Abstract
Human induced pluripotent stem cell-derived cardiomyocytes (hiPSC-CMs) have become an important cell source to address the lack of primary cardiomyocytes available for basic research and translational applications. To differentiate hiPSCs into cardiomyocytes, various protocols including embryoid body (EB)-based differentiation and growth factor induction have been developed. However, these protocols are inefficient and highly variable in their ability to generate purified cardiomyocytes. Recently, a small molecule-based protocol utilizing modulation of Wnt/β-Catenin signaling was shown to promote cardiac differentiation with high efficiency. With this protocol, greater than 50%-60% of differentiated cells were cardiac troponin-positive cardiomyocytes were consistently observed. To further increase cardiomyocyte purity, the differentiated cells were subjected to glucose starvation to specifically eliminate non-cardiomyocytes based on the metabolic differences between cardiomyocytes and non-cardiomyocytes. Using this selection strategy, we consistently obtained a greater than 30% increase in the ratio of cardiomyocytes to non-cardiomyocytes in a population of differentiated cells. These highly purified cardiomyocytes should enhance the reliability of results from human iPSC-based in vitro disease modeling studies and drug screening assays.
Keywords: Developmental Biology, Issue 97, Human induced pluripotent stem cells, cardiac differentiation, small molecule compounds, cardiomyocytes, glucose starvation
Introduction
Primary human cardiomyocytes are difficult to obtain because of the requirement for invasive cardiac biopsies, difficulty in dissociating to single cells, and because of poor long-term cell survival in culture. Given this lack of primary human cardiomyocytes, patient-specific human induced pluripotent stem cell-derived cardiomyocyte (hiPSC-CM) technology has been regarded as a powerful alternative cardiomyocyte source for basic research as well as clinical and translational applications such as disease modeling and drug discovery1. Early efforts in differentiating pluripotent stem cells into cardiomyocytes employed differentiation protocols using embryoid bodies (EBs), but this method is inefficient in producing cardiomyocytes because often less than 25% of cells in an EB are beating cardiomyocytes2,3. Comparatively, a monolayer-based differentiation protocol using the cytokines activin A and BMP4 displayed a higher efficiency than EBs, but this protocol is still relatively inefficient, requires expensive growth factors, and only functions in a limited number of human pluripotent stem cell lines4. Recently, a highly efficient, hiPSC monolayer-based cardiomyocyte differentiation protocol was developed by modulating Wnt/β-Catenin signaling5. These hiPSC-CMs express cardiac troponin T and alpha actinin, two sarcomeric proteins that are standard markers of cardiomyocytes6. The protocol describe here is an adaptation of this small molecule-based, feeder cell-free, monolayer differentiation method5,7. We are able to obtain beating cardiomyocytes from hiPSCs after 7-10 days (Figure 1). However, following a cardiomyocyte differentiation resulting in 50% beating cells, immunostaining consistently shows the existence of a population of non-cardiomyocytes that are negative for cardiomyocyte-specific markers such as cardiac-specific troponin T and alpha-actinin. To further purify cardiomyocytes and eliminate non-cardiomyocytes, heterogeneous differentiated cell populations were subjected to glucose starvation by treating them with an extremely low glucose culture medium for multiple days (Figure 2). This treatment selectively eliminates non-cardiomyocytes due to the ability of cardiomyocytes, but not non-cardiomyocytes, to metabolize lactate as the primary energy source in order to survive in a low glucose environment8. After this purification step, a 40% increase in the ratio of cardiomyocytes to non-cardiomyocytes is observed, (Figure 3, Figure 4) and these cells can be used for downstream gene expression analysis, disease modeling, and drug screening assays.
Protocol
NOTE: Vendor information for all reagents used in this protocol has been listed in Table 1 and Materials List. All solutions and equipment coming into contact with cells must be sterile, and aseptic technique should be used accordingly. Perform all culture incubations in a humidified 37 °C, 5% CO2 incubator unless otherwise specified. In this protocol, all differentiations are performed in 6-well plates, in which the hiPSCs are seeded. Following differentiation and purification, cells can be dissociated and replated for downstream use.
1. Medium Preparation
For the E8 medium, make a stock solution for each medium component (NaHCO3, L-ascorbic acid 2-phosphate, sodium selenite, transferrin, insulin, FGF2, TGFB1) and store the solutions at -20 °C. Add appropriate amount of stock solutions to the bottle of DMEM/F12 and filter to sterilize. The solution concentration and reaction setup for the stock components and E8 medium can be found in Table 1.
- For the extracellular matrix solution, thaw the basement membrane matrix (commonly supplied at 10 mg/ml) at 4 °C O/N. Matrigel is commonly used for pluripotent stem cell culture because human pluripotent stem cells can adhere well to this material using the alpha-6-beta-1 integrin9. Once thawed, make 2.25 mg (250 µl) aliquots on ice using ice cold pipet tips and tubes and store the aliquots at −80 °C.
- When precoating plates with ECMS, thaw and aliquot the extracellular matrix solution (ECMS) on ice. Mix the ECMS and ice-cold DMEM/F12 medium in a ratio of 1:200 on ice. Keep cool to prevent premature ECMS solidification.
For the RPMI/B27 without insulin medium, add 10 ml of B27 Minus Insulin into 500 ml of RPMI media.
For the RPMI/B27 (with insulin) medium, add 10 ml of B27 Supplement (with insulin) and 5 ml of Pen-strep antibiotic into 500 ml of RPMI media.
For the low glucose medium, add 10 ml of B27 Supplement and 5 ml of Pen-strep antibiotic into 500 ml of glucose-free RPMI media.
For the freezing/cryopreservation medium, mix fetal bovine serum together with dimethylsulfoxide (DMSO) at a 9:1 ratio. The freezing medium can be stored up to 1 month at 4 °C. NOTE: All media should be filtered and stored in 4 °C and used within 2 weeks unless otherwise specified. All reagent/solution volumes used below are intended for a single well in a 6-well plate, unless otherwise specified.
2. Pre-coating 6-well Plates with ECMS
Make ECMS by mixing basement membrane matrix (e.g., Matrigel) with ice-cold DMEM/F12 media in a ratio of 1:200, then apply 2 ml of freshly made ECMS to each well for a 6-well plate. Each well in a 6-well plate will receive approximately 90 mg ECMS.
Incubate the ECMS coated plates for 1 hr at 37 °C. Immediately prior to plating cells, aspirate DMEM/F12 solution from each well. The plates will then be ready to receive cells.
3. Thawing the Frozen hiPSCs
NOTE: Closely follow this procedure as it may lead to optimal culturing and downstream differentiation of hiPSCs into hiPSC-CMs following the freeze/thaw cycle.
Thaw one vial of hiPSCs for each well of a 6-well plate. Prepare a 15 ml conical tube with 9 ml of cold E8 medium with ROCK inhibitor (10 μM) for each vial. ROCK inhibitor improves hiPSCs survival following cryopreservation10. Keep the vials in 37 °C water bath to thaw the cells until an approximately 5 mm diameter ice crystal is left.
Spray the exterior of the vials with 70% ethanol. Wipe the vials with tissue paper before moving them to the sterile laminar hood.
Transfer the cells into the prepared conical tube. Rinse the vial once with 500 μl of E8 medium and transfer the medium containing the thawed cells to the same conical tube. Centrifuge at RT for 4 min at 200 x g.
Aspirate the supernatant after centrifugation and gently resuspend the cell pellet with 2 ml of E8 medium with ROCK inhibitor (10 μM). Transfer the resuspended cells to one well of a 6-well plate pre-coated with ECMS.
Replace the medium with E8 medium without ROCK inhibitor after 24 hr of culturing. Cells should have adhered by this time.
4. Passaging of hiPSCs
- Typically, passage hiPSCs after reaching 75%-80% confluency in a 6-well plate. Aspirate the culture medium. Quickly wash wells with 1 ml of sterile, RT PBS. Aspirate PBS.
- Add 500 μl of 0.5 mM EDTA or 1x cell detachment solution (e.g., Accutase) and incubate for 1-7 min at RT. Appropriate time for EDTA incubation varies with the hiPSC line. Expect to see visible curling or thickening of colonies around the edges, as this will indicate that EDTA or cell detachment solution is ready to be removed.
Aspirate the EDTA or the cell detachment solution. Add 1 ml of E8 medium supplemented with 10 μM ROCK inhibitor. Displace hiPSC colonies in the well by repeatedly pipetting up and down and spraying colonies off with a P1000 pipette. Colonies should be broken into 5-10 cell clumps after completion.
Transfer the colonies that are suspended in 1 ml of E8 medium with ROCK inhibitor into a 15 ml conical tube.
- Disrupt the large cell clumps into small clumps in the E8 medium. Add an appropriate volume of E8 media with inhibitor to the cell suspension to dilute the cells. The media volume to add for dilution depends on the cell density and the number of wells to be seeded. Ideally, a single, 80% confluent well of hiPSCs should be diluted 1:12.
- Add 11 ml E8 medium with ROCK inhibitor to the existing 1 ml of cell solution in aforementioned 15 ml conical tube to obtain a 1:12 dilution.
Further triturate the cell clumps into small cell fragments (around 50-200 cells in each fragment is best). Avoid over-triturating as this reduces cell survival.
Aspirate ECMS from the pre-coated plates and add 2 ml of resuspended cells into each well. Approximately 100,000 cells per well of a 6-well plate is ideal. Aim to dispense the cells evenly around the well to avoid clustering of cells in the center of the well.
After 24 hr of culturing, replace the medium with E8 medium without ROCK inhibitor. Change the culture medium every 24 hr until cells become 80% confluent. Then, the cells are ready to passage again. It usually takes 3-6 days between passages to reach 80% confluency.
5. Freezing hiPSCs
NOTE: Closely follow this procedure as it may lead to optimal culturing and downstream differentiation of hiPSCs into hiPSC-CMs following the freeze/thaw cycle.
Label cryogenic tubes with the cell line name, cell type, passage number, and freezing date. As a general guideline, use one vial for each well of a 6-well plate. Prepare a 15 ml conical tube filled with 9 ml of E8 medium with ROCK inhibitor (10 μM). Prepare freezing medium with 90% FBS and 10% DMSO, and keep the medium at 4 °C until ready to use.
Aspirate the culture media, add 500 μl of 0.5 mM EDTA or 1x cell detachment solution and incubate for 2-7 min at RT. The duration of EDTA or cell detachment solution exposure varies from cell line to cell line.
Aspirate EDTA or cell detachment solution. Add 1 ml of E8 medium.
Displace hiPSC colonies in the well by pipetting up and down and spraying colonies off with a P1000 pipette. Colonies should not be broken into smaller than 100 cell clumps. Transfer the suspended cells to the prepared conical tube containing E8. Centrifuge at RT for 4 min at 200 x g.
Aspirate the supernatant and add 500 μl of cold freezing medium to the pellet. Resuspend the pellet by pipetting one to two times. Cell survival is improved by keeping cells in large clumps.
Transfer the resuspended cells into a labeled cryogenic tube. Quickly move the vials into a freezing container containing isopropanol, which will allow for gradual cooling. Keep the container with cells at -80 °C for 24 hr, then transfer the cells to liquid nitrogen for long-term storage.
6. Cardiac Differentiation of Human iPSC
NOTE: All media should be at least at RT when added.
After dissociation with EDTA or 1x cell detachment solution, seed approximately 100,000 human iPSCs on ECMS coated 6-well culture plates for differentiation (same steps as cell passaging procedures). When the cells reach 85% confluency, change the medium to RPMI/B27 without insulin medium with 6 µM GSK3-beta inhibitor CHIR99021 (CHIR) and maintain for 48 hr.
After 48 hr, replace the CHIR-containing culture medium with RPMI/B27 without insulin medium and leave alone for 24 hr (until day 3).
At day 3, change the media to RPMI/B27 without insulin with 5 µM Wnt inhibitor IWR1 and maintain for 48 hr (until day 5). NOTE: Wnt inhibition can also be attempted using other small molecule compounds, as described in earlier studies11. IWR1 was selected over other small molecule Wnt inhibitors due to the increased range in which it has been shown to be effective in inhibiting Wnt signaling11.
At day 5, change the medium back to RPMI/B27 without insulin medium and leave for 48 hr (until day 7).
At day 7, replace the medium with RPMI/B27 medium (with insulin) and replace medium every 3 days thereafter with the same medium. Spontaneous beating of cardiomyocytes should first be visible at approximately day 8 to day 10.
7. Purification of Human Cardiomyocytes through Glucose Starvation
At day 10 post-differentiation, change the medium in each well of the 6-well plate to 2 ml low glucose medium and maintain the cells in this medium for 3 days (until day 13).
At day 13, return cells to RPMI/B27 medium (with insulin).
- Optionally, replate cardiomyocytes prior to the second round of glucose starvation to help loosen non-cardiomyocytes from the culture plate, allowing for easier dissociation of non-cardiomyocytes during glucose starvation.
- At day 13, aspirate medium, wash once with PBS, and dissociate the cells into single cells using 500 μl of cell disassociation enzyme for 5 min at 37 °C. Specifically, after 5 min of enzyme treatment, use a 1,000 µl pipette to manually dissociate cardiomyocytes from the 6-well plate by repeatedly pulling up the cell disassociation enzyme and spraying it against the cardiomyocyte monolayer. Up to 30 pipetting repetitions may be required to dissociate the cardiomyocytes into single cells.
- After cells are dissociated and are in single-cell form, collect all cells into a 15 ml conical tube filled with 5 ml of RPMI/B27 medium with insulin to dilute out the cell disassociation enzyme and centrifuge for 4 min at 200 x g. Aspirate and discard the supernatant.
- Re-suspend the cells with 2 ml RPMI/B27 medium and plate onto a new ECMS-coated 6-well plate. Typically, higher confluency of cardiomyocytes helps with cell survival during replating. Aim to replate 2 million cells per new 6-well dish for optimal survival during replating.
At day 14, change the medium back to 2 ml of low glucose medium for a second glucose deprivation cycle. Culture the cells in this low glucose state for 3 more days. Most of the non-cardiomyocytes will die in this low-glucose culture condition.
At day 17, change the medium to 2 ml of RPMI/B27 medium with insulin. The remaining cells will be highly purified cardiomyocytes. These cardiomyocytes can be used for gene expression analysis, drug screening, metabolic analysis, and various other downstream assays.
Representative Results
The morphological changes during hiPSC differentiation.
The hiPSC cultured in feeder-free plates grew as flat, two-dimensional colonies. Upon reaching around 85% confluency, hiPSCs were treated with 6 µM CHIR for differentiation (Figure 1A). Substantial amounts of cell death, a normal and common phenomenon, were observed after 24 hr of CHIR treatment. After two days of CHIR treatment, the hiPSCs continued to differentiate towards a mesodermal fate. In comparison to cells in hiPSC colonies, cell size for these day 2 cells increased. Cell number also increased through cell division (Figure 1B). At day 5, the mesodermal cells were directed towards the cardiac lineage (cardiac mesoderm). At this time, cells begin coalescing and forming characteristic, branch-like structures (Figure 1C). At day 10, branch-like structures were highly pronounced, and cardiomyocytes started spontaneously beating (Figure 1D). Terminally-differentiated cardiomyocytes will not proliferate beyond this point. A timeline of the differentiation protocol is shown in Figure 2.
Immunostaining and flow cytometry results showed cardiomyocytes were purified with glucose starvation.
After 10 days of differentiation, more than 50% of cell areas are beating. However, beating cell sheets sometimes are also intermingled with non-cardiomyocytes. To examine cardiomyocyte purity, cells with immunostaining against cardiomyocyte marker cardiac Troponin-T (cTnT) were characterized to find that most cells were cTnT positive. However, in an unpurified population of differentiated cells, a number of cells also lacked expression of cTnT, suggesting the presence of non-cardiomyocytes in this differentiated population at day 13 (Figure 3A). However, with glucose starvation, almost all remaining cells were cTnT positive at day 13 (Figure 3B), indicating successful purification of the cardiomyocytes following glucose starvation. These data were further corroborated using flow cytometry analysis. A population of differentiated cells at day 13 post-differentiation contained approximately 50% TNNT2+ cells when not glucose starved (Figure 4A). A parallel differentiation was also conducted but with a 3 day glucose deprivation beginning at day 10 after differentiation. In contrast to the unstarved differentiation, glucose deprivation led to a purified population containing 90% TNNT2+ cells at day 13 (Figure 4B).
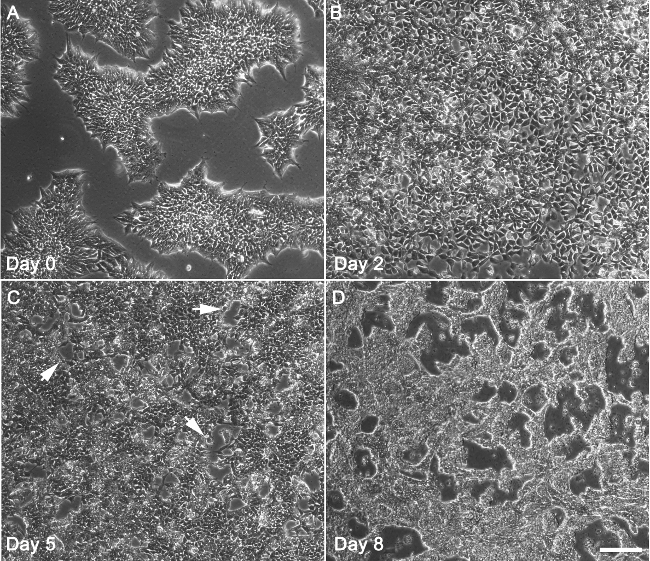 Figure 1. The sequential morphological changes during hiPSC differentiation towards the cardiac lineage. (A) Undifferentiated hiPSCs at day 0 exhibit typical colony morphology and reached around 85% confluency. (B) At day 2, cells after treatment with CHIR for two days reached 100% confluency and entered the mesodermal lineage. (C) At day 5, the cells after treatment with IWR1 for 2 days entered the cardiac mesoderm stage. Some cells began to fuse and form characteristic branch-like morphologies (indicated by arrows). (D) At day 7-10, branch-like structures are highly evident, and cardiomyocytes start spontaneously beating. Bar = 200 μm. Please click here to view a larger version of this figure.
Figure 1. The sequential morphological changes during hiPSC differentiation towards the cardiac lineage. (A) Undifferentiated hiPSCs at day 0 exhibit typical colony morphology and reached around 85% confluency. (B) At day 2, cells after treatment with CHIR for two days reached 100% confluency and entered the mesodermal lineage. (C) At day 5, the cells after treatment with IWR1 for 2 days entered the cardiac mesoderm stage. Some cells began to fuse and form characteristic branch-like morphologies (indicated by arrows). (D) At day 7-10, branch-like structures are highly evident, and cardiomyocytes start spontaneously beating. Bar = 200 μm. Please click here to view a larger version of this figure.
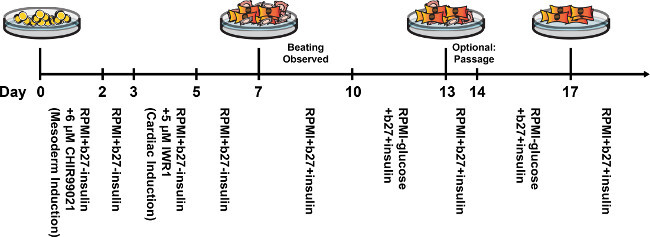 Figure 2. Timeline of hiPSC-CM differentiation protocol and subsequent glucose starvation process. Beating cardiomyocytes are typically first observed at approximately days 7-10. Two rounds of glucose starvation will result in a purified population of cardiomyocytes by day 17. Please click here to view a larger version of this figure.
Figure 2. Timeline of hiPSC-CM differentiation protocol and subsequent glucose starvation process. Beating cardiomyocytes are typically first observed at approximately days 7-10. Two rounds of glucose starvation will result in a purified population of cardiomyocytes by day 17. Please click here to view a larger version of this figure.
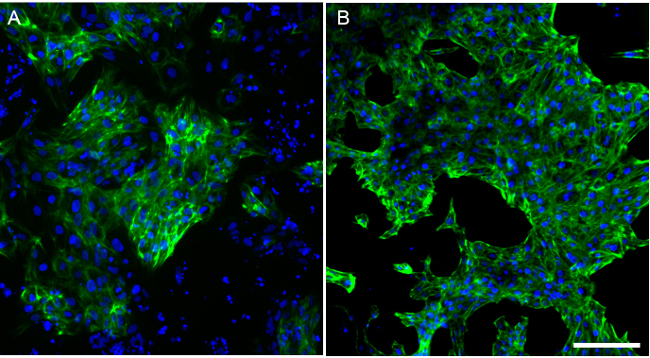 Figure 3. Immunofluorescence reveals purification of cardiomyocytes following glucose starvation. (A) Unpurified cells after 13 days of differentiation were stained with Cardiac Troponin T (cTnT). Most cells have differentiated into cardiomyocytes and were cTnT positive, but a number of non-cardiac, cTnT-negative cells are also present. (B) In contrast, after a 3 day glucose starvation beginning at day 10, almost all of the surviving cells were cTnT positive by day 13. Scale bar = 200 μm. Please click here to view a larger version of this figure.
Figure 3. Immunofluorescence reveals purification of cardiomyocytes following glucose starvation. (A) Unpurified cells after 13 days of differentiation were stained with Cardiac Troponin T (cTnT). Most cells have differentiated into cardiomyocytes and were cTnT positive, but a number of non-cardiac, cTnT-negative cells are also present. (B) In contrast, after a 3 day glucose starvation beginning at day 10, almost all of the surviving cells were cTnT positive by day 13. Scale bar = 200 μm. Please click here to view a larger version of this figure.
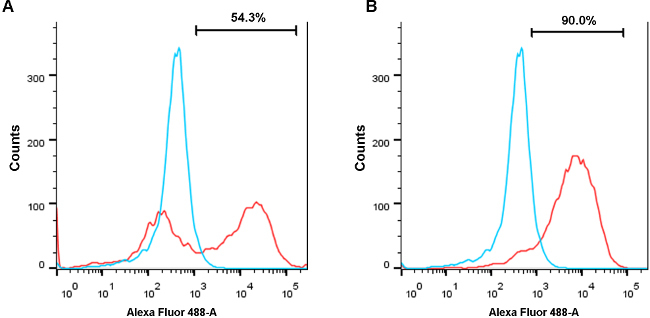 Figure 4. Flow cytometry reveals purification of cardiomyocytes following glucose starvation. (A) After 13 days of cardiac differentiation without glucose starvation, a population of cells exhibited roughly 50% TNNT2+ cardiomyocytes. Red indicates the differentiated cell population and blue indicates undifferentiated hiPSCs as a negative control. (B) After 10 days of cardiac differentiation and followed by 3 days of glucose starvation, a population of cells from the same batch of differentiations was purified to contain 90% TNNT2+ cardiomyocytes. Please click here to view a larger version of this figure.
Figure 4. Flow cytometry reveals purification of cardiomyocytes following glucose starvation. (A) After 13 days of cardiac differentiation without glucose starvation, a population of cells exhibited roughly 50% TNNT2+ cardiomyocytes. Red indicates the differentiated cell population and blue indicates undifferentiated hiPSCs as a negative control. (B) After 10 days of cardiac differentiation and followed by 3 days of glucose starvation, a population of cells from the same batch of differentiations was purified to contain 90% TNNT2+ cardiomyocytes. Please click here to view a larger version of this figure.
| The compositions for E8 medium | |||
| E8 | Volume: 1 L | Company | Catalog number |
| DMEM/F12 with Glutamine and HEPES | 1,000 ml | Invitrogen | 11330-032 |
| NaHCO3 (7.5%, 75 mg/ml) | 7.24 ml | Invitrogen | 25080-094 |
| L-Ascorbic acid 2-phosphate (64 mg/ml) | 1 ml | Sigma | A8960 |
| sodium selenite (70 µg/ml) | 200 µl | Sigma | S5261 |
| transferrin (50 mg/ml) | 214 µl | Sigma | T3705 |
| insulin (4 mg/ml) | 5 ml | Invitrogen | 12585-014 |
| FGF2 (200 ng/µl) | 500 µl | Peprotech | 100-18B |
| TGFB1 (100 ng/µl) | 20 µl | Peprotech | 100-21 |
| The stock solutions for E8 compositions | |
| L-Ascorbic acid-2-phosphate (64 mg/ml) | 3.2 g in 50 ml Ultrapure water, store 500 µl aliquots at -20 °C |
| Transferrin (50 mg/ml) | 500 mg in 10 ml of Ultrapure water, store 107 µl aliquots at -20 °C |
| Sodium selenite (70 µg/ml) | 35 mg sodium selenite into 500 ml ultrapure water, store 100 µl aliquots at -20 °C |
| FGF2 (200 ng/µl) | 1 mg in 5 ml cold D-PBS, store 250 µl aliquots at -20 °C |
| TGFB1 (100 ng/µl) | 100 µg in 1 ml cold 10 mM citric acid, pH 3, store 10 µl aliquots at -20 °C |
Table 1. Composition of E8 Medium.
Discussion
Obtaining a large amount of highly purified hiPSC-derived cardiomyocytes is critical for basic cardiac research as well as clinical and translational applications. Cardiac differentiation protocols have undergone tremendous improvements in recent years, transitioning from embryoid body-based methods utilizing cardiogenic growth factors2, to matrix sandwich methods12, and finally to small molecule-modulated and monolayer-based methods5. Of the aforementioned protocols, the protocol described here displayed the highest and the most reproducible cardiac differentiation efficiency for different hiPSC cell lines by modulating Wnt/β-catenin signaling using the small molecules CHIR and IWR5,7. Specifically, the undifferentiated hiPSCs were induced to undergo mesodermal differentiation with the GSK3-beta inhibitor CHIR and subsequent cardiac differentiation with the Wnt signaling inhibitor IWR1. These sequential steps of Wnt signaling modulation, aided by insulin depletion during the cardiac lineage induction phase, led to highly efficient cardiac differentiation. CHIR is a common small molecule GSK3-beta inhibitor used for mesodermal induction, but other small molecules for GSK3-beta inhibition have been tested in cardiomyocyte differentiation protocols11. Multiple options are also used for the subsequent small molecule Wnt inhibitor. For example, a recent publication focusing on chemically-defined differentiation of pluripotent stem cells to cardiomyocytes uses 2 µM Wnt-C59 for effective Wnt inhibition and cardiac mesoderm induction11.
In addition, the differentiated cardiomyocytes with a glucose starvation method were further purified, which takes advantage of the distinct glucose metabolic flow existing between cardiomyocytes and non-cardiomyocytes8. It is essential to note that the effectiveness of the glucose starvation method is density-dependent (i.e., differentiations that yielded higher percentages of cardiomyocytes are more likely to achieve greater cardiomyocyte survival). However, the transition to the low glucose medium is also a stressful condition for the cardiomyocytes. It is important to change the cells back to the regular RPMI/B27 with insulin medium after 3 days of glucose starvation. In this protocol, a feeder-free growth system was utilized, in which pluripotent stem cells were not grown on feeder mouse embryonic fibroblasts (MEFs). However, prior cardiac differentiation protocols have utilized feeder-based systems to produce cardiomyocytes from pluripotent stem cells5. Likewise, prior protocols have also used mTeSR1 medium for stem cell maintenance, but the E8 media and feeder-free system utilized here is superior due to its simplicity and lack of excess xenogeneic components such as bovine serum albumin and mouse embryonic fibroblasts.
Currently, pluripotent stem cell differentiation towards cardiomyocyte lineages is largely robust, but still suffers from hiPSC line-to-line variability in terms of efficiency. This remains a major issue in the cardiac differentiation field and will require further study. The differentiation efficiency is improved by proper maintenance of hiPSC lines, and in particular, preventing overconfluency during hiPSC maintenance. Additional variability arises from cell seeding during the passaging process, as an evenly seeded monolayer of hiPSCs tends to give rise to the best overall differentiations. Here, a mouse-derived, Matrigel-based ECMS for cell seeding during cell culture was successfully utilized, but an eventual transition to xeno-free substrates is recommended. In regards to cell seeding on ECMS, uneven seeding often results in uneven distribution of cardiomyocytes within a particular well. For example, if hiPSCs are seeded unevenly, the edges of a well in a six-well plate may give rise to higher numbers of cardiomyocytes than the center of the well. These uneven seeding conditions may give rise to low efficiency differentiations and a larger number of non-cardiomyocyte, mesodermal derivatives such as fibroblasts and smooth muscle cells. We have also observed that low efficiency differentiations (below 50%) are much more difficult to purify using the glucose deprivation process mentioned here. Another side effect of long-term glucose starvation is a potential loss in cardiomyocyte viability. Although the cardiomyocytes are able to metabolize lactate in the absence of glucose, we find that this switch to a low-glucose environment is stressful for the cells. Cells may stop spontaneously beating, and some cell loss may be observed if glucose starvation is prolonged beyond the recommended time. This enhanced sensitivity to glucose deprivation may be reflective of the well-established developmental, functional, and electrophysiological immaturity of stem cell-derived cardiomyocytes in comparison to true adult cardiomyocytes13.
In summary, the protocol described here combines the small molecule-based, hiPSC monolayer cardiac differentiation method with a cardiomyocyte-purifying glucose starvation method. This protocol allows for the reproducible generation of highly purified cardiomyocytes and should facilitate various downstream assays relevant to cardiovascular disease modeling and drug screening.
Disclosures
The authors do not declare competing interests.
Acknowledgments
This work was supported in part by the NIH/NHBI (U01 HL099776-5), the NIH Director’s New Innovator Award (DP2 OD004411-2), the California Institute of Regenerative Medicine (RB3-05129), the American Heart Association (14GRNT18630016) and the Endowed Faculty Scholar Award from the Lucile Packard Foundation for Children and the Child Health Research Institute at Stanford (to SMW). We also acknowledge funding support from the American Heart Association Predoctoral Fellowship 13PRE15770000, and National Science Foundation Graduate Research Fellowship Program DGE-114747 (AS).
References
- Sharma A, Wu JC, Wu SM. Induced pluripotent stem cell-derived cardiomyocytes for cardiovascular disease modeling and drug screening. Stem Cell Research & Therapy. 2013;4(6):150. doi: 10.1186/scrt380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kehat I, et al. Human embryonic stem cells can differentiate into myocytes with structural and functional properties of cardiomyocytes. The Journal of Clinical Investigation. 2001;108(3):407–414. doi: 10.1172/JCI12131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, et al. Functional cardiomyocytes derived from human induced pluripotent stem cells. Circulation Research. 2009;104(4):e30–e41. doi: 10.1161/CIRCRESAHA.108.192237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kattman SJ, et al. Stage-specific optimization of activin/nodal and BMP signaling promotes cardiac differentiation of mouse and human pluripotent stem cell lines. Cell Stem Cell. 2011;8(2):228–240. doi: 10.1016/j.stem.2010.12.008. [DOI] [PubMed] [Google Scholar]
- Lian X, et al. Robust cardiomyocyte differentiation from human pluripotent stem cells via temporal modulation of canonical Wnt signaling. Proceedings of the National Academy of Sciences of the United States of America. 2012;109(27):E1848–E1857. doi: 10.1073/pnas.1200250109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma A, et al. Human induced pluripotent stem cell-derived cardiomyocytes as an in vitro model for coxsackievirus B3-induced myocarditis and antiviral drug screening platform. Circulation Research. 2014;115(6):556–566. doi: 10.1161/CIRCRESAHA.115.303810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lian X, et al. Directed cardiomyocyte differentiation from human pluripotent stem cells by modulating Wnt/beta-catenin signaling under fully defined conditions. Nature Protocols. 2013;8(1):162–175. doi: 10.1038/nprot.2012.150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tohyama S, et al. Distinct metabolic flow enables large-scale purification of mouse and human pluripotent stem cell-derived cardiomyocytes. Cell Stem Cell. 2013;12(12):127–137. doi: 10.1016/j.stem.2012.09.013. [DOI] [PubMed] [Google Scholar]
- Rodin S, et al. Long-term self-renewal of human pluripotent stem cells on human recombinant laminin-511. Nature Biotechnology. 2010;28(6):611–615. doi: 10.1038/nbt.1620. [DOI] [PubMed] [Google Scholar]
- Li X, Meng G, Krawetz R, Liu S, Rancourt DE. The ROCK inhibitor Y-27632 enhances the survival rate of human embryonic stem cells following cryopreservation. Stem Cells And Development. 2008;17(6):1079–1085. doi: 10.1089/scd.2007.0247. [DOI] [PubMed] [Google Scholar]
- Burridge PW, et al. Chemically defined generation of human cardiomyocytes. Nature Methods. 2014;11(8):855–860. doi: 10.1038/nmeth.2999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, et al. Extracellular matrix promotes highly efficient cardiac differentiation of human pluripotent stem cells: the matrix sandwich method. Circulation Research. 2012;111(9):1125–1136. doi: 10.1161/CIRCRESAHA.112.273144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burridge PW, Keller G, Gold JD, Wu JC. Production of de novo cardiomyocytes: human pluripotent stem cell differentiation and direct reprogramming. Cell Stem Cell. 2012;10(1):16–28. doi: 10.1016/j.stem.2011.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]


