Abstract
Mutations in ALK are a common mechanism of acquired resistance to small molecule ALK inhibitors in ALK-rearranged lung cancer. Different mutants exhibit differential sensitivity to ALK inhibitors. Matching the mutational profile of the tumor with the appropriate ALK inhibitor is likely to be important to maximize benefit for patients.
In this issue of Clinical Cancer Research, Katayama and colleagues describe the identification and characterization of two mutations that confer resistance to the novel small molecule ALK (anaplastic lymphoma kinase) inhibitor, alectinib (1). Since the discovery of rearrangements in the ALK gene in non–small cell lung cancers (NSCLC) in 2007, there has been rapid clinical development of small molecule ALK inhibitors for the 3% to 5% of NSCLCs driven by an ALK fusion oncogene (2). Crizotinib was the first ALK inhibitor to receive FDA approval for patients with ALK-rearranged (ALK+) NSCLC. Although the majority of patients with ALK+ NSCLC who are treated with crizotinib achieve dramatic radiographic and/or clinical improvement (3), resistance inevitably develops, generally within 1 year of starting crizotinib. Resistance to crizotinib emerges by a variety of mechanisms (4). In ∼30% of cases, point mutations in ALK or amplification of the fusion gene can be identified, suggesting that such tumors may still be dependent on ALK for their survival. Another third of crizotinib-resistant tumors exhibit activation of signaling pathways that bypass the requirement for ALK (via EGFR activation or KIT amplification). The mechanisms of resistance in the remaining ∼30% of cases are unknown.
To counter ALK-dependent mechanisms of resistance to crizotinib, multiple next-generation ALK inhibitors have been identified and are currently in clinical development, with FDA approval granted to ceritinib in 2014 for the treatment of advanced ALK+ NSCLC previously treated with crizotinib (5). Encouraging activity has also been observed with the ALK inhibitors alectinib and AP26113, both currently being evaluated in registrational clinical trials. (6, 7). As new agents receive FDA approval, clinicians will be faced with the challenge of deciding how to choose initial therapy and sequence subsequent therapies to maximize benefit for their patients. Knowledge of the common and unique mechanisms of resistance to the different agents will be critical to inform these decisions.
Eight different mutations in the ALK tyrosine kinase (TK) domain have been described in crizotinib-resistant NSCLCs, with the L1196M “gatekeeper” and G1269A mutations being the most common (gatekeeper residues are found in multiple kinases and play a role in binding of ATP-competitive inhibitors; mutations at these residues are frequently causes of resistance to these drugs, e.g., EGFRT790M and BCR-ABLT315I; ref. 8). Ceritinib, alectinib, and AP-26113 are potent ALK inhibitors that have lower IC50s than crizotinib for ALK and additionally suppress the kinase activity of several mutations associated with crizotinib-resistance including L1196M and G1296A. In vitro studies have demonstrated that some crizotinib-resistant mutants are cross-resistant to ceritinib (e.g., C1156Y, G1202R, 1151T-ins, and F1174C) and/or alectinib (G1202R). Indeed, analysis of ceritinib-resistant tumors from 10 patients revealed the presence of either the F1174C or G1202R mutations in 4 cases; in 2 of the cases, these mutations replaced either G1269A or S1206Y point mutations in ALK that had been identified following crizotinib resistance (9).
A limited number of studies to date have been conducted to understand mechanisms of resistance to alectinib. Molecular analysis from one alectinib-resistant tumor has been reported identifying the G1202R mutation (10). In this issue of Clinical Cancer Research, Katayama and colleagues integrate evidence from ALK-rearranged cell lines cultured long-term in alectinib and analysis of an alectinib-resistant tumor from a patient to identify two novel ALK mutations (V1180L and I1171T) that confer resistance to alectinib. Interestingly, the V1180L mutation—identified in the H3122 cell line following exposure to alectinib—is near the L1196 gatekeeper residue, and the authors predict that the methyl group present on the leucine interferes with alectinib binding. Although, to date, the V1180L mutation has not been identified in crizotinib-resistant tumors, the authors demonstrate that this mutation does confer resistance to crizotinib. Surprisingly, however, the V1180L mutant retains sensitivity to ceritinib and AP26113, highlighting potential therapeutic avenues should this mutation eventually be identified in patient samples. The I1171T mutation was identified in a liver biopsy specimen from a patient with crizotinib-resistant disease treated with alectinib. This patient initially responded to alectinib, and disease progression was noted after 4 months of therapy. Molecular modeling of the mutation suggests that it causes a shift in the position of the C-helix and disrupts the formation of a hydrogen bond between E1167 of ALK and alectinib, leading to a reduced affinity of the mutant kinase for alectinib. It is possible that the I1171T mutation was present, at low frequency, upon crizotinib resistance, because the same mutation has been found in crizotinib-resistant ALK+ neuroblastomas and was isolated in vitro in mutagenesis screens for mutations that confer crizotinib resistance (11, 12). Similar to the V1180L mutant, the I1171T mutation was sensitive to ceritinib (and partially to AP26113) in cell line experiments. Further confirming these observations, the patient described in this article exhibited a partial response to ceritinib following alectinib resistance.
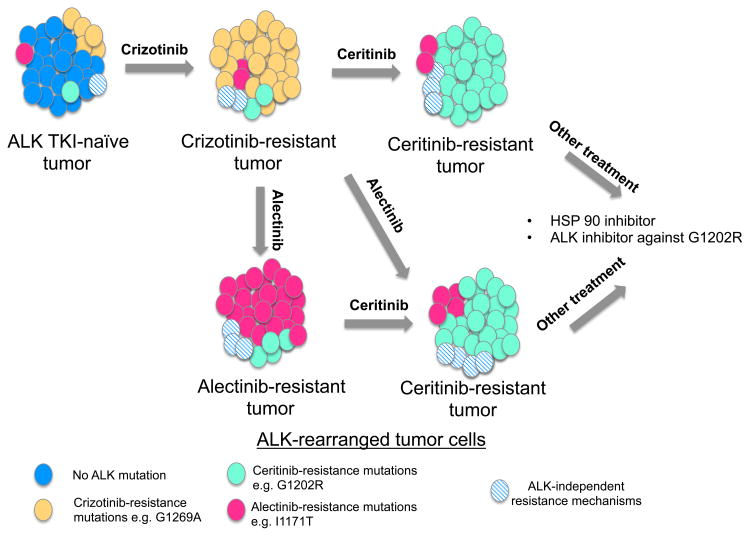
Results from studies like those described by Katayama and colleagues (1) suggest that the spectrum of resistance-conferring ALK mutations is different for each ALK inhibitor, although some of the mutations confer resistance to one or more agents. Moreover, the findings indicate that multiple distinct mutations can emerge, even after exposure to the most potent ALK inhibitors. Finally, data from studies of crizotinib and ceritinib (9) indicate that ALK inhibitor– resistant tumors are heterogeneous, with several resistance mutations being present in individual tumors (although one may dominate). Depending on which ALK inhibitor is used, subclones of cells harboring specific resistance mutations emerge while others remain suppressed (Fig. 1). These results highlight the need for additional studies to comprehensively catalog and characterize the mutations that emerge in patients upon treatment with different ALK inhibitors.
Figure 1.
Schematic representation showing the possible evolution of an ALK-rearranged lung cancer following sequential treatment with ALK inhibitors. This ALK inhibitor–naïve tumor is composed mainly of sensitive cells (blue) interspersed with rare cells harboring ALK point mutations (pink, orange, and green) or other ALK-independent alterations (blue hatched cells). During treatment with crizotinib, clones with mutations that confer resistance to crizotinib are positively selected. In this example, the more abundant clone harboring a G1269A mutation (orange) emerges, whereas clones harboring other resistance mutations, such as G1202R and I1171T, persist at low concentrations. Clones that have alterations other than ALK mutations that resist crizotinib will also persist (blue hatched cells). Upon treatment with ceritinib, cells with mutations that confer resistance to this drug (e.g., G1202R mutation, green cells) eventually dominate. Similarly, tumors with acquired resistance to alectinib are mostly composed of resistant clones that dominate after prolonged exposure to alectinib, for example, those with an I1171T ALK mutation or the G1202R mutation that also confers resistance to ceritinib. *For simplicity, these rare clones are depicted as present in the untreated tumor. This may be the case or, alternatively, they may also emerge during treatment. Further, the number and spectrum of clones present are likely to be unique for each tumor/patient.
Findings from this study and previous investigations have implications for the management of patients with ALK-rearranged lung cancer. Indeed, these data highlight the need for repeat tumor biopsies at the time of resistance to each individual agent to determine if ALK mutations are present in the tumor, and if so, which ones. This practice will allow subsequent treatment to be tailored to the most current mutational state of the tumor. In the study performed by Katayama and colleagues, for example, both of the alectinib-resistance mutations described retain sensitivity to ceritinib (1).
Despite our increasing knowledge of how to treat ALK-rearranged lung cancer, several challenges remain. First, only ∼30% of crizotinib-resistant tumors harbor ALK-mediated resistance mechanisms. Although activity of the next- generation ALK inhibitors has been observed in a subset of crizotinib-resistant tumors without known ALK mutations or amplification, further research to define the mechanisms of resistance and vulnerabilities of this subset of tumors is essential. Second, the G1202R mutation has been found in tumors resistant to crizotinib, ceritinib, and alectinib. The identification of an agent that can effectively counter this mutation will be important. Third, clinical trials investigating next- generation ALK inhibitors in both the first-line and refractory settings will be critical to establish whether use of more potent ALK inhibitors at diagnosis is superior to sequencing of therapies as resistance emerges. Progress in the treatment and development of therapies for ALK-rearranged lung cancer has been exponential in the past 5 years. Studies, like the one presented here, that define biomarkers predictive of response to respective ALK inhibitors and other potential therapies will be essential to refine treatment paradigms for patients with ALK-rearranged lung cancer.
Acknowledgments
Grant Support: This work was supported by the NIH/National Cancer Institute grants R01CA120247 (K.Politi) and R01-CA121210 (K. Politi), the Lung Cancer Research Foundation (K. Politi), the Department of Defense Lung Cancer Research Program (K. Politi) and the Yale Cancer Center (K. Politi and S. Gettinger).
Footnotes
Disclosure of Potential Conflicts of Interest: S. Gettinger is a consultant/advisory board member for ARIAD pharmaceuticals. K. Politi is a consultant/advisory board member for Takeda and has a patent relating to EGFR T790M mutation testing that was licensed by Memorial Sloan Kettering Cancer Center to MolecularMD. No other potential conflict of interest was disclosed.
Authors' Contributions: Conception and design: S. Gettinger, K. Politi
Writing, review, and/or revision of the manuscript: S. Gettinger, K. Politi
References
- 1.Katayama R, Friboulet L, Koike S, Lockerman E, Khan TM, Gainor JF, et al. Two novel ALK mutations mediate acquired resistance to the next generation ALK inhbitor alectinib. Clin Cancer Res. 2014;20:XXX–XXX. doi: 10.1158/1078-0432.CCR-14-1511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Shaw AT, Engelman JA. ALK in lung cancer: past, present, and future. J Clin Oncol. 2013;31:1105–11. doi: 10.1200/JCO.2012.44.5353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Shaw AT, Kim DW, Nakagawa K, Seto T, Crino L, Ahn MJ, et al. Crizotinib versus chemotherapy in advanced ALK-positive lung cancer. N Engl J Med. 2013;368:2385–94. doi: 10.1056/NEJMoa1214886. [DOI] [PubMed] [Google Scholar]
- 4.Katayama R, Shaw AT, Khan TM, Mino-Kenudson M, Solomon BJ, Halmos B, et al. Mechanisms of acquired crizotinib resistance in ALK-rearranged lung Cancers. Sci Transl Med. 2012;4:120ra17. doi: 10.1126/scitranslmed.3003316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shaw AT, Kim DW, Mehra R, Tan DS, Felip E, Chow LQ, et al. Ceritinib in ALK-rearranged non-small-cell lung cancer. N Engl J Med. 2014;370:1189–97. doi: 10.1056/NEJMoa1311107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gettinger SN, Bazhenova L, Salgia R, Langer CJ, Gold KA, Rosell R, et al. Updated efficacy and safety of the ALK inhibitor AP26113 in patients (pts) with advanced malignancies, including ALK+ non-small cell lung cancer (NSCLC) J Clin Oncol. 2014;32:5s. suppl; abstr 8047. [Google Scholar]
- 7.Ou SH, Gadgeel S, Chiappori A, Riely G, Lee R, Garcia L, et al. Safety and efficacy analysis of RO5424802/CH5424802 in anaplastic lymphoma kinase (ALK)-positive non-small cell lung cancer (NSCLC) patients who have failed crizotinib in a dose-finding phase I study (AF-002JG, NCT01588028) European J Cancer. 2013;49(3) [Google Scholar]
- 8.Lovly CM, Pao W. Escaping ALK inhibition: mechanisms of and strategies to overcome resistance. Sci Transl Med. 2012;4:120ps2. doi: 10.1126/scitranslmed.3003728. [DOI] [PubMed] [Google Scholar]
- 9.Friboulet L, Li N, Katayama R, Lee CC, Gainor JF, Crystal AS, et al. The ALK inhibitor ceritinib overcomes crizotinib resistance in non-small cell lung cancer. Cancer Discov. 2014;4:662–73. doi: 10.1158/2159-8290.CD-13-0846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ignatius Ou SH, Azada M, Hsiang DJ, Herman JM, Kain TS, Siwak-Tapp C, et al. Next-generation sequencing reveals a Novel NSCLC ALK F1174V mutation and confirms ALK G1202R mutation confers high-level resistance to alectinib (CH5424802/RO5424802) in ALK-rearranged NSCLC patients who progressed on crizotinib. J Thorac Oncol. 2014;9:549–53. doi: 10.1097/JTO.0000000000000094. [DOI] [PubMed] [Google Scholar]
- 11.Mosse YP, Laudenslager M, Longo L, Cole KA, Wood A, Attiyeh EF, et al. Identification of ALK as a major familial neuroblastoma predisposition gene. Nature. 2008;455:930–5. doi: 10.1038/nature07261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang S, Wang F, Keats J, Zhu X, Ning Y, Wardwell SD, et al. Crizotinib-resistant mutants of EML4-ALK identified through an accelerated mutagenesis screen. Chem Biol Drug Design. 2011;78:999–1005. doi: 10.1111/j.1747-0285.2011.01239.x. [DOI] [PMC free article] [PubMed] [Google Scholar]