Abstract
Similar to healthy tissues, many blood and solid malignancies are now thought to be organised hierarchically, with a subset of stem-like cancer cells that self-renew while giving rise to more differentiated progeny. Understanding and targeting these cancer stem cells in breast cancer, which may possess enhanced chemo- and radio-resistance compared to the non-stem tumor bulk, has become an important research area. Markers including CD44, CD24, and ALDH activity can be assessed using fluorescence activated cell sorting (FACS) to prospectively isolate cells that display enhanced tumorigenicity when implanted into immunocompromised mice: the mammosphere assay has also become widely used for its ability to retrospectively identify sphere-forming cells that develop from single stem cell-like clones. Here we outline approaches for the appropriate culturing of mammospheres from cell lines or primary patient samples, their passaging, and calculations to estimate sphere forming efficiency (SFE). First we discuss key considerations and pitfalls in the appropriate planning and interpretation of mammosphere experiments.
Keywords: Medicine, Issue 97, breast cancer, cancer stem cell, CSC, BCSC, tumorsphere, mammosphere, tumorigenesis, self-renewal.
Introduction
The existence of tumor cell lineages headed by stem-like cancer stem cells has greatly added to our understanding of tumor heterogeneity. While some phenotypic diversity in tumors does arise from the clonal outgrowth of genetically distinct clones, a substantial component appears to result from epigenetic differences: cancer cells can transition (sometimes reversibly) between stem, progenitor, and differentiated states via activation or repression of specific gene expression programs1–3. This may reflect cell intrinsic or extrinsic factors, reflecting the gene expression program currently being expressed in a cell with its resultant autocrine signalling in conjunction with paracrine signalling from neighboring cancer, stromal or immune cells delivering modulatory factors, and microenviromental conditions such as the degree of hypoxia2,4,5.
Although innovative lineage tracing approaches are advancing our ability to study putative cancer stem cells in their in vivo niche6–8, sphere-forming assays remain a popular and convenient approach to estimate breast cancer cells’ potential to behave like stem cells, at least under the assay conditions used. It is often used alongside retrospective methods for purifying cancer stem cells, by their expression of membrane markers CD44 and CD249, and activity levels of the enzyme ALDH (aldehyde dehydrogenase)10, markers that have been proposed to correspond to more mesenchymal- and epithelial-like cancer stem cells respectively11. The sphere formation approach was first developed as the neurosphere assay, enabling the growth of putative stem cells from single clones in non-adherent, serum free conditions with the addition of epithelial growth factor (EGF)12, later being usefully applied to normal and cancerous breast tissues.
The identity of the sphere-forming founder cell, and the mixed cell types making up the sphere mass, are relevant for the inferences that can be made from mammo-, or other-sphere forming assays. Long-term quiescent bone fide stem cells, thought to rest in G0 phase, will not experience the precise combination of factors that would favor activation in vivo. The mammosphere assay instead enables the growth of cells either poised for mitotic division or already dividing13. These progenitors, although not a truly quiescent cell, may be the cell stage that proliferates with the EGF and basic fibroblast growth factor (bFGF) mitogens used in the assay. Nevertheless, they contain a range of activated stem cell-associated signalling pathways14. In addition, the rate of their formation relates to the tumorigenicity of the tissue they were taken from, when measured by their potency in limited dilution assays in mouse xenografts2,15,16.
Here we provide detailed protocols to isolate single cells and generate primary mammospheres from both human breast cancer cell lines and clinical samples of breast tumors. We also describe how to perform serial passages of primary mammospheres to assess self-renewal, and how to calculate sphere forming efficiency that allows comparison across different seeding densities (see scheme in Figure 1).
Protocol
The procedures below have been ethically approved by Imperial College, London.
1. Generation of Primary Mammospheres from Human Breast Cancer Cell Lines
NOTE: Perform the following steps under a sterile culture hood.
Prepare Mammosphere Media containing DMEM/F12 supplemented with 2 mM L-glutamine, 100 U/ml penicillin, 100 U/ml streptomycin. Prepare complete media immediately before use by adding 20 ng/ml recombinant human epidermal growth factor (EGF; Sigma), 10 ng/ml recombinant human basic fibroblast growth factor (bFGF; R&D Systems) and 1x B27 supplement.
Aspirate media from flask containing adherent MCF-7 or MDA-MB-231 cells (or a breast cancer cell line of your choice; 70-80% confluence), wash twice in 1x PBS, and trypsinize cells.
Centrifuge cells at 200 x g at room temperature for 5 min. Decant supernatant and resuspend cells in 1-5 ml of mammosphere media. Pipette up and down and if necessary use a 40 µm cell strainer cap filter to obtain single-cell suspension. Use a hemocytometer to ensure a single cell suspension has formed (if not, use a 25 G needle to syringe the cell suspension up to 3 times).
If necessary isolate CD44+CD24- cellular subset via FACS using anti-CD24-phycoerythrin (PE) and anti-CD44-fluorescein isothiocyanate (FITC) monoclonal antibodies9. Alternatively, isolate such population using a magnetic-activated cell sorting (MACS) system with anti-CD44 and anti-CD24-biotin combined microbeads as per manufacturer’s instructions. Perform positive selection using LS columns, and negative selection using LD columns and confirm the phenotypes of all isolated cells by flow cytometry17.
- Calculate the number of viable cells per ml using trypan blue. NOTE: Cell viability is calculated as the number of viable cells divided by the total number of cells within the grids on the hemocytometer. Cells that take up trypan blue are considered non-viable. The following procedure is used to accurately determine proportion of viable cells.
- Prepare a 0.4% solution of trypan blue in PBS. Add 0.1 ml of trypan blue stock solution to 1 ml of cells. Load a hemocytometer and examine immediately under a microscope at low magnification. Count the number of total cells and the number of blue staining cells. Viable cells = [1.00 – (Number of blue cells ÷ Number of total cells)] × 100.
- To calculate the number of viable cells per ml of culture, use the formula below. Remember to correct for the dilution factor. Number of viable cells × 104 × 1.1 = cells/ml culture
Resuspend a pre-determined amount of cells in 2 ml of complete mammosphere media in each well of a 6-well ultra-low adherent plate. The seeding density is normally 500-4,000 cells/cm2 cell per well. Optionally use low attachment 24-well plates while seeding cells at the same density in 0.5 ml of media. NOTE: We recommend optimization of seeding density and time of culture for each cell lines of interest.
Incubate ultra low-attachment 6-well plates at 37 °C and 5% CO2 for 5-10 days (depending on the cell line and the size of mammospheres) without disturbing the plates, particularly during the growth phase of the first 5 days.
2. Generation of Primary Mammospheres from Human Breast Cancer Clinical Samples
NOTE: Human breast cancer tissue can be obtained from patients undergoing surgery for the removal of breast tumors. Store tissue on ice for up to 24 hr in 50 ml sterile tubes in culture media containing 100 U/ml penicillin and 100 U/ml streptomycin. Perform the following steps under a sterile culture hood.
Transfer the sample into a 100 mm tissue Petri dish with a small volume of media. Remove the adipose tissue using sterile scissors, scapel and tweezers.
Add 2-3 ml of DMEM/F12 and mince the sample into 1 mm3 pieces with a sterile scapel or razor blade until no large pieces remain.
Resuspend the tissue pieces in pre-warmed 10 ml DMEM containing proteolytic enzymes (3,000 U/ml collagenase and 1,000 U/ml hyaluronidase) and incubate at 37 °C in a rotary shaker until all the tissue fragments are digested. Usually complete digestion takes from 1-3 hr. Adjust the digestion time according to pathological characters of tumors. (For example breast adenocarcinoma is usually harder to digest compared to mucinous carcinoma which is more fragile and needs shorter digestion time). Assess the degree of digestion every ½ hour by observing 20 µl suspension under hemocytometer.
Allow the fragments to sediment for 5 min, then transfer the supernatant in a 15 ml conical polypropylene tube and centrifuge at 200 x g for 10 min at room temperature. Carefully decant off the supernatant and resuspend cells in 1 - 5 ml of mammosphere media.
Follow the steps 1.1-1.4 described above for cell lines to obtain and plate single cell suspensions. Pass cells through the 25 G syringe a maximum of 2 times if necessary to limit damaging cells.
3. Mammosphere Forming Efficiency (%) Calculation
After the culture period, count the mammospheres (greater than 40 µm) diameter under a microscope at 40X magnification. Digitally image 5 random fields using a digital camera on a light microscope and determine the mammospheres size using the acquisition software. Use any Image Analysis Software.
Calculate Mammosphere Forming Efficiency (MFE%) using the following equation: MFE (%) = (# of mammospheres per well) / (# of cells seeded per well) x 100
4. Serial Passaging of Mammospheres for Assessment of Self-renewal
Pipette the media containing the mammospheres from each well into a 15 ml tube. Wash the wells with PBS and add to the collected media. Centrifuge at 115 x g for 10 min at room temperature.
Discard supernatant and resuspend the pellet in 500 µl of pre-warmed 0.5% trypsin/0.2% EDTA. Incubate for 2-3 min.
Add 500 µl of FCS to neutralize trypsin. Centrifuge at 500 x g for 5 min. Discard supernatant and resuspend pellet in 100 µl of mammospheres media, pipetting up and down to disaggregate spheres.
Count cells with hemocytometer and determine if cells are in single cell suspension. If not pass through a 25 G syringe up to 3 times to obtain single cells. Seed the cells into a new ultra low-attachment 6-well plate at the same density used in the primary generation.
After 5-10 days, count spheres >40 µm and calculate sphere-forming efficiency using the formula reported before.
Representative Results
Different samples or those subjected to different treatments may vary in the number of mammospheres >40 µm that form after normalizing for initial cells seeded. Calculate mammosphere forming efficiency (MFE) for each treatment grown in triplicate. This enables experiments with different seeding densities to be compared. Data is best displayed on a bar graph, ideally with positive and negative controls, and displaying the standard deviation across the triplicate wells. Consistently transfer adherent cells into non-adherent plates at between 60-80% confluence. Careful cell counting is essential to accurately quantifying the effects of treatments. Estimate the degree of variability that this key step is contributing to your results by preparing multiple suspensions separately from the same treatment well. If the cell concentration measured differs significantly from the same well, consider refining your counting approach (Figure 2).
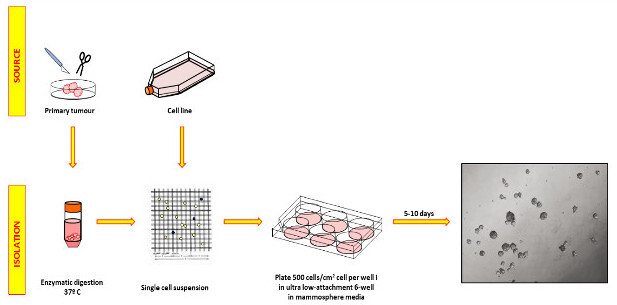 Figure 1: Processing of primary breast cancers and cell line samples to obtain mammosphere cultures. Samples are enzymatically digested, validated as single cells, and plated on non-adherent plates in serum-free mammosphere medium with growth factors. Plates should not be handled during the growth phase to avoid sphere fusion. The picture on the right shows representative MCF-7 spheres photographed after 5 days. Please click here to view a larger version of this figure.
Figure 1: Processing of primary breast cancers and cell line samples to obtain mammosphere cultures. Samples are enzymatically digested, validated as single cells, and plated on non-adherent plates in serum-free mammosphere medium with growth factors. Plates should not be handled during the growth phase to avoid sphere fusion. The picture on the right shows representative MCF-7 spheres photographed after 5 days. Please click here to view a larger version of this figure.
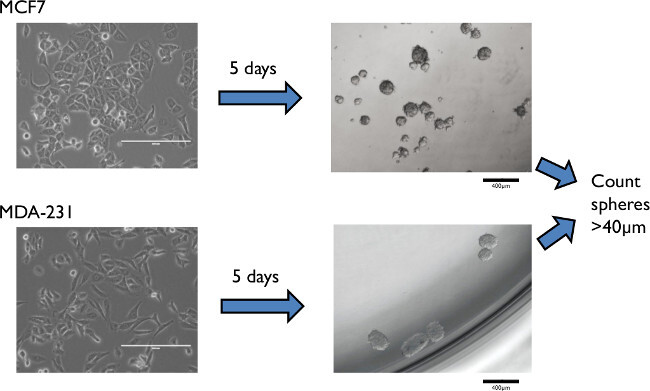 Figure 2: Representative results of mammospheres formed from an epithelial estrogen positive (MCF-7), and a mesenchymal triple negative (MDA-MB-231) cell line. Minimal cell fusion/aggregation was attained by low density plating, here at 500 cells/cm2 for MCF-7, and 1,000 cells/cm2 for MDA-MB-231. Please click here to view a larger version of this figure.
Figure 2: Representative results of mammospheres formed from an epithelial estrogen positive (MCF-7), and a mesenchymal triple negative (MDA-MB-231) cell line. Minimal cell fusion/aggregation was attained by low density plating, here at 500 cells/cm2 for MCF-7, and 1,000 cells/cm2 for MDA-MB-231. Please click here to view a larger version of this figure.
Discussion
Successful assessment of primary and secondary mammospheres relies on cells being plated at sufficiently low densities that mammospheres form from single clones, with minimal sphere aggregation. However at densities that are too low, too few mammospheres may form to distinguish the effects of treatments statistically. Seeding density should be optimised for each cell line used since they can vary considerably in their sphere forming efficiencies (those expressing low E-cadherin may form less stable and shorter term mammosphere cultures18). This optimization stage should ensure most primary and secondary sphere formation results from the clonal growth of single cells. Plating one cell per well is the only way to entirely ensure clonal mammosphere formation, and can be recommended for quantifying the self-renewal/differentiation rates of freshly derived samples13. Yet this approach also suffers from complicating factors: a mammosphere forming efficiency of 1% will yield only a single mammosphere per 96-well plate, and efficiency may be underestimated with fewer mammospheres being formed in the absence of paracrine growth factor signalling between suspended cells. Finally, the frequency of secondary spheres after passaging can estimate levels of self-renewal, and higher efficiency of secondary than primary sphere formation should be expected if truly selecting for cells with cancer stem cell properties. Reaggregation is a particular problem when deriving secondary spheres and cell-type appropriate very low cell densities should be optimized towards.
The time taken for mammospheres to grow to >40 µm will also vary, and mammospheres from clinical samples may need to be counted between 5-12 days depending on sphere growth speed. Aggregation and fusion of spheres inevitably occurs due to inherent locomotion of cells around the media, but the process appears to be substantially enhanced by experimenters moving the plates during the growth phase19, something therefore to be avoided.
While large spheres are routinely thought to come from stem cells with higher long-term replicative capacity this assumption should be treated with caution. Larger spheres may reflect the proliferative capacity of progenitors, the responsiveness to growth factors, or be a product of aggregation or fusion. Both stem cells and their transit amplifying daughter cells are capable of giving rise to mammospheres20, although the combined frequency of both cell types may still reflect the tumorigenicity of the source tissue.
The mammosphere assay is limited by a failure to capture the complexity of cancer stem cell formation and behavior in their in vivo niche. To truly assess the long-term in vitro and in vivo self-renewal and differentiation of prospectively isolated breast cancer stem cells, mammosphere assays should therefore ideally be performed in conjunction with lineage tracing and xenotransplantation experiments. While lineage tracing relies on markers that accurately reflect the cancer stem cell subset, they benefit greatly from retaining the in vivo conditions that contribute to the dynamic formation of ‘bone fide’ cancer stem cells. Assessing tumor formation in xenograft experiments in immunocompromised mice will also provide many niche components missing from mammosphere assays, while still lacking human immune and stromal components21. Though more technically demanding, these techniques should be attempted for a more complete and informative view of breast cancer stem cell properties and tumorigenicity.
The mammosphere assay does provide an informative and convenient tool, provided it is carried out carefully and the above limitations considered: indeed as our knowledge of the in vivo conditions experienced by cancer stem cells advances, mammosphere assays may incorporate an increasing range of molecular and environmental factors. Using the approach outlined above, sphere-formation rates correspond to tumor-initiation rates in xenografts2,15,16, and self-renewal of spheres does increase in more aggressive breast cancers22 and following stem cell-increasing interventions such as chemotherapy23. Meanwhile the spheres themselves can provide a valuable resource with which to study expression differences and novel factors involved in the stem cell gene expression architecture.
Disclosures
The authors declare that there is no conflict of interest that could be perceived as prejudicing the impartiality of this article.
Acknowledgments
This work is supported by the Imperial BRC, the National Institute for Health Research, and Action Against Cancer with special mention to Hilary Craft and Sir Douglas Myers.
References
- Iliopoulos D, Hirsch Ha, Struhl K. An epigenetic switch involving NF-kappaB, Lin28, Let-7 MicroRNA, and IL6. Cell. 2009;139(4):693–706. doi: 10.1016/j.cell.2009.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iliopoulos D, Hirsch Ha, Wang G, Struhl K. Inducible formation of breast cancer stem cells and their dynamic equilibrium with non-stem cancer cells via IL6 secretion. Proc Natl Acad Sci U S A. 2011;108(4):1397–1402. doi: 10.1073/pnas.1018898108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visvader JE, Lindeman GJ. Cancer stem cells: current status and evolving complexities. Cell stem cell. 2012;10(6):717–728. doi: 10.1016/j.stem.2012.05.007. [DOI] [PubMed] [Google Scholar]
- Rosen JM, Jordan CT. The increasing complexity of the cancer stem cell paradigm. Science. 2009;324(5935):1670–1673. doi: 10.1126/science.1171837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rokavec M, Wu W, Luo J-L. IL6-mediated suppression of miR-200c directs constitutive activation of inflammatory signaling circuit driving transformation and tumorigenesis. Mol Cell. 2012;45(6):777–789. doi: 10.1016/j.molcel.2012.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, Li Y, et al. A restricted cell population propagates glioblastoma growth after chemotherapy. Nature. 2012;488(7412):522–526. doi: 10.1038/nature11287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Driessens G, Beck B, Caauwe A, Simons BD, Blanpain C. Defining the mode of tumour growth by clonal analysis. Nature. 2012;488(7412):527–530. doi: 10.1038/nature11344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schepers AG, Snippert HJ, et al. Lineage tracing reveals Lgr5+ stem cell activity in mouse intestinal adenomas. Science. 2012;337(6095):730–735. doi: 10.1126/science.1224676. [DOI] [PubMed] [Google Scholar]
- Sheridan C, Kishimoto H, et al. CD44+/CD24- breast cancer cells exhibit enhanced invasive properties: an early step necessary for metastasis. Breast cancer research: BCR. 2006;8(5):R59. doi: 10.1186/bcr1610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ginestier C, Hur MH, et al. ALDH1 is a marker of normal and malignant human mammary stem cells and a predictor of poor clinical outcome. Cell stem cell. 2007;1(5):555–567. doi: 10.1016/j.stem.2007.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S, Cong Y, et al. Breast Cancer Stem Cells Transition between Epithelial and Mesenchymal States Reflective of their Normal Counterparts. Stem cell reports. 2014;2(1):78–91. doi: 10.1016/j.stemcr.2013.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reynolds BA, Weiss S. Generation of neurons and astrocytes from isolated cells of the adult mammalian central nervous system. Science. 1992;255(5052):1707–1710. doi: 10.1126/science.1553558. http://www.ncbi.nlm.nih.gov/pubmed/1553558 . [DOI] [PubMed] [Google Scholar]
- Pastrana E, Silva-Vargas V, Doetsch F. Eyes wide open: a critical review of sphere-formation as an assay for stem cells. Cell stem cell. 2011;8(5):486–498. doi: 10.1016/j.stem.2011.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dontu G, Abdallah WM, et al. In vitro propagation and transcriptional profiling of human mammary stem / progenitor cells. Genes Dev. 2003. pp. 1253–1270. [DOI] [PMC free article] [PubMed]
- Ponti D, Costa A, et al. Isolation and in vitro propagation of tumorigenic breast cancer cells with stem/progenitor cell properties. Cancer Res. 2005;65(13):5506–5011. doi: 10.1158/0008-5472.CAN-05-0626. [DOI] [PubMed] [Google Scholar]
- Grimshaw MJ, Cooper L, et al. Mammosphere culture of metastatic breast cancer cells enriches for tumorigenic breast cancer cells. Breast Cancer Res. 2008;10(3):R52. doi: 10.1186/bcr2106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pham PV, Phan NLC, et al. Differentiation of breast cancer stem cells by knockdown of CD44: promising differentiation therapy. J Transl Med. 2011;9(1):209. doi: 10.1186/1479-5876-9-209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manuel Iglesias J, Beloqui I, et al. Mammosphere formation in breast carcinoma cell lines depends upon expression of E-cadherin. PloS one. 2013;8(10):e77281. doi: 10.1371/journal.pone.0077281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coles-Takabe BLK, Brain I, et al. Don’t look: growing clonal versus nonclonal neural stem cell colonies. Stem Cells. 2008;26(11):2938–2944. doi: 10.1634/stemcells.2008-0558. [DOI] [PubMed] [Google Scholar]
- Stingl J. Detection and analysis of mammary gland stem cells. J Pathol. 2009;217(2):229–241. doi: 10.1002/path.2457. [DOI] [PubMed] [Google Scholar]
- Kreso A, Dick JE. Evolution of the cancer stem cell model. Cell stem cell. 2014;14(3):275–291. doi: 10.1016/j.stem.2014.02.006. [DOI] [PubMed] [Google Scholar]
- Al-Hajj M, Clarke MF. Self-renewal and solid tumor stem cells. Oncogene. 2004;23(43):7274–7282. doi: 10.1038/sj.onc.1207947. [DOI] [PubMed] [Google Scholar]
- Yu F, Yao H, et al. let-7 regulates self renewal and tumorigenicity of breast cancer cells. Cell. 2007;131(6):1109–1123. doi: 10.1016/j.cell.2007.10.054. [DOI] [PubMed] [Google Scholar]


