Abstract
Here we report the crystal structures of the Thermus thermophilus ribosome at 2.3-2.5Å-resolution, which have enabled a comprehensive modeling of rRNA modifications. The structures reveal contacts of modified nucleotides with mRNA and tRNAs or protein pY, and contacts within the ribosome interior stabilizing the functional fold of rRNA. Our work provides a resource to explore the roles of rRNA modifications and yields the most complete atomic model of bacterial ribosome.
Current models suggest that the high fidelity of protein synthesis by the ribosome requires numerous modifications of rRNA1-3. In bacteria, there are three major types of rRNA modifications: conversion of uridines to pseudouridines, methylation of the 2’-hydroxyl groups of ribose, and methylation of nucleotide bases1. Modified nucleotides comprise only a small fraction of all rRNA residues (23 out of ~4400 total in T. thermophilus ribosomes4) and cluster mainly near the ribosome functional centers. Numerous studies show that the loss of rRNA modifications results in the alteration of the structures of the active sites5-7, which causes slower rates and lower accuracy of translation8-10, as well as impaired responses to metabolites and antibiotics11-13. Despite the functional importance of rRNA modifications, the resolution of previous structural studies of the 70S ribosomes was insufficient to unambiguously model modified nucleotides, which were instead modeled as regular RNA nucleotides.
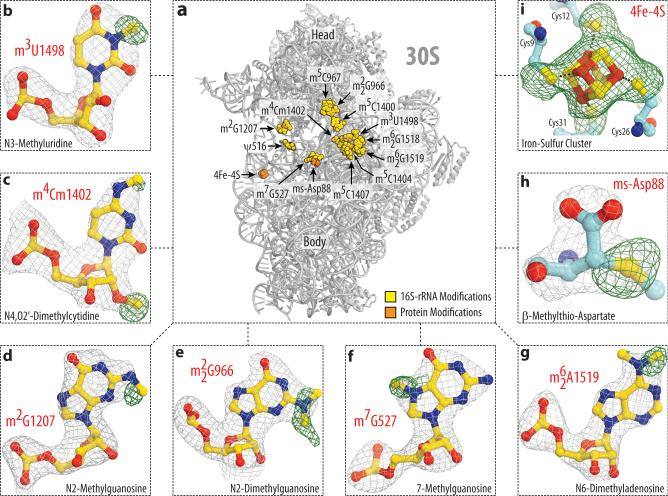
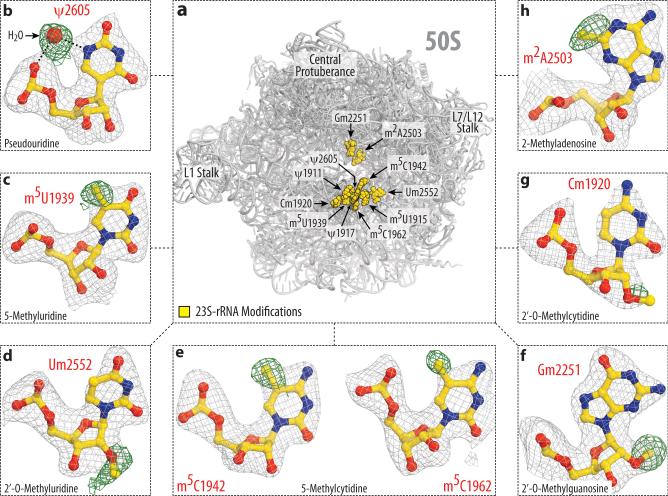
Here, we set out to apply our recent advances in crystal treatment to reach diffraction resolution suitable for visualization of modified nucleotides within the 70S ribosome (see Online Methods). As a result, we report two crystal structures of the T. thermophilus (Tth) 70S ribosome reflecting two functional states – hibernating ribosomes, in complex with the hibernation factor pY, and translating ribosomes, in complex with mRNA and tRNAs, – determined at 2.3Å and 2.5Å resolution, respectively (I/σI=1) (Supplementary Table 1). The resolution at which I/σI=2 is 2.5Å and 2.7Å, respectively. At this resolution methylation of ribose and nucleotide bases can be directly visualized in the unbiased electron density maps (Fig. 1, panels d-j and m-q). Pseudouridines cannot be distinguished from uridines and, therefore, their modeling was guided by biochemical data4. In one particular case, however, the electron density map confirmed the presence of pseudouridine (Ψ2605), consistent with hydrogen bonding between the base and the phosphate via a water molecule, which is impossible for uridine base (Fig. 2b). Apart from rRNA modifications, the maps also confirmed the presence of a modification in ribosomal protein S12 – β-methyl-thiolation of residue Asp88 (Fig. 1h, Supplementary Fig. 1a, b), – and a prosthetic group in ribosomal protein S4 – the 4Fe-4S iron-sulfur cluster, coordinated by cysteine residues (Fig. 1i, Supplementary Fig. 1c, d). In total, each of our models contains 23 modified RNA nucleotides – all known modifications of Tth rRNA, – among which 5 are conserved across bacteria, archaea and eukaryotes, 18 are common between Tth and E. coli, and 5 are specific to Tth (Fig. 1a, Fig. 2a, Supplementary Table 1).
Figure 1. Electron density maps allow comprehensive modeling of rRNA and ribosomal protein modifications of the 30S subunit.
(a) Spatial distribution of modified nucleotides in the structure of the small ribosomal subunit from T. thermophilus. Here, in Fig. 2 and throughout the text rRNA nucleotides are named according to the E. coli numbering system. Panels (b-i) show unbiased difference Fo-Fc electron density maps for eight types of modifications present in 16S rRNA and ribosomal proteins S12 and S4. Grey mesh shows the Fo-Fc map after refinement with the entire modified nucleotides or amino acid residues omitted (contoured at 2.7-3.0σ). Green mesh shows the Fo-Fc electron density map after refinement with unmodified nucleotides (contoured at 1.5-2.5σ). The modifications shown are: (b) N3-methyluridine 1498, (c) N4,O2’-dimethylcytidine 1402, (d) N2-methylguanosine 1207, (e) N2-dimethylguanosine 966, (f) N7-methylguanosine 527, (g) N6-dimethyladenosine 1519; (h) β-methylthioaspartate of protein S12, (i) 4Fe-4S iron-sulfur cluster of protein S4.
Figure 2. Modeling of rRNA modifications of the 50S subunit.
(a) Spatial distribution of modified nucleotides in the structure of the large ribosomal subunit from T. thermophilus. Panels (b-h) show unbiased difference Fo-Fc electron density maps for eight types of modifications present in 23S rRNA. The color scheme is the same as in Fig.1. The modifications shown are: (b) pseudouridine 2605, (c) 5-methyluridine 1939, (d) 2’-O-methyluridine 2552, (e) 5-methylcytidines 1942 and 1962, (f) 2’-O-methylguanosine 2251; (g) 2’-O-methylcytidine 1920, (h) 2-methyladenosine 2503.
The structures revealed three major types of contacts formed by rRNA modifications: (1) with the ribosome ligands; (2) between the ribosomal subunits; and (3) within the interior of the ribosomal RNA (Fig. 3a). The first group comprises nucleotides m22G966, m5C1400, m4Cm1402 of the 16S rRNA, which are all located within the P site of the small ribosomal subunit. In the complex of 70S ribosomes with mRNA and tRNAs, the methyl groups of these residues surround the mRNA-tRNA duplex and contact the mRNA codon (m4Cm1402) and the wobble base-pair (m22G966, m5C1400), as predicted earlier14,15 (Fig. 3b). These contacts rationalize why loss of m22G966 methylation affects recruitment of the initiator tRNA and the initiation of translation16-18. One of these modifications, 5-C-methylation of C1400, is absent in E. coli ribosomes and thereby may reflect adaptation of Tth ribosomes to high temperature via the extended stacking between m5C1400 base and the wobble-pair (Supplementary Table 1). In the complex of 70S ribosomes with protein pY, the methyl groups of m22G966, m5C1400, and m4Cm1402 form hydrophobic contacts with pY (Fig. 3c). These contacts suggest that loss of the rRNA modifications could not only alter the ribosome affinity to the P-site tRNA but also could reduce its affinity to the hibernation factor pY and, thus, impair translational response to stress and stationary phase.
Figure 3. Modified nucleotides form numerous molecular contacts with the ribosome ligands and within the ribosome interior.
(a) Categorization of modified rRNA nucleotides into three groups based on their molecular contacts: (i) interacting with the ribosome ligands (orange), (ii) forming the inter-subunit bridges (light blue), and (iii) strengthening RNA-RNA and RNA-protein interactions within ribosomal interior (yellow). The actual sites of modifications are shown as red spheres. Modifications specific only to T. thermophilus are labeled in red (see Supplementary Table 2). (b-c)rRNA modifications that directly interact with either mRNA and tRNAs (b), or with protein pY (c). (d) rRNA modifications that maintain structure of the helix 44 in the mRNA channel (mRNA is not shown for clarity). In panels (b-d), modified residues of 16S rRNA are shown in orange, unmodified residues are in in light yellow.
The second group comprises the 23S rRNA modifications of m5U1915 and Cm1920, which are located at the interface between the large and the small ribosomal subunits (Supplementary Fig. 2a). As previously shown, these modification are among the most variable across the species19. While U1915 is 5-C-methylated in Tth, it is 3-N-methylated and pseudouridylated in E. coli, and unmethylated but pseudouridylated in archaea (H. marismortui) and eukaryotes (S. cerevisiae and H. sapiens)19. The second residue, C1920, is 2’-O-methylated in Tth, unmethylated in E. coli, and pseudouridylated in eukaryotic ribosomes. In the classical state of the ribosome in both of our complexes, 2’-O-methyl group forms hydrophobic interactions between the large and the small subunits (Supplementary Fig. 2a). Therefore, these two modifications might adjust the strength of the inter-subunit contacts to meet the requirements of translation at different temperatures or compensate for the interspecies variability in the inter-subunit bridges.
The third and the most abundant group comprises the remaining nucleotides that are buried inside and reside within the single-stranded rRNA segments or in highly distorted helices, where rRNA adopts energetically unfavorable folds. Our structures revealed that, apart from pseudouridines, most of these modifications form or extend the hydrophobic interactions that further stabilize rRNA structure. Perhaps, the most unusual example of structural motifs involving rRNA modifications is the one observed in a cluster of modified nucleotides in the mRNA channel (Fig. 3d). In the middle part of this cluster, nucleotides C1403, m3U1498 and A1499 form a base-triple, whose structure is supported by 3-N-methylation of U1498 and by stacking with the flanking bases of m4Cm1402 and m5C1404. These interactions occur at the interface of the mRNA channel and, thus, might be important for the proper accommodation of the mRNA. As predicted by previous studies, similar motifs are found in other functional sites of the ribosome, such as the peptide-exit tunnel or the tRNA-binding sites on the large ribosomal subunit3. In the peptide exit tunnel, the 2-C-methyl group of A2503 (m2A2503) extends the stacking between A2059 and A2503. This stacking supports the fold of the two single-stranded rRNA segments of the peptidyl transferase loop, which form the wall of the peptide exit tunnel (Supplementary Fig. 2b). In the A-loop of the 23S rRNA, the 2’-O-methyl group of Um2552 intercalates between the bases of G2553 and U2554. Apparently, these contacts are maintaining the active conformation of the G2553 base, which is directly involved in accommodation of the aminoacyl-tRNA (Supplementary Fig. 2c). Similarly, the P-loop of the 23S rRNA carries 2’-O-methylated Gm2251, which is the key residue required for accommodation of the P-site tRNA via Watson-Crick base pairing with the 3’-end of tRNA. The 2’-O-methyl group of Gm2251 forms additional hydrophobic contacts with the sugar of C2065 and the base of U2449 (Supplementary Fig. 2d). Since the m2A2503, Gm2251 and Um2552 modifications are universally conserved across kingdoms of life, similar or identical interactions are expected to occur in the ribosomes from other species, including human. Overall, Tth rRNA carries eighteen rRNA modifications, which are buried or oriented towards the interior of rRNA helices and loops, where they form additional hydrophobic contacts with neighboring nucleotides and, in a few cases, with ribosomal proteins (Supplementary Table 1). Therefore, the major role of modified nucleotides within the ribosome is possibly related to maintenance of distorted rRNA folds at the functional interfaces of the ribosome.
In summary, we have visualized previously unseen modifications of ribosomal RNA. This allowed us to create the most comprehensive models of bacterial ribosome and provide a resource to explore unique properties of modified nucleotides in the context of translating or hibernating ribosome. Since RNA modifications are present in most non-coding RNAs and mRNAs, our structures pave the way to understanding the general principles of how nucleotide modifications control RNA structure and function.
ONLINE METHODS
Purification of Thermus thermophilus ribosomes and preparation of tRNAs
70S ribosomes from T. thermophilus and unmodified tRNAiMet and tRNAPhe from E. coli were purified as previously described20. Synthetic mRNA with the sequence 5’-GGC AAG GAG GUA AAA AUG UUC UAA-3’ was obtained from Integrated DNA Technologies (Coralville, IA).
Crystallization and data collection
The ribosome complex with protein pY was formed essentially as described previously21. The ribosome complex with mRNA and tRNAs was formed by programming 5 μM Tth 70S ribosomes with 10 μM mRNA and 20 μM P- and A-site tRNA substrates. All Tth 70S ribosome complexes were formed in the buffer containing 5 mM HEPES-KOH (pH 7.6), 50 mM KCl, 10 mM NH4Cl, and 10 mM Mg(CH3COO)2, and then crystallized in the buffer containing 100 mM Tris-HCl (pH 7.6), 2.9% (w/v) PEG-20K, 7-12% (v/v) MPD, 100-200 mM arginine, 0.5 mM β-mercaptoethanol. Crystals were grown by the vapor diffusion method in sitting drops at 19°C and stabilized as described20. Diffraction data were collected using beamline 24IDC at the Advanced Photon Source (Argonne, IL). A complete dataset for each ribosome complex was collected using 0.979Å wavelength at 100K from multiple regions of the same crystal using 0.2° oscillations.
Crystallographic data processing and model building
The raw data were integrated and processed using the XDS software package22. All crystals belonged to the primitive orthorhombic space group P212121 with approximate unit cell dimensions of 210Å × 450Å × 620Å and contained two copies of the 70S ribosome per asymmetric unit. The initial molecular replacement solutions were refined by rigid body refinement, followed by positional and individual B-factor refinement. Location of rRNA modifications within the rRNA was facilitated by the use of the Fournier'S Lab database of ribosomal RNA modifications19 and biochemical data for T. thermophilus4. The electron density maps allowed location of the previously unknown position of methyl groups in the nucleotides C1915 (m5C1915) and C1940 (m5C1940). All modified nucleotides were also modeled in both tRNAiMet and tRNAPhe. The final models of the 70S ribosome in complex with either pY or mRNA and tRNAs were generated by multiple rounds of model building in COOT23, followed by refinement in PHENIX24. The improved quality of the electron density maps allowed us to build a more accurate model of the 70S ribosome with more residues of the ribosomal proteins falling into the favored and allowed regions of the Ramachandran plot: 95.79% favored, 3.73% allowed, 0.47% disallowed for the 70S-pY complex, and 94.80% favored, 4.29% allowed, 0.91% disallowed for the 70S-mRNA-tRNAs complex. The statistics of data collection and refinement for each complex are compiled in Table 1. In both structures, all rRNA nucleotides were renumbered according to E. coli numbering system.
Table 1.
Data collection and refinement statistics
| 70S-pYa | 70S-mRNA-tRNAsa | |
|---|---|---|
| Data collection | ||
| Space group | P212121 | P212121 |
| Cell dimensions | ||
| a, b, c (Å) | 209.16, 448.37, 618.12 | 209.70, 450.05, 624.09 |
| α, β, γ (°) | 90.0, 90.0, 90.0 | 90.0, 90.0, 90.0 |
| Resolution (Å) | 309-2.30 (2.36-2.30) | 312-2.50 (2.56-2.50) |
| R merge | 15.1 (186.6) | 14.1 (111.6) |
| I / σI | 8.50 (0.85)b | 8.27 (0.90)c |
| Completeness (%) | 97.6 (81.4) | 97.9 (88.5) |
| Redundancy | 5.60 (4.70) | 3.28 (2.52) |
| Refinement | ||
| Resolution (Å) | 173.22-2.32 | 122.01-2.50 |
| No. reflections | 2.467.089 | 1.962.003 |
| Rwork / Rfree | 20.69/24.67 | 23.07-28.07 |
| No. atoms | ||
| Protein | 93.016 | 90.976 |
| Ligand/ion | 194.892 | 203.092 |
| Water | 9.516 | 5.058 |
| B factors | ||
| Protein | 59.5 | 62.8 |
| Ligand/ion | 54.9 | 59.1 |
| Water | 43.8 | 46.3 |
| r.m.s. deviations | ||
| Bond lengths (Å) | 0.006 | 0.006 |
| Bond angles (°) | 1.151 | 1.063 |
Values in parentheses are for highest-resolution shell.
Single crystal was used to obtain the structure
I/σI = 2 at 2.5Å resolution
I/σI = 2 at 2.7Å resolution
Supplementary Material
ACKNOWLEDGMENTS
We thank J. Lin, C.A. Innis and N.V. Sumbatyan for bringing this project to our attention, R.L. Grodzicki for preparation of the unmodified tRNAs and for critical reading of the manuscript, and members of the T.A.S. and D.S. laboratories for discussions. We also thank the staff at the Advanced Photon Source (beamline 24ID), supported by award GM103403 from the National Center for Research Resources at the National Institutes of Health, the staff at the National Synchrotron Light Source (beamline X25) for help during data collection and the staff at the Richards Center at Yale University for computational support. This work was supported by US National Institutes of Health grants GM022778 (T.A.S.) and GM022854 (D.S.).
Footnotes
AUTHOR CONTRIBUTIONS
Y.S.P. and S.V.M. devised experiments; Y.S.P. crystallized ribosomal complexes and performed crystallographic data collection and processing, S.V.M. and Y.S.P. analyzed the data, and S.V.M., Y.S.P., D.S. and T.A.S. wrote the manuscript.
ACCESSION CODES
Coordinates and structure factors were deposited in the RCSB Protein Data Bank with accession codes 4Y4O for the Tth 70S ribosome in complex with protein pY, and 4Y4P for the Tth 70S ribosome in complex with mRNA and tRNA ligands.
REFERENCES
- 1.Decatur WA, Fournier MJ. rRNA modifications and ribosome function. Trends Biochem. Sci. 2002;27:344–351. doi: 10.1016/s0968-0004(02)02109-6. [DOI] [PubMed] [Google Scholar]
- 2.Chow CS, Lamichhane TN, Mahto SK. Expanding the nucleotide repertoire of the ribosome with post-transcriptional modifications. ACS Chem. Biol. 2007;2:610–619. doi: 10.1021/cb7001494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sergiev PV, et al. Modifications of ribosomal RNA: From enzymes to function. Ribosomes: Structure, Function, and Dynamics. 2011:97–110. [Google Scholar]
- 4.Mengel-Jorgensen J, et al. Modifications in Thermus thermophilus 23 S ribosomal RNA are centered in regions of RNA-RNA contact. J Biol Chem. 2006;281:22108–17. doi: 10.1074/jbc.M600377200. [DOI] [PubMed] [Google Scholar]
- 5.Demirci H, et al. Modification of 16S ribosomal RNA by the KsgA methyltransferase restructures the 30S subunit to optimize ribosome function. RNA. 2010;16:2319–2324. doi: 10.1261/rna.2357210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Desaulniers JP, et al. Pseudouridines in rRNA helix 69 play a role in loop stacking interactions. Org. Biomol. Chem. 2008;6:3892–3895. doi: 10.1039/b812731j. [DOI] [PubMed] [Google Scholar]
- 7.Sumita M, Jiang J, SantaLucia J, Jr., Chow CS. Comparison of solution conformations and stabilities of modified helix 69 rRNA analogs from bacteria and human. Biopolymers. 2011;97:94–106. doi: 10.1002/bip.21706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Liang XH, Liu Q, Fournier MJ. rRNA modifications in an intersubunit bridge of the ribosome strongly affect both ribosome biogenesis and activity. Mol. Cell. 2007;28:965–977. doi: 10.1016/j.molcel.2007.10.012. [DOI] [PubMed] [Google Scholar]
- 9.Baudin-Baillieu A, et al. Nucleotide modifications in three functionally important regions of the Saccharomyces cerevisiae ribosome affect translation accuracy. Nucleic Acids Res. 2009;37:7665–7677. doi: 10.1093/nar/gkp816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Baxter-Roshek JL, Petrov AN, Dinman JD. Optimization of ribosome structure and function by rRNA base modification. PLoS One. 2007;2:e174. doi: 10.1371/journal.pone.0000174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Helser TL, Davies JE, Dahlberg JE. Change in methylation of 16S ribosomal RNA associated with mutation to kasugamycin resistance in Escherichia coli. Nat. New Biol. 1971;233:12–14. doi: 10.1038/newbio233012a0. [DOI] [PubMed] [Google Scholar]
- 12.Doi Y, Arakawa Y. 16S ribosomal RNA methylation: emerging resistance mechanism against aminoglycosides. Clin. Infect. Dis. 2007;45:88–94. doi: 10.1086/518605. [DOI] [PubMed] [Google Scholar]
- 13.Prokhorova IV, et al. Modified nucleotides m(2)G966/m(5)C967 of Escherichia coli 16S rRNA are required for attenuation of tryptophan operon. Sci. Rep. 2013;3:3236. doi: 10.1038/srep03236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Prince JB, Taylor BH, Thurlow DL, Ofengand J, Zimmermann RA. Covalent crosslinking of transfer Rna1val to 16s Rna at the ribosomal-P Site - identification of crosslinked residues. Proc. Natl. Acad. Sci. USA. 1982;79:5450–5454. doi: 10.1073/pnas.79.18.5450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kimura S, Suzuki T. Fine-tuning of the ribosomal decoding center by conserved methyl-modifications in the Escherichia coli 16S rRNA. Nucleic Acids Res. 2009;38:1341–1352. doi: 10.1093/nar/gkp1073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Burakovsky DE, et al. Impact of methylations of m(2)G966/m(5)C967 in 16S rRNA on bacterial fitness and translation initiation. Nucleic Acids Res. 2012;40:7885–7895. doi: 10.1093/nar/gks508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jemiolo DK, Taurence JS, Giese S. Mutations in 16S rRNA in Escherichia coli at methyl-modified sites: G966, C967, and G1207. Nucleic Acids Res. 1991;19:4259–4265. doi: 10.1093/nar/19.15.4259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Arora S, et al. Distinctive contributions of the ribosomal P-site elements m2G966, m5C967 and the C-terminal tail of the S9 protein in the fidelity of initiation of translation in Escherichia coli. Nucleic Acids Res. 2013;41:4963–4975. doi: 10.1093/nar/gkt175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Piekna-Przybylska D, Decatur WA, Fournier MJ. The 3D rRNA modification maps database: with interactive tools for ribosome analysis. Nucleic Acids Res. 2008;36:D178–183. doi: 10.1093/nar/gkm855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Polikanov YS, Steitz TA, Innis CA. A proton wire to couple aminoacyl-tRNA accommodation and peptide-bond formation on the ribosome. Nat. Struct. Mol. Biol. 2014;21:787–793. doi: 10.1038/nsmb.2871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Polikanov YS, Blaha GM, Steitz TA. How Hibernation Factors RMF, HPF, and YfiA Turn Off Protein Synthesis. Science. 2012;336:915–918. doi: 10.1126/science.1218538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kabsch W. Xds. Acta Cryst. Sect. D Biol. Cryst. 2010;66:125–132. doi: 10.1107/S0907444909047337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallographica Section D-Biological Crystallography. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Adams PD, et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallographica Section D-Biological Crystallography. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.