Abstract
Comprehensive genomic analysis has uncovered surprisingly large numbers of genetic alterations in various types of cancers. To robustly and efficiently identify oncogenic “drivers” among these tumors and define their complex relationships with concurrent genetic alterations during tumor pathogenesis remains a daunting task. Recently, zebrafish have emerged as an important animal model for studying human diseases, largely because of their ease of maintenance, high fecundity, obvious advantages for in vivo imaging, high conservation of oncogenes and their molecular pathways, susceptibility to tumorigenesis and, most importantly, the availability of transgenic techniques suitable for use in the fish. Transgenic zebrafish models of cancer have been widely used to dissect oncogenic pathways in diverse tumor types. However, developing a stable transgenic fish model is both tedious and time-consuming, and it is even more difficult and more time-consuming to dissect the cooperation of multiple genes in disease pathogenesis using this approach, which requires the generation of multiple transgenic lines with overexpression of the individual genes of interest followed by complicated breeding of these stable transgenic lines. Hence, use of a mosaic transient transgenic approach in zebrafish offers unique advantages for functional genomic analysis in vivo. Briefly, candidate transgenes can be coinjected into one-cell-stage wild-type or transgenic zebrafish embryos and allowed to integrate together into each somatic cell in a mosaic pattern that leads to mixed genotypes in the same primarily injected animal. This permits one to investigate in a faster and less expensive manner whether and how the candidate genes can collaborate with each other to drive tumorigenesis. By transient overexpression of activated ALK in the transgenic fish overexpressing MYCN, we demonstrate here the cooperation of these two oncogenes in the pathogenesis of a pediatric cancer, neuroblastoma that has resisted most forms of contemporary treatment.
Keywords: Developmental Biology, Issue 97, zebrafish, animal model, mosaic transgenesis, coinjection, functional genomics, tumor initiation
Introduction
Cancers are progressive diseases marked by the accumulation of pathologic mutations, deletions and chromosome gains over time. These genetic abnormalities can affect multiple cellular processes ranging from the cell cycle, cell death, energetic metabolism and assembly of the cytoskeleton to stress responses such as hypoxia. Hence, tumorigenesis reflects the collective actions of multiple genetic aberrations across a spectrum of biological processes. Recent integrative genomic research efforts, including whole genome sequencing, exome sequencing, targeted sequencing, deep sequencing and genome-wide association studies, have identified a growing number of novel genetic alterations in essentially all types of tumors 1-4. In many instances, the genetic lesions occur together in a nonrandom manner 5-8, suggesting their cooperation in disease pathogenesis. Dissecting the oncogenic roles of the large array of aberrantly expressed genes resulting from these genomic lesions is necessary to devise new therapeutic strategies and to understand the responses of tumor cells to these agents, but this has proved to be a daunting task, requiring very robust animal model systems for the conduct of high-throughput functional genomic analysis in vivo.
Although mammals, especially rodents, are favored models in cancer biology, the zebrafish has begun to attract considerable attention. The teleost zebrafish (Dario rerio) has been used as a model organism for development study since the 1960s and was first applied to the study of tumor pathogenesis in 1982 9-11. Ease of maintenance, small body size, and high fecundity make the zebrafish a robust model for large-scale forward genetic screens to identify mutations that confer abnormal and pathological phenotypes 10. The optical transparency of zebrafish embryos is another key feature supporting wider use of this cancer model, as it allows in vivo imaging to be conducted to locate tumor development in real time 9, an application that is relatively difficult in rodents 12. Recent comparative genomics analysis of the zebrafish reference genome (Zv9) revealed 26,206 protein-coding genes, with 71% having human orthologues, of which 82% are correlated with disease-associated genes in the Online Mendelian Inheritance in Man (OMIM) database 13,14. Consequently, the zebrafish has been used to model diverse types of human cancers, including neuroblastoma 8, T-cell acute lymphoblastic leukemia (T-ALL) 15,16, melanoma 17,18, Ewing’s sarcoma 19, rhabdomyosarcoma 20,21, pancreatic carcinoma 22, hepatocellular carcinoma 23 and myeloid malignancies 24,25, and has been selected as a cancer model for xenotransplantation studies 11,26.
A stable transgenic approach in zebrafish is commonly used to study the effect of gain-of-function of genes in normal development or disease pathogenesis 27,28. To develop such a model (Figure 1A), one injects a DNA construct containing the gene of interest driven by a tissue-specific promoter into one-cell wild-type embryos. Three to four months after injection, when the injected embryos reach sexual maturity, they are outcrossed with wild-type fish to screen for the ones showing integration of the DNA construct in their germline, which licenses them as founder fish. Many factors, such as the copy number and integration site of the transgene, affect expression of the transgene in stable transgenic lines. Thus, to develop a transgenic tumor model, multiple stable transgenic lines overexpressing a single oncogene have to be generated first and screened for the line expressing the transgene at a level that might lead to tumor induction. However, if overexpression of a candidate oncogene is toxic to germ cells, it is difficult to generate a stable transgenic line by directly overexpressing the transgene 29. Hence, this approach can be time-consuming, with a high risk of failure to generate a suitable cancer model.
Here, we illustrate an alternative strategy based on mosaic transient transgenesis (Figure 1B) that provides unique advantages over traditional stable transgenesis for functional genomic study in vivo. In this approach, one or more transgene constructs are injected into the one-cell stage of transgenic or wild-type embryos. The injected DNA constructs containing transgenes are then mosaically and randomly integrated into the primary injected fish, resulting in mixed genotypes within multiple cell populations in individual fish 30. Moreover, coinjection of multiple DNA constructs in one-cell embryos leads to co-integration into the same cell at random sites, allowing one to trace the cells with expression of transgenes and explore the interactions of different genes during disease pathogenesis in the mosaic animals 31. As proof of principle, we transiently overexpressed mutationally activated ALK (F1174L) with mCherry reporter gene in the peripheral sympathetic nervous system (PSNS) under control of the dopamine beta hydroxylase (dβh) promoter in wild-type fish and transgenic fish overexpressing MYCN. ALK, which encodes a receptor tyrosine kinase, is the most frequently mutated gene in high-risk neuroblastoma 5-7,32,33. ALK (F1174L), as one of the most frequent and potent somatic activating mutations, is over-represented in MYCN-amplified high-risk neuroblastoma patients and synergizes with MYCN overexpression to accelerate neuroblastoma tumorigenesis in both stable transgenic mice and transgenic zebrafish models 8,34,35. By mosaic transient overexpression of ALK (F1174L) with mCherry in the MYCN transgenic fish, we recapitulated the acceleration of tumor onset observed in the stable transgenic fish overexpressing both ALK (F1174L) and MYCN, suggesting that the mosaic transgenesis strategy can be used to rapidly and efficiently assess the relative contributions of multiple oncogenes in tumor initiation in vivo.
Protocol
NOTE: All zebrafish studies and maintenance of the animals were done in accord with Mayo Clinic Institute IACUC-approved protocol # A41213.
1. DNA Constructs for Transgenesis
Amplify a 5.2-kb dopamine beta hydroxylase (dβh) promoter region8 using the CH211-270H11 BAC clone (from BACPAC resources center (BPRC)) as a DNA template. Use a PCR system appropriate for long and accurate PCR amplification of long DNA templates and the following cycle programs for PCR: 94 °C for 2 min, 10 cycles of (94 °C, 15 sec, 50 °C, 30 sec, 68 °C, 8 min), followed by 30 cycles of (94 °C, 15 sec, 53 °C, 30 sec, 68 °C, 8 min), 68 °C, 4 min (forward primer 5’-GGGGACAACTTTGTATAGAAAAGTTGGCGTACTCCCCCTTTTTAGG-3’ and reverse primer 5’- GGGGACTGCTTTTTTGTACAAACTTGTGTTGCTTTGTCGTCTTTTGA-3’).
Create the dβh-pDONRP4-P1R entry clone 8 using commercial recombinant cloning system such as multisite gateway. Mix 1 μl of purified dβh PCR product (172 ng/μl), 1 μl (150 ng/μl) pDONRP4-P1R donor vector (together with other gateway vectors are generous gifts from Dr. Chi-Bin Chien, Univ. of Utah), 4 μl of TE buffer (pH 8.0) with 2 µl of BP clonase enzyme mix, incubate for 1 hr at 25 °C and then transform to One Shot TOP10 competent E. coli according to the manufacturer's protocol.
Generate the dβh:EGFP-MYCN transgenic construct 8 using recombinant cloning system mentioned in step 1.2. Mix 1 μl of each entry clone (150 ng/μl ) including a 5.2-kb dβh promoter (dβh-pDONR P4-P1R construct), EGFP lacking a stop codon (pME-EGFP construct), and human MYCN cDNA (a generous gift from Dr. Hogarty at the Children’s Hospital of Pennsylvania, MYCN-pDONRP2R-P3 construct), 1 μl (150 ng/μl ) of the modified destination vector containing I-SceI recognition sites (a generous gift from Dr. C. Grabher, Karlsruhe Institute of Technology, Karlsruhe, Germany), 4 μl of TE buffer (pH 8.0) with 2 µl of LR Clonase enzyme mix, incubate for 1 hr at 25 °C and then transform to chemically competent E. coli such as One shot top 10 and follow the manufacturer's protocol.
Make mitf:mitf transgenic construct 8 by digesting the PNP-mitf vector (a generous gift from Dr. D. Raible, Univ. of Washington) with NotI and SalI restriction enzymes for 2 h at RT, and by subcloning the released 2.65-kb DNA fragment that contains the mitf promoter and the coding sequence of the zebrafish mitf gene into the NotI and MluI sites of a modified pBluescript vector containing flanking I-SceI recognition sites (a generous gift from Dr. Hui Feng, Boston University), using T4 DNA ligase according to the manufacturer’s protocol. NOTE: To increase the efficiency of generating stable MYCN-expressing lines, dβh:EGFP-MYCN and the mitf:mitf DNA constructs are coinjected into one-cell stage nacre embryos. Nacre designates a type of mutant fish lacking a neural crest-derived melanophore during development 36. Thus, the appearance of pigment cells in the injected embryos suggests the integration of mitf:mitf DNA constructs into the genome. It has been demonstrated that two or three coinjected DNA constructs can be cointegrated into the fish genome 31. Thus, pigmentation caused by mitf expression can serve as a marker for the integration of the dβh:EGFP-MYCN transgene and for easier identification of MYCN stable transgenic line.
Mix the dβh:EGFP-MYCN and mitf:mitf DNA constructs at a 3:1 ratio in a total volume of a 15 μl reaction with 1 μl of I-SceI enzyme and 0.75 μl of buffer. Ensure that the total amount of DNA in the reaction does not exceed 750 ng.
Carry out the I-SceI digestion at RT for 4 hr or O/N. On the second day, the I-SceI-digested DNAs are ready for microinjection or can be stored at -20 °C for injection in the future. Store the I-SceI enzyme at -80 °C in small aliquots to maintain its enzyme efficiency.
Subclone the human ALKF1174L and wild-type ALK gene from the PCDNA3 vector (a generous gift from Dr. George at Dana-Farber Cancer Institute) 7 into EcoRI and NotI sites of a pENTRY1A vector (a generous gift from Dr. C. Grabher, Karlsruhe Institute of Technology, Karlsruhe, Germany) using T4 DNA ligase according to the manufacturer’s protocol.
Generate the dβh:ALKF1174L or dβh:ALKWT transgenic constructs using the recombinant system as mentioned in Protocol 1.2. Briefly, combine three entry clones, dβh-pDONRP4-P1R, ALKF1174L-pENTRY1A (or ALKWT-pENTRY1A) and p3E-polyA, into the modified destination vector containing I-SceI recognition sites (a generous gift from Dr. C. Grabher, Karlsruhe Institute of Technology, Karlsruhe, Germany), using LR Clonase enzyme Mix, according to manufacturer's protocol.
Mix the dβh: ALKF1174L (or dβh:ALKWT) and dβh:mCherry DNA constructs at a 3:1 ratio and linearize them with I-SceI enzyme as described in PROTOCOL 1.5-1.6.
2. Microinjection
Use glass micropipettes with 1.0 mm diameter for all injections 37. Break off the tip of the glass micropipettes with a razor blade before injection. Calibrate the injection volume by injecting H2O into a drop of mineral oil. Measure the diameter of the resulting droplets and adjust the microinjector (pressure or duration of pressure pulse) to ensure that the injection volume is less than 10% of the total cell volume of one cell-stage embryos.
Add an additional 0.5 μl of fresh I-SceI enzyme (5 unit/μl) in 5 μl of injection solution containing DNA constructs right before the injection to increase the efficiency of transgenesis 38. Inject 50-80 pg of linearized DNA constructs into the cytoplasm of one-cell stage embryos. To ensure the success of injection, mix the sample with 0.25% phenol red for visualization. If cells turn red after injection, it indicates the microinjection is successful. To ensure a high success rate for generating transgenic fish, we typically inject as many as 500 embryos.
To generate transgenic fish stably expressing MYCN, coinject the linearized dβh:EGFP-MYCN and mitf:mitf DNA constructs (at a 3:1 ratio) into embryos of the pigmentation mutant nacre zebrafish at the one-cell stage. The expression of Mitf serves as a reporter for the integration of transgenic constructs in the fish genome.
For mosaic overexpression of ALK in the wild-type or MYCN transgenic embryos, co-inject the linearized dβh:ALK F1174L (or dβh:ALKWT) with dβh:mCherry DNA constructs (at a 3:1 ratio) into the one-cell embryos resulting from breeding of F1 heterozygous Tg(dβh:EGFP-MYCN) transgenic fish with wild-type AB fish. Thus, half of offspring are transgenic for MCYN and half are wild-type.
3. Screen for Stable or Mosaic Transgenic Fish
To increase the efficiency of identification of stable MYCN-expressing lines, anesthetize primarily injected nacre embryos with tricaine (0.02%) at days 3-5 of postfertilization and screen for pigmentation. Transfer the embryos with pigmentation to a new petri dish with fresh egg water and raise them to sexual maturity for further screening for the founder fish, which carry the dβh:EGFP-MYCN transgene in the germ cells.
To identify the founder with germline transmission of EGFP-MYCN, outcross pigmented F0 adult fish with wild-type AB fish and screen for EGFP-positive embryos at 1-2 days post fertilization for further genotyping. Place a single EGFP-positive F1 embryo into a PCR tube and remove all liquid. Add 50 μl of gDNA extraction buffer which contains 12.5 μl of 4x lysis buffer, 35 μl H2O and 2.5 μl proteinase K (10 mg/ml) to single embryo. Incubate the reaction for 2-3 hr at 55 °C, followed by 10 min of incubation at 98ºC to inactivate proteinase K. To make 50 ml of 4x lysis buffer, add 500 μl of 1M Tris (pH 8.4), 2.5 ml of 1M KCl to 47 ml of H2O. NOTE: After F0 MYCN stable transgenic fish are bred with wild-type AB fish, all of the offspring are pigmented. Thus, pigmentation cannot serve as marker for the identification of MYCN-positive fish. While in the MYCN transgenic fish, the EGFP-MYCN fusion protein is expressed in the PSNS, including the sympathetic neurons of the superior cervical ganglia and sequential segmental ganglion of the sympathetic chain and the non-PSNS dopaminergic neurons, such as the medulla oblongata and cranial ganglia 8. Thus, the expression of EGFP can serve as a marker for identification of the MYCN stable transgenic embryos.
Use 2 μl of gDNA extracted from the pigmented F1 embryo as template, primers MYCN-test F1: 5’-CTG CTT GAG AAC GAG CTG TG-3’; MYCN-R3: 5’-AGG CAT CGT TTG AGG ATC AG-3’, and the following program with the GC-RICH PCR System: 1 cycle of 95 °C for 3 min, 25 cycles of 95 °C for 30 sec, 58 °C for 30 sec, and 72 °C for 3 min. We typically genotype 14-16 pigmented embryos from a single mating to confirm the presence of integrated MYCN transgene in the pigmented embryos, more than 6 founder fish overexpressing mitf and MYCN were identified by this method.
Raise up the remainder of the EGFP-positive F1 embryos. Fin clip and genotype them at 2-3 month of age using the above protocol to further confirm the integration of MYCN transgene into the fish. Breed F1 MYCN stable transgenic fish with wild-type AB fish. Co-inject the linearized dβh:ALK F1174L (or dβh:ALKWT) with dβh:mCherry DNA constructs into the one-cell embryos as described in PROTOCOL 2.4.
Sort the MYCN-expressing embryos in the experiment described in PROTOCOL 2.4 using a stereoscopic fluorescence microscope and screen the primarily injected embryos at days 1-3 of postfertilization for the expression of EGFP-MYCN in the PSNS. Then, during days 2-5 of postfertilization, anesthetize the embryos with tricaine (0.02%) and sort those MYCN-positive or negative embryos again based on the expression of mCherry in the PSNS using a stereoscopic fluorescence microscope. The expression of mCherry serves as a marker for the coexpression of ALK in tissues of the mosaic primary injected animals. NOTE: ~600 offspring resulting from the breeding of MYCN stable transgenic fish with wild-type fish were injected per group with the linearized DNA constructs, including dβh:ALK F1174L and dβh:mCherry, dβh:ALKWT and dβh:mCherry, or dβh:mCherry alone, respectively. Mosaic expression of mCherry can be observed in 70-90% of the primarily injected fish. One-half to one-third of the injected fish survived through the larvae stage for tumor watch.
Raise all of the mCherry+MYCN+ and mCherry+MYCN- embryos according to the standard protocols from the zebrafish book 39 and monitor tumor onset beginning at 5 weeks postfertilization.
4. Tumor Watch in Mosaic Transgenic Fish
Monitor mCherry-positive primarily injected fish every 2 weeks starting at 5 weeks postfertilization for evidence of tumor onset.
Anesthetize fish with tricaine (0.02%) and screen for the presence of mCherry- and EGFP-expressing tumors in the PSNS under the Nikon SMZ-1500 stereoscopic fluorescence microscope equipped with a digital sight DS-U1 camera.
To confirm whether the tumors are expressing the ALK transgene, isolate the mCherry- and EGFP-positive masses for ALK genotyping PCR.
Using PCR, amplify genomic DNA extracted from developed tumors with the following primers: ALK P7: 5’-AGG CCA GGT GTC CGG AAT GC-3’ and ALK P18: 5’-TGT CTT CAG GCT GAT GTT GC-3’ and the following PCR reaction: 1 cycle of 94 °C for 5 min, 30 cycles of (94 °C for 30 sec, 55 °C for 30 sec, and 72 °C for 60 sec). Then, sequence the PCR product with ALK P7 primer to further confirm the existence of mutant or wild-type ALK in the mCherry-positive tumors.
Representative Results
To investigate whether overexpression of mutationally activated ALKF1174L or wild-type ALK could collaborate with MYCN in neuroblastoma induction, we overexpressed either activated human ALK or wild-type human ALK under control of the dβh promoter in the PSNS of transgenic fish overexpressing MYCN. Either of the following constructs, dβh-ALKF1174L or dβh-ALKWT, were coinjected with dβh-mCherry into one-cell wild-type or MYCN transgenic embryos (Figure 2A) 8. The expression of mCherry served as a marker for the coexpression of ALK in tissues of the mosaic primary injected animals. All of the expected genotypes were represented in the injected fish: (1) MYCN-expressing fish with mosaic coexpression of activated ALKF1174L and mCherry, designated MYCN;mosaic ALKmut; (2) MYCN-expressing fish with mosaic coexpression of wild-type ALK and mCherry, designated MYCN;mosaic ALKWT; (3) MYCN-expressing fish with mosaic expression of mCherry, designated MYCN;mosaic mCherry; (4) wild-type fish with mosaic coexpression of activated ALKF1174L and mCherry, designated WT;mosaic ALKmut; and (5) wild-type fish with mosaic coexpression of wild-type ALK and mCherry, designated WT;mosaic ALKWT. A tumor watch was then performed every 2 weeks from 5 weeks postfertilization (wpf) on a total of 492 injected animals.
Eight GFP+/mCherry+ tumors arose in the interrenal gland, the human adrenal gland equivalent, by 9 wpf in the MYCN-expressing fish coinjected with dβh-ALKF1174L and dβh-mCherry (Figures 2B and 3). These tumors were histologically, immunohistochemically, and ultrastructurally comparable to human neuroblastoma (data can be found in reference 8). By contrast, no tumors were observed by 9 weeks of age in the MYCN-expressing fish coinjected with dβh-ALKWT and dβh-mCherry (Figures 2C and 3, p = 0.002) or injected with dβh-mCherry alone (Figures 2D and 3, p = 0.007). Tumors arose in both the MYCN;mosaic ALKWT and MYCN;mosaic mCherry fish with the same rate of induction after 12 weeks of age. In addition, neuroblastomas were not detected in any wild-type fish coinjected with either the wild-type ALK and mCherry or ALKF1174L and mCherry transgenes during the 15 weeks of monitoring. Taken together, these findings show that mosaic overexpression of mutationally activated ALK accelerates MYCN-induced tumor onset, regardless of the integration site in individual mosaic animals, and that overexpression of wild-type ALK at the levels driven by the dβh promoter does not appear to collaborate with MYCN overexpression to induce neuroblastoma in this model system.
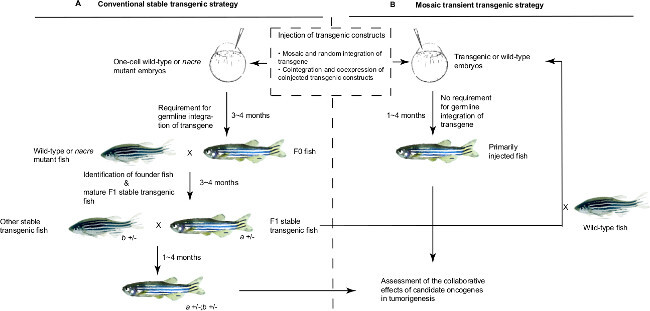 Figure 1: Schematic outline of conventional stable and mosaic transient zebrafish transgenic strategies in the study of the Cooperative Contributions of Candidate Oncogenes to Tumorigenesis. (A) Stable transgenic strategy. Linearized transgenic DNA construct containing candidate oncogene is injected into one-cell stage wild-type or nacre mutant zebrafish embryos where the transgene can be integrated into the fish genome at random genomic loci. Integration of transgene into the germ cells is required for the generation of stable transgenic line. Primarily injected embryos take 3 to 4 months to reach sexual maturity (F0 generation of fish), which will be outcrossed with wild-type or nacre mutant fish to screen for the founder fish with the transgene germline transmission. The resultant offspring of founder fish, termed the F1 generation, carry the heterozygous alleles of the transgene in their genome. To assess the collaborative effects of two candidate oncogenes, such as gene “a” and gene “b”, in tumorigenesis, we bred the F1 progeny of two stable transgenic lines overexpressing these transgenes, with only a quarter of offspring carrying the compound transgenes. Because of the relatively low efficiency of generating stable transgenic lines, this approach is not feasible for high-throughput functional genomic analyses. (B) Mosaic transient transgenic strategy. Similar to the stable transgenesis, linearized transgenic DNA constructs containing candidate genes can be injected into one-cell stage wild-type embryos. Because the transgene does not have to be integrated into the genome of germ cells, much time and effort can be saved in the process of identifying founder fish. Alternatively, if the transgenic fish line overexpressing one of the candidate oncogenes is available (in our case, MYCN transgenic fish were developed), linearized transgenic DNA constructs containing other candidate genes can be injected into one-cell stage transgenic embryos to study their cooperation in tumorigenesis. Up to three transgene constructs can be coinjected and coexpressed in the primarily injected fish 31; thus, the interaction of candidate oncogenes in tumorigenesis can be assessed in the primarily injected fish. Please click here to view a larger version of this figure.
Figure 1: Schematic outline of conventional stable and mosaic transient zebrafish transgenic strategies in the study of the Cooperative Contributions of Candidate Oncogenes to Tumorigenesis. (A) Stable transgenic strategy. Linearized transgenic DNA construct containing candidate oncogene is injected into one-cell stage wild-type or nacre mutant zebrafish embryos where the transgene can be integrated into the fish genome at random genomic loci. Integration of transgene into the germ cells is required for the generation of stable transgenic line. Primarily injected embryos take 3 to 4 months to reach sexual maturity (F0 generation of fish), which will be outcrossed with wild-type or nacre mutant fish to screen for the founder fish with the transgene germline transmission. The resultant offspring of founder fish, termed the F1 generation, carry the heterozygous alleles of the transgene in their genome. To assess the collaborative effects of two candidate oncogenes, such as gene “a” and gene “b”, in tumorigenesis, we bred the F1 progeny of two stable transgenic lines overexpressing these transgenes, with only a quarter of offspring carrying the compound transgenes. Because of the relatively low efficiency of generating stable transgenic lines, this approach is not feasible for high-throughput functional genomic analyses. (B) Mosaic transient transgenic strategy. Similar to the stable transgenesis, linearized transgenic DNA constructs containing candidate genes can be injected into one-cell stage wild-type embryos. Because the transgene does not have to be integrated into the genome of germ cells, much time and effort can be saved in the process of identifying founder fish. Alternatively, if the transgenic fish line overexpressing one of the candidate oncogenes is available (in our case, MYCN transgenic fish were developed), linearized transgenic DNA constructs containing other candidate genes can be injected into one-cell stage transgenic embryos to study their cooperation in tumorigenesis. Up to three transgene constructs can be coinjected and coexpressed in the primarily injected fish 31; thus, the interaction of candidate oncogenes in tumorigenesis can be assessed in the primarily injected fish. Please click here to view a larger version of this figure.
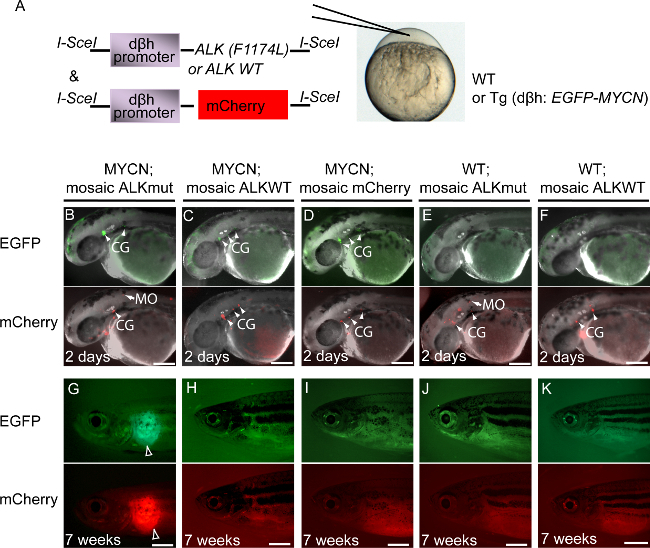 Figure 2: Mosaic Expression of Activated ALK Accelerates the Onset of MYCN-induced Neuroblastoma 8. Modified figure with permission from Elsevier (reference 3466510609819). (A) Approach for constructing mosaic transgenesis. (B-F) 2-day-old larvae of primarily injected embryos. Lateral views of stable EGFP expression in the dopaminergic neurons, including cranial ganglia (CG, arrowheads) in merged fluorescence-brightfield images (upper panels). Lateral views of mosaic mCherry expression (lower panels) in the cranial ganglia (CG, arrowheads), medulla oblongata (MO, arrows). Some ectopic mCherry expression is observed in the non-PSNS and non-dopaminergic neurons. (G-K) 7-week-old primarily injected fish. (B, G) MYCN-expressing fish coinjected with dβh-ALKF1174L and dβh-mCherry constructs (MYCN;mosaic ALKmut). EGFP- and mCherry-positive tumors arose at 7 weeks (white empty arrows in G). (C, H) MYCN-expressing fish coinjected with dβh-ALKWT and dβh-mCherry constructs (MYCN;mosaic ALKWT). (D, I) MYCN-expressing fish injected with dβh-mCherry construct alone (MYCN; mosaic mCherry). (E, J) Wild-type (WT) fish coinjected with dβh-ALKF1174L and dβh-mCherry constructs (WT; mosaic ALKmut). (F, K) Wild-type (WT) fish coinjected with dβh-ALKWT and dβh-mCherry constructs (WT;mosaic ALKWT). Neuroblastomas were not observed in the MYCN-expressing fish coinjected with dβh-ALKWT and dβh-mCherry or dβh-mCherry alone at 7 wpf, or in any of the siblings that did not inherit the MYCN transgene and were injected with either the ALKWT gene or the ALKF1174L gene. Scale bars, 100 μm in B-F and 1 mm in G-K. A Nikon SMZ-1500 stereoscopic fluorescence microscope equipped with a digital sight DS-U1 camera and a Leica MZ10F stereoscopic fluorescence microscope equipped with a Leica DFC 345FX digital camera were used to capture the fluorescent images.The acquired images were processed and compiled with Adobe Photoshop and Illustrator CS3 (Adobe) software. Please click here to view a larger version of this figure.
Figure 2: Mosaic Expression of Activated ALK Accelerates the Onset of MYCN-induced Neuroblastoma 8. Modified figure with permission from Elsevier (reference 3466510609819). (A) Approach for constructing mosaic transgenesis. (B-F) 2-day-old larvae of primarily injected embryos. Lateral views of stable EGFP expression in the dopaminergic neurons, including cranial ganglia (CG, arrowheads) in merged fluorescence-brightfield images (upper panels). Lateral views of mosaic mCherry expression (lower panels) in the cranial ganglia (CG, arrowheads), medulla oblongata (MO, arrows). Some ectopic mCherry expression is observed in the non-PSNS and non-dopaminergic neurons. (G-K) 7-week-old primarily injected fish. (B, G) MYCN-expressing fish coinjected with dβh-ALKF1174L and dβh-mCherry constructs (MYCN;mosaic ALKmut). EGFP- and mCherry-positive tumors arose at 7 weeks (white empty arrows in G). (C, H) MYCN-expressing fish coinjected with dβh-ALKWT and dβh-mCherry constructs (MYCN;mosaic ALKWT). (D, I) MYCN-expressing fish injected with dβh-mCherry construct alone (MYCN; mosaic mCherry). (E, J) Wild-type (WT) fish coinjected with dβh-ALKF1174L and dβh-mCherry constructs (WT; mosaic ALKmut). (F, K) Wild-type (WT) fish coinjected with dβh-ALKWT and dβh-mCherry constructs (WT;mosaic ALKWT). Neuroblastomas were not observed in the MYCN-expressing fish coinjected with dβh-ALKWT and dβh-mCherry or dβh-mCherry alone at 7 wpf, or in any of the siblings that did not inherit the MYCN transgene and were injected with either the ALKWT gene or the ALKF1174L gene. Scale bars, 100 μm in B-F and 1 mm in G-K. A Nikon SMZ-1500 stereoscopic fluorescence microscope equipped with a digital sight DS-U1 camera and a Leica MZ10F stereoscopic fluorescence microscope equipped with a Leica DFC 345FX digital camera were used to capture the fluorescent images.The acquired images were processed and compiled with Adobe Photoshop and Illustrator CS3 (Adobe) software. Please click here to view a larger version of this figure.
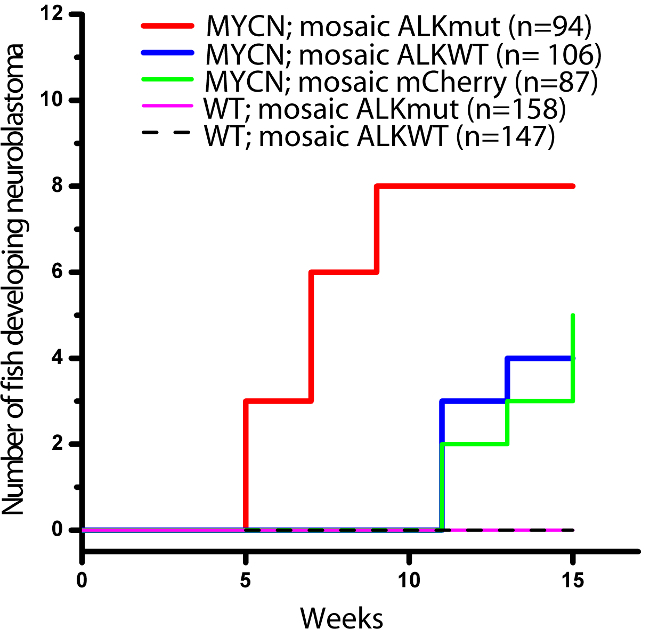 Figure 3:Onset of Neuroblastoma in MYCN Transgenic Fish or Wild-type (WT) Fish as Mosaics Coinjected with the DNA Constructs. Reprinted with permission from Elsevier (reference 3466510609819). dβh-ALKF1174L and dβh-mCherry (mosaic ALKmut); dβh-ALKWT and dβh-mCherry (mosaic ALKWT); or dβh-mCherry (mosaic mCherry) alone 8. The difference between tumor onset by 9 wpf in the MYCN-expressing fish coinjected with dβh-ALKF1174L and dβh-mCherry (MYCN;mosaic ALKmut) and that in the MYCN line coinjected with dβh-ALKWT and dβh-mCherry (MYCN;mosaic ALKWT) or dβh-mCherry alone (MYCN; mosaic mCherry) is significant at p = 0.002 and p = 0.007, respectively, by two-tailed Fisher’s exact test.
Figure 3:Onset of Neuroblastoma in MYCN Transgenic Fish or Wild-type (WT) Fish as Mosaics Coinjected with the DNA Constructs. Reprinted with permission from Elsevier (reference 3466510609819). dβh-ALKF1174L and dβh-mCherry (mosaic ALKmut); dβh-ALKWT and dβh-mCherry (mosaic ALKWT); or dβh-mCherry (mosaic mCherry) alone 8. The difference between tumor onset by 9 wpf in the MYCN-expressing fish coinjected with dβh-ALKF1174L and dβh-mCherry (MYCN;mosaic ALKmut) and that in the MYCN line coinjected with dβh-ALKWT and dβh-mCherry (MYCN;mosaic ALKWT) or dβh-mCherry alone (MYCN; mosaic mCherry) is significant at p = 0.002 and p = 0.007, respectively, by two-tailed Fisher’s exact test.
Discussion
In this representative study, we used transient coinjection and coexpression of activated ALK with the mCherry reporter gene in MYCN-expressing transgenic fish to show that these genes cooperate to markedly accelerate the onset of neuroblastoma, consistent with our previous finding in compound stable transgenic fish coexpressing both activated ALK and MYCN 8. This mosaic transgenic approach possesses several distinct advantages over the conventional method. Most important, it enables rapid examination of the effects of coexpressing candidate oncogenes in primarily injected animals (F0 generation) without the requirement for stable transgenic animals, such as eliminating the excessive time and labor typically involved in the breeding and identification of ALK-expressing stable transgenic fish. We also examined the effect of overexpression of wild-type ALK, with or without MYCN overexpression, on tumor initiation. The results show that transient overexpression of wild-type ALK in our fish model is not sufficient to initiate neuroblastoma tumorigenesis, regardless of the status of the MYCN oncogene. It would be interesting to test in the future whether transient coexpression of both activated ALK and MYCN in wild-type fish could induce earlier onset of tumorigenesis in the primarily injected fish. This study would more faithfully mimic the actual disease context, in which somatic alterations of both genes co-occur in a subset of high-risk neuroblastoma patients 5.
An important design element of the mosaic transient transgenic strategy is the inclusion of a reporter gene for coinjection with any candidate oncogenes of interest, which aids in screening for animals with successful integration of transgenes among the primarily injected zebrafish embryos and to monitor the positive fish for tumor onset and progression. To increase the efficiency of transgenesis, we have applied the I-SceI meganuclease-mediated transgenic approach, which significantly increases the uniform promoter-dependent expression of transgenes from 26% of the primarily injected embryos without meganuclease to 76% of injected embryos with meganuclease 38. Although use of the I-SceI meganuclease-mediated transgenic approach has improved the efficiency of transgenesis remarkably, the percentage of injected embryos positively for the transgenes still remains well below 100%. Thus, the expression of a coinjected reporter gene can serve as a marker to identify injected embryos with successful integration of candidate transgenes before further monitoring the fish for the onset of tumors. In contrast, a Tol2 transposon-mediated transgenic approach has become a popular genetic tool in model vertebrates and has been successfully used for transgenesis, insertional mutagenesis, gene trapping, and enhancer trapping 40,41. Coinjection of Tol2 transposons with transposase mRNA into fertilized embryos can facilitate early integration events that increases the efficiency of chromosomal integration and potentiates successful germline transmission of the transgene42. However, due to the “cut-and-paste” mechanism of DNA transposition, a single copy of transgene is integrated into the chromosome per insertion locus42. Thus, coinjected candidate genes are most likely integrated individually into the fish genome at different sites, which could lead to the expression of candidate genes in different cells and a more complicated transient and mosaic expression pattern of coinjected genes in the primarily injected fish.
Overall, the current study supports the future use of mosaic transgenesis as a rapid means to evaluate the collaborative, perhaps synergistic, relationships among multiple oncogenes in a high-throughput manner, a challenge that is difficult to meet with other animal models, including rodents. Thus far, this strategy has also been successfully applied in transgenic zebrafish to identify genes that suppress the activated RAS-induced initiation of rhabdomyosarcoma 31 and modify the radiation sensitivity of MYC-induced T-cell acute lymphoblastic leukemia 31. Furthermore, Langenau et al, 15 have demonstrated that combining the coinjection approach with a heat-shock-inducible transgenic approach can induce transgene expression by heat shock in the primarily injected fish, which would be very useful in exploring the cooperative roles of oncogenes in tumor progression, as it permits gene expression to be turned on after the primary tumor is established. Very recently, Langenau and colleagues developed the first immunocompromised zebrafish model by targeting rag2 gene with engineered zinc-finger nucleases 43. Loss of function of rag2 permitted robust and long-term engraftment of multiple tissues and cancer cells 43, an advantage that could be useful in transplanting the heterogeneous cancer cells that overexpress candidate transgenes and assessing the effect of mosaic transgene expression on self-renewal capacity, therapeutic responses and the growth rate of these cancer cells.
Finally, mosaic transgenesis offers another unique advantage over stable transgenesis. In stable transgenic animals, the transgenes are integrated into the genome of every single cell in the body and are heritable from generation to generation 27. However, many cancers arise from genetic alterations in somatic cells instead of inherited mutations in germ cells 44. Thus, the mosaic pattern of transgene integration in the primary injected fish and the mixed genotypes within cell populations of individual mosaic transgenic fish would better recapitulate the somatic defects found in patients and would avoid the artificial positional effect caused by a single insertion site in a stable transgenic line. On the other hand, the mosaicism and small population of cells overexpressing the transgenes in the primarily injected fish makes mechanistic study more difficult.
In summary, the mosaic transgenic strategy provides a robust tool for rapid and effective assessment of the cooperative contribution of oncogenes to tumor initiation and spread. This advance is expected to generate seminal information on the interplay among candidate oncogenes, which is urgently needed for further productive investigation of oncogenic mechanisms and pathways and for developing effective targeted therapy. Increasing the efficiency of cointegration of more than three coinjected DNA constructs would open the opportunity for examination of the complex relationships among concurrently expressed multigene combinations in various types of cancers. Challenges for the future will be to modify the mosaic transgenic strategy to accommodate any genomic or epigenetic alteration that could play a role in the neoplastic process, rather than limit its use to mutations that change only protein-coding or to genes are overexpressed, gained or amplified. Recently, improved genome editing techniques, such as transcription activator-like effector nucleases (TALENS) 45 and clustered regularly interspaced short palindromic repeats (CRISPR)/CRISPR-associated (Cas) systems 46, have become widely used to more rapidly and efficiently modify endogenous genes in various types of cells and organisms 46. Such techniques would allow us to perform targeted, highly efficient modifications of genome sequence and gene expression in the zebrafish model system and to better understand the mechanism(s) underlying the role of genomic or epigenetic alterations in tumor pathogenesis. These, in turn, are likely to spur the development of novel molecular therapeutics for a range of cancers.
Disclosures
The authors declare that they have no competing financial interests.
Acknowledgments
We appreciate Dr. Jeong-Soo Lee for sharing the Tg(dbh:EGFP-MYCN) transgenic fish with us in our study. This work was supported by a grant 1K99CA178189-01 from the National Cancer Institute, a fellowship from the Pablove Foundation and the Friends for Life, and young investigator awards from the Alex's Lemonade Stand Foundation and the CureSearch for Children's Cancer Foundation.
References
- Tenesa A, Dunlop MG. New insights into the aetiology of colorectal cancer from genome-wide association studies. Nat Rev Genet. 2009;10(6):353–358. doi: 10.1038/nrg2574. [DOI] [PubMed] [Google Scholar]
- Maher B. Exome sequencing takes centre stage in cancer profiling. Nature. 2009;459(7244):146–147. doi: 10.1038/459146b. [DOI] [PubMed] [Google Scholar]
- Meyerson M, Gabriel S, Getz G. Advances in understanding cancer genomes through second-generation sequencing. Nat Rev Genet. 2010;11(10):685–696. doi: 10.1038/nrg2841. [DOI] [PubMed] [Google Scholar]
- Chung CC, Chanock SJ. Current status of genome-wide association studies in cancer. Hum Genet. 2011;130(1):59–78. doi: 10.1007/s00439-011-1030-9. [DOI] [PubMed] [Google Scholar]
- Mosse YP, et al. Identification of ALK as a major familial neuroblastoma predisposition gene. Nature. 2008;455(7215):930–935. doi: 10.1038/nature07261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janoueix-Lerosey I, et al. Somatic and germline activating mutations of the ALK kinase receptor in neuroblastoma. Nature. 2008;455(7215):967–970. doi: 10.1038/nature07398. [DOI] [PubMed] [Google Scholar]
- George RE, et al. Activating mutations in ALK provide a therapeutic target in neuroblastoma. Nature. 2008;455(7215):975–978. doi: 10.1038/nature07397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu S, et al. Activated ALK Collaborates with MYCN in Neuroblastoma Pathogenesis. Cancer Cell. 2012;21(3):362–373. doi: 10.1016/j.ccr.2012.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White R, Rose K, Zon L. Zebrafish cancer: the state of the art and the path forward. Nat Rev Cancer. 2013;13(9):624–636. doi: 10.1038/nrc3589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amatruda JF, Patton EE. Genetic models of cancer in zebrafish. Int Rev Cell Mol Biol. 2008;271:1–34. doi: 10.1016/S1937-6448(08)01201-X. [DOI] [PubMed] [Google Scholar]
- Konantz M, et al. Zebrafish xenografts as a tool for in vivo studies on human cancer. Ann N Y Acad Sci. 2012;1266:124–137. doi: 10.1111/j.1749-6632.2012.06575.x. [DOI] [PubMed] [Google Scholar]
- Ellenbroek SI, van Rheenen J. Imaging hallmarks of cancer in living mice. Nat Rev Cancer. 2014;14(6):406–418. doi: 10.1038/nrc3742. [DOI] [PubMed] [Google Scholar]
- Kettleborough RN, et al. A systematic genome-wide analysis of zebrafish protein-coding gene function. Nature. 2013;496(7446):494–497. doi: 10.1038/nature11992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howe K, et al. The zebrafish reference genome sequence and its relationship to the human genome. Nature. 2013;496(7446):498–503. doi: 10.1038/nature12111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langenau DM, et al. Myc-induced T cell leukemia in transgenic zebrafish. Science. 2003;299(5608):887–890. doi: 10.1126/science.1080280. [DOI] [PubMed] [Google Scholar]
- Feng H, et al. T-lymphoblastic lymphoma cells express high levels of BCL2, S1P1, and ICAM1, leading to a blockade of tumor cell intravasation. Cancer Cell. 2010;18(4):353–366. doi: 10.1016/j.ccr.2010.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patton EE, et al. BRAF mutations are sufficient to promote nevi formation and cooperate with p53 in the genesis of melanoma. Current biology : CB. 2005;15(3):249–254. doi: 10.1016/j.cub.2005.01.031. [DOI] [PubMed] [Google Scholar]
- Santoriello C, Anelli V, Alghisi E, Mione M. Highly penetrant melanoma in a zebrafish model is independent of ErbB3b signaling. Pigment Cell Melanoma Res. 2012;25(2):287–289. doi: 10.1111/j.1755-148X.2012.00973.x. [DOI] [PubMed] [Google Scholar]
- Leacock SW, et al. A zebrafish transgenic model of Ewing's sarcoma reveals conserved mediators of EWS-FLI1 tumorigenesis. Dis Model Mech. 2012;5(1):95–106. doi: 10.1242/dmm.007401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le X, et al. Heat shock-inducible Cre/Lox approaches to induce diverse types of tumors and hyperplasia in transgenic zebrafish. Proceedings of the National Academy of Sciences of the United States of America. 2007;104(22):9410–9415. doi: 10.1073/pnas.0611302104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langenau DM, et al. Effects of RAS on the genesis of embryonal rhabdomyosarcoma. Genes & development. 2007;21(11):1382–1395. doi: 10.1101/gad.1545007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park SW, et al. Oncogenic KRAS induces progenitor cell expansion and malignant transformation in zebrafish exocrine pancreas. Gastroenterology. 2008;134(7):2080–2090. doi: 10.1053/j.gastro.2008.02.084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng W, et al. Xmrk, kras and myc transgenic zebrafish liver cancer models share molecular signatures with subsets of human hepatocellular carcinoma. PLoS One. 2014;9(3):e91179. doi: 10.1371/journal.pone.0091179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forrester AM, et al. NUP98-HOXA9-transgenic zebrafish develop a myeloproliferative neoplasm and provide new insight into mechanisms of myeloid leukaemogenesis. British journal of haematology. 2011;155(2):167–181. doi: 10.1111/j.1365-2141.2011.08810.x. [DOI] [PubMed] [Google Scholar]
- Alghisi E, et al. Targeting oncogene expression to endothelial cells induces proliferation of the myelo-erythroid lineage by repressing the Notch pathway. Leukemia. 2013;27(11):2229–2241. doi: 10.1038/leu.2013.132. [DOI] [PubMed] [Google Scholar]
- Veinotte CJ, Dellaire G, Berman JN. Hooking the big one: the potential of zebrafish xenotransplantation to reform cancer drug screening in the genomic era. Dis Model Mech. 2014;7(7):745–754. doi: 10.1242/dmm.015784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patton EE, Zon LI. The art and design of genetic screens: zebrafish. Nat Rev Genet. 2001;2(12):956–966. doi: 10.1038/35103567. [DOI] [PubMed] [Google Scholar]
- Lieschke GJ, Currie PD. Animal models of human disease: zebrafish swim into view. Nat Rev Genet. 2007;8(5):353–367. doi: 10.1038/nrg2091. [DOI] [PubMed] [Google Scholar]
- Igoucheva O, Alexeev V, Yoon K. Differential cellular responses to exogenous DNA in mammalian cells and its effect on oligonucleotide-directed gene modification. Gene Ther. 2006;13(3):266–275. doi: 10.1038/sj.gt.3302643. [DOI] [PubMed] [Google Scholar]
- Koster RW, Fraser SE. Tracing transgene expression in living zebrafish embryos. Dev Biol. 2001;233(2):329–346. doi: 10.1006/dbio.2001.0242. [DOI] [PubMed] [Google Scholar]
- Langenau DM, et al. Co-injection strategies to modify radiation sensitivity and tumor initiation in transgenic Zebrafish. Oncogene. 2008;27(30):4242–4248. doi: 10.1038/onc.2008.56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, et al. Oncogenic mutations of ALK kinase in neuroblastoma. Nature. 2008;455(7215):971–974. doi: 10.1038/nature07399. [DOI] [PubMed] [Google Scholar]
- Pugh TJ, et al. The genetic landscape of high-risk neuroblastoma. Nature genetics. 2013. [DOI] [PMC free article] [PubMed]
- Berry T, et al. The ALK(F1174L) mutation potentiates the oncogenic activity of MYCN in neuroblastoma. Cancer Cell. 2012;22(1):117–130. doi: 10.1016/j.ccr.2012.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heukamp LC, et al. Targeted expression of mutated ALK induces neuroblastoma in transgenic mice. Sci Transl Med. 2012;4(141):141ra191. doi: 10.1126/scitranslmed.3003967. [DOI] [PubMed] [Google Scholar]
- Lister JA, Robertson CP, Lepage T, Johnson SL, Raible DW. nacre encodes a zebrafish microphthalmia-related protein that regulates neural-crest-derived pigment cell fate. Development. 1999;126(17):3757–3767. doi: 10.1242/dev.126.17.3757. [DOI] [PubMed] [Google Scholar]
- Rosen JN, Sweeney MF, Mably JD. Microinjection of zebrafish embryos to analyze gene function. J Vis Exp. 2009. [DOI] [PMC free article] [PubMed]
- Thermes V, et al. I-SceI meganuclease mediates highly efficient transgenesis in fish. Mech Dev. 2002;118(1-2):91–98. doi: 10.1016/s0925-4773(02)00218-6. [DOI] [PubMed] [Google Scholar]
- Kimmel CB, Ballard WW, Kimmel SR, Ullmann B, Schilling TF. Stages of embryonic development of the zebrafish. Dev Dyn. 1995;203(3):253–310. doi: 10.1002/aja.1002030302. [DOI] [PubMed] [Google Scholar]
- Urasaki A, Asakawa K, Kawakami K. Efficient transposition of the Tol2 transposable element from a single-copy donor in zebrafish. Proc Natl Acad Sci U S A. 2008;105(50):19827–19832. doi: 10.1073/pnas.0810380105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caneparo L, Pantazis P, Dempsey W, Fraser SE. Intercellular bridges in vertebrate gastrulation. PLoS One. 2011;6(5):e20230. doi: 10.1371/journal.pone.0020230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivics Z, Izsvak Z. The expanding universe of transposon technologies for gene and cell engineering. Mob DNA. 2010;1(1):25. doi: 10.1186/1759-8753-1-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang Q, et al. Optimized cell transplantation using adult rag2 mutant zebrafish. Nat Methods. 2014;11(8):821–824. doi: 10.1038/nmeth.3031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watson IR, Takahashi K, Futreal PA, Chin L. Emerging patterns of somatic mutations in cancer. Nat Rev Genet. 2013;14(10):703–718. doi: 10.1038/nrg3539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sander JD, et al. Targeted gene disruption in somatic zebrafish cells using engineered TALENs. Nat Biotechnol. 2011;29(8):697–698. doi: 10.1038/nbt.1934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sander JD, Joung JK. CRISPR-Cas systems for editing, regulating and targeting genomes. Nat Biotechnol. 2014;32(4):347–355. doi: 10.1038/nbt.2842. [DOI] [PMC free article] [PubMed] [Google Scholar]


