Abstract
Extracellular vesicles (EVs), membranous particles released from various types of cells, hold a great potential for clinical applications. They contain nucleic acid and protein cargo and are increasingly recognized as a means of intercellular communication utilized by both eukaryote and prokaryote cells. However, due to their small size, current protocols for isolation of EVs are often time consuming, cumbersome, and require large sample volumes and expensive equipment, such as an ultracentrifuge. To address these limitations, we developed a paper-based immunoaffinity platform for separating subgroups of EVs that is easy, efficient, and requires sample volumes as low as 10 μl. Biological samples can be pipetted directly onto paper test zones that have been chemically modified with capture molecules that have high affinity to specific EV surface markers. We validate the assay by using scanning electron microscopy (SEM), paper-based enzyme-linked immunosorbent assays (P-ELISA), and transcriptome analysis. These paper-based devices will enable the study of EVs in the clinic and the research setting to help advance our understanding of EV functions in health and disease.
Keywords: Bioengineering, Issue 98, extracellular vesicles, exosomes, cellulose paper, microfluidics, paper ELISA, aqueous humor, chemical conjugation
Introduction
Extracellular vesicles (EVs) are heterogeneous membranous particles that range in size from 40 nm to 5,000 nm and are released actively by many cell types via different biogenesis routes1-9. They contain unique and selected subsets of DNA, RNA, proteins, and surface markers from parental cells. Their involvement in a variety of cellular processes, such as intercellular communication10, immunity modulation11, angiogenesis12, metastasis12, chemoresistance13, and the development of eye diseases9, is increasingly recognized and has spurred a great interest in their utility in diagnostic, prognostic, therapeutic, and basic biology applications.
EVs can be classically categorized as exosomes, microvesicles, apoptotic bodies, oncosomes, ectosomes, microparticles, telerosomes, prostatosomes, cardiosomes, and vexosomes, etc., based on their biogenesis or cellular origin. For example, exosomes are formed in multivesicular bodies, whereas microvesicles are generated by budding directly from plasma membrane and apoptotic vesicles are from apoptotic or necrotic cells. However, the nomenclature is still under refined, partly due to a lack of thorough understanding and characterization of EVs. Several methods have been developed to purify EVs, including ultracentrifugation14, ultrafiltration15, magnetic beads16, polymeric precipitation17-19, and microfluidic techniques20-22. The most common procedure to purify EVs involves a series of centrifugations and/or filtration to remove large debris and other cellular contaminants, followed by a final high-speed ultracentrifugation, a process that is expensive, tedious, and nonspecific14,23,24. Unfortunately, technological need for rapid and reliable isolation of EVs with satisfactory purity and efficiency is not yet met.
We have developed a paper-based immunoaffinity device that provides a simple, time- and cost-saving, yet efficient way to isolate and characterize subgroups of EVs22. Cellulose paper cut into a defined shape can be arranged and laminated using two plastic sheets with registered through-holes. In contrast to the general strategy to define the fluid boundary in paper-based devices by printing hydrophobic wax or polymers25-27, these laminated paper patterns are resistant to many organic liquids, including ethanol. Paper test zones are chemically modified to provide stable and dense coverage of capture molecules (e.g., target-specific antibodies) that have high affinity to specific surface markers on EV subgroups. Biological samples can be pipetted directly onto the paper test zones, and purified EVs are retained after rinse steps. Characterization of isolated EVs can be performed by SEM, ELISA, and transcriptomic analysis.
Protocol
A general diagram of the operation procedure is provided in Figure 1. Using ethical practices, we collected blood samples from healthy subjects, and obtained aqueous humor samples from patients through the Taichung Veterans General Hospital (TCVGH), Taichung, Taiwan under IRB approved protocols (IRB TCVGH No. CF11213-1).
1. Fabrication of Paper Devices
Cut chromatography paper into circles of 5 mm in diameter to provide the same layout as a 96-well microtiter plate. Sandwich these paper pieces with two polystyrene sheets with registered through-holes, and laminate.
- Modify paper devices using the chemical conjugation method described in the following steps28.
- Treat paper devices with oxygen plasma (100 mW, 1% oxygen, 30 sec) in a plasma chamber. CAUTION: Special caution must be taken when working with 3-mercaptopropyl trimethoxysilane and N-γ-maleimidobutyryloxy succinimide ester (GMBS), since they are both moisture sensitive and toxic. Wear protective gloves and avoid inhalation or contact with skin and eyes. Keep the stock bottle tightly closed and allow it to equilibrate to RT before opening to avoid water condensation. Alternatively, open the stock bottle inside a nitrogen-filled glove bag or box. Aliquot into small vials to avoid frequent opening of the stock bottle.
- Immediately incubate treated paper devices in a 4% (v/v) solution of 3-mercaptopropyl trimethoxysilane in ethanol (200 proof) for 30 min.
- Rinse paper devices with ethanol and incubate them with 0.01 μmol/ml GMBS in ethanol for 15 min.
- Rinse with ethanol (200 proof) and incubate paper devices with 10 µg/ml avidin solution in phosphate buffered saline (PBS) for 1 hr at 4 °C. Store at 4 °C if needed, and use within 4 weeks.
Wet each paper test zones with 10 μl PBS containing 1% (w/v) bovine serum albumin (BSA) for 3 × 10 min before spotting 10 μl biotinylated capture molecules for 3 × 10 min. Use anti-CD63 antibody (20 μg/ml in PBS containing 1% (w/v) BSA and 0.09% (w/v) sodium azide) or annexin V (1:20, v/v in annexin V binding buffer) as capture molecules.
Rinse off unbound anti-CD63 antibody or annexin V molecules using 10 μl corresponding PBS containing 1% (w/v) BSA or annexin V binding buffer solutions for 3 × 1 min, respectively.
2. Serum and Aqueous Humor Sample Collection and Processing
- Serum collection.
- Collect 10 ml of peripheral blood by venipuncture in serum separation tubes and gently invert the tube five times. Set the tube in a vertical position and wait for 30 min.
- Centrifuge at 1,200 × g for 15 min. Transfer the serum from the top layer to a clean tube, and centrifuge again at 3,000 × g for 30 min.
- Pass the supernatant through a 0.8 μm filter. Keep the sample at -80 °C until use.
Aqueous humor collection: collect aqueous humor samples from patients diagnosed by an ophthalmologist directly through invasive procedures and store samples at -80 °C until use.
3. Isolation of Extracellular Vesicles
Spot samples onto each paper test zone at a rate of 5 μl/min.
Rinse off unbound EVs with 10 μl corresponding PBS containing 1% (w/v) BSA or annexin V binding buffer solutions for 3 × 1 min for paper devices functionalized with anti-CD63 antibody or annexin V molecules, respectively. Perform the following downstream assays.
4. Downstream Assay Example 1: Scanning Electromicrographs
Fix EVs captured on functionalized paper test zone using 10 μl 0.5× Karnovsky’s fixative for 10 min.
Rinse the samples with ample PBS for 2 × 5 min.
Dehydrate the samples with subsequent 35% ethanol for 10 min, 50% ethanol for 2 × 10 min, 70% ethanol for 2 × 10 min, 95% ethanol for 2 × 10 min, and 100% ethanol for 4 × 10 min.
Critical dry and sputter coat the samples with palladium/gold, and examine using an scanning electron microscope operated at low electron acceleration voltage (~ 5 kV).
5. Downstream Assay Example 2: Paper-based ELISA
Add a 5 μl solution containing anti-CD9 primary antibody at 1:1,000 dilution in PBS to each test zone containing captured EVs. Wait 1 min.
Rinse the samples with 30 ml PBS shaken at 100 rpm for 30 sec.
Add a 5 μl solution containing horseradish peroxidase (HRP)-linked secondary antibody at 1:1,000 dilution in PBS and wait 1 min.
Rinse the samples with 30 ml PBS shaken at 100 rpm for 30 sec.
Add a 5 μl colorimetric substrate containing 1:1 volume ratio of hydrogen peroxide and 3,3’,5,5’-tetramethylbenzidine (TMB) and scan using a desktop scanner.
6. Downstream Assay Example 3: RNA Isolation
Immerse the samples in 35 μl of polyvinylpyrrolidone-based RNA isolation aid and 265 μl of lysis/binding buffer to lyse EVs captured. Vortex vigorously.
Add 30 μl homogenate provided in the isolation kit to the lysate. Vortex vigorously and put on ice for 10 min.
- Extract the RNA using acid phenol-chloroform separation described in the following steps.
- Take 330 μl of phenol-chloroform from the bottom layer of the bottle and add to the homogenate/lysate. Vortex vigorously.
- Centrifuge at 10,000 x g for 5 min. Transfer the aqueous (upper) phase to a clean tube and note the volume removed.
Precipitate the total RNA with 100% ethanol of the volume that is 1.25 times the volume of the aqueous phase removed in the previous step and collect the RNA in the elution solution preheated to 95 °C.
Concentrate and further purify the RNA using an RNA cleanup kit according to the manufacture’s protocol.
Representative Results
The ability of the paper device to isolate subgroups of EVs efficiently relies upon its sensitive and specific recognition of EV surface markers. The stable modification of paper fibers with capture molecules is achieved by using avidin-biotin chemistry as described elsewhere28-30. The effectiveness of chemical conjugation and that of the physisorption method is assessed using fluorescence-based readouts. The paper test zones are prepared following the protocol step 1) except the capture molecule is replaced with 20 μg/ml fluorophore R-phycoerythrin-conjugated biotin molecules (PE-biotin) in step 1.3. The paper test zones are then rinsed with PBS to remove unbound PE-biotin molecules. In contrast, 10 μl 20 μg/ml PE-biotin in PBS containing 1% (w/v) BSA and 0.09% (w/v) sodium azide is directly spotted on the paper test zone for 3 × 10 min, followed by PBS rinse for conducting the physisorption of PE-biotin dye molecules. Chemically modified paper devices ("paper+crosslinker+dye") along with unprocessed paper devices ("paper"), paper devices modified with the same protocol without the application of PE-biotin molecules ("paper+crosslinker") and paper devices with physisorbed PE-biotin molecules ("physisorbed-dye") are scanned using an image-record system set to 540-560 nm excitation, 575 nm-long-pass emission, and the data is saved in the form of 16-bit files and analyzed using ImageJ software. As shown in Figure 2A, fluorescence intensity of test zones prepared by the chemical conjugation method is substantially higher, as compared to physically coated paper test zones (p-value < 0.0005, n = 5, Student’s t-test). The capturing of EVs is also specific as exemplified in Figure 2B, C, where few EVs are retained on the paper device when the capture molecules are replaced by PBS solution.
EVs are heterogeneous in sizes, specific contents, and biogenesis routes. To bind them, paper devices coated with two different EV capture molecules, anti-CD63 antibodies and annexin V, were prepared. CD63, a member of the tetraspanin family (which includes CD9, CD63, CD81 and CD82 molecules) is enriched on EV membrane31, and annexin V has a strong affinity for phosphatidylserine (PS)32,33. It is found that EVs are released constitutively during early apoptosis or under stress conditions, in which process normal cell phospholipid asymmetry is lost, leading to increased exposure of PS on the outer leaflet of the EV membrane. Analyses of SEM images show that the size distributions of CD63+ EVs and PS+ EVs are different, with mean diameters of 80 and 43 nm, respectively (Figure 3).The two-tailed Student’s t-test is used to determine the statistical p-value (p-value < 0.0001). Surprisingly, CD63+ EVs appear rounder and larger than PS+ EVs on average, which biological significance remains to be elucidated.
RNA from EVs captured on the paper device can be extracted. Analyses with a bioanalyzer showed similar overall profiles for total RNAs extracted from CD63+ and PS+ EVs (Figure 4A). Compared to the amounts of RNA extracted using the microfluidic and ultracentrifugation techniques, respectively about twice and 39 times more RNAs per unit volume of serum sample were extracted utilizing the paper device, albeit using a smaller sample volume (Table 1). P-ELISA can be carried out for the detection of EVs. Anti-CD9 primary antibody and HRP-conjugated secondary antibody are used in this study. Figure 4B displays grey value changes produced by the enzymatic reaction of HRP for paper devices applied with 10 μl PBS, 50% serum (diluted with PBS), or 100% serum, respectively. The grey value increases linearly within the range of serum samples tested.
Paper devices are especially advantageous when the sample amount is limited such as in the case of aqueous humor, when only < 200 μl/eye can be collected. Multiple samples are often needed and pooled together if other isolation methods, e.g., ultracentrifugation, are to be used. EVs from patient aqueous humor samples isolated with paper devices coated with anti-CD63 antibodies can be seen in SEM images as showed in Figure 5.
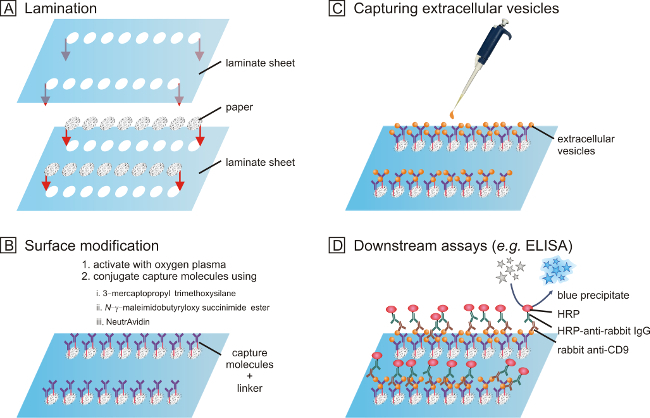 Figure 1. The fabrication and operation procedure of paper-based assays aimed at isolation and characterization of extracellular vesicles. (A) Filter paper is cut into the desired shape and laminated. (B) The surface of the paper is modified with a brief treatment of oxygen plasma and reacted with 3-mercaptopropyl trimethoxysilane, N-γ-maleimidobutyryloxy succinimide ester, and NeutrAvidin, in that order. Biotinylated capture molecules, e.g., antibodies, can then be immobilized via the strong affinity between biotin and NeutrAvidin molecules. (C) Biosamples can be pipette onto the functionalized paper device. Extracellular vesicles can be isolated. (D) Downstream assays, such as SEM, transcriptome, and ELISA (shown), can be performed. Please click here to view a larger version of this figure.
Figure 1. The fabrication and operation procedure of paper-based assays aimed at isolation and characterization of extracellular vesicles. (A) Filter paper is cut into the desired shape and laminated. (B) The surface of the paper is modified with a brief treatment of oxygen plasma and reacted with 3-mercaptopropyl trimethoxysilane, N-γ-maleimidobutyryloxy succinimide ester, and NeutrAvidin, in that order. Biotinylated capture molecules, e.g., antibodies, can then be immobilized via the strong affinity between biotin and NeutrAvidin molecules. (C) Biosamples can be pipette onto the functionalized paper device. Extracellular vesicles can be isolated. (D) Downstream assays, such as SEM, transcriptome, and ELISA (shown), can be performed. Please click here to view a larger version of this figure.
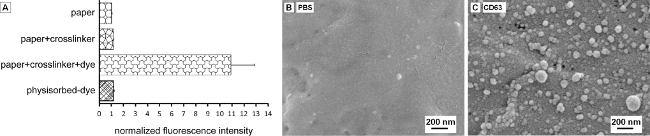 Figure 2. Extracellular vesicles (EVs) can be captured on functionalized paper devices with good specificity and sensitivity. (A) Fluorescently labeled biotin molecules are highly immobilized onto the paper device via chemical conjugation process ("paper+crosslinker+dye") in contrast to physical adsorption ("physisorbed-dye"). Unprocessed paper ("paper") and conjugation molecules ("paper+crosslinker") contribute little to the fluorescence intensity. (B) A representative SEM image showing few EVs are captured on the paper device non-specifically when the capture molecules is replaced by PBS containing 1% (w/v) BSA. (C) Many EVs are retained on paper devices coated with anti-CD63 antibodies. Please click here to view a larger version of this figure.
Figure 2. Extracellular vesicles (EVs) can be captured on functionalized paper devices with good specificity and sensitivity. (A) Fluorescently labeled biotin molecules are highly immobilized onto the paper device via chemical conjugation process ("paper+crosslinker+dye") in contrast to physical adsorption ("physisorbed-dye"). Unprocessed paper ("paper") and conjugation molecules ("paper+crosslinker") contribute little to the fluorescence intensity. (B) A representative SEM image showing few EVs are captured on the paper device non-specifically when the capture molecules is replaced by PBS containing 1% (w/v) BSA. (C) Many EVs are retained on paper devices coated with anti-CD63 antibodies. Please click here to view a larger version of this figure.
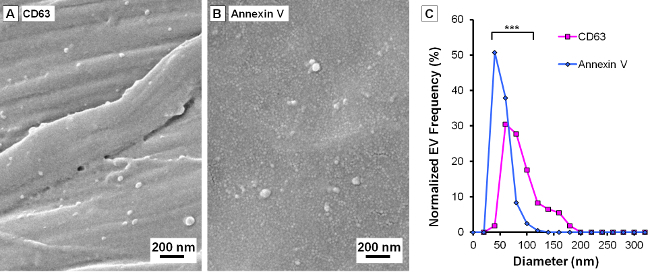 Figure 3. Subgroups of Extracellular vesicles (EVs) can have different size distributions. (A) and (B) are representative SEM images of EVs captured on paper devices coated with anti-CD63 antibodies and annexin V molecules, respectively. (C) CD63+ EVs appear larger than PS+ EVs under experimental conditions used in this study (p-value < 0.0001, n = 124 and 203, Student’s t-test). Please click here to view a larger version of this figure.
Figure 3. Subgroups of Extracellular vesicles (EVs) can have different size distributions. (A) and (B) are representative SEM images of EVs captured on paper devices coated with anti-CD63 antibodies and annexin V molecules, respectively. (C) CD63+ EVs appear larger than PS+ EVs under experimental conditions used in this study (p-value < 0.0001, n = 124 and 203, Student’s t-test). Please click here to view a larger version of this figure.
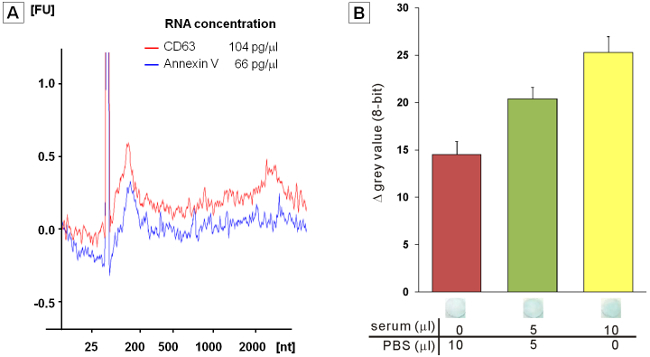 Figure 4. Downstream transcriptome and ELISA assays. (A) Bioanalyzer data showing the intensities (y-axis, in arbitrary units) and size (x-axis, in nucleotides (nt)) distributions of fluorescently-labeled total RNA extracted from EVs captured on anti-CD63 antibody- or annexin V-coated filter papers. The migrating peak at ~25 nt represents an internal standard. (B) Grey value changes produced by the enzymatic reaction of horseradish peroxidase (HRP) in the P-ELISA assay for paper devices applied with 10 μl samples of PBS, 50% serum (diluted with PBS) and 100% serum samples. Please click here to view a larger version of this figure.
Figure 4. Downstream transcriptome and ELISA assays. (A) Bioanalyzer data showing the intensities (y-axis, in arbitrary units) and size (x-axis, in nucleotides (nt)) distributions of fluorescently-labeled total RNA extracted from EVs captured on anti-CD63 antibody- or annexin V-coated filter papers. The migrating peak at ~25 nt represents an internal standard. (B) Grey value changes produced by the enzymatic reaction of horseradish peroxidase (HRP) in the P-ELISA assay for paper devices applied with 10 μl samples of PBS, 50% serum (diluted with PBS) and 100% serum samples. Please click here to view a larger version of this figure.
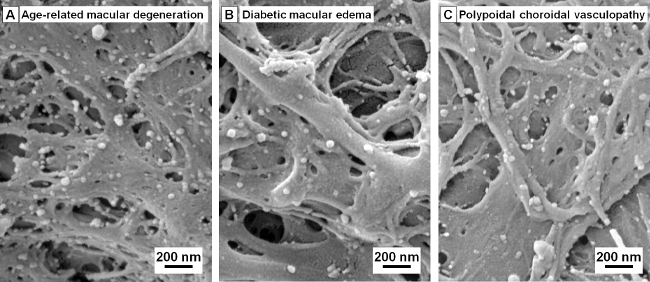 Figure 5. Extracellular vesicles (EVs) in aqueous humor samples can be captured on functionalized filter paper without sample pooling. SEM images of isolated CD63+ EVs in aqueous humor samples from patients with (A) age-related macular degeneration, (B) diabetic macular edema, and (C) polypoidal choroidal vasculopathy, respectively. Please click here to view a larger version of this figure.
Figure 5. Extracellular vesicles (EVs) in aqueous humor samples can be captured on functionalized filter paper without sample pooling. SEM images of isolated CD63+ EVs in aqueous humor samples from patients with (A) age-related macular degeneration, (B) diabetic macular edema, and (C) polypoidal choroidal vasculopathy, respectively. Please click here to view a larger version of this figure.
| isolation method | capture molecule | serum vol. (µl) | average RNA extracted (ng) | average extracted RNA/serum (pg/µl) | ratio of average RNA/serum | ref |
| microfluidic device | anti-human CD63 | 400 | 4.14 | 10.4 | 20 | 20 |
| paper device | anti-human CD63 | 72 | 1.49 ± 0.08 | 20.7 | 39 | 22 |
| paper device | annexin V | 72 | 0.99 ± 0.26 | 13.8 | 26 | 22 |
| ultracentrifugation | - | 2,000 | 1.06 ± 0.64 | 0.53 | 1 | |
| serum | - | 72 | 1.23 ± 0.58 | 17 | 32 | 22 |
Table 1. Total RNA extracted from serum samples from healthy volunteers analyzed with a bioanalyzer (n = 4 for all conditions).
Discussion
The most critical steps for successful isolation of subgroups of extracellular vesicles are: 1) a good choice of paper; 2) stable and high coverage of capture molecules on the surface of the paper fibers; 3) proper handling of samples; and 4) general laboratory hygiene practice.
Porous materials have been utilized in many inexpensive and equipment-free assays. They may have tunable pore size, versatile functionality, low cost and high surface-to-volume ratio permitting passive wicking of fluids. We chose chromatography cellulose paper grade 1 mainly for its proper pore size and low protein adsorption. Compared to another commonly used porous materials, nitrocellulose, cellulose paper has much lower nonspecific protein adsorption34. We also find little nonspecific capture of EVs in this study. In addition, its particle retention level, 11 μm, is larger than the size of EVs35, allowing for specific capture in contrast to physical trapping of EVs. Here, we focus on EVs of a smaller size by passing serum samples through a 0.8 μm filter, which step may be modified when larger EVs are to be studied.
Chemical surface modification can be a key to achieve a stable and high coverage of capture molecules on the surface as a means of improving the efficiency of EV isolation. Surface modifications of cellulose have been reviewed36,37. There are high numbers of hydroxyl (-OH) groups on the surface of cellulose and they can react with silane to give stable chemical Si-O-C bonds after being activated with oxygen plasma or other condensation process. The silane used in this study is (3-mercaptopropyl) trimethoxysilane, (MeO)3Si-(CH2)3-SH. The thiol (-SH) group can react with GMBS to provide a short space arm (0.73 nm) and a crosslinking group that couples with primary amines. Capture molecules with amine groups can be conjugated to GMBS directly. In this study, however, we conjugated NeutrAvidin instead to provide a general scheme to conjugate various capture molecules that are biotin-labeled. However, consensus on EV subtype-specific markers has yet to be reached17. NeutrAvidin has a very strong affinity to biotin and a near-neutral isoelectric point (pH 6.3), minimizing nonspecific interactions with negatively charged objects, e.g. the surface of EVs. In contrast, avidin with an isoelectric point of 10.5 carries positive charges at neutral pH. A higher concentration of anti-CD63 antibody is used than that in Chen et al.20, mainly due to the small volume applied and the larger surface-to-volume ratio of paper device. However, the amount and compositions of proteins adsorbed onto the paper fibers have not been characterized, which may be sample-dependent. Hence, care must be taken when interpreting the results of protein analysis.
Collected biological samples should be handled gently and processed rapidly to prevent artifactually increased EV levels. It is recommended to remove cells and platelets, if any, before storage at -80 °C. A current standardization of sample handling in EV research can be found in this review17.
Proper laboratory hygiene practice should be employed. Always wear gloves and change gloves frequently to minimize the introduction of dust particles and ribonucleases. Isolation of RNA from EVs in cell culture-conditioned media can yield more than 70 times greater concentrations using paper devices as compared to that extracted from EVs concentrated using the ultracentrifuge method (unpublished).
The protocol described herein uses cellulose paper devices modified chemically with capture molecules to isolate different subgroups of EVs. The method is simple, efficient, and especially valuable when the sample volume is limited.
Disclosures
The authors declare that they have no competing financial interests.
Acknowledgments
This work was supported in part by the Taiwan National Science Council grants- NSC 99-2320-B-007-005-MY2 (CC) and NSC 101-2628-E-007-011-MY3 (CMC), and the Veterans General Hospitals and University System of Taiwan Joint Research Program (CC).
References
- Caby MP, Lankar D, Vincendeau-Scherrer C, Raposo G, Bonnerot C. Exosomal-like vesicles are present in human blood plasma. Int. Immunol. 2005;17:879–887. doi: 10.1093/intimm/dxh267. [DOI] [PubMed] [Google Scholar]
- Lasser C, et al. Human saliva, plasma and breast milk exosomes contain RNA: uptake by macrophages. J. Transl. Med. 2011;9:9. doi: 10.1186/1479-5876-9-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raj DAA, Fiume I, Capasso G, Pocsfalvi G. A multiplex quantitative proteomics strategy for protein biomarker studies in urinary exosomes. Kidney Int. 2012;81:1263–1272. doi: 10.1038/ki.2012.25. [DOI] [PubMed] [Google Scholar]
- Wiggins RC, Glatfelter A, Kshirsagar B, Brukman J. Procoagulant activity in normal human-urine associated with subcellular particles. Kidney Int. 1986;29:591–597. doi: 10.1038/ki.1986.39. [DOI] [PubMed] [Google Scholar]
- Admyre C, et al. Exosomes with immune modulatory features are present in human breast milk. J. Immunol. 2007;179:1969–1978. doi: 10.4049/jimmunol.179.3.1969. [DOI] [PubMed] [Google Scholar]
- Keller S, Ridinger J, Rupp AK, Janssen JWG, Altevogt P. Body fluid derived exosomes as a novel template for clinical diagnostics. J. Transl. Med. 2011;9:86. doi: 10.1186/1479-5876-9-86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asea A, et al. Heat shock protein-containing exosomes in mid-trimester amniotic fluids. J. Reprod. Immunol. 2008;79:12–17. doi: 10.1016/j.jri.2008.06.001. [DOI] [PubMed] [Google Scholar]
- Bard MP, et al. Proteomic analysis of exosomes isolated from human malignant pleural effusions. Am. J. Resp. Cell Mol. Biol. 2004;31:114–121. doi: 10.1165/rcmb.2003-0238OC. [DOI] [PubMed] [Google Scholar]
- Perkumas KM, Hoffman EA, McKay BS, Allingham RR, Stamer WD. Myocilin-associated exosomes in human ocular samples. Exp. Eye Res. 2007;84:209–212. doi: 10.1016/j.exer.2006.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson HC, Mulhall D, Garimella R. Role of extracellular membrane vesicles in the pathogenesis of various diseases, including cancer, renal diseases, atherosclerosis, and arthritis. Lab. Invest. 2010;90:1549–1557. doi: 10.1038/labinvest.2010.152. [DOI] [PubMed] [Google Scholar]
- Montecalvo A, et al. Mechanism of transfer of functional microRNAs between mouse dendritic cells via exosomes. Blood. 2012;119:756–766. doi: 10.1182/blood-2011-02-338004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grange C, et al. Microvesicles released from human renal cancer stem cells stimulate angiogenesis and formation of lung premetastatic niche. Cancer Res. 2011;71:5346–5356. doi: 10.1158/0008-5472.CAN-11-0241. [DOI] [PubMed] [Google Scholar]
- Jaiswal R, et al. Microparticle-associated nucleic acids mediate trait dominance in cancer. Faseb. J. 2012;26:420–429. doi: 10.1096/fj.11-186817. [DOI] [PubMed] [Google Scholar]
- Thery C, Clayton A, Amigorena S, Raposo G. In: Current protocols in cell biology. UNIT 3.22. Morgan KK, editor. New York, NY: John Wiley; 2006. [DOI] [PubMed] [Google Scholar]
- Rood IM, et al. Comparison of three methods for isolation of urinary microvesicles to identify biomarkers of nephrotic syndrome. Kidney Int. 2010;78:810–816. doi: 10.1038/ki.2010.262. [DOI] [PubMed] [Google Scholar]
- Taylor DD, Gercel-Taylor C. MicroRNA signatures of tumor-derived exosomes as diagnostic biomarkers of ovarian cancer. Gynecol. Oncol. 2008;110:13–21. doi: 10.1016/j.ygyno.2008.04.033. [DOI] [PubMed] [Google Scholar]
- Witwer KW, et al. Standardization of sample collection, isolation and analysis methods in extracellular vesicle research. J. Extracellular Vesicles. 2013;2:20360. doi: 10.3402/jev.v2i0.20360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez-Llama P, et al. Tamm-Horsfall protein and urinary exosome isolation. Kidney Int. 2010;77:736–742. doi: 10.1038/ki.2009.550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alvarez ML, Khosroheidari M, Ravi RK, DiStefano JK. Comparison of protein, microRNA, and mRNA yields using different methods of urinary exosome isolation for the discovery of kidney disease biomarkers. Kidney Int. 2012;82:1024–1032. doi: 10.1038/ki.2012.256. [DOI] [PubMed] [Google Scholar]
- Chen C, et al. Microfluidic isolation and transcriptome analysis of serum microvesicles. Lab. Chip. 2010;10:505–511. doi: 10.1039/b916199f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies RT, et al. Microfluidic filtration system to isolate extracellular vesicles from blood. Lab. Chip. 2012;12:5202–5210. doi: 10.1039/c2lc41006k. [DOI] [PubMed] [Google Scholar]
- Chen C, et al. Paper-based immunoaffinity devices for accessible isolation and characterization of extracellular vesicles. Microfluid. Nanofluid. 2014;16:849–856. [Google Scholar]
- Lamparski HG, et al. Production and characterization of clinical grade exosomes derived from dendritic cells. J. Immunol. Methods. 2002;270:211–226. doi: 10.1016/s0022-1759(02)00330-7. [DOI] [PubMed] [Google Scholar]
- Cantin R, Diou J, Belanger D, Tremblay AM, Gilbert C. Discrimination between exosomes and HIV-1: Purification of both vesicles from cell-free supernatants. J. Immunol. Methods. 2008;338:21–30. doi: 10.1016/j.jim.2008.07.007. [DOI] [PubMed] [Google Scholar]
- Carrilho E, Martinez AW, Whitesides GM. Understanding wax printing: a simple micropatterning process for paper-based microfluidics. Anal. Chem. 2009;81:7091–7095. doi: 10.1021/ac901071p. [DOI] [PubMed] [Google Scholar]
- Dungchai W, Chailapakul O, Henry CS. Electrochemical detection for paper-based microfluidics. Anal. Chem. 2009;81:5821–5826. doi: 10.1021/ac9007573. [DOI] [PubMed] [Google Scholar]
- Glavan AC, et al. Omniphobic 'R-F paper' produced by silanization of paper with fluoroalkyltrichlorosilanes. Adv. Funct. Mater. 2014;24:60–70. [Google Scholar]
- Usami S, Chen HH, Zhao YH, Chien S, Skalak R. Design and construction of a linear shear-stress flow chamber. Ann. Biomed. Eng. 1993;21:77–83. doi: 10.1007/BF02368167. [DOI] [PubMed] [Google Scholar]
- Murthy SK, Sin A, Tompkins RG, Toner M. Effect of flow and surface conditions on human lymphocyte isolation using microfluidic chambers. Langmuir. 2004;20:11649–11655. doi: 10.1021/la048047b. [DOI] [PubMed] [Google Scholar]
- Cras JJ, Rowe-Taitt CA, Nivens DA, Ligler FS. Comparison of chemical cleaning methods of glass in preparation for silanization. Biosens. Bioelectron. 1999;14:683–688. [Google Scholar]
- Thery C, Zitvogel L, Amigorena S. Exosomes: composition, biogenesis and function. Nat. Rev. Immunol. 2002;2:569–579. doi: 10.1038/nri855. [DOI] [PubMed] [Google Scholar]
- Scanu A, et al. Stimulated T cells generate microparticles, which mimic cellular contact activation of human monocytes: differential regulation of pro- and anti-inflammatory cytokine production by high-density lipoproteins. J. Leukocyte Biol. 2008;83:921–927. doi: 10.1189/jlb.0807551. [DOI] [PubMed] [Google Scholar]
- Inal JM, et al. Microvesicles in health and disease. Arch. Immunol. Ther. Ex. 2012;60:107–121. doi: 10.1007/s00005-012-0165-2. [DOI] [PubMed] [Google Scholar]
- Fridley GE, Holstein CA, Oza SB, Yager P. The evolution of nitrocellulose as a material for bioassays. Mrs Bull. 2013;38:326–330. [Google Scholar]
- Gyorgy B, et al. Membrane vesicles, current state-of-the-art: emerging role of extracellular vesicles. Cell. Mol. Life Sci. 2011;68:2667–2688. doi: 10.1007/s00018-011-0689-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Missoum K, Belgacem MN, Bras J. Nanofibrillated cellulose surface modification: a review. Materials. 2013;6:1745–1766. doi: 10.3390/ma6051745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalia S, Boufi S, Celli A, Kango S. Nanofibrillated cellulose: surface modification and potential applications. Colloid Polym. Sci. 2014;292:5–31. [Google Scholar]


