Abstract
Conventional methods for experimental manipulation of microalgae have employed large volumes of culture (20 ml to 5 L), so that the culture can be subsampled throughout the experiment1–7. Subsampling of large volumes can be problematic for several reasons: 1) it causes variation in the total volume and the surface area:volume ratio of the culture during the experiment; 2) pseudo-replication (i.e., replicate samples from the same treatment flask8) is often employed rather than true replicates (i.e., sampling from replicate treatments); 3) the duration of the experiment is limited by the total volume; and 4) axenic cultures or the usual bacterial microbiota are difficult to maintain during long-term experiments as contamination commonly occurs during subsampling.
The use of microtiter plates enables 1 ml culture volumes to be used for each replicate, with up to 48 separate treatments within a 12.65 x 8.5 x 2.2 cm plate, thereby decreasing the experimental volume and allowing for extensive replication without subsampling any treatment. Additionally, this technique can be modified to fit a variety of experimental formats including: bacterial-algal co-cultures, algal physiology tests, and toxin screening9–11. Individual wells with an alga, bacterium and/or co-cultures can be sampled for numerous laboratory procedures including, but not limited to: WATER-Pulse-Amplitude-Modulated (WATER-PAM) fluorometry, microscopy, bacterial colony forming unit (cfu) counts and flow cytometry. The combination of the microtiter plate format and WATER-PAM fluorometry allows for multiple rapid measurements of photochemical yield and other photochemical parameters with low variability between samples, high reproducibility and avoids the many pitfalls of subsampling a carboy or conical flask over the course of an experiment.
Keywords: Environmental Sciences, Issue 97, Phytoplankton, microalgae, co-culture, bacteria, WATER-Pulse-Amplitude-Modulated (WATER-PAM) fluorometry, photosynthetic yield, chlorophyll, microtiter plate assay, high-throughput bioassay
Introduction
Phytoplankton physiology has traditionally been studied in meso-scale experiments ranging from 20 ml in conical flasks to 5 L in carboys1–7. This experimental scale requires subsampling for experimental monitoring, as sacrificing replicate samples for each time point creates an unmanageable experimental setup.
The ability to increase the number of independent experiments while using the same diurnal incubator space by miniaturizing the experimental volume for algal physiology experiments will reduce or eliminate the limitations of subsampling and pseudo-replication from large volumes. A microtiter plate format has been developed for algal bioassays using a 1 ml culture volume for experimentally manipulating algae in variable conditions. This small experimental volume allows for the number of replicates to be increased, increases experimental reproducibility due to a decreased variability between replicate samples and experiments, and allows true replication while maintaining experimental controls (i.e., axenic algal cultures) for 140 days (Figure 2)12.
This microtiter plate format is easily adapted for a variety of experimental questions, such as: does a bacterium have a symbiotic, neutral or pathogenic interaction with its algal host? Is the addition of a compound stimulating or toxic to an alga? These and other questions can be addressed in a rapid high-throughput manner using this new format9–11.
A 48-well microtiter culture plate allows each 1 ml well to be an independent experimental setup that is sampled at a single time-point. Various parameters can be sampled from this 1 ml volume including, but not limited to: chlorophyll fluorescence and photochemical parameters using WATER-Pulse-Amplitude-Modulated (WATER-PAM) fluorometry (see Materials and Equipment table)13. WATER-PAM fluorometry is a rapid and non-invasive technique that can be used to monitor experiments performed with algae13. It allows measurement of photosynthetic efficiency and PSII health from a small culture volume (150 - 300 μl of culture diluted in medium to a 2 - 4 ml volume for WATER-PAM)14,15. In addition to WATER-PAM fluorometry, this setup can be used to measure a variety of other parameters including, but not limited to: microscopy to visualize the bacteria attached to algal cells and changes in the algal cell morphology; bacterial colony forming unit (cfu) counts; and flow cytometry for algal cell counts and identifying subpopulations.
Protocol
1. Calculations for Experimental Setup
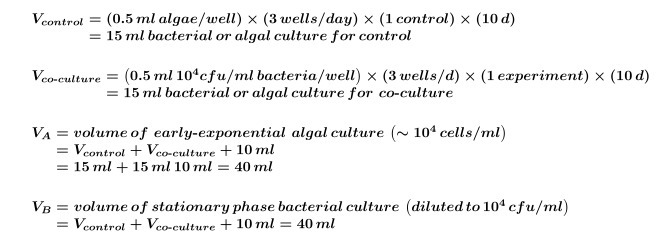
Calculate the volume of algal and/or bacterial cultures needed for controls that will be required for the entire experiment by using Equation 1:
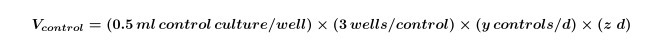 Where y equals the number of controls needed per day and z equals the number of days.
Where y equals the number of controls needed per day and z equals the number of days.Calculate the volume of algal and/or bacterial cultures that are needed for co-cultures for the experiment by using Equation 2:
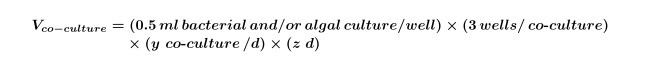 NOTE: It is possible to replace ‘co-culture’ experiments with any compound screen; just adjust the final compound, solvent and algal concentrations to fit the experimental design.
NOTE: It is possible to replace ‘co-culture’ experiments with any compound screen; just adjust the final compound, solvent and algal concentrations to fit the experimental design.Use Equations 1 and 2 to calculate the final volume of early-exponential algal culture (commonly 5 days for ~104 cells/ml but this should be determined by performing an algal growth curve) required for the experiment (this volume is needed for step 2.3) using Equation 3:

Use Equations 1 and 2 to calculate the final volume of 104 cfu/ml bacterial culture (or desired inoculation concentration) required for the experiment (this volume is needed for step 4.2) using Equation 4:
 NOTE: Use the additional 10 ml in steps 1.3 and 1.4 to account for pipetting error and any tests that are performed (e.g., WATER-PAM, flow cytometry, microscopy, etc.) on 0 d. Increase this volume if needed to fit the experimental design. NOTE: For an example calculation of an 8-day experiment see Section 10. See supplemental table for media recipes.
NOTE: Use the additional 10 ml in steps 1.3 and 1.4 to account for pipetting error and any tests that are performed (e.g., WATER-PAM, flow cytometry, microscopy, etc.) on 0 d. Increase this volume if needed to fit the experimental design. NOTE: For an example calculation of an 8-day experiment see Section 10. See supplemental table for media recipes.
2. Growing Algal Cells for Experimental Setup
Isolate or obtain an actively growing axenic algal culture.
Aseptically transfer 10% of the final volume of the algal culture into sterile algal medium (e.g., L1 or similar marine algal medium, see Materials and Equipment table), ensuring a 1:9 dilution of algal culture in fresh medium. Grow the diluted alga in a diurnal incubator using previously determined growth conditions for that strain (e.g., 18 °C with a 16:8 hr light:dark cycle is commonly used).
When the culture has reached early-exponential phase, re-culture the alga in the same sterile algal medium (e.g., L1 or similar marine algal medium), ensure the final concentration is a 1:9 dilution and that the final volume of algae is equal to the VA calculated (step 1.3). NOTE: It is important to ensure these cultures are axenic. Test all algal media and algal stock bottles for contamination by plating a 20 μl aliquot onto a general marine bacterial medium (e.g., Marine Broth 2216 supplemented with 1.5% agar or similar).
Grow algal culture to early-exponential phase (~104 cells/ml). NOTE: Every algal strain has a unique growth curve depending on culturing conditions. Early-exponential phase (~104 cells/ml) can be determined by performing an algal growth curve based on cell density (i.e. using flow cytometry or microscopy).
3. Preparing Bacterial Cells for Inoculation
Pre-determine the bacterial concentration (cfu/ml) and optical density (OD) of the bacterium at stationary phase by doing a growth curve in the chosen bacterial medium and growth conditions (e.g., Marine Broth 2216 at 25 °C and 160 rpm, or similar). Use this information to grow the cells (step 3.3) and later to dilute the cells correctly in step 3.7.
Aseptically transfer an isolated and freshly grown bacterial colony from a 1.5% agar plate (e.g., Marine Broth 2216 supplemented with 1.5% agar or similar marine bacterial solid medium) into 5 ml of bacterial liquid medium (e.g., Marine Broth 2216 or similar marine bacterial medium).
Grow the bacterium to stationary phase on a rolling drum or shaker (~12 - 36 hr, depending on the bacterium). Plan the experiment so that the bacterium reaches stationary phase (~108 - 109 cfu/ml) at the same time that the algal cells have reached the early-exponential phase (~104 cells/ml).
Using a pipette, wash down any biofilm attached to the test tube into the medium. Pipette 1 ml of well-mixed bacterial culture into a sterile 1.5 ml microtube. Centrifuge for 1 min at 14,000 x g.
Remove and dispose of the supernatant (bacterial media) without disrupting the pellet (bacterial cells). Add 1 ml of sterile algal media (e.g., L1 or similar) to the microtube (with pellet). Vortex microtube to resuspend pellet in algal media.
Second wash: repeat step 3.5. NOTE: It is critical to wash the bacterial cells with algal media in order to thoroughly remove all of the bacterial media, cell detritus, excreted proteins and small molecules from the cells prior to inoculating the algae with them as this could change the nutrient composition of the algal media or introduce bioactive molecules to the screen.
Serially dilute the washed bacterial cells in algal media, to a final concentration that is 100-fold more concentrated than the desired final bacterial concentration (cfu/ml). Save the microtube containing cells that have been washed and diluted in algal media for step 4.2. NOTE: Plan the experiment based on having a 1:1 ratio of algae to bacteria on 0 d. To do this the desired initial bacterial concentration for the experiment is 104 cfu/ml, so in step 3.7 the initial cells should be serially diluted to 106 cfu/ml.
4. Preparing Bacteria for Experimental Setup
Prepare 4 sterile autoclaved glass conical flasks and label them: a) ‘algal control flask’, b) ‘diluted bacterial stock flask’, c) ‘bacterial control flask’, and d) ‘co-culture flask’.
Dilute the bacterial suspension from step 3.7 1:99 with sterile algal media to a final volume = VB (step 1.4). Make the dilution in the flask labeled diluted bacterial stock (step 4.1). NOTE: In the previous example (step 4.2), this 1:99 dilution gives a final concentration of 104 cfu/ml. Swirl flask to mix cells.
Pipette Vcontrol (step 1.1) from the diluted bacterial stock and put it in the bacterial control flask.
Pipette Vcontrol of sterile algal medium into the bacterial control flask (this is a 1:1 dilution). Swirl flask to mix cells and set aside for step 7.3.
Pipette Vco-culture from the diluted bacterial stock flask to the co-culture flask, set flask aside for step 6.1. NOTE: When doing multiple co-cultures on the same alga at once (i.e., a co-culture of two different bacterial isolates with one control) it is necessary to re-calculate the Vco-culture for each strain individually and then repeat step 4.5 for each co-culture separately. It is also necessary to increase the VA calculated in step 1.3 to include both co-cultures shown in Equation 5:
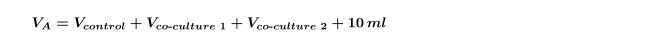
5. Preparing Algae for Experimental Setup
Gently mix the early-exponential algal culture (from step 2.4) with a wide-mouth pipette tip until cells appear well mixed.
Pipette Vcontrol from the algal stock bottle to the algal control flask. Gently pipette using a 10 ml pipette. Then return the algal stock bottle in the diurnal incubator.
Pipette Vcontrol of sterile algal medium to the algal control flask (this is a 1:1 dilution). Swirl to mix flask and place in the diurnal incubator, until needed for step 7.4.
6. Preparing Experimental Co-culture
Gently pipette Vco-culture from the algal stock flask into the bacterial co-culture flask from step 4.5. Return co-culture flask to the diurnal incubator until needed for step 7.5. NOTE: The concentration of bacteria in the bacterial control should equal the bacterial concentration in the experimental co-culture. Similarly, the concentration of algae in the algal control should equal the concentration of algae in the experimental co-culture. By having the same initial bacterial and algal concentrations, the population density can be compared throughout the experiment.
7. Setting up Microtiter Plates
Divide a sterile 48-well microtiter plate as per Figure 1. Label above outer wells containing either sterile diluent or non-photosynthetic samples. NOTE: Randomization of the plate wells can be done for samples taken on the same day. For example, quadrant 1 will be sampled at 1 d in the experiment; the wells that should be randomized are B2 - 4 and C2 - 4. Leave the perimeter of the plate filled with sterile solution as shown in Figure 1.
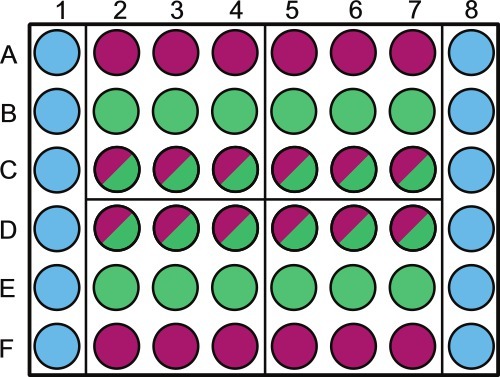 Figure 1. Schematic representation of sample placement in a 48-well microtiter plate. Wells are to be filled as follows: columns 1 and 6, wells A through F (
Figure 1. Schematic representation of sample placement in a 48-well microtiter plate. Wells are to be filled as follows: columns 1 and 6, wells A through F (![]() ) are filled with 1 ml 1x PBS (or other sterile solution/media). Rows A and F, wells 2 - 8 (
) are filled with 1 ml 1x PBS (or other sterile solution/media). Rows A and F, wells 2 - 8 (![]() ) are filled with 1 ml bacterial control; rows B and E wells 2 - 8 (
) are filled with 1 ml bacterial control; rows B and E wells 2 - 8 (![]() ) are filled with 1 ml algal control; rows C and D, wells 2 - 8 (
) are filled with 1 ml algal control; rows C and D, wells 2 - 8 (![]() ) are filled with 1 ml co-culture. The plate is divided into 4 quadrants (A2, A5, D2, and D5) these quadrants are each specific sampling days 1 - 4, this should be randomized throughout plates and labeled accordingly. Within each day we advise randomizing the algal control (
) are filled with 1 ml co-culture. The plate is divided into 4 quadrants (A2, A5, D2, and D5) these quadrants are each specific sampling days 1 - 4, this should be randomized throughout plates and labeled accordingly. Within each day we advise randomizing the algal control (![]() ) and co-culture (
) and co-culture (![]() ) wells using a random number generator. Label the lid over the 1x PBS and/or bacterial control wells to prevent shading of algal cultures.
) wells using a random number generator. Label the lid over the 1x PBS and/or bacterial control wells to prevent shading of algal cultures.
Pipette 1 ml 1x phosphate buffer solution (PBS, pH 7.4) or other sterile solution in the appropriate wells as indicated in Figure 1. Perform this slowly and with care. PBS will change the ionic strength of the algal and/or co-culture medium if it splatters in other wells so handle carefully.
Pipette 1 ml of bacterial control culture into wells labeled bacterial control (Figure 1). Swirl the bacterial flask before pipetting each plate to avoid bacterial settling.
Pipette 1 ml of algal control culture into wells labeled algal control using a wide mouth pipette tip (Figure 1). Swirl the algal flask as regularly as the bacterial flask. If the alga tends to sink or float, swirl as regularly as is needed to maintain a visually uniform culture.
Using a wide mouth pipette tip, pipette 1 ml of co-culture in the appropriate wells (Figure 1). Swirling the co-culture flask as regularly as the algal flask.
Seal each plate with parafilm and place in a diurnal incubator at the desired temperature and diurnal light cycle (18 °C with a 16:8 hr light:dark cycle is commonly used). Leave the plates in the diurnal incubator until ready to take PAM readings (Section 8). Ensure all plates are oriented in the same direction to allow for consistent light exposure.
Take a 20 μl aliquot from the PBS, algal media, algal stock and algal control and drop plate onto an appropriate non-selective medium (e.g., Marine Broth 2216 supplemented with 1.5% agar) and incubate plates at 18 - 25 °C for 72 hr to test for contamination. If growth appears in any of the solutions that should not contain bacteria, then these data should not be used.
Take PAM fluorometry readings using the remaining sample from the algal control and co-culture flasks for the experimental 0 d PAM fluorometry reading (see step 8.1). Any other 0 d measurements should also be performed with the remaining algal control, bacterial control and co-culture samples.
8. Taking PAM Fluorometry Readings from Stock Samples
Zero the WATER-PAM with sterile algal media in a clean cuvette before taking readings8.
Pipette 300 µl from algal control or co-culture flasks into a clean cuvette containing 2.7 ml of the same algal medium used in the experiment. Mix the sample and diluent gently with a wide mouth pipette tip.
Wipe-off all fingerprints from outside of cuvette with a tissue before placing the cuvette in the WATER-PAM.
Place cuvette into WATER-PAM. Cover the sample with the cap and allow it to dark-adapt for 3 min. Dark adaptation times vary depending on the species of algae and must be determined for the specific alga used in the experiment. Avoid lengthy dark adaptation times (>20 min) by taking WATER-PAM readings during the middle of the dark cycle of the algal diurnal incubators16,17.
After dark adaption, hit F0 button. If fluorescence readings are above 3,900, dilute sample 1:1 in algal medium. Dark-adapt for an additional 3 min and take a new reading. If the F0 or Fm readings are still above 3,900, continue to dilute the sample 1:1 in algal medium until the F0 and Fm readings are below 3,900. NOTE: Make sure to account for these dilutions when recording final fluorescence: for instance if the algal sample is diluted 1:9 during the initial transfer from the well to the dilution tube, then the algal fluorescence reading of 500 should be multiplied by the inverse of the dilution factor (in this case 10) and the actual fluorescence of the tube is then 5,000.
After setting F0, take a saturating pulse (SAT-Pulse) reading every 90 sec by hitting the SAT button to take Fm readings. The time interval between readings may be adjusted depending on algal strain. Discard sample.
Repeat steps 8.1 - 8.6 for the remaining samples.
9. Taking PAM Fluorometry Readings from Microtiter Plates
Label 6 sterile sample tubes of >3 ml volume for wells B2 - 4 for the algal control and wells C2 - 4 for the bacterial co-culture (see plate layout in Figure 1).
Aliquot 2.7 ml of sterile algal medium into each tube.
Place tubes in diurnal incubator and allow them to acclimate to the temperature the alga was grown at for 30 min.
Before removing the microtiter plate from the incubator ensure that light penetration into the dark acclimated wells is limited by covering the plate with aluminum foil (or similar), only remove the foil while actively transferring culture from the wells to the dilution tubes (steps 9.5 - 9.7).
Aseptically mix the first microtiter plate well (well B2) with a wide mouth pipette tip by slowly pipetting up and down.
Obtain the dilution tubes from the incubator and aseptically transfer 300 µl from well B2 (Figure 1) to its corresponding sample tube with a wide mouth pipette tip (this is a 1:9 dilution of the sample in algal medium).
Repeat steps 9.5 - 9.6 for the remaining wells (B3,4 and C2 - 4).
Cover sample tubes with aluminum foil and return them to the diurnal incubator until ready for WATER-PAM.
Perform WATER-PAM readings as described in steps 8.1 - 8.7. Seal the microtiter plate with parafilm before returning it to the incubator.
Repeat readings at planned time intervals for the duration of the experiment. The frequency and length of sampling should be planned at the beginning of the experiment.
10. Sample Experiment
The sample experiment is a 10 day co-culture of a bacterium (Phaeobacter gallaeciensis BS107) and a microalga (Emiliania huxleyi strain (CCMP3266)). It includes an algal control, bacterial control, and a bacterial-algal experimental co-culture.
Calculate volumes of bacterial and algal stocks required (steps 1.1 - 1.4)
Prepare the algal stock, a VA of 40 ml, by inoculating 4 ml of alga in 36 ml medium and incubated until early-exponential growth is achieved with a cell density of ~104 cells/ml (steps 2.1 - 2.4).
Prepare the bacterial stock flask by diluting 400 μl of the 106 cfu/ml bacteria obtained in Section 3 to a VB of 40 ml with algal media (final concentration of bacteria will be 104 cfu/ml).
Transfer half (20 ml) of the bacterial stock to the bacterial control flask and add 20 ml of algal media to make the bacterial control. Transfer half (20 ml) the algal stock to the algal control flask and add 20 ml of algal media to make up the algal control (Section 5). Transfer the remaining 20 ml of bacterial stock to the co-culture flask and mix with the remaining 20 ml of algal stock to establish the co-culture (Section 6).
Pipette the 1x PBS (pH 7.4), controls, and co-culture in two pre-labeled microtiter plates (Figure 1), 1 ml per well. Use 3 ml of the remaining controls and co-cultures to make 0 d measurements (cfu counts, WATER-PAM (Section 8), microscopic observation of algal cell morphology, and algal cell fixation for flow cytometry).
Take WATER-PAM readings for the algal control and co-culture once a day for 10 d, and always at the same time as the 0 d measurements were taken (Section 8).
11. Other Parameters of Interest
Determine bacterial cfu concentration: perform a serial dilution of the bacterial control and co-culture in sterile 1x PBS (or similar), then drop plate onto Marine Broth 2216 supplemented with 1.5% agar (or similar) and observe the number of cfu that grow to determine the bacterial cfu/ml for each well18.
To evaluate the algal cell concentration: fix algal control and co-culture with a 0.15% final concentration of glutaraldehyde. Incubate for 10 min in the dark, then flash freeze in liquid nitrogen, and store at -80 °C. Process all samples on a flow cytometer (FACS Calibur or similar) to count algal cells.
Observe algal cell morphology: algal cultures and bacterial-algal co-culture using microscopy (i.e., light microscopy, epifluorescence, or similar).
Representative Results
WATER-PAM fluorometry readings.
WATER-Pulse-Amplitude-Modulated (PAM) fluorometry is a quick and efficient method to determine the fluorescence (a proxy for chlorophyll content) and photosynthetic yield (PSII health) of algal cultures. The PAM WinControl software generates a spreadsheet of raw data values for (the following are the basic parameters for dark adapted algal samples):
F0 = fluorescence of dark-adapted cells
Fm = maximum fluorescence after saturating light-emitting diodes (LED) pulse
Fv/Fm = (Fm-F0)/Fm = potential quantum yield of a dark adapted sample19
PAM fluorometry has many uses and can give a lot of information about the photosystem and health of the algae. In addition, there are other parameters that are important when testing light adapted samples. For further reading these are discussed in detail in these reviews13,16,20–23. These data can be transferred to a spreadsheet or graphing software to generate graphs of the initial algal fluorescence (F0), the maximum algal fluorescence (Fm), and the potential quantum yield (Fv/Fm; Figures 2 and 3). The graphs in Figure 3 depict how the algal fluorescence and Fv/Fm are influenced by the bacterium by comparing the alga grown alone (control) to it being grown with the bacterium in co-culture throughout the 10-day co-culture experiment (Figure 3C). In this example, a two-sample t-test was used to compare the parameters statistically between the two treatments on each day. Similarly, for experiments in which more than two treatments are present, an ANOVA could be used. The Fm reading (Figure 3B) is taken directly after the saturating LED pulse, which means the primary PSII electron acceptor QA is fully reduced and cannot accept any more electrons from the PSII reaction centre P680, and as such, all reaction centers are 'closed'17. This hinders the photochemical use of light energy, bringing fluorescence emission to a maximum. This then gives the maximum fluorescence (Fm) reading. Figure 3C illustrates a dramatic decline in PSII health between 5 d and 10 d when co-cultured with the bacterium compared to growing alone (control). The standard error bars are derived from triplicate microtiter wells, which are independent experiments from the same parental bacterial and algal cultures. The consistently small standard error confirms the robustness and reproducibility of the microtiter plate format for algal bioassays as it allows independent experiments to be used as replicates and eliminates the need to subsample the experimental unit over the duration of the experiment.
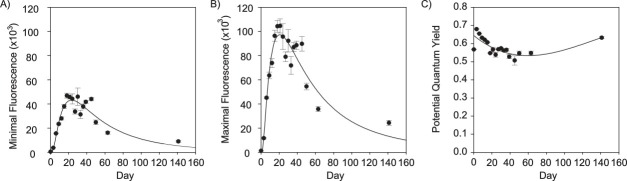 Figure 2. Representative WATER-PAM fluorometry graphs of a 140 d growth curve of axenic Emiliania huxleyi (CCMP3266). (A) Readings for the initial algal fluorescence (F0), (B) maximum algal fluorescence (Fm), and (C) potential quantum yield (Fv/Fm) for E. huxleyi are shown as black circles. The line of best fit for F0 and Fm was normal log 3 parameter (SigmaPlot) R2 = 0.94, 0.95 respectively. Potential quantum yield (Fv/Fm) is a dimensionless expression of photosynthetic health, which is calculated as (Fm – F0) / Fm. The line of best fit for the Fv/Fm curve was a 3 factor polynomial R2 = 0.6. Error bars represent the standard error between triplicate wells. Please click here to view a larger version of this figure.
Figure 2. Representative WATER-PAM fluorometry graphs of a 140 d growth curve of axenic Emiliania huxleyi (CCMP3266). (A) Readings for the initial algal fluorescence (F0), (B) maximum algal fluorescence (Fm), and (C) potential quantum yield (Fv/Fm) for E. huxleyi are shown as black circles. The line of best fit for F0 and Fm was normal log 3 parameter (SigmaPlot) R2 = 0.94, 0.95 respectively. Potential quantum yield (Fv/Fm) is a dimensionless expression of photosynthetic health, which is calculated as (Fm – F0) / Fm. The line of best fit for the Fv/Fm curve was a 3 factor polynomial R2 = 0.6. Error bars represent the standard error between triplicate wells. Please click here to view a larger version of this figure.
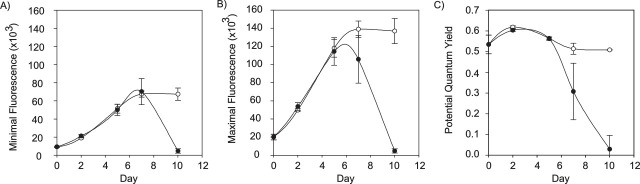 Figure 3. Representative WATER-PAM fluorometry graphs of a 10 d co-culturing experiment of Emiliania huxleyi (CCMP3266) with Phaeobacter gallaeciensis BS107. (A) The initial algal fluorescence (F0), (B) maximum algal fluorescence (Fm), and (C) potential quantum yield (Fv/Fm) are graphed for control algal (open circles) and alga co-cultured with a bacterium (black circles). Potential quantum yield (Fv/Fm) is a dimensionless expression of photosynthetic health, which is calculated as (Fm - F0) / Fm. Error bars represent the standard error between triplicate wells. An asterisk (*) denotes days on which the parameters for the control and co-culture treatments differed significantly. Differences between treatments for all days were non-significant except for 10 d in all three parameters (10 d – F0: t-test, df = 2.5, t-ratio = -15, p = 0.0017*; Fm: t-test, df = 2.1, t-ratio = -16.15, p = 0.003*; Fv/Fm: t-test, df = -18.68, t-ratio = 2.0, p = 0.0028*). Please click here to view a larger version of this figure.
Figure 3. Representative WATER-PAM fluorometry graphs of a 10 d co-culturing experiment of Emiliania huxleyi (CCMP3266) with Phaeobacter gallaeciensis BS107. (A) The initial algal fluorescence (F0), (B) maximum algal fluorescence (Fm), and (C) potential quantum yield (Fv/Fm) are graphed for control algal (open circles) and alga co-cultured with a bacterium (black circles). Potential quantum yield (Fv/Fm) is a dimensionless expression of photosynthetic health, which is calculated as (Fm - F0) / Fm. Error bars represent the standard error between triplicate wells. An asterisk (*) denotes days on which the parameters for the control and co-culture treatments differed significantly. Differences between treatments for all days were non-significant except for 10 d in all three parameters (10 d – F0: t-test, df = 2.5, t-ratio = -15, p = 0.0017*; Fm: t-test, df = 2.1, t-ratio = -16.15, p = 0.003*; Fv/Fm: t-test, df = -18.68, t-ratio = 2.0, p = 0.0028*). Please click here to view a larger version of this figure.
Discussion
Algal growth in a miniaturized format.
The miniaturization of algal cultures to a 1 ml culture volume in a microtiter plate allows for the replication within an experiment to be increased. It is important to ensure the alga is healthy throughout an experiment; perform a growth curve (Figure 2), using the microtiter plate format to assess various algal media, to ensure the nutritional requirements of the alga are met. Additionally, it may be important to optimize the diurnal cycle (light and dark periods) and temperature. Proper optimization for a given alga can allow for maintaining healthy algal cultures at peak fluorescence for 26 d and for detecting potential quantum yield after 140 days (Figure 2).
Minimizing evaporative effects.
It is important to minimize the evaporative effects of liquid based assays as an ‘edge effect’ is commonly observed where there is greater evaporation in wells at the edge of the microtiter plate than in wells located towards the middle of the plate. While evaporation has been observed at the edge of plates, the rate of evaporation does not limit the experimental duration as healthy algal cultures, with peak fluorescence at 26 d has been maintained in this format (Figure 2). To minimize any potential ‘edge effect’, 1x PBS (pH 7.4), or other sterile solution, is aliquoted into wells along all four edges (columns A and H, rows 1 and 6, Figure 1).
Lighting within a diurnal incubator.
Microtiter plates should all have the same orientation in the diurnal incubator. For instance, lengthwise orientation of microtiter plates in an incubator means that the plates are placed along the short axis. If this orientation is used, algal cultures along the long axis B-G (Figure 1) can experience a slight light and temperature gradient due to the variation in distance from the light source (the light bulb is a source of both light and heat). This has been observed to influence temperature assays on algae at the extreme of their temperature range. Consequently this is unlikely to affect most algae grown at their optimum temperature. To minimize the impact of this light and temperature gradient ensure that larger bottles are not creating shading, place plates at a consistent distance from the lights, use shade cloth to reduce the light level if necessary and check the light intensity across the long axis of the diurnal incubator to ensure that it travels evenly.
Dark adaptation of algal samples for WATER-PAM fluorometry.
Before conducting WATER-PAM readings it is important to dark-adapt the algal samples so that the PSII reaction centers are fully open and the light-induced transthylakoidal pH gradient is fully dissipated, thus giving true Fo and Fm values from which to calculate Fv/Fm. Sampling the alga from the assay for WATER-PAM measurements in the middle of the dark phase of the diurnal cycle (i.e., for a 16:8 hr light:dark cycle, a 2 hr sampling session would be performed from T(dark) = 3 - 5 hr) makes dark adaptation time shorter (3 - 5 min) compared with the middle of the light cycle (T(light) = 7 - 9 hr) when dark adaptation is longer (>20 min)17. The alga’s dark adaption time will also vary depending on the algal species, growth conditions and the light conditions of its natural habitat range.
Sensitivity of WATER-PAM fluorometry readings.
The WATER-PAM was designed to be an ultrasensitive fluorometer capable of detecting F, F0 and Fm from low chlorophyll samples such as ocean surface water16. Consequently it is ideally suited to a miniaturized bioassay where samples can be diluted. Due to this sensitivity it is important to understand that the WATER-PAM machine has an upper limit of fluorescence readings (i.e., F, F0 and/or Fm) if the sample is not sufficiently diluted (Figure 4). Within the WinControl software the maximum values are first noticeable after pressing F0 and reading the F0 (directly after dark adaptation). Consequently the F and Fm are at the maximum value 4,056 (Figure 4, no. 2286). The maximum values of F and Fm depend on the type of algal medium that is used to calibrate the WATER-PAM, but samples above the detection limit can be readily identified because repeated readings with the same maximum value occur until the sample is sufficiently diluted. After a 1:1 dilution in algal media, the less concentrated sample is dark-adapted again and the F0 measurement was repeated. The F value appears to be measured correctly, but the Fm is still reading the maximum value 4,056 (Figure 4, no. 2287). This sample was again diluted 1:1 in algal media again and dark adapted for an additional 3 min before taking another F0 reading (Figure 4, no. 2288) and valid readings were obtained, so the next measurement (F) is taken and the Fv/Fm calculation is valid. In this example, the sample was diluted twice in a 1:1 ratio with medium, which needs to be factored into the calculation of Fm and F0. It is important to note that Fv/Fm, as a direct function of F0 and Fm, is incorrectly calculated if the samples are too concentrated despite being a dimensionless expression of photosynthetic health.
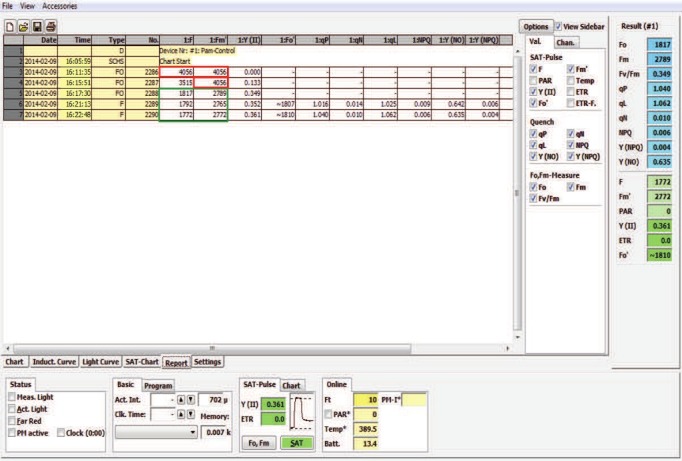 Figure 4. WinControl display of several WATER-PAM fluorometry readings of an algal sample being sequentially diluted to achieve the correct dilution. This figure displays algal fluorescence readings reaching the upper limit of the WATER-PAM machine (red box) and those where a suitable dilution of the sample has been achieved (green box). In addition, this figure depicts some of the other variables that this method calculates, but these are not discussed in detail here (see reviews13,16,20–22). Please click here to view a larger version of this figure.
Figure 4. WinControl display of several WATER-PAM fluorometry readings of an algal sample being sequentially diluted to achieve the correct dilution. This figure displays algal fluorescence readings reaching the upper limit of the WATER-PAM machine (red box) and those where a suitable dilution of the sample has been achieved (green box). In addition, this figure depicts some of the other variables that this method calculates, but these are not discussed in detail here (see reviews13,16,20–22). Please click here to view a larger version of this figure.
Bacterial contamination.
In all algal experiments, algal controls are necessary as a baseline of algal health, so it is imperative that it remains free of bacterial contamination throughout the experiment. Bacterial contamination occurs easily as there is no selection (such as antibiotics) and photosynthesis constantly produces new organic carbon for bacterial growth. The two most important ways to avoid contamination is to ensure sterility of all solutions (e.g., algal media, 1x PBS, pH 7.4) and equipment at T = 0 d of the experiment and to maintain aseptic technique while handling the microtiter plates. The experiment should be monitored for contamination at each time point by plating a 20 μl aliquot from all algal control wells. If contamination is observed from any of the algal control wells then those WATER-PAM fluorometry readings should be noted and excluded from data analysis. If a solution used to set up the experiment causes the entire experiment to be contaminated, then discard the experiment and all contaminated reagents and start again. The bacterial controls and bacterial-algal co-cultures can also become contaminated and agar plates used for bacterial cfu counts should be monitored for alternate colony morphologies. Well-to-well cross contamination can occur when removing the microtiter plate lid, but is easily avoided by not tilting or shaking of the plates and employing aseptic technique in a laminar flow hood or near a flame.
Future Applications.
This small volume bioassay provides a rapid screening method for microalgae by combining a microtiter plate format with WATER-PAM fluorometry. Examples of future applications are various, and could include Imaging PAM fluorometry, which provides insight into cell-cell variation of PSII health within a population as it performs PAM fluorometry on individual cells24. The bioassay can also be combined with microscopy and flow cytometry as previously discussed. Another combination with the potential to provide further insight is cell staining for flow cytometry and microscopy to elucidate morphological variation within subpopulations of the algal culture.
Disclosures
The authors have nothing to disclose.
Acknowledgments
This work was supported by Natural Sciences and Engineering Research Council of Canada (grant 402105), Canadian Foundation for Innovation (grant 129087) and Alberta Education and Training (grant AAETRCP-12-026-SEG) to RJC.
References
- Scarratt MG, Marchetti A. Assessing microbial responses to iron enrichment in the Subarctic Northeast Pacific: Do microcosms reproduce the in situ condition? Deep Sea Res Part II Top. Stud. Oceanogr. 2006;53(20-22):2182–2200. [Google Scholar]
- Bidle KD, Haramaty L, Barcelos E Ramos J, Falkowski P. Viral activation and recruitment of metacaspases in the unicellular coccolithophore, Emiliania huxleyi. Proc. Natl. Acad. Sci. U. S. A. 2007;104(14):6049–6054. doi: 10.1073/pnas.0701240104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore LR, Goericke R, Chisholm SW. Comparative physiology of Synechococcus and Prochlorococcus: influence of light and temperature on growth, pigments, fluorescence and absorptive. Mar. Ecol. Prog. Ser. 1995;116 [Google Scholar]
- Iglesias-Rodriguez MD, Halloran PR. Phytoplankton calcification in a high-CO2 world. Science. 2008;320(5874):336–340. doi: 10.1126/science.1154122. [DOI] [PubMed] [Google Scholar]
- Chen M, Tang H, Ma H, Holland TC, Ng KYS, Salley SO. Effect of nutrients on growth and lipid accumulation in the green algae Dunaliella tertiolecta. Bioresour. Technol. 2011;102(2):1649–1655. doi: 10.1016/j.biortech.2010.09.062. [DOI] [PubMed] [Google Scholar]
- Lv J-M, Cheng L-H, Xu X-H, Zhang L, Chen H-L. Enhanced lipid production of Chlorella vulgaris by adjustment of cultivation conditions. Bioresour. Technol. 2010;101(17):6797–6804. doi: 10.1016/j.biortech.2010.03.120. [DOI] [PubMed] [Google Scholar]
- Geider R, Graziano L, McKay RM. Responses of the photosynthetic apparatus of Dunaliella tertiolecta (Chlorophyceae) to nitrogen and phosphorus limitation. Eur. J. Phycol. 1998;33(4):315–332. [Google Scholar]
- MacIntyre HL, Cullen JJ. Using Cultures to Investigate the Physiological Ecology of Microalgae. Algal Cult. Tech. 2005. pp. 287–326.
- Blaise C, Vasseur P. Algal microplate toxicity test. Small-scale Freshw. Toxic. Investig. Vol. 1 Toxic. Test Methods. 2005. pp. 137–179.
- Skjelbred B, Edvardsen B, Andersen T. A high-throughput method for measuring growth and loss rates in microalgal cultures. J. Appl. Phycol. 2012;24:1589–1599. [Google Scholar]
- Nagai T, Taya K, Annoh H, Ishihara S. Application of a fluorometric microplate algal toxicity assay for riverine periphytic algal species. Ecotoxicol. Environ. Saf. 2013;94:37–44. doi: 10.1016/j.ecoenv.2013.04.020. [DOI] [PubMed] [Google Scholar]
- Seyedsayamdost MR, Case RJ, Kolter R, Clardy J. The Jekyll-and-Hyde chemistry of Phaeobacter gallaeciensis. Nat. Chem. 2011;3(4):331–335. doi: 10.1038/nchem.1002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schreiber U, Schliwa U, Bilger W. Continuous recording of photochemical and non-photochemical chlorophyll fluorescence quenching with a new type of modulation fluorometer. Photosynth. Res. 1986;10(1-2):51–62. doi: 10.1007/BF00024185. [DOI] [PubMed] [Google Scholar]
- Jones RJ, Ward S, Amri AY, Hoegh-Guldber O. Changes in quantum efficiency of photosystem II of symbiotic dinoflagellates of corals after heat stress, and of bleached corals sampled after the 1998 Great Barrier Reef mass bleaching event. Mar. Freshw. Res. 1998;51(345):659–668. [Google Scholar]
- Beer S, Larsson C, Poryan O, Axelsson L. Photosynthetic rates of Ulva (Chlorophyta) measured by pulse amplitude modulated fluorometry. Eur. J. Phycol. 2000;35(1):69–74. [Google Scholar]
- WATER-PAM Chlorophyll Fluorometer. Instrument Description and Information for Users. Germany: Heinz Walz GmbH; 2013. Available at: http://www.walz.com/downloads/manuals/water-pam/waterp3e.pdf. [Google Scholar]
- Maxwell K, Johnson GM, Heers J. Chlorophyll fluorescence--a practical guide. J. Exp. Bot. 2000;51(345):659–668. doi: 10.1093/jxb/51.345.659. [DOI] [PubMed] [Google Scholar]
- Herigstad B, Hamilton M, Heersink J. How to optimize the drop plate method for enumerating bacteria. J. Microbiol. Methods. 2001;44(2):121–129. doi: 10.1016/s0167-7012(00)00241-4. Available at: http://www.ncbi.nlm.nih.gov/pubmed/11165341. [DOI] [PubMed] [Google Scholar]
- Kooten O, Snel J. The use of chlorophyll fluorescence nomenclature in plant stress physiology. Photosynth. Res. 1990;25(3):147–150. doi: 10.1007/BF00033156. Available at: http://www.springerlink.com/index/r3689k5w72782r50.pdf. [DOI] [PubMed] [Google Scholar]
- Maxwell K, Johnson GN. Chlorophyll fluorescence--a practical guide. J. Exp. Bot. 2000;51(345):659–668. doi: 10.1093/jxb/51.345.659. [DOI] [PubMed] [Google Scholar]
- Schreiber U. Pulse-Amplitude-Modulation (PAM) Fluorometry and Saturation Pulse Method: An Overview. Chlorophyll a Fluoresc. A Signat. Photosynth. 2004. pp. 279–319.
- Roháček K, Barták M. Technique of the modulated chlorophyll fluorescence: basic concepts, useful parameters, and some applications. Photosynthetica. 1999;37(3):339–363. [Google Scholar]
- Da Silva JM, da Silva AB, Pádua M. Modulated chlorophyll a fluorescence: a tool for teaching photosynthesis. J. Biol. Educ. 2007;41(4):178–183. [Google Scholar]
- Vieira S, Ribeiro L, Jesus B, Cartaxana P, da Silva JM. Photosynthesis assessment in microphytobenthos using conventional and imaging pulse amplitude modulation fluorometry. Photochem. Photobiol. 2013;89(1):97–102. doi: 10.1111/j.1751-1097.2012.01224.x. [DOI] [PubMed] [Google Scholar]


