partial cDNA sequence contains a 5’end similar to that of CR749411.
3.
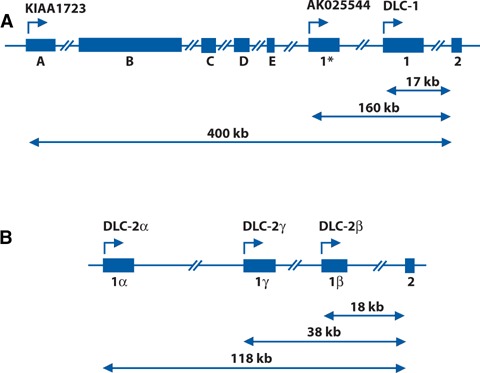
Origin of human DLC-1 and DLC-2 transcriptional variants. (A). Diagram of the genomic region at the 5′ end of DLC1, with exons represented by boxes. The five exons comprising the novel 5′sequence of the KIAA1723 transcript are labelled A–E, and the alternative first exon of the AK025544 transcript is denoted as 1*. The three transcripts share exons 2–14, and the distances between the putative transcription start sites (marked with arrows) and exon 2 are indicated. (B). Diagram of the 5’end of the DLC-2 gene, showing the first exons of the DLC-2α (1α), DLC-2β (1β) and DLC-2γ (1γ) isoforms. Exons 2–14 are common to all three transcripts, and the distances between exon 2 and the putative transcription start sites are indicated. The genomic DNA distances in A and B were obtained from the human genome sequence compilation (NCBI Build 36) and are not drawn to scale. The structures of the human DLC-3 gene and its transcripts were described in Ref 22.
DLC-2
DLC-2 was identified by analysis of genomic and cDNA sequences as a gene on chromosome 13q13 that encoded a RhoGAP domain protein related to DLC-1 [19, 20]. The full-length DLC-2 cDNA is ≍6 kb, and on Northern blots an additional 4 kb transcript was detected in some tissues [19], possibly due to alternative polyadenylation site usage. The DLC-2 gene (STARD13) also appears to have more than one transcription start site, potentially yielding three DLC-2 isoforms with different 5'-untranslated regions and different N-termini (Fig. 3B). The DLC-2α transcript encodes an 1113-aa polypeptide of 125 kb that is 58% identical to DLC-1 and has the same SAM-RhoGAP-START domain organization [19]. The DLC-2 transcript contains a different first exon and encodes an 1105 aa polypeptide, in which 46 aa replace the first 56 aa of the α form [27]. In the third transcript, DLC-2, translation is predicted to initiate at an AUG codon equivalent to aa 119 of DLC-2α, yielding a 995-aa protein missing the SAM domain. The biological significance, the relative abundance and the tissue distribution of the three DLC-2 isoforms remain to be determined. The mouse Stard13 gene maps to chromosome 5 G3, and full-length mouse cDNAs have been identified that encode a protein 90% identical the human DLC-2α isoform.
DLC-3
The KIAA0189 cDNA clone was isolated from a human myeloid cell line library and noted to encode a protein related to p122RhoGAP [21]. Due to its homology with DLC-1 and-2, we have now termed this protein DLC-3. The gene encoding DLC-3 (STARD8) is present on chromosome Xq13, and the organization of exons 2–14 of STARD8 is similar to that of DLC1 and STARD13 [22]. The DLC-3 mRNA has a size of ≍5 kb, and the gene appears to have two transcription start sites that would produce polypeptides with different N-termini. A transcript originating from a distal promoter encodes the 122-kDa DLC-3α isoform, an 1103-aa polypeptide containing the SAM, RhoGAP and START domains that is 44% identical to DLC-1 and 52% identical to DLC-2 [22]. The original KIAA0189 cDNA apparently initiates from a proximal promoter and encodes the DLC-3β isoform, predicted to encode a 1023-aa protein that lacks the SAM domain. As with the DLC-2 isoforms, there is a lack of information concerning the nature of the endogenous DLC-3 protein. The mouse STARD8 gene is found on chromosome XC2, and there are mouse transcripts corresponding to both the DLC-3α and β variants [22].
Invertebrate DLC-1-like proteins
In the Drosophila genome sequence there is a single DLC-1-like protein, RhoGAP88 C, encoded by the gene crossveinless-c (cv-c) [11, 28]. The 1017-aa RhoGAP88 C polypeptide has the characteristic SAM -GAP-START domain organization and has 40% identity with DLC-1 and DLC-2 and 36% identity with DLC-3. The C.elegans DLC-1 orthologue is the product of the gene gei-1 (gut on exterior-interacting) [29]. There are two gei-1 transcripts (F45H7.2a and F45H7.2b) that encode hypothetical proteins of 842 and 722 aa, respectively (WormBase; http://www. wormbase.org). Both of the predicted gene products lack the SAM domain; however, further characterization of the 5′ ends of the gei-1 transcripts will be needed to confirm the absence of a SAM domain. The 842-aa protein is slightly more similar to DLC-2 (33% identity) than to DLC-1 or DLC-3 (30% identity).
Expression of DLC family proteins
DLC-1 mRNA is widely expressed in human and mouse tissues, as shown by Northern blot hybridization [16, 24, 30] and by the large number of expressed sequence tags (ESTs) isolated from many tissues and at various stages of development (http://www.ncbi.nlm.nih.gov, unigene database). By in situ hybridization, the DLC-1 transcript was detected in several tissues in 10-day mouse embryos [31]. The transcription start sites of the human and mouse DLC-1 genes have been identified, and the promoter regions of both genes are GC-rich with characteristics of CpG islands [24, 32–34]. Genomic DNA upstream of the translation start site was found to stimulate transcription of a luciferase reporter gene in several human cancer cell lines [34]. DLC-1 expression could also be subject to control at the post-transcriptional level, as the 5′-untranslated region of the human, mouse, and rat transcripts have upstream AUG codons, which might inhibit initiation of translation at the start codon of the major open reading frame [24]. The distribution of the DLC-2 and DLC-3 mRNAs appears to overlap with that of DLC-1 in many adult tissues [19, 21, 22]. Fewer ESTs have been obtained for DLC-3, suggesting that it may be synthesized by a more limited number of cell types or that the transcript is present at lower levels.
Features of DLC family protein domains
SAM domain
The N-terminus of the DLC-1 polypeptide contains a SAM domain (aa 11–78), an ≍70 amino acid motif that occurs in more than 200 human proteins, including transcription factors and signalling proteins [35]. Most SAM domains are thought to be involved in protein–protein interactions, although certain SAM modules have been reported to bind RNA and lipids [35]. Some SAM domain proteins interact with other SAM domains in a homo- or heterotypic fashion to form oligomers or multi-protein complexes, respectively. Recent data indicated that the SAM domains of the DLC family proteins may have unique features. Determination of the 3D structure of the SAM domain of DLC-2 showed that it formed a four δ– helical bundle, instead of the usual five helices typical of other SAM domains [36, 37]. The DLC-2 SAM domain was found to exist in solution as a monomer and may interact with a lipid ligand [36, 37]. The absence of a SAM domain in the DLC-2γ and DLC-3β isoforms could alter their biological properties; consistent with this possibility, HCC cells could be stably transfected with vectors encoding DLC-2γ but not DLC-2δ[27].
RhoGAP domain
Residues 639–847 of DLC-1 comprise the RhoGAP domain, which is the most highly conserved domain among the three DLC family members, with around 70% sequence identity. The intrinsic GTP hydrolysis rates of Rho proteins are low, and GAP domains enhance their GTPase activity by positioning the catalytic glutamine residue in the proper conformation for aligning the nucleophilic water molecule that attacks the γ-phosphate of GTP [8]. All of the DLC proteins have a conserved arginine residue that is essential for RhoGAP activity, corresponding to Arg677 of human DLC-1. The ‘arginine finger’ is present in a loop and introduces a positive charge into the catalytic site that stabilizes the transition state of the GTP hydrolysis reaction [38]. Rat p122RhoGAP and human DLC-1, DLC-2 and DLC-3 were found to have GAP activity for RhoA in vivo and in vitro [19, 39–41].Human DLC-1 and DLC-2 have also been reported to increase the hydrolysis of Cdc42-GTP in vitro, although less efficiently than that of RhoA-GTP, and both had little effect on the GTPase activity of Rac1 [22, 40].
START domain
The START domain at the C-terminus of DLC-1 (aa 878–1081) is a domain first identified in several proteins with a role in lipid transport or lipid metabolism, and subsequently START domains have been found in 15 human proteins [18, 42]. The 3D structures obtained for several START domains show that they exist as a hydrophobic tunnel formed by a curved, 9-stranded β sheet gripped by N-terminal and C-terminal helices [42]. Different lipid-binding modules are found in other RhoGAP proteins, such as the Sec14 domain in Cdc42GAP and C1 domain in 2-chimaerin, and they are thought to regulate the subcellular localization of the protein and/or its GAP activity [10, 43].However, it has been suggested that START domains may not be involved in membrane targeting due to buried nature of lipid-binding region [44]. Determination of the identity of the ligand(s) for the START domains of the DLC family proteins will be essential for understanding the function of this region.
Serine-rich, unstructured middle region
The region between the SAM and RhoGAP domains of DLC-1 (aa 86–638), encoded primarily by the unusually long exon 5 (1424 bp in humans), displays little overall sequence similarity with known protein domains. This region has the least sequence conservation with DLC-2 and DLC-3, although alignment of the sequences reveals short stretches of high similarity separated by gaps and insertions of variable length [22]. One of the conserved sequence elements (LDDILYHV, residues 469–476 in human DLC-1) is similar to the consensus LD motif (LDXLLXXL) found in paxillin and other signalling proteins, which mediates the binding of paxillin to vinculin and focal adhesion kinase [45]. This middle region is rich in serine residues, and computer analysis shows that there are a number of potential sites for phosphorylation by ser-ine-threonine protein kinases (ELM server, http://elm.eu.org). There are also several proline-rich segments in the serine-rich (SR) region that could bind to proline recognition domains, such as the SH3 (src homology 3) and WW (Trp-Trp motif) modules that are present in many signalling proteins [46].
Secondary structure analysis using the programs FoldIndex (http://bioportal.weizmann.ac.il/fldbin/findex) and PONDR (http://www.pondr.com) predicts that large stretches of the SR region are not likely to adopt a globular conformation (Fig. 4). Instead, this domain has several features in common with a class of proteins that have been termed intrinsically unstructured (or disordered) proteins [47, 48]. Unstructured protein domains are characteristically enriched in amino acids that promoter disorder (S, P, Q, E, K) and deficient in hydrophobic amino acids that form the core of globular proteins. Compared to globular domains, unstructured domains tend to have a lower degree of sequence conservation, due to a faster rate of evolution. This category includes many regulatory and signalling proteins, and their lack of secondary structure has been recognized as an important factor in the functions of these proteins. Their open, extended conformation confers flexibility and the ability to harbour multiple sites for interactions with other molecules. The unfolded structure can also impart increased susceptibility to proteolysis, providing a means for regulated turnover of the protein.
4.
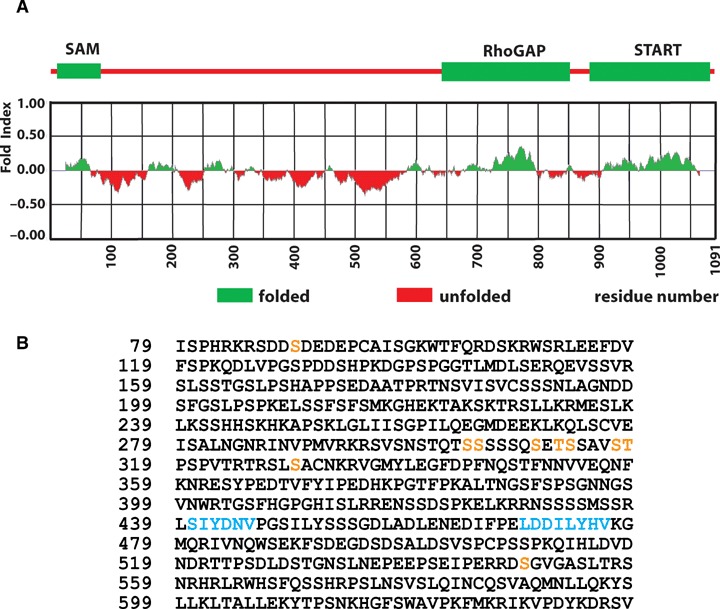
Structural and functional features of the serine-rich domain of DLC-1. (A). Graphical representation of the predicted secondary structure of the human DLC-1 protein, performed using the FoldIndex program (http://bioportal.weizmann.ac.il/fldbin/findex). The serine-rich region between the SAM and RhoGAP domains is predicted to have a largely unfolded conformation. (B). Features of the serine-rich region. The amino acid sequence of the region between the SAM and RhoGAP domains (residues 79–638) of human DLC-1 is shown, and residues that were found to be phosphorylated in the mouse [51] and rat [49,50] DLC-1 proteins are indicated in yellow. Phosphorylation of Ser549 was presumed, since the equivalent serine in a highly conserved region of mouse DLC-2 was phosphorylated [51]. The tensin-binding site (SIYDNV) and the LD motif (LDDILYHV) are shown in blue.
Unstructured domains are also preferred sites for post-translational modifications such as phosphory-lation [47], which serves as a reversible means of regulating the activity and/or subcellular localization of many signalling proteins, including several RhoGAPs [43]. This domain appears to be undergo phosphorylation at multiple sites in the DLC family proteins. Ser322 of rat p122RhoGAP (equivalent to Ser329 in human DLC-1) was phosphorylated in rat adipocytes after insulin treatment, and this was blocked by wortmannin, a phosphatidylinositol 3-kinase inhibitor [49]. Recombinant p122RhoGAP was phosphorylated on Ser322 in vitro by protein kinase B and ribosomal S6 kinase. A cluster of 7 phosphorylation sites in rat DLC-1, equivalent to Ser304, Ser305, Ser310, Thr312, Ser313, Ser 317 and Thr318 in the human sequence, was found in a large-scale characterization of phosphopeptides in vasopressin-treated rat kidney cells [50]. A proteomic analysis of phosphorylated peptides in mouse liver identified Ser89 of DLC-1 and an equivalent residue in DLC-2 (Ser133 in the human sequence) as phosphorylation sites [51]. An additional site was found in DLC-2 (corresponding to Ser572 in the human protein), and the sequence surrounding Ser572 (RRDSGVGASLTR) is identical in DLC-1 and DLC-3 and is similar to the RRXS protein kinase A phosphorylation site consensus sequence [52]. Phosphorylation of these sites has not yet been shown to have an effect on the activities of DLC-1 or DLC-2.
Biological functions of DLC-1
Cytoskeletal organization
The assembly of actin filaments into various structures is initiated at sites of cell-cell and cell-extracellular matrix contact and involves the formation of large multi-protein complexes that include structural and regulatory proteins [53]. These complexes are dynamic structures, and their re-modelling coincides with changes in morphology and cell movement during development, and in adults with processes, such as wound healing and tumour metastasis. The identification of p122RhoGAP as a regulator of RhoA activity prompted studies of its effect on the formation of two RhoA-dependent structures: actin stress fibres, long thick bundles of actin filaments associated with myosin, and focal adhesions, which are multi-protein complexes at the end of stress fibres that provide a link between the actin cytoskeleton and the integrin family of extracellular matrix receptors [54]. Over-expression of DLC-1/p122RhoGAP in cultured cells resulted in a rounded morphology associated with the disruption of actin stress fibres and focal adhesions [39, 55, 56] (Fig 1D and E). Using overlapping deletion mutants, the ability of DLC-1 to disrupt the cytoskeleton was shown to require the presence of the RhoGAP domain, and point mutations that abolished GAP activity reduced this inhibitory effect [39, 55, 56]. DLC-1 over-expression led to the de-phosphorylation of the focal adhesion proteins FAK, paxillin and Cas [56], pointing to a possible mechanism by which DLC-1 may interfere with focal adhesion formation. cDLC-1 may have additional effects in cells cultured on different substrata, as it was found to promote the extension of long membrane protrusions in cells plated on laminin-1 [56].
DLC-1 localizes to focal adhesions via binding to tensin family proteins
While DLC-1 ectopically expressed in cultured cells can have a diffuse cytoplasmic localization [39, 55, 57], in certain cell types DLC-1 may be present in focal adhesions. In normal rat kidney (NRK) fibroblasts, endogenous p122 RhoGAP/DLC-1 and a GFP-tagged N-terminal fragment (aa 1–534) were detected in focal adhesions, as shown by their co-localization with the tips of actin stress fibres and with vin-culin, a focal adhesion protein [58]. Recently, yeast two-hybrid screenings identified DLC-1 as a binding partner for members of the tensin family of focal adhesion proteins [41, 59, 60]. The four tensin proteins (tensin1, tensin2, tensin3 and cten) bind to the cytoplasmic tails of β integrins [61]. The middle region of DLC1 was found to interact with the C-ter-mini of the tensins, which consist of SH2 (src homology 2) and phosphotyrosine-binding (PTB) domains [41, 59, 60]. DLC-1 bound to the SH2 domains of cten and tensin1, and the binding site was mapped to aa 440–445 (SIYDNV) of DLC-1 [41, 60]. Mutation of Tyr442 of DLC-1 abolished the SH2 domain binding, but phosphorylation of Tyr442 was not required for the interaction, unlike most ligands for SH2 domains. [41, 60]. Binding of DLC-1 to the PTB domains of tensin1 and tensin2 was also detected [41, 59], and DLC-1 competed with integrin β3 for binding to the tensin1 PTB domain [41]. Tensin and DLC-1 co-localize (Fig. 5), and the localization of ectopically expressed DLC-1 to focal adhesion-like structures was dependent on the co-expression of tensin proteins [41, 59, 60].
5.
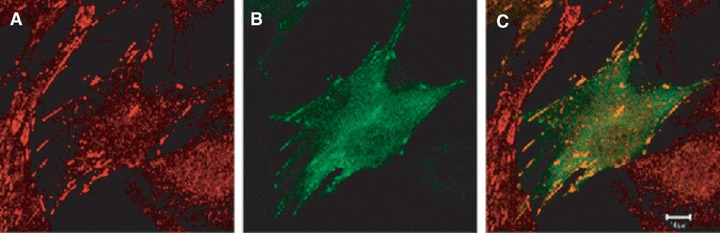
Co-localization of DLC-1 and tensin. Confocal photomicrographs showing endogenous tensin (red, A) and transfected GFP-DLC1 fusion protein (green, B) in human fibroblasts. The merged image (C) shows co-localization of tensin and DLC-1 in focal adhesions, indicated in yellow. The bar represents 10 μm. (Images courtesy of Dr. Guorong Li, Laboratory of Cellular Oncology, National Cancer Institute).
Interaction of DLC-1 with caveolin-1
Caveolin-1 is a 22 kD protein that is an essential component of caveolae, plasma membrane domains that appear as flask-shaped invaginations at the cell surface and are enriched in cholesterol and sphingolipids [62]. Endogenous DLC-1 was found to sediment with caveolin-1 in low-density cholesterol-rich membrane fractions [63], and both endogenous and myc-tagged DLC-1 were co-immunoprecipitated with caveolin-1 [59, 63]. Since multiple functions have been proposed for caveolae [62, 64], and caveolin-1 has been found in membrane subdomains other than caveolae, including focal adhesions [64, 65], the physiological significance of the interaction between DLC-1 and caveolin-1 requires further investigation.
DLC-1 and phosphoinositide signalling
The isolation of the rat p122RhoGAP cDNA by screening an expression library with anti-serum raised against PLC- 1 suggested that the two proteins can interact, and recombinant p122RhoGAP was found to bind PLC- 1 and to stimulate the hydrolysis of phosphatidylinositol 4,5-bisphosphate (PIP2) by PLC- 1 [17].PIP2 hydrolysis generates two second messengers, diacylglycerol, which activates proteins with C1 domains, such as protein kinase C, and inositol 1,4,5-trisphosphate, which promotes the release of Ca2+ from internal stores to regulate calcium-responsive proteins [66]. p122RhoGAP did not bind PLC- 1 or PLC- 1 [17], which are activated by receptor-coupled heterotrimeric G proteins and receptor tyrosine kinases, respectively [66]. The PLC- 1 binding activity of p122RhoGAP appeared to reside in the C-terminal half of the polypeptide, as a fragment consisting of aa 617–1083 promoted the release of intracellular Ca2+[39]. Whether DLC-2 and DLC-3 also interact with PLC- 1 has not been deter-mined. In addition to serving as a precursor for other lipid second messengers, PIP2 also has an important role in regulating cytoskeleton assembly by inducing conformational changes in actin-binding proteins, such as vinculin and talin [67]. Thus DLC-1 could influence cytoskeletal dynamics by altering local PIP2 levels as well as by regulating Rho GTPase activity.
Biological activities of DLC-2 and DLC-3
Consistent with its similarity to DLC-1, over-expression of DLC-2 has been found to induce cell rounding and disrupt actin filaments in liver and breast cancer cells [20, 27], and this effect was eliminated by mutation of critical residues in the RhoGAP domain [27]. When recombinant DLC-2 was expressed at lower levels to reduce cytotoxicity, the protein was found to co-localize with mitochondrial markers, using immunofluorescence and cell fractionation [68]. DLC-2 was also detected in structures that resembled lipid droplets, and targeting to mitochondria and lipid droplets was mapped to the START domain [68].Recombinant DLC-2 was found to form homodimers in solution, and residues 120–672 were responsible for self-association [36]. It will be of interest to determine whether dimerization is a property of the other DLC family proteins. Studies on the biological functions of DLC-3 have just begun, but the amino acid sequence contains a segment similar to the DLC-1 tensin-binding site (STYDNL, aa 353–358) and full-length DLC-3 was shown to bind the SH2 and PTB domains of tensin1 [41].
Genetic analysis of DLC-1 function
Mouse DLC-1 gene knockout
Evidence that DLC-1 has a crucial role in cytoskeletal organization and morphogenesis in vivo has come from the discovery that null mutations in the gene result in embryonic lethality in mice and flies. We used gene targeting to disrupt the mouse DLC-1 gene and found that DLC-1 was essentialfor viability, as embryos homozygous for the mutated allele did not survive beyond 10.5 days gestation [31]. Histological examination of 9.5-day DLC-1−/− embryos revealed an open anterior neural tube, abnormalities in the branchial arches, brain neuroepithelium and heart, and defective foetal blood vessels in the placental labyrinth [31]. The related DLC-2 and DLC-3 gene products were apparently unable to compensate for DLC-1 deficiency during development. Fibroblasts from 9.5 day DLC-1−/−embryos were able to proliferate in culture, but displayed a reduction in actin stress fibres and vinculin-containing focal adhesions, suggesting that loss of DLC-1 may have interfered with development by adversely affecting the assembly of the cytoskeleton and cell adhesion complexes [31]. The phenotype of DLC-1-deficient mouse embryo fibroblasts may seem inconsistent with studies showing that over-expression of DLC-1 in cultured cells disrupts focal adhesions and actin stress fibres [39, 55]. On one hand, the response of primary cell cultures may differ from that of immortalized cell lines that have accumulated genetic alterations. However, the deleterious consequences of both the absence and over-expression of DLC-1 may signify that it serves as an adaptor or scaffolding protein that must be present at an optimal level to interact with other components of a signalling complex. Either a deficiency or an excess of a scaffolding protein can impair formation of the complex and reduce signal propagation [69].
RhoGAP88 C in Drosophila
The Drosophila DLC-1 orthologue RhoGAP88 C was found to be essential for viability in a study using RNA interference to inactivate Drosophila RhoGAP genes [70]. Phenotypic analysis of mutant alleles of the cv-c gene encoding RhoGAP88 C has provided striking confirmation of the essential role of the DLC proteins in actin cytoskeleton dynamics during mor-phogenesis. Viable hypomorphic alleles of cv-c lead to loss of the posterior crossvein and detachment of the anterior crossvein in the wings of adult flies [28]. Molecular characterization of embryonic lethal alleles showed that two are apparently null alleles resulting from nonsense mutations, and a third (cv-c7) has a missense mutation (Arg601Gln) that targets the ‘arginine finger’ of the RhoGAP domain. The lethal alleles caused abnormalities in tissues undergoing morphogenetic movements, including delayed dorsal closure of the epidermis and defects in head involution, midgut constriction and Malpighian tubule elongation. In Malpighian tubules, either RhoGAP88 C deficiency or over-expression resulted in actin cytoskeleton disorganization. Genetic analysis of the interactions between cv-c and several RhoGTPase mutants indicated that RhoGAP88C regulated different RhoGTPase substrates in a tissue-specific fashion [28].
Two recent studies indicated that cv-c is required for the cell shape changes that take place during epithelial invagination in Drosophila spiracle and trachea morphogenesis. The apical constriction of the invaginating cells of the trachea and the posterior spiracle depended on the activation of Rho1 at the apical membrane and the consequent apical accumulation of F-actin and myosin II [71, 72]. In cv-c mutants, the invagination defects in tracheal and spiracle cells were associated with the abnormal subcellular distribution of F-actin and myosin II [71, 72]. RhoGAP88C was present in the basolateral domain of invaginating spiracle cells in normal embryos, where it may down-regulate Rho activity and cooperate with two apically localized RhoGEFs, RhoGEF64C and RhoGEF2, in restricting Rho1 activation and actomyosin contraction to the apical surface to generate cell shape changes during morpho-genesis [72]. Expression of cv-c in the developing trachea was reduced in mutants with defective EGF signalling [71], thus placing the DLC family proteins downstream of the EGF receptor in some tissues during development.
No mutations in the C.elegans DLC-1 homologue gei-1 have been reported, but there is indirect evidence that it may also be involved in cytoskeletal organization. By yeast two hybrid screening, the gei-1 gene product was identified as a protein that interacts with gex-2 (gut on exterior 2) [29], a gene essential for normal morphogenesis in C. elegans that encodes the nematode homologue of mammalian Sra-1 (specifically Rac associated protein 1, also known as PIR121) [73], a Rac1 effector and component of the WAVE/Scar protein complex that regulates branching actin filament assembly [53].
The DLC family proteins in cancer
Decreased expression of DLC-1 in human cancers
Rho GTPases are involved in the regulation of cellular processes, such as proliferation, motility and apoptosis that are typically altered during oncogenesis [74]. Rho GTPase activity is increased in a variety of human cancers, but this has not been associated with the occurrence of activating mutations in Rho protein genes, in contrast with Ras [5, 75]. Some RhoGAPs could potentially serve as tumour suppressors, if their loss resulted in a dysregulation of Rho protein activity that facilitated the growth and metastasis of cancer cells. Considerable evidence has accumulated that DLC-1 can act as a tumour suppressor in a wide spectrum of human cancers. While DLC-1 mRNA is expressed in many adult human tissues, it is down-regulated or absent in a number of common human cancers, including HCC [16, 40, 76], breast [77–79] colon [79], ovarian [79, 80], uterine [79], gastric [33], lung [79, 81–83], pancreatic [84], prostate [85], renal [79] and nasopharyngeal tumours [86].Reduced mRNA levels have also been found in non-malignant conditions, such as uterine fibroids [87] and benign prostatic hyperplasia [85]. There had been little data on the relative abundance of the DLC-1 protein in normal and neoplastic tissues, but recently decreased DLC-1 protein levels were reported in prostate tumour samples compared to normal prostate tissue [85].
Deletions of DLC1 in tumours
The 8p22 region-containing DLC1 is commonly under-represented in a variety of solid tumours and haematological malignancies [88], and deletions at the DLC1 locus were detected in HCC cell lines and primary HCC using Southern blot hybridization and loss of heterozygosity (LOH) analysis [16, 40, 76]. Subsequently DLC1 copy number losses were found in other common human cancers, including breast [77, 89], lung [81], nasopharyngeal [86] and prostate cancer [85].Although the 8p21–22 regions does not correspond to a fragile site (FS), its propensity for deletion is similar to that of the most unstable and vulnerable FSs [90, 91]. The proximity of DLC1 to a break in the mouse/human synteny [24] could predispose it to deletions, since these evolutionary breakpoints often coincide with regions of instability in tumours [92].
Epigenetic inactivation of DLC-1 expression
Epigenetic mechanisms also lead to silencing of tumour suppressor genes, and hypermethylation of cytosine bases within CpG dinucleotides in GC-rich promoter regions is the best characterized chromatin modification associated with transcriptional repression in human malignancies [93]. The first evidence implicating aberrant promoter methylation in the down-regulation of the DLC-1 gene was obtained by screening several cell lines derived from HCC, breast, colon and prostate tumours [32]. Subsequent analyses showed that promoter methylation is the principal mechanism responsible for inactivation of the DLC-1 gene in a number of solid tumours (Table 2). Hypermethylation of the DLC-1 promoter may be an early event in the development of prostate neo-plasms, as it was also observed in a majority of benign prostatic hyperplasia cases [85]. Aberrant DLC1 methylation has been detected in several haematological malignancies (Table 3), which is of interest, since DLC-1 mRNA levels are low in normal peripheral blood leucocytes but can be detected in lymph node, thymus and spleen [22, 97], suggesting that DLC-1 may be expressed at certain stages of lymphocyte development. The relative ease of PCR-based techniques for detecting methylation may make DLC1 promoter hypermethylation a useful biomarker for diagnosis, staging, prediction and monitoring for recurrence of certain malignancies. Reactivation of DLC-1 expression in cancer cell lines with promoter hypermethylation can be achieved in some cases by treatment with the DNA methyltrans-ferase inhibitor 5-aza-2′-deoxycytidine [32, 33, 40, 85, 94–96, 98].
2.
Methylation of the DLC1 promoter region in human cancers:solid tumours
| Tumour type | Fraction of samples methylated | Percent methylated | Reference |
|---|---|---|---|
| Primary hepatocellular carcinomas | 6/25 | 24 | [40] |
| Hepatocellular carcinoma cell lines | 3/7 | 43 | [40] |
| Non-small cell carcinoma cell lines | 10/19 | 53 | [81] |
| Non-small cell carcinomas | 11/18 | 61 | [82] |
| Matching normal tissue | 2/8 | 25 | [82] |
| Non-small cell carcinoma cell lines | 2/11 | 18 | [82] |
| Gastric carcinomas | 29/97 | 30 | [33] |
| Gastric cancer cell lines | 5/7 | 71 | [33] |
| Prostate carcinomas | 13/27 | 48 | [85] |
| Matching normal tissue | 2/27 | 7 | [85] |
| Benign prostate hyperplasia | 15/21 | 71 | [85] |
| Normal prostate tissue | 0/10 | 0 | [85] |
| Breast carcinomas | 5/14 | 36 | [34] |
| Breast carcinoma cell lines | 3/9 | 33 | [34] |
| Breast carcinomas | 4/39 | 10 | [94] |
| Breast carcinoma cell lines | 5/20 | 25 | [94] |
| Nasopharyngeal carcinomas | 31/39 | 79 | [86] |
| Nasopharyngeal carcinomas | 64/72 | 89 | [34] |
| Nasopharyngeal carcinoma cell lines | 11/12 | 91 | [34] |
| Oesophageal carcinomas | 48/94 | 51 | [34] |
| Oesophageal carcinoma cell lines | 6/15 | 40 | [34] |
| Cervical carcinomas | 7/8 | 87 | [34] |
| Cervical carcinoma cell lines | 5/8 | 63 | [34] |
| Renal cell carcinomas | 12/34 | 35 | [95] |
| Matching normal tissue | 1/34 | 3 | [95] |
| Renal carcinoma cell lines | 1/7 | 14 | [95] |
3.
Methylation of the DLC1 promoter region in human cancers:Haematological malignancies
| Cancer type | Fraction of Samples methylated | Percent methylated | Reference |
|---|---|---|---|
| Multiple myeloma | 11/14 | 78 | [96] |
| Normal bone marrow | 0/4 | 0 | [96] |
| Multiple myeloma cell lines | 3/4 | 75 | [96] |
| Non-Hodgkin's lymphoma | 65/75 | 87 | [97] |
| Normal lymphocytes | 0/5 | 0 | [97] |
| Non-Hodgkin's lymphoma cell lines | 6/6 | 100 | [97] |
| Acute lymphoblastic leukaemia | 14/16 | 88 | [98] |
| Leukaemia cell lines | 3/4 | 75 | [98] |
Alterations in histone modifications also occur in cancer and may contribute to silencing of DLC-1. Transcriptional repression has been linked with his-tone hypoacetylation, which is promoted by the recruitment of histone deacetylases to chromatin [99]. DLC-1 expression was increased after treatment with the histone deacetylase inhibitor trichostatin A (TSA) in several human gastric cancer [33], prostate cancer [85], multiple myeloma [96] and leukaemia [98] cell lines. In some cell lines DLC-1 was not induced by TSA unless the cells were co-treated with 5-aza-2′-deoxycytidine [33, 97, 98]. Since epigenetic changes are potentially reversible, compounds that reactivate tumour suppressor genes silenced by DNA methylation or histone de-acetylation may be useful anti-cancer agents. Drugs that bring about DNA de-methylation or histone acetylation have shown promise in clinical trials, and combination therapy with inhibitors of both processes is under investigation [100].
Cancer-associated epigenetic changes may have their origin in transcriptional mechanisms that control developmentally regulated genes. The Polycomb group (PcG) of transcriptional repressors help to maintain the multi-potent state of stem cells by reversibly repressing genes involved in differentiation and inhibition of cell proliferation [101]. Recently DLC1 and several other genes silenced by methyla-tion in cancer were found to be targets of PcG proteins in human embryonic fibroblasts [102] and human embryonic stem cells [103]. PcG binding to tumour suppressor genes, such as DLC1 may be a mark that predisposes them to persistent silencing during the development of neoplasia by aberrant recruitment of DNA methyl transferases [102, 104].
DLC-1 sequence variants in human cancers
Mutational analysis of the DLC-1 gene in tumour DNA samples revealed that somatic mutations in the coding region are uncommon in HCC [76, 105] and col-orectal [30], ovarian [30], brain [106], head and neck [107] and prostate cancers [108]. A number of single nucleotide polymorphisms (SNPs) have been identified in the DLC-1 genomic DNA sequence that could be used for determining whether DLC-1 variants are associated with altered susceptibility to cancer or other human disorders. A familial prostate cancer susceptibility gene was mapped near the DLC-1 locus on 8p22 [109], but a mutation screening and association study did not reveal significant differences in the DLC1 SNP frequencies in prostate cancer cases and controls [108]. However, in a large-scale genomic screen, DLC1 was identified as a highly significant breast cancer susceptibility gene [110, 111].
DLC-1 suppresses tumour cell growth
Evidence that DLC-1 can inhibit the growth of tumour cells has come from experiments in which the cDNA was transfected into human cancer cell lines that do not express the endogenous gene. Restoration of DLC-1 expression inhibited cell proliferation and colony formation in HCC [55–57, 76] and in breast [77, 89], lung [41, 81, 83], nasopharyngeal [34], oesophageal [34] and epithelial ovarian cancer cells [112] (Fig. 1B). DLC-1-mediated inhibition of HCC and ovarian cancer cell proliferation was associated with the induction of apoptosis (Fig. 1G), marked by the cleavage of procaspase-3 [57, 112]. DLC-1 expression reduced anchorage-independent growth of HCC [55], ovarian cancer [112] and lung cancer [41] cells in soft agar colony forming assays, an in vitro measure of tumourigenicity. Over-expression of DLC-1 was also able to suppress the growth of tumours in athymic nude mouse xenograft models. Tumour formation was abolished in DLC-1-transfect-ed human breast cancer cells [89] and non-small cell lung cancer cells [81], and the size of tumours was reduced in HCC cells [55, 57] (Fig. 1F). Inhibition of tumour cell growth by DLC-1 has been attributed to its ability to down-regulate Rho protein activity, as RhoGAP-deficient mutants were less active in suppressing cell growth [41, 55]. Mutation of the tensin-binding site also reduced growth inhibition by DLC-1 [41, 60], without affecting its overall RhoGAP activity in vivo[41], indicating that either the tumour suppressor function of DLC-1 may require targeting of the RhoGAP activity by tensin or that DLC-1 has additional functions that do not involve the GAP domain.
DLC-1 as a metastasis suppressor gene
Losses at chromosome 8p have been linked to increased metastasis, poorer prognosis and higher tumour grade in several human cancers [113], and the DLC-1 gene at 8p22 has been implicated in the metastatic process. DLC-1 was found to be consistently down-regulated in highly invasive HCC cell lines and metastatic subclones compared to non-metastatic ones [114]. In assays using cultured cells, re-expression of DLC-1 inhibited the motility and invasiveness of HCC [55–57] (Fig. 1C), breast cancer [115], ovarian cancer [112] and lung cancer cell lines [41].Both the RhoGAP and tensin-binding activities have been implicated in the inhibition of migration [41]. Transcriptional profiling of two subclones of the MDA-MB-435 cell line showed that DLC-1 was one of the genes down-regulated in metastatic M4A4 cells relative to non-metastatic NM2C5 cells [115].Restoration of DLC-1 expression in M4A4 cells to the levels observed in the non-metastatic cells decreased the size and abundance of pulmonary metastases formed in nude mice [115] (Fig. 1H). In another metastasis model system, DLC-1 expression was reduced in human MDA-MB-231 breast cancer cells that had been selected for increased metastasis to bone [116]. Lower DLC-1 levels in primary tumours could serve as a marker for those with a greater potential for metastasis and a worse prognosis. Gene expression profiling analyses have found that DLC-1 expression was higher in doxorubicin-sensitive breast tumours compared to doxorubicin-resistant ones [117], and down-regulation of DLC-1 was associated with reduced survival of patients with high-grade gliomas [118] and of hepatitis C virus-positive HCC patients awaiting liver transplantation [119]. Microarray studies have also shown that DLC-1 expression can be used to distinguish lymphoma and leukaemia subtypes with different clinical behaviours [120,121].
Regulation of DLC-1 by anti-oncogenic factors
Several studies using microarray gene expression profiling have shown that DLC-1 expression was up-regulated in cultured cells after treatment with agents that inhibit cell proliferation and/or promote differentiation. DLC-1 mRNA levels were increased in HT-29 colon cancer cells treated with flavone, a dietary flavonoid that inhibits growth and induces apoptosis [122]. A recent study confirmed that flavone caused a nearly fivefold increase in the amount of DLC-1 mRNA in HT-29 colon cancer cells and also found that flavone treatment induces DLC-1 expression in several human breast cancer cell lines [123]. DLC-1 levels were increased in cultured Wilm's tumour cells treated with all-trans retinoic acid [124], a vitamin A metabolite well-known for stimulating differentiation and suppressing carcinogenesis [125]. DLC-1 was one of the genes up-regulated in ovarian cancer cells treated with high concentrations of progesterone that inhibited cell growth [112]. Activators of the peroxi-some proliferator-activated receptorγ (PPARγ), a member of the steroid hormone receptor family of transcription factors, promote differentiation and inhibit the growth of several epithelial types of cancer [126]. Over-expression of PPAR in lung cancer cells decreased invasivesness and tumourigenesis and induced expression of DLC-1 [127]. Further investigations may reveal how DLC-1 contributes to the phenotype of cells treated with anti-oncogenic compounds.
Animal models of DLC-1 in cancer
The embryonic lethality of the DLC-1−/− mice precluded attempts to investigate the effect of constitutive DLC-1 deficiency on tumour development. The heterozygous animals were phenotypically normal despite lower levels of DLC-1 mRNA, and they did not have an increased incidence of spontaneous tumour formation [31], suggesting that haploinsuffi-ciency of Dlc1 was unable to initiate neoplastic development. Alternative technologies may be needed to generate tissue-specific inactivation of the gene to study the effect on neoplasia in various organs. The rat may also be a promising model for studying DLC-1 in tumourigenesis. The CpG island at the 5′ end of the Dlc1 gene was found to be hypermethylated in rat liver tumour cell lines that did not express DLC-1 RNA [128]. A gastric cancer resistance locus (Gcr3) that reduced the size and depth of invasion of N-methyl-N’-nitro-N-nitrosoguanidine-induced stomach carcinomas was mapped to the vicinity of the Dlc1 gene on rat chromosome 16q [129].
Evidence for roles of DLC-2 and DLC-3 in neoplasia
In addition to their structural and functional similarities with DLC-1, the other two human DLC family genes may also be involved in oncogenesis. The STARD13/DLC2 locus at chromosome 13q13 is another frequent target of genomic DNA deletions in several human cancers [88].LOH at the 13q13 locus and under-expression of DLC-2 were observed in primary HCC tissues [19]. Down-regulation of DLC-2 mRNA was also detected in a significant fraction of primary lung, ovary, kidney, breast and uterine tumours [79]. The GAP domain of DLC-2 was able to suppress Ras signalling and inhibit the transformation of NIH3T3 cells when co-transfected with oncogenic Ras [19]. DLC-2 over-expression was found to inhibit the growth of MCF-7 breast cancer cells [20]. Transfection of the DLC-2 isoform into the HepG2 cell line decreased proliferation, RhoA activity, motility and the ability to form colonies in soft agar [27].
The STARD8/DLC3 locus on chromosome Xq13 is near a site of LOH in ovarian cancers [130], but this region has not been reported to be frequently deleted in other cancer types [131]. Reduced DLC-3 mRNA levels were observed in a majority of human prostate, kidney, lung, breast, uterine and ovarian cancer tissues analysed [22]. Over-expression of DLC-3 inhibited the proliferation, colony forming ability, and anchorage-independent growth of human breast and prostate cancer cell lines [22]. In a largescale analysis of mutations in human breast and colon cancers, two missense mutations in STARD8/DLC3 were found in breast tumours [132]. Whether these alterations, both located in the SR region (Gly268Ser and Glu322Lys, in the DLC-3 amino acid sequence), have an effect on DLC-3 activity remains to be determined.
Conclusions and future directions
Both in vitro and in vivo studies have provided firm evidence that DLC-1 has a role in regulating actin cytoskeleton organization. The Drosophila DLC-1 orthologue participates in localizing Rho GTPase activity to sites undergoing cytoskeleton rearrangements during morphogenesis. The RhoGAP activity of DLC-1 may be targeted to particular subcellular domains by binding to the tensins and other molecules. Further efforts using mass-spectrometry-based proteomic analyses and yeast two-hybrid screening will reveal additional interaction partners of DLC-1 and help to elucidate its role in the signalling pathways that regulate cell adhesion, migration and prolif-eration. This knowledge will also aid in understanding how the loss of DLC-1 in neoplasia contributes to tumour cell growth and metastasis. More information is required on the mechanisms responsible for regulating DLC-1 at various levels, including transcription, translation, and post-translational processes, such as phosphorylation and proteolytic degradation. Determining the biological functions specific to each of the DLC proteins will shed light on the raison d'être for having three DLC genes in vertebrates.
The anti-cancer properties of DLC-1 may have clinical applications. Since cells derived from different types of cancer are highly sensitive to re-expression of DLC-1, the potential for effective therapy based on DLC-1 transfer appears high. Screening for dietary and pharmacological agents that up-regulate DLC-1 gene expression could lead to the development of drugs that might be useful for chemoprevention and therapy. Identification of the pathways in which DLC-1 operates may aid in the screening or design of small molecules that can mimic the effects of DLC-1 and inhibit tumour progression.
Acknowledgments
This research was supported by the Intramural Research Program of the National Cancer Institute, NIH.
The authors would like to thank Steve Goodison, Ming Guan, Mark Miller, Alex Papageorge, Sang-Won Park, Xiaolan Qian, Steven Reynolds, Demetrios Spandidos, Nikolaus Soulitzis, Veronika Ullmannova, Virginia Urquidi and Catherine Keck-Waggoner for their significant contribution to the study of DLC-1.
References
- 1.Van Aelst L, D'Souza-Schorey C. Rho GTPases and signaling networks. Genes Dev. 1997;11:2295–322. doi: 10.1101/gad.11.18.2295. [DOI] [PubMed] [Google Scholar]
- 2.Ridley AJ. Rho family proteins: coordinating cell responses. Trends Cell Biol. 2001;11:471–7. doi: 10.1016/s0962-8924(01)02153-5. [DOI] [PubMed] [Google Scholar]
- 3.Jaffe AB, Hall A. Rho-GTPases: biochemistry and biology. Annu Rev Cell Dev Biol. 2005;21:247–69. doi: 10.1146/annurev.cellbio.21.020604.150721. [DOI] [PubMed] [Google Scholar]
- 4.Sahai E, Marshall CJ. Rho-GTPases and cancer. Nat Rev Cancer. 2002;2:133–42. doi: 10.1038/nrc725. [DOI] [PubMed] [Google Scholar]
- 5.Gomez del Pulgar T, Benitah SA, Valeron PF, Espina C, Lacal JC. Rho GTPase expression in tumourigenesis: evidence for a significant link. Bioessays. 2005;27:602–13. doi: 10.1002/bies.20238. [DOI] [PubMed] [Google Scholar]
- 6.Wennerberg K, Der CJ. Rho-family GTPases:it's not only Rac and Rho (and I like it) J Cell Sci. 2004;117:1301–12. doi: 10.1242/jcs.01118. [DOI] [PubMed] [Google Scholar]
- 7.Boureux A, Vignal E, Faure S, Fort P. Evolution of the Rho family of Ras-like GTPases in eukaryotes. Mol Biol Evol. 2007;24:203–16. doi: 10.1093/molbev/msl145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bos JL, Rehmann H, Wittinghofer A. GEFs and GAPs:critical elements in the control of small G proteins. Cell. 2007;129:865–77. doi: 10.1016/j.cell.2007.05.018. [DOI] [PubMed] [Google Scholar]
- 9.Rossman KL, Der CJ, Sondek J. GEF means go: turning on Rho GTPases with guanine nucleotide-exchange factors. Nat Rev Mol Cell Biol. 2005;6:167–80. doi: 10.1038/nrm1587. [DOI] [PubMed] [Google Scholar]
- 10.Moon SY, Zheng Y. Rho GTPase-activating proteins in cell regulation. Trends Cell Biol. 2003;13:13–22. doi: 10.1016/s0962-8924(02)00004-1. [DOI] [PubMed] [Google Scholar]
- 11.Bernards A. GAPs galore! A survey of putative Ras superfamily GTPase activating proteins in man and Drosophila. Biochim Biophys Acta. 2003;1603:47–82. doi: 10.1016/s0304-419x(02)00082-3. [DOI] [PubMed] [Google Scholar]
- 12.Tcherkezian J, Lamarche-Vane N. Current knowledge of the large RhoGAP family of proteins. Biol Cell. 2007;99:67–86. doi: 10.1042/BC20060086. [DOI] [PubMed] [Google Scholar]
- 13.Marinissen MJ, Gutkind JS. Scaffold proteins dictate Rho GTPase-signaling specificity. Trends Biochem Sci. 2005;30:423–6. doi: 10.1016/j.tibs.2005.06.006. [DOI] [PubMed] [Google Scholar]
- 14.García-Mata R, Burridge K. Catching a GEF by its tail. Trends Cell Biol. 2007;17:36–43. doi: 10.1016/j.tcb.2006.11.004. [DOI] [PubMed] [Google Scholar]
- 15.Wang L, Zheng Y. Cell type-specific functions of Rho GTPases revealed by gene targeting in mice. Trends Cell Biol. 2007;17:58–64. doi: 10.1016/j.tcb.2006.11.009. [DOI] [PubMed] [Google Scholar]
- 16.Yuan BZ, Miller MJ, Keck CL, Zimonjic DB, Thorgeirsson SS, Popescu NC. Cloning, characterization, and chromosomal localization of a gene frequently deleted in human liver cancer (DLC-1) homologous to rat RhoGAP. Cancer Res. 1998;58:2196–9. [PubMed] [Google Scholar]
- 17.Homma Y, Emori Y. A dual functional signal mediator showing RhoGAP and phosopholipase C- stimulating activities. EMBO J. 1995;14:286–91. doi: 10.1002/j.1460-2075.1995.tb07002.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ponting CP, Aravind L. START: a lipid-binding domain in StAR, HD-ZIP and signalling proteins. Trends Biochem Sci. 1999;24:130–2. doi: 10.1016/s0968-0004(99)01362-6. [DOI] [PubMed] [Google Scholar]
- 19.Ching YP, Wong CM, Chan SF, Leung TH, Ng DC, Jin DY, Ng IO. Deleted in liver cancer (DLC) 2 encodes a RhoGAP protein with growth suppressor function and is underexpressed in hepatocellular carcinoma. J Biol Chem. 2003;278:10824–30. doi: 10.1074/jbc.M208310200. [DOI] [PubMed] [Google Scholar]
- 20.Nagaraja GM, Kandpal RP. Chromosome 13q12 encoded Rho GTPase activating protein suppresses growth of breast carcinoma cells, and yeast two-hybrid screen shows its interaction with several proteins. Biochem Biophys Res Commun. 2004;313:654–65. doi: 10.1016/j.bbrc.2003.12.001. [DOI] [PubMed] [Google Scholar]
- 21.Nagase T, Seki N, Ishikawa K, Tanaka A, Nomura N. Prediction of the coding sequences of unidentified human genes. V. The coding sequences of 40 new genes (KIAA0161-KIAA0200) deduced by analysis of cDNA clones from human cell line KG-1. DNA Res. 1996;3:17–24. doi: 10.1093/dnares/3.1.17. [DOI] [PubMed] [Google Scholar]
- 22.Durkin ME, Ullmannova V, Guan M, Popescu NC. Deleted in Liver Cancer 3 (DLC-3), a novel Rho GTPase activating protein, is downregulated in cancer and inhibits tumor cell growth. Oncogene. 2007;26:4580–9. doi: 10.1038/sj.onc.1210244. [DOI] [PubMed] [Google Scholar]
- 23.Schimdt A, Schmelzle T, Hall MN. The RHO1-GAPs SAC7, BEM2 and BAG7 control distinct RHO1 functions in Saccharomyces cerevisiae. Mol Microbiol. 2002;45:1433–41. doi: 10.1046/j.1365-2958.2002.03110.x. [DOI] [PubMed] [Google Scholar]
- 24.Durkin ME, Yuan BZ, Thorgeirsson SS, Popescu NC. Gene structure, tissue expression, and linkage mapping of the mouse DLC-1 gene (Arhgap7. Gene. 2002;288:119–27. doi: 10.1016/s0378-1119(02)00462-6. [DOI] [PubMed] [Google Scholar]
- 25.Yuan BZ, Yang Y, Keck-Waggoner CL, Zimonjic DB, Thorgeirsson SS, Popescu NC. Assignment and cloning of mouse Arhgap7 to chromosome 8A4-B2, a conserved syntenic region of human chromosome 8p22®p21. Cytogenet Cell Genet. 1999;87:189–90. doi: 10.1159/000015462. [DOI] [PubMed] [Google Scholar]
- 26.Nagase T, Kikuno R, Hattori A, Kondo Y, Okumura K, Ohara O. Prediction of the coding sequences of unidentified human genes. XIX. The complete sequences of 100 new cDNA clones from brain which code for large proteins in vitro. DNA Res. 2000;7:347–55. doi: 10.1093/dnares/7.6.347. [DOI] [PubMed] [Google Scholar]
- 27.Leung TH, Ching YP, Yam JW, Wong CM, Yau TO, Jin DY, Ng IO. Deleted in liver cancer 2 (DLC2) suppresses cell transformation by means of inhibition of RhoA activity. Proc Natl Acad Sci USA. 2005;102:15207–12. doi: 10.1073/pnas.0504501102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Denholm B, Brown S, Ray RP, Ruiz-Gomez M, Skaer H, Hombria JC. Crossveinless-c is a RhoGAP required for actin reorganisation during morphogene-sis. Development. 2005;132:2389–400. doi: 10.1242/dev.01829. [DOI] [PubMed] [Google Scholar]
- 29.Tsuboi D, Qadota H, Kasuya K, Amano M, Kaibuchi K. Isolation of the interacting molecules with GEX-3 by a novel functional screening. Biochem Biophys Res Commun. 2002;292:697–701. doi: 10.1006/bbrc.2002.6717. [DOI] [PubMed] [Google Scholar]
- 30.Wilson PJ, McGlinn E, Marsh A, Evans T, Arnold J, Wright K, Biden K, Young J, Wainwright B, Wicking C, Chenevix-Trench G. Sequence variants of DLC1 in colorectal and ovarian tumours. Hum Mutat. 2000;15:156–65. doi: 10.1002/(SICI)1098-1004(200002)15:2<156::AID-HUMU4>3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]
- 31.Durkin ME, Avner MR, Huh CG, Yuan BZ, Thorgeirsson SS, Popescu NC. DLC-1, a Rho GTPase-activating protein with tumor suppressor function, is essential for embryonic development. FEBS Lett. 2005;579:1191–6. doi: 10.1016/j.febslet.2004.12.090. [DOI] [PubMed] [Google Scholar]
- 32.Yuan BZ, Durkin ME, Popescu NC. Promoter hyper-methylation of DLC-1, a candidate tumor suppressor gene, in several common human cancers. Cancer Genet Cytogenet. 2003;140:113–7. doi: 10.1016/s0165-4608(02)00674-x. [DOI] [PubMed] [Google Scholar]
- 33.Kim TY, Jong HS, Song SH, Dimtchev A, Jeong SJ, Lee JW, Kim TY, Kim NK, Jung M, Bang YJ. Transcriptional silencing of the DLC-1 tumor suppressor gene by epigenetic mechanism in gastric cancer cells. Oncogene. 2003;22:3943–51. doi: 10.1038/sj.onc.1206573. [DOI] [PubMed] [Google Scholar]
- 34.Seng TJ, Low JS, Li H, Cui Y, Goh HK, Wong ML, Srivastava G, Sidransky D, Califano J, Steenbergen RD, Rha SY, Tan J, Hsieh WS, Ambinder RF, Lin X, Chan AT, Tao Q. The major 8p22 tumor suppressor DLC1 is frequently silenced by methylation in both endemic and sporadic nasopharyngeal, esophageal, and cervical carcinomas, and inhibits tumor cell colony formation. Oncogene. 2007;26:934–44. doi: 10.1038/sj.onc.1209839. [DOI] [PubMed] [Google Scholar]
- 35.Qiao F, Bowie JU. The many faces of SAM. Sci STKE. 2005;2005:re7. doi: 10.1126/stke.2862005re7. [DOI] [PubMed] [Google Scholar]
- 36.Li H, Fung KL, Jin DY, Chung SS, Ching YP, Ng IO, Sze KH, Ko BC, Sun H. Solution structures, dynamics, and lipid-binding of the sterile alpha-motif domain of the deleted in liver cancer 2. Proteins. 2007;67:1154–66. doi: 10.1002/prot.21361. [DOI] [PubMed] [Google Scholar]
- 37.Kwan JJ, Donaldson LW. The NMR structure of the murine DLC2 SAM domain reveals a variant fold that is similar to a four-helix bundle. BMC Struct Biol. 2007;7:34. doi: 10.1186/1472-6807-7-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Li G, Zhang XC. GTP hydrolysis mechanism of Ras-like GTPases. J Mol Biol. 2004;340:921–32. doi: 10.1016/j.jmb.2004.06.007. [DOI] [PubMed] [Google Scholar]
- 39.Sekimata M, Kabuyama Y, Emori Y, Homma Y. Morphological changes and detachment of adherent cells induced by p122, a GTPase-activating protein for Rho. J Biol Chem. 1999;274:17757–62. doi: 10.1074/jbc.274.25.17757. [DOI] [PubMed] [Google Scholar]
- 40.Wong CM, Lee JM, Ching YP, Jin DY, Ng IO. Genetic and epigenetic alterations of DLC-1 gene in hepatocellular carcinoma. Cancer Res. 2003;63:7646–51. [PubMed] [Google Scholar]
- 41.Qian X, Li G, Asmussen HK, Asnaghi L, Vass WC, Braverman R, Yamada KM, Popescu NC, Papageorge AG, Lowy DR. Oncogenic inhibition by a deleted in liver cancer gene requires cooperation between tensin binding and Rho-specific GTPase-activating protein activities. Proc Natl Acad Sci USA. 2007;104:9012–7. doi: 10.1073/pnas.0703033104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Alpy F, Tomasetto C. Give lipids a START: the StAR-related lipid transfer (START) domain in mammals. J Cell Sci. 2005;118:2791–801. doi: 10.1242/jcs.02485. [DOI] [PubMed] [Google Scholar]
- 43.Bernards A, Settleman J. GAP control: regulating the regulators of small GTPases. Trends Cell Biol. 2004;14:377–85. doi: 10.1016/j.tcb.2004.05.003. [DOI] [PubMed] [Google Scholar]
- 44.Tsujishita Y, Hurley JH. Structure and lipid transport mechanism of a StAR-related domain. Nat Struct Biol. 2000;7:408–14. doi: 10.1038/75192. [DOI] [PubMed] [Google Scholar]
- 45.Brown MC, Curtis MS, Turner CE. Paxillin LD motifs may define a new family of protein recognition domains. Nat Struct Biol. 1998;5:677–8. doi: 10.1038/1370. [DOI] [PubMed] [Google Scholar]
- 46.Li SS. Specificity and versatility of SH3 and other proline-recognition domains: structural basis and implications for cellular signal transduction. Biochem J. 2005;390:641–53. doi: 10.1042/BJ20050411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tompa P. Intrinsically unstructured proteins. Trends Biochem Sci. 2002;27:527–33. doi: 10.1016/s0968-0004(02)02169-2. [DOI] [PubMed] [Google Scholar]
- 48.Radivojac P, Iakoucheva LM, Oldfield CJ, Obradovic Z, Uversky VN, Dunker AK. Intrinsic disorder and functional proteomics. Biophys J. 2007;92:1439–56. doi: 10.1529/biophysj.106.094045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hers I, Wherlock M, Homma Y, Yagisawa H, Tavaré JM. Identification of p122RhoGAP (deleted in liver cancer-1) Serine 322 as a substrate for protein kinase B and ribosomal S6 kinase in insulin-stimulated cells. J Biol Chem. 2006;281:4762–70. doi: 10.1074/jbc.M511008200. [DOI] [PubMed] [Google Scholar]
- 50.Hoffert JD, Pisitkun T, Wang G, Shen RF, Knepper MA. Quantitative phosphoproteomics of vasopressin-sensitive renal cells:regulation of aquaporin-2 phos-phorylation at two sites. Proc Natl Acad Sci USA. 2006;103:7159–64. doi: 10.1073/pnas.0600895103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Villén J, Beausoleil SA, Gerber SA, Gygi SP. Large-scale phosphorylation analysis of mouse liver. Proc Nat Acad Sci USA. 2007;104:1488–93. doi: 10.1073/pnas.0609836104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Songyang Z, Blechner S, Hoagland N, Hoekstra MF, Piwnica-Worms H, Cantley LC. Use of an oriented peptide library to determine the optimal substrates of protein kinases. Curr Biol. 1994;4:973–82. doi: 10.1016/s0960-9822(00)00221-9. [DOI] [PubMed] [Google Scholar]
- 53.Brunton VG, MacPherson IRJ, Frame MC. Cell adhesion receptors, tyrosine kinases and actin modulators: a complex three-way circuitry. Biochim Biophys Acta. 2004;1692:121–44. doi: 10.1016/j.bbamcr.2004.04.010. [DOI] [PubMed] [Google Scholar]
- 54.Burridge K, Wennerberg K. Rho and Rac take center stage. Cell. 2004;116:167–79. doi: 10.1016/s0092-8674(04)00003-0. [DOI] [PubMed] [Google Scholar]
- 55.Wong CM, Yam JW, Ching YP, Yau TO, Leung TH, Jin DY, Ng IO. Rho GTPase-activating protein deleted in liver cancer suppresses cell proliferation and invasion in hepatocellular carcinoma. Cancer Res. 2005;65:8861–8. doi: 10.1158/0008-5472.CAN-05-1318. [DOI] [PubMed] [Google Scholar]
- 56.Kim TY, Lee JW, Kim HP, Jong HS, Kim TY, Jung M, Bang YJ. DLC-1, a GTPase-activating protein for Rho, is associated with cell proliferation, morphology and migration in human hepatocellular carcinoma. Biochem Biophys Res Commun. 2007;355:72–7. doi: 10.1016/j.bbrc.2007.01.121. [DOI] [PubMed] [Google Scholar]
- 57.Zhou X, Thorgeirsson SS, Popescu NC. Restoration of DLC-1 gene expression induces apop-tosis and inhibits both cell growth and tumorigenicity in human hepatocellular carcinoma cells. Oncogene. 2004;23:1308–13. doi: 10.1038/sj.onc.1207246. [DOI] [PubMed] [Google Scholar]
- 58.Kawai K, Yamaga M, Iwamae Y, Kiyota M, Kamata H, Hirata H, Homma Y, Yagisawa H. A PLCδ 1-binding protein, p122RhoGAP, is localized in focal adhesions. Biochem Soc Trans. 2004;32:1107–9. doi: 10.1042/BST0321107. [DOI] [PubMed] [Google Scholar]
- 59.Yam JW, Ko FC, Chan CY, Jin DY, Ng IO. Interaction of deleted in liver cancer 1 with tensin2 in caveolae and implications in tumor suppression. Cancer Res. 2006;66:8367–72. doi: 10.1158/0008-5472.CAN-05-2850. [DOI] [PubMed] [Google Scholar]
- 60.Liao YC, Si L, DeVere White RW, Lo SH. The phos-photyrosine-independent interaction of DLC-1 and the SH2 domain of cten regulates focal adhesion localization and growth suppression activity of DLC-1. J Cell Biol. 2007;176:43–9. doi: 10.1083/jcb.200608015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lo SH. Tensin. Int J Biochem Cell Biol. 2004;36:31–4. doi: 10.1016/s1357-2725(03)00171-7. [DOI] [PubMed] [Google Scholar]
- 62.Cohen AW, Hnasko R, Schubert W, Lisanti MP. Role of caveolae and caveolins in health and disease. Physiol Rev. 2004;84:1341–79. doi: 10.1152/physrev.00046.2003. [DOI] [PubMed] [Google Scholar]
- 63.Yamaga M, Sekimata M, Fujii M, Kawai K, Kamata H, Hirata H, Homma Y, Yagisawa H. A PLC 1-binding protein, p122/RhoGAP, is localized in caveolin-enriched membrane domains and regulates caveolin internalization. Genes Cells. 2004;9:25–37. doi: 10.1111/j.1356-9597.2004.00698.x. [DOI] [PubMed] [Google Scholar]
- 64.Parton RG, Simons K. The multiple faces of caveo-lae. Nat Rev Mol Cell Biol. 2007;8:185–94. doi: 10.1038/nrm2122. [DOI] [PubMed] [Google Scholar]
- 65.Swaney JS, Patel HH, Yokoyama U, Head BP, Roth DM, Insel PA. Focal adhesions in (myo)fibroblasts scaffold adenylyl cyclase with phosphorylated cave-olin. J Biol Chem. 2006;281:17173–9. doi: 10.1074/jbc.M513097200. [DOI] [PubMed] [Google Scholar]
- 66.Rhee SG. Regulation of phosphoinositide-specific phospholipase C. Annu Rev Biochem. 2001;70:281–312. doi: 10.1146/annurev.biochem.70.1.281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Niggli V. Regulation of protein activities by phospho-inositide phosphates. Annu Rev Cell Dev Biol. 2005;21:57–79. doi: 10.1146/annurev.cellbio.21.021704.102317. [DOI] [PubMed] [Google Scholar]
- 68.Ng DC, Chan SF, Kok KH, Yam JW, Ching YP, Ng IO, Jin DY. Mitochondrial targeting of growth suppressor protein DLC2 through the START domain. FEBS Lett. 2006;580:191–8. doi: 10.1016/j.febslet.2005.11.073. [DOI] [PubMed] [Google Scholar]
- 69.Ferrell JE., Jr What do scaffold proteins really do? Sci STKE. 2000;2000:PE1. doi: 10.1126/stke.2000.52.pe1. [DOI] [PubMed] [Google Scholar]
- 70.Billuart P, Winter GG, Maresh A, Zhao X, Luo L. Regulating axon branch stability: the role of p190 RhoGAP in repressing a retraction signaling pathway. Cell. 2001;107:195–207. doi: 10.1016/s0092-8674(01)00522-0. [DOI] [PubMed] [Google Scholar]
- 71.Brodu V, Casanova J. 2006. The RhoGAP crossvein-less-c links trachealess and EGFR signaling to cell shape remodeling in Drosophila tracheal invagina-tion. Genes Dev. 2006;20:1817–28. doi: 10.1101/gad.375706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Simões S, Denholm B, Azevedo D, Sotillos S, Martin P, Skaer H, Hombria JC, Jacinto A. Compartmentalisation of Rho regulators directs cell invagination during tissue morphogenesis. Development. 2006;133:4257–67. doi: 10.1242/dev.02588. [DOI] [PubMed] [Google Scholar]
- 73.Soto MC, Qadota H, Kasuya K, Inoue M, Tsuboi D, Mello CC, Kaibuchi K. The GEX-2 and GEX-3 proteins are required for tissue morphogenesis and cell migrations in C. elegans. Genes Dev. 2002;16:620–32. doi: 10.1101/gad.955702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Hanahan D, Weinberg RA. The hallmarks of cancer. Cell. 2000;100:57–70. doi: 10.1016/s0092-8674(00)81683-9. [DOI] [PubMed] [Google Scholar]
- 75.Jaffe AB, Hall A. Rho GTPases in transformation and metastasis. Adv Cancer Res. 2002;84:57–80. doi: 10.1016/s0065-230x(02)84003-9. [DOI] [PubMed] [Google Scholar]
- 76.Ng IO, Liang ZD, Cao L, Lee TK. DLC-1 is deleted in primary hepatocellular carcinoma and exerts inhibitory effects on the proliferation of hepatoma cell lines with deleted DLC-1. Cancer Res. 2000;60:6581–4. [PubMed] [Google Scholar]
- 77.Plaumann M, Seitz S, Frege R, Estevez-Schwarz L, Scherneck S. Analysis of DLC-1 expression in human breast cancer. J Cancer Res Clin Oncol. 2003;129:349–54. doi: 10.1007/s00432-003-0440-z. [DOI] [PubMed] [Google Scholar]
- 78.Gatalica Z, Velagaleti G, Kuivaniemi H, Tromp G, Palazzo J, Graves KM, Guigneaux M, Wood T, Sinha M, Luxon B. Gene expression profile of an adenomyoepithelioma of the breast with a reciprocal translocation involving chromosomes 8 and 16. Cancer Genet Cytogenet. 2005;156:14–22. doi: 10.1016/j.cancergencyto.2004.04.024. [DOI] [PubMed] [Google Scholar]
- 79.Ullmannova V, Popescu NC. Expression profile of the tumor suppressor genes DLC-1 and DLC-2 in solid tumors. Int J Oncol. 2006;29:1127–32. [PubMed] [Google Scholar]
- 80.Zhang X, Feng J, Cheng Y, Yao Y, Ye X, Fu T, Cheng H. Characterization of differentially expressed genes in ovarian cancer by cDNA microarrays. Int J Gynecol Cancer. 2005;15:50–7. doi: 10.1111/j.1048-891X.2005.15007.x. [DOI] [PubMed] [Google Scholar]
- 81.Yuan BZ, Jefferson AM, Baldwin TK, Thorgeirsson SS, Popescu NC, Reynolds SH. DLC-1 operates as a tumor suppressor gene in human non-small cell lung carcinomas. Oncogene. 2004;23:1405–11. doi: 10.1038/sj.onc.1207291. [DOI] [PubMed] [Google Scholar]
- 82.Dammann R, Strunnikova M, Schagdarsurengin U, Rastetter M, Papritz M, Hattenhorst UE, Hofmann HS, Silber RE, Burdach S, Hansen G. CpG island methylation and expression of tumour-associated genes in lung carcinoma. Eur J Cancer. 2005;41:1223–36. doi: 10.1016/j.ejca.2005.02.020. [DOI] [PubMed] [Google Scholar]
- 83.Healy KD, Kim TY, Shutes AT, Bang YJ, Juliano RL, Der CJ. RhoGAP DLC-1 tumor suppression and aberrant Rho GTPase activation in lung cancer. Proc Am Assoc Cancer Res. 2006;47:970. [Google Scholar]
- 84.Nakamura T, Furukawa Y, Nakagawa H, Tsunoda T, Ohigashi H, Murata K, Ishikawa O, Ohgaki K, Kashimura N, Miyamoto M, Hirano S, Kondo S, Katoh H, Nakamura Y, Katagiri T. Genome-wide cDNA microarray analysis of gene expression profiles in pancreatic cancers using populations of tumor cells and normal ductal epithelial cells selected for purity by laser microdissection. Oncogene. 2004;23:2385–400. doi: 10.1038/sj.onc.1207392. [DOI] [PubMed] [Google Scholar]
- 85.Guan M, Zhou X, Soulitzis N, Spandidos DA, Popescu NC. Aberrant methylation and deacetyla-tion of deleted in liver cancer-1 gene in prostate cancer: potential clinical applications. Clin Cancer Res. 2006;12:1412–9. doi: 10.1158/1078-0432.CCR-05-1906. [DOI] [PubMed] [Google Scholar]
- 86.Peng D, Ren CP, Yi HM, Zhou L, Yang XY, Li H, Yao KT. Genetic and epigenetic alterations of DLC-1, a candidate tumor suppressor gene, in nasopharyngeal carcinoma. Acta Biochim Biophys Sin. 2006;38:349–55. doi: 10.1111/j.1745-7270.2006.00164.x. [DOI] [PubMed] [Google Scholar]
- 87.Hoffman PJ, Milliken DB, Gregg LC, Davis RR, Gregg JP. Molecular characterization of uterine fibroids and its implication for underlying mechanisms of pathogenesis. Fertil Steril. 2004;82:639–49. doi: 10.1016/j.fertnstert.2004.01.047. [DOI] [PubMed] [Google Scholar]
- 88.Struski S, Doco- Fenzy M, Cornillet-Lefebvre P. Compilation of published comparative genomic hybridization studies. Cancer Genet Cytogenet. 2002;135:63–90. doi: 10.1016/s0165-4608(01)00624-0. [DOI] [PubMed] [Google Scholar]
- 89.Yuan BZ, Zhou X, Durkin ME, Zimonjic DB, Gumundsdottir K, Eyfjord JE, Thorgeirsson SS, Popescu NC. DLC-1 gene inhibits human breast cancer cell growth and in vivo tumorigenicity. Oncogene. 2003;22:445–50. doi: 10.1038/sj.onc.1206064. [DOI] [PubMed] [Google Scholar]
- 90.Popescu NC. Genetic alterations in cancer as a result of breakage at fragile sites. Cancer Lett. 2003;92:1–17. doi: 10.1016/s0304-3835(02)00596-7. [DOI] [PubMed] [Google Scholar]
- 91.Popescu NC. Fragile sites and cancer genes on the short arm of chromosome 8. Lancet Oncol. 2004;5:77. doi: 10.1016/S1470-2045(04)01377-4. [DOI] [PubMed] [Google Scholar]
- 92.Kost-Alimova M, Imreh S. Modeling non-random deletions in cancer. Semin Cancer Biol. 2007;17:19–30. doi: 10.1016/j.semcancer.2006.11.003. [DOI] [PubMed] [Google Scholar]
- 93.Herman JG, Baylin SB. Gene silencing in cancer in association with promoter hypermethylation. N Eng J Med. 2003;349:2042–54. doi: 10.1056/NEJMra023075. [DOI] [PubMed] [Google Scholar]
- 94.Teramoto A, Tsukuda K, Yano M, Toyooka S, Dote H, Doihara H, Shimizu N. Less frequent promoter hypermethylation of DLC-1 gene in primary breast cancers. Oncol Rep. 2004;12:141–4. [PubMed] [Google Scholar]
- 95.Zhang Q, Ying J, Zhang K, Li H, Ng KM, Zhao Y, He Q, Yang X, Xin D, Liao SK, Tao Q, Jin J. Aberrant methylation of the 8p22 tumor suppressor gene DLC1 in renal cell carcinoma. Cancer Lett. 2007;249:220–6. doi: 10.1016/j.canlet.2006.08.019. [DOI] [PubMed] [Google Scholar]
- 96.Song YF, Xu R, Zhang XH, Chen BB, Chen Q, Chen YM, Xie Y. High-frequency promoter hypermethyla-tion of the deleted in liver cancer-1 gene in multiple myeloma. J Clin Pathol. 2006;59:947–51. doi: 10.1136/jcp.2005.031377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Shi H, Guo J, Duff DJ, Rahmatpanah F, Chitima-Matsiga R, Al-Kuhlani M, Taylor KH, Sjahputera O, Andreski M, Wooldridge JE, Caldwell CW. Discovery of novel epigenetic markers in non-Hodgkin's lymphoma. Carcinogenesis. 2007;28:60–70. doi: 10.1093/carcin/bgl092. [DOI] [PubMed] [Google Scholar]
- 98.Taylor KH, Pena-Hernandez KE, Davis JW, Arthur GL, Duff DJ, Shi H, Rahmatpanah FB, Sjahputera O, Caldwell CW. Large-scale CpG methylation analysis identifies novel candidate genes and reveals methylation hotspots in acute lymphoblastic leukemia. Cancer Res. 2007;67:2617–25. doi: 10.1158/0008-5472.CAN-06-3993. [DOI] [PubMed] [Google Scholar]
- 99.Marks P, Rifkind RA, Richon VM, Breslow R, Miller T, Kelly WK. Histone deacetylases and cancer:caus-es and therapies. Nat Rev Cancer. 2001;1:194–202. doi: 10.1038/35106079. [DOI] [PubMed] [Google Scholar]
- 100.Gore SD, Baylin S, Sugar E, Carraway H, Miller CB, Carducci M, Grever M, Galm O, Dauses T, Karp JE, Rudek MA, Zhao M, Smith BD, Manning J, Jiemjit A, Dover G, Mays A, Zwiebel J, Murgo A, Weng LJ, Herman JG. Combined DNA methyltrans-ferase and histone deacetylase inhibition in the treatment of myeloid neoplasms. Cancer Re. 2006;66:6361–9. doi: 10.1158/0008-5472.CAN-06-0080. [DOI] [PubMed] [Google Scholar]
- 101.Sparmann A, Van Lohuizen M. Polycomb silencers control cell fate, development and cancer. Nat Rev Cancer. 2006;6:846–56. doi: 10.1038/nrc1991. [DOI] [PubMed] [Google Scholar]
- 102.Bracken A, Dietrich N, Pasini D, Hansen KH, Helin K. Genome-wide mapping of Polycomb target genes unravels their roles in cell fate transitions. Genes Dev. 2006;20:1123–36. doi: 10.1101/gad.381706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Lee TI, Jenner RG, Boyer LA, Guenther MG, Levine SS, Kumar RM, Chevalier B, Johnstone SE, Cole MF, Isono K, Koseki H, Fuchikami T, Abe K, Murray HL, Zucker JP, Yuan B, Bell GW, Herbolsheimer E, Hannett NM, Sun K, Odom DT, Otte AP, Volkert TL, Bartel DP, Melton DA, Gifford DK, Jaenisch R, Young RA. Control of developmental regulators by Polycomb in human embryonic stem cells. Cell. 2006;125:301–13. doi: 10.1016/j.cell.2006.02.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Widschwendter M, Fiegl H, Egle D, Mueller-Holzner E, Spizzo G, Marth C, Weisenberger DJ, Campan M, Young J, Jacobs I, Laird PW. Epigenetic stem cell signature in cancer. Nat Genet. 2007;39:157–8. doi: 10.1038/ng1941. [DOI] [PubMed] [Google Scholar]
- 105.Park SW, Durkin ME, Thorgeirsson SS, Popescu NC. DNA variants of DLC-1, a candidate tumor suppressor gene in human hepatocellular carcinoma. Int J Oncol. 2003;23:133–7. [PubMed] [Google Scholar]
- 106.Pang JC, Chang Q, Chung YF, Teo JG, Poon WS, Zhou LF, Kong X, Ng HK. Epigenetic inactivation of DLC-1 in supratentorial primitive neuroectodermal tumor. Hum Pathol. 2005;36:36–43. doi: 10.1016/j.humpath.2004.09.021. [DOI] [PubMed] [Google Scholar]
- 107.Hewitt C, Wilson P, McGlinn E, MacFarlane G, Papageorgiou A, Woodwards RT, Sloan P, Gollin SM, Paterson I, Parkinson KK, Read AP, Thakker N. DLC1 is unlikely to be a primary target for deletions on chromosome arm 8p22 in head and neck squamous cell carcinoma. Cancer Lett. 2004;209:207–13. doi: 10.1016/j.canlet.2003.12.018. [DOI] [PubMed] [Google Scholar]
- 108.Zheng SL, Mychaleckyj JC, Hawkins GA, Isaacs SD, Wiley KE, Turner A, Chang BL, Von Kap-Herr C, Carpten JD, Pettenati M, Bleecker ER, Walsh PC, Trent JM, Meyers DA, Isaacs WB, Xu J. Evaluation of DLC1 as a prostate cancer susceptibility gene: mutation screen and association study. Mutat Res. 2003;528:45–53. doi: 10.1016/s0027-5107(03)00081-2. [DOI] [PubMed] [Google Scholar]
- 109.Chang BL, Liu W, Sun J, Dimitrov L, Li T, Turner AR, Zheng SL, Isaacs WB, Xu J. Integration of somatic deletion analysis of prostate cancers and germline linkage analysis of prostate cancer families reveals two small consensus regions for prostate cancer genes at 8p. Cancer Res. 2007;67:4098–103. doi: 10.1158/0008-5472.CAN-06-4570. [DOI] [PubMed] [Google Scholar]
- 110.Tang K, Oeth P, Kammerer S, Denissenko MF, Ekblom J, Jurinke C, Van Den Boom D, Braun A, Cantor CR. Mining disease susceptibility genes through SNP analyses and expression profiling using MALDI-TOF mass spectrometry. J Proteome Res. 2004;3:218–27. doi: 10.1021/pr034080s. [DOI] [PubMed] [Google Scholar]
- 111.Van Den Boom D, Beaulieu M, Oeth P, Roth R, Honisch C, Nelson MR, Jurinke C, Cantor C. MALDI-TOF MS: a platform technology for genetic discovery. Int J Mass Spectrom. 2004;238:173–88. [Google Scholar]
- 112.Syed V, Mukherjee K, Lyons-Weiler J, Lau KM, Mashima T, Tsuruo T, Ho SM. Identification of ATF-3, caveolin-1, DLC-1, and NM23-H2 as putative antitumori-genic, progesterone-regulated genes for ovarian cancer cells by gene profiling. Oncogene. 2005;24:1774–87. doi: 10.1038/sj.onc.1207991. [DOI] [PubMed] [Google Scholar]
- 113.Qin LX. Chromosomal aberrations related to metastasis of human solid tumors. World J Gastroenterol. 2002;8:769–76. doi: 10.3748/wjg.v8.i5.769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Song LJ, Ye SL, Wang KF, Weng YQ, Liang CM, Sun RX, Zhao Y, Liu YK, Tang ZY. Relationship between DLC-1 expressions and metastasis in hepa-tocellular carcinoma. Zhonghua Gan Zang Bing Za Zhi. 2005;13:428–31. [PubMed] [Google Scholar]
- 115.Goodison S, Yuan J, Sloan D, Kim R, Li C, Popescu NC, Urquidi V. The RhoGAP protein DLC-1 functions as a metastasis suppressor in breast cancer cells. Cancer Res. 2005;65:6042–53. doi: 10.1158/0008-5472.CAN-04-3043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Kang Y, Siegel PM, Shu W, Drobnjak M, Kakonen SM, Cordón-Cardo C, Guise TA, Massagué J. A multigenic program mediating breast cancer metastasis to bone. Cancer Cell. 2003;3:537–49. doi: 10.1016/s1535-6108(03)00132-6. [DOI] [PubMed] [Google Scholar]
- 117.Folgueira MA, Carraro DM, Brentani H, Patrão DF, Barbosa EM, Netto MM, Caldeira JR, Katayama ML, Soares FA, Oliveira CT, Reis LF, Kaiano JH, Camargo LP, Vêncio RZ, Snitcovsky IM, Makdissi FB, E Silva PJ, Góes JC, Brentani MM. Gene expression profile associated with response to dox-orubicin-based therapy in breast cancer. Clin Cancer Res. 2005;11:7434–43. doi: 10.1158/1078-0432.CCR-04-0548. [DOI] [PubMed] [Google Scholar]
- 118.Czernicki T, Zegarska J, Paczek L, Cukrowska B, Grajkowska W, Zajaczkowska A, Brudzewski K, Ulaczyk J, Marchel A. Gene expression profile as a prognostic factor in high-grade gliomas. Int J Oncol. 2007;30:55–64. [PubMed] [Google Scholar]
- 119.Mas VR, Fisher RA, Archer KJ, Yanek KC, Williams B, Dumur CI, Maluf DG. Genes associated with progression and recurrence of hepatocellular carcinoma in hepatitis C patients waiting and undergoing liver transplantation: preliminary results. Transplantation. 2007;83:973–81. doi: 10.1097/01.tp.0000258643.05294.0b. [DOI] [PubMed] [Google Scholar]
- 120.Basso K, Liso A, Tiacci E, Benedetti R, Pulsoni A, Foa R, Di Raimondo F, Ambrosetti A, Califano A, Klein U, Dalla Favera R, Falini B. Gene expression profiling of hairy cell leukemia reveals a phenotype related to memory B cells with altered expression of chemokine and adhesion receptors. J Exp Med. 2004;199:59–68. doi: 10.1084/jem.20031175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Ross ME, Mahfouz R, Onciu M, Liu HC, Zhou X, Song G, Shurtleff SA, Pounds S, Cheng C, Ma J, Ribeiro RC, Rubnitz JE, Girtman K, Williams WK, Raimondi SC, Liang DC, Shih LY, Pui CH, Downing JR. Gene expression profiling of pediatric acute myel-ogenous leukaemia. Blood. 2004;104:3679–87. doi: 10.1182/blood-2004-03-1154. [DOI] [PubMed] [Google Scholar]
- 122.Herzog A, Kindermann B, Döring F, Daniel H, Wenzel U. Pleiotropic molecular effects of the pro-apoptotic dietary constituent flavone in human colon cancer cells identified by protein and mRNA expression profiling. Proteomics. 2004;4:2455–64. doi: 10.1002/pmic.200300754. [DOI] [PubMed] [Google Scholar]
- 123.Ullmannova V, Popescu NC. Inhibition of cell proliferation, induction of apoptosis, reactivation of DLC1, and modulation of other gene expression by dietary flavone in breast cancer cell lines. Cancer Detect Prev. 2007;31:110–8. doi: 10.1016/j.cdp.2007.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Zirn B, Samans B, Spangenberg C, Graf N, Eilers M, Gessler M. All-trans retinoic acid treatment of Wilms tumor cells reverses expression of genes associated with high risk and relapse in vivo. Oncogene. 2005;24:5246–51. doi: 10.1038/sj.onc.1208725. [DOI] [PubMed] [Google Scholar]
- 125.Hansen LA, Sigman CC, Andreola F, Ross SA, Kelloff GJ, De Luca LM. Retinoids in chemopreven-tion and differentiation therapy. Carcinogenesis. 2000;21:1271–9. [PubMed] [Google Scholar]
- 126.Grommes C, Landreth GE, Heneka MT. Antineoplastic effects of peroxisome proliferator-activated receptor gamma agonists. Lancet Oncol. 2004;5:419–29. doi: 10.1016/S1470-2045(04)01509-8. [DOI] [PubMed] [Google Scholar]
- 127.Bren-Mattison Y, Van Putten V, Chan D, Winn R, Geraci MW, Nemenoff RA. Peroxisome proliferator-activated receptor- (PPAR) inhibits tumorigenesis by reversing the undifferentiated phenotype of metastatic non-small-cell lung cancer cells (NSCLC) Oncogene. 2005;24:1412–22. doi: 10.1038/sj.onc.1208333. [DOI] [PubMed] [Google Scholar]
- 128.Asada K, Asada R, Yoshiji H, Fukui H, Floyd RA, Kotake Y. DNA cytosine methylation profile in various cancer-related genes is altered in cultured rat hepa-tocyte cell lines as compared with primary hepato-cytes. Oncol Rep. 2006;15:1241–8. [PubMed] [Google Scholar]
- 129.Ushijima T, Yamamoto M, Suzui M, Kuramoto T, Yoshida Y, Nomoto T, Tatematsu M, Sugimura T, Nagao M. Chromosomal mapping of genes controlling development, histological grade, depth of invasion, and size of rat stomach carcinomas. Cancer Res. 2000;60:1092–6. [PubMed] [Google Scholar]
- 130.Edelson MI, Lau CC, Colitti CV, Welch WR, Bell DA, Berkowitz RS, Mok SC. A one centimorgan deletion unit on chromosome Xq12 is commonly lost in borderline and invasive epithelial ovarian tumors. Oncogene. 1998;16:197–202. doi: 10.1038/sj.onc.1201479. [DOI] [PubMed] [Google Scholar]
- 131.Spatz A, Borg C, Feunteun J. X-chromosome genetics and human cancer. Nat Rev Cancer. 2004;4:617–29. doi: 10.1038/nrc1413. [DOI] [PubMed] [Google Scholar]
- 132.Sjöblom T, Jones S, Wood LD, Parsons DW, Lin J, Barber TD, Mandelker D, Leary RJ, Ptak J, Silliman N, Szabo S, Buckhaults P, Farrell C, Meeh P, Markowitz SD, Willis J, Dawson D, Willson JK, Gazdar AF, Hartigan J, Wu L, Liu C, Parmigiani G, Park BH, Bachman KE, Papadopoulos N, Vogelstein B, Kinzler KW, Velculescu VE. The consensus coding sequences of human breast and col-orectal cancers. Science. 2006;314:268–74. doi: 10.1126/science.1133427. [DOI] [PubMed] [Google Scholar]


