Abstract
Background and Objective
Porphyromonas gingivalis has been implicated as one of the major pathogens in chronic periodontitis, an infectious disease affecting the majority of the adult population. We have previously demonstrated that a surface protein, arginine deiminase (ArcA), of Streptococcus cristatus represses production of P. gingivalis long fimbriae and interrupts the formation of P. gingivalis biofilms in vitro. Our in vivo studies have also shown that the distribution of P. gingivalis and S. cristatus in human subgingival plaque is negatively correlated. The objective of this study is to determine if S. cristatus ArcA inhibits P. gingivalis colonization and attenuates its subsequent pathogenesis in alveolar bone loss in the murine oral cavity.
Material and Methods
A wild-type strain of S. cristatus, CC5A and its arcA knock-out mutant ArcAE were used as initial colonizers in the oral cavity of BALB/cByJ mice. Colonization of P. gingivalis on the existing S. cristatus biofilms was assessed by qPCR and P. gingivalis-induced alveolar bone loss was measured 6 weeks after P. gingivalis infection.
Results
The presence of S. cristatus CC5A, but not its arcA mutant, attenuated P. gingivalis colonization in the murine oral cavity. In addition, P. gingivalis-induced alveolar bone loss was significantly less in mice initially infected with S. cristatus CC5A than in those infected with the arcA mutant.
Conclusions
This study provides direct evidence that S. cristatus ArcA has an inhibitory effect on P. gingivalis colonization, which may in turn attenuate the pathogenecity of P. gingivalis.
Keywords: Porphyromonas gingivalis, Streptococcus cristatus ArcA, mouse, pathogenesis
INTRODUCTION
Chronic periodontitis is among the most common infectious diseases in humans and affects not only the periodontium but also systemic conditions. Associations have been demonstrated between periodontitis and several systemic diseases such as cardiovascular diseases, preterm labor, and respiratory diseases (1-3). It has been generally accepted that the initiation of periodontitis depends on the existence of certain gram-negative species of bacteria and, Porphyromonas gingivalis is the predominant species implicated in periodontitis (4, 5).
P. gingivalis has been studied extensively in vitro and in vivo for its role in chronic periodontitis. The majority of P. gingivalis clinical isolates are fimbriated, especially when isolated from deep periodontal pockets (6, 7) and different FimA genotypes have been demonstrated (8-10). The major fimbriae (long fimbriae) of P. gingivalis composed of polymeric FimA subunit proteins are a well-studied virulence factor contributing to colonization, biofilm formation, cell invasion, bone resorption, and evasion of host defenses (11-19). Thus, reduction of FimA production in P. gingivalis has been considered as an attractive strategy for preventing the bacterial colonization and subsequent periodontal pathogenesis. We previously reported that arginine deiminase (ArcA) of Streptococcus cristatus is capable of selectively repressing FimA expression in P. gingivalis (20). As a surface protein of S. cristatus, ArcA was identified as the signaling molecule to which P. gingivalis responds by repressing expression of the fimA gene and production of the FimA protein. Our recent results also showed that the distribution of S. cristatus and P. gingivalis in dental plaque in periodontitis patients was negatively correlated (21). These studies suggested that an early colonizer of dental plaque, S. cristatus, may serve as a beneficial bacterium to block P. gingivalis accumulation in dental plaque in sufficient numbers as to reduce periodontal inflammation.
In the present study, we investigated the role of ArcA in P. gingivalis colonization and its pathogenesis in alveolar bone loss using a mouse model. Utilizing a wild-type S. cristatus strain CC5A and its arcA knockout mutant (22) as initial colonizers, we report here direct evidence that S. cristatus ArcA attenuates sequential P. gingivalis colonization and subsequent P. gingivalis-induced alveolar bone loss in vivo.
MATERIALS & METHODS
Bacterial Strains and Media
S. cristatus CC5A and its arcA knockout mutant, ArcAE, (erythromycin-resistant) (22) were grown in Trypticase peptone broth (TPB) supplemented with 0.5% glucose at 37°C under aerobic conditions. P. gingivalis 33277 was grown from frozen stocks in Trypticase Soy broth (TSB) or on TSB blood agar plates supplemented with yeast extract (1 mg/ml), hemin (5 μg/ml), and menadione (1 μg/ml) at 37°C in an anaerobic chamber (85% N2, 10% H2, 5% CO2).
Infection of Mice
Specific-pathogen-free BALB/cByJ male mice (Jackson Laboratory, Bar Harbor, ME, USA) were maintained in the Laboratory Animal Facility of the University at Buffalo, State University of New York. The experimental protocols were approved by the Institutional Animal Care and Use Committee of the University at Buffalo. Infection timeline and the experimental groups are listed in Table 1. Briefly, 6 week old mice (8 mice per group) were treated with kanamycin (Sigma-Aldrich, Saint Louis, MO) at 1 mg/ml in water ad libitum for 7 days, followed by a three-day antibiotic-free period (23). Mice were infected orally with micropipettes using 2 × 109 cfu of live S. cristatus CC5A or its arcA knock out mutant (ArcAE) twice a day for 5 days in 50 μl of PBS with 2% carboxymethyl cellulose (PBS-CMC). P. gingivalis 33277 cells in late log phase were pelleted and re-suspended in PBS-CMC. Inoculation of P. gingivalis (5 × 109 cfu in 50 μl PBS-CMC intraorally by micropipettes, three inoculations at two day intervals) was carried out 7 days after the last inoculation of S. cristatus and was repeated two weeks after the last P. gingivalis inoculation. Control (sham-infected) mice received the antibiotic treatment and the intraoral PBS-CMC inoculation without the bacteria. Mice were kept away from food and water for 1 h after each inoculation of bacteria.
Table 1.
Timeline and experimental groups.
| Days | 1-7 | 8-10 | 11-16 | 23 | 23, 25, 27 41, 43, 45 | 41 | 69 |
|---|---|---|---|---|---|---|---|
| Antibiotics | Antibiotics | S. cristatus infection | qPCR & Colony morphology | P. gingivalis infection | qPCR | Assessment of bone loss | |
| Group 1 | Yes | No | CC5A | No | Yes | Yes | Yes |
| Group 2 | Yes | No | ArcAE | No | Yes | Yes | Yes |
| Group 3 | Yes | No | CC5A | Yes | No | No | Yes |
| Group 4 | Yes | No | ArcAE | Yes | No | No | Yes |
| Group 5 | Yes | No | PBS-CMC | Yes | No | Yes | Yes |
Bacterial Quantitation
The colonization of S. cristatus or P. gingivalis in the murine oral cavities was examined at 7 or 14 days after the final inoculation of bacteria, respectively (see Table 1). The plaque samples were taken from tooth surfaces and surrounding gingival mucosa using cotton applicators and immersed in 1 ml of Tris-EDTA (TE) buffer (pH 7.5). The suspensions were dispersed by vortex at full speed for 30 seconds and bacteria were harvested by centrifugation. The samples obtained were boiled for 20 minutes to release chromosomal DNA. To minimize experimental error, plaque samples from the oral cavity were collected by a single investigator (JH). The final volume per sample before DNA extraction by boiling was fixed at 50 μl in TE buffer. A fixed volume of 5 μl DNA per sample was utilized in qPCR. P. gingivalis and S. cristatus cells were enumerated by using a QuantiTect SYBR Green PCR Kit (Qiagen, CA) with P. gingivalis species-specific 16S rDNA gene primers, (TGTAGATGACTGATGGTGAAA and ACTGTTAGCAACTACCGATGT) (24) or S. cristatus 23S rDNA gene primers (ACTGCAATGTGGACTCAGAATTTAT and TACAGAATCTATTTAAAATACGAGGCTCT). Standards used to determine numbers of P. gingivalis or S. cristatus cells were prepared using genomic DNAs from the wild type strain 33277 or CC5A (21). A fresh culture of bacteria was serially diluted in PBS and plated to enumerate colony forming units at each dilution. Chromosomal DNA was isolated from the dilutions and a qPCR assay was run to determine cell numbers. S. cristatus colonization in the oral cavity was confirmed by colony morphology. The colonization of S. cristatus in the murine oral cavities was examined 7 days after the final inoculation of the bacteria. The plaque samples were taken from tooth surfaces and surrounding gingival mucosa using cotton applicators (from 3 mice per group that have not be sampled for qPCR) and smeared on Mitis Salivarius (MS) agar plates. Blue colonies representing S. cristatus were counted after 36-48 h incubation at 37°C in candle jars.
Alveolar Bone Loss Measurement
To examine alveolar bone loss, the mice were sacrificed on day 69 (see Table 1). The maxillae from the sacrificed mice were removed, autoclaved, and mechanically defleshed to remove all the soft tissue. The maxillae was then immersed overnight in 3% hydrogen peroxide and stained with 1% methylene blue. The distance between the cementoenamel junction (CEJ) and alveolar bone crest (ABC) were measured for a total of 14 buccal sites on the right and left maxillary molars [3 sites on first molar, 2 sites on second molar and another 2 sites on third molar, as described by Sharma et al (25)]. Measurement was made under a dissecting microscope with an Aquinto imaging measurement system. Alveolar bone loss per experimental group was calculated as the average of 14-site total CEL-ABC distances for each group.
Statistical Analysis
ANOVA analysis and Student's t-test were performed to determine statistically significant difference in colonization and alveolar bone loss among different experimental groups. A p value < 0.05 was considered significant.
RESULTS
Colonization of S. cristatus in the Murine Oral Cavity
To establish the feasibility of S. cristatus colonization in the murine oral cavity, wild-type S. cristatus CC5A and its arcA knockout mutant ArcAE were introduced into the murine oral cavities. Quantification of colonized S. cristatus was carried out using qPCR, with in vitro cultured S. cristatus CC5A as a standard control. Both the wild-type CC5A and the knockout mutant consistently colonized the murine oral cavity (positive qPCR was obtained from every mouse infected, data not shown). There was no significant difference in colonization efficiencies between the two groups [(13.0 ± 3.4) x 103 vs. (14.2 ± 2.9) x 103); p > 0.05] (Fig. 1).
Figure 1.
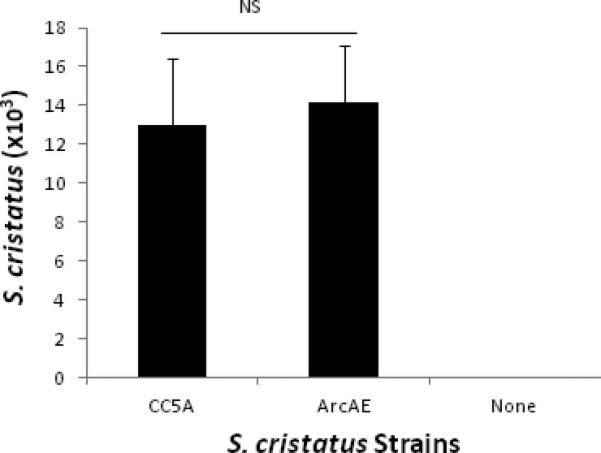
Colonization of S. cristatus in murine oral cavities. The plaque samples were taken from tooth surfaces and surrounding gingival mucosa 7 days after the last inoculation of S. cristatus in the oral cavities. DNA extracted from these plaque samples (50 μl final volume/sample in TE buffer) was examined for quantitation of S. cristatus using qPCR with primers corresponding to 23S rDNA as probes. ANOVA analysis revealed no significant difference in colonization efficiencies between the wild-type CC5A and the knockout mutant ArcAE (p > 0.05). Data represent the means ± S.D. from 5 mice per group in Experimental Groups 3, 4, and 5.
Colonization of S. cristatus strains in the murine oral cavity was also determined by culturing the bacteria on MS agar plates; streptococcal colonies were observed in the samples retrieved from CC5A or its arcA mutant infected mice, but not from sham-infected mice. This result further confirmed the colonization of S. cristatus in the murine oral cavity (Fig. 2). We also confirmed that the streptococcal colonies on MS agar plates were indeed the S. cristatus strains, using qPCR with the S. cristatus 23S rDNA primers (data not shown).
Figure 2.
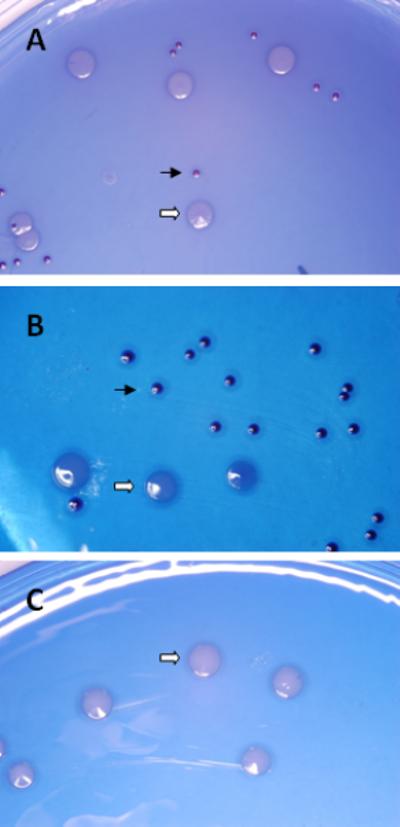
S. cristatus morphology on MS agar plates. The plaque samples were taken from tooth surfaces and surrounding gingival mucosa 7 days after the last intraoral inoculation of S. cristatus in the oral cavities. The bacteria on the cotton applicator were streaked on the MS plates and incubated for 36-48 hours anaerobically. Photographs are the representatives of each experimental group (3 mice/group sampled from Experimental Groups 3, 4, and 5). A, CC5A-infected; B, ArcAE-infected; and C, sham-infected. The small blue colonies represent colonized S. cristatus (solid arrows) and large white colonies were residual endogenous murine bacteria (open arrows).
S. cristatus ArcA Attenuated the Colonization of P. gingivalis in the Murine Oral Cavity
Formation of dental plaque on human enamel chips has been shown to be a sequential process with streptococci as the dominating initial colonizers (26, 27). Based on our earlier observations that ArcA of S. cristatus inhibits formation of P. gingivalis biofilms in vitro, we examined the role of S. cristatus biofilms on P. gingivalis colonization in the murine oral cavities. Colonization of P. gingivalis was determined using qPCR, with 16S rDNA primers as probes. As shown in Fig. 3A, at 2 weeks post P. gingivalis inoculation, the colonization of P. gingivalis in the wild-type S. cristatus CC5A colonized mice was significantly less efficient than that in the arcA mutant ArcAE colonized mice [(4.48 ± 2.54) x 103 vs. (24.93 ± 2.08) x 103; p = 0.0062], indicating that ArcA of S. cristatus interferes with P. gingivalis colonization in the murine oral cavity.
Figure 3.
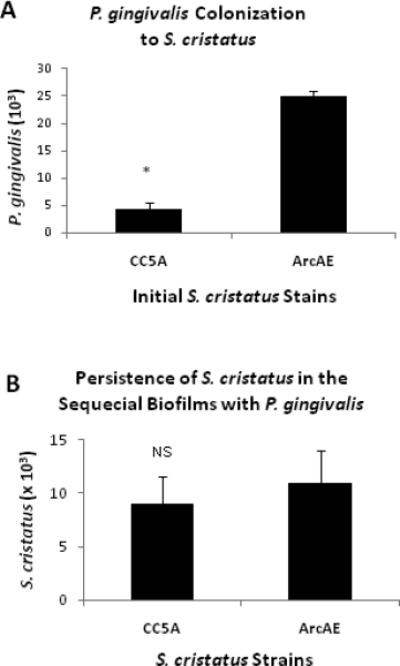
Sequential P. gingivalis colonization on preexisting S. cristatus biofilms in the murine oral cavity. The plaque samples were taken from tooth surfaces and surrounding gingival mucosa 2 weeks post P. gingivalis inoculation. DNA extracted from these plaque samples (50 μl final volume/sample in TE buffer) was examined for quantitation of P. gingivalis and S. cristatus, using qPCR with primers corresponding to 16S or 23S rDNA as probes. Data represent the means ± S.D. from 8 mice per group. The variances between the groups were analyzed by ANOVA. A, P. gingivalis colonization on S. cristatus biofilms; B, Persistence of S. cristatus in the sequential biofilms with P. gingivalis.
We also examined the persistence of initial S. cristatus colonization in the mixed S. cristatus-P. gingivalis biofilms. As shown in Fig. 3B, S. cristatus colonization persisted for 4 weeks after the last S. cristatus inoculation and 2 weeks after the sequential P. gingivalis inoculation. There was no statistically significant difference in quantity between the wild-type S. cristatus CC5A and its arcA knockout counterpart [(8.94 ± 2.65) x 103 vs. (10.93 ± 3.04) x 103; p = 0.2245].
Our results demonstrated that S. cristatus and P. gingivalis are able to co-exist in the murine oral cavity and the expression of ArcA by S. cristatus inhibits P. gingivalis colonization.
S. cristatus ArcA Prevented P. gingivalis-induced Alveolar Bone Loss in Mice
We next examined P. gingivalis-induced alveolar bone loss in the presence of S. cristatus CC5A or S. cristatus ArcAE mutant on day 69 (6 weeks after P. gingivalis infection). As shown in Fig. 4, no significant bone loss was observed in the mice infected only with S. cristatus CC5A or ArcAE, relative to the sham-infected mice (0.076 ± 0.019 mm, 0.077 ± 0.028 mm, 0.073 ± 0.018 mm; p > 0.05). P. gingivalis-induced alveolar bone loss was significantly more prominent in the S. cristatus ArcAE group than in the comparable CC5A group (0.120 ± 0.037 mm, 0.092 ± 0.033 mm; p = 0.011). These results indicated that ArcA serves as a potent inhibiter for P. gingivalis pathogenesis.
Figure 4.
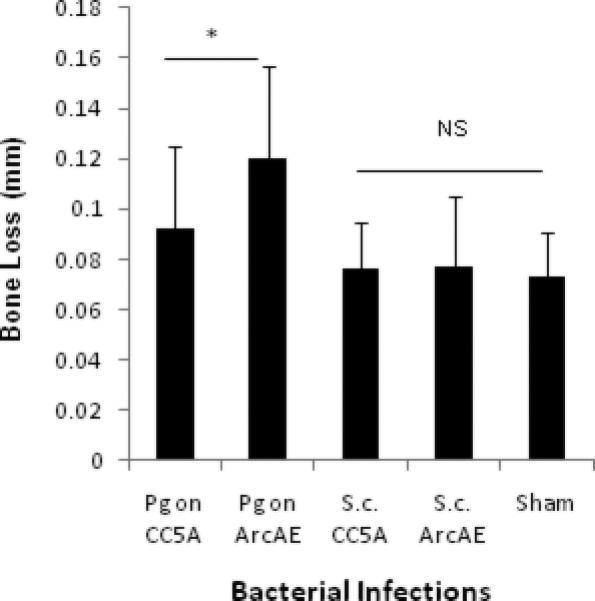
Effect of S. cristatus ArcA on P. gingivalis-induced alveolar bone loss in BALB/cByJ mice. Data represent the means ± S.D. from 8 mice per group. Alveolar bone loss was calculated as the average of 14-site (3 sites on first molar, 2 sites on second molar, and 2 sites on third molar, at both right and left sides of the maxilla) total CEJ-ABC distances for each group.
NS, no significant difference in alveolar bone loss when compared with the sham-infected group *, the difference in alveolar bone loss was significant (p < 0.05) between the two P. gingivalis infected groups
DISCUSSION
Dental plaque formation is generally programmed beginning with the initial colonization by gram-positive aerobic bacteria, followed by a succession of gram-negative anaerobic bacteria. The transition from commensal bacterial accumulation to periodontopathic plaque involves the colonization of certain species of gram-negative anaerobic bacteria such as P. gingivalis. Evidence is accumulating that some early colonizers of dental plaque provide a favorable environment for P. gingivalis, at least with regard to attachment sites and lower oxygen tension. This in turn facilitates retention and multiplication of P. gingivalis, ultimately leading to the progression of adult periodontitis (14). Although less well understood, antagonistic relationships have been reported between/among oral bacteria. P. gingivalis, for example, does not grow with Streptococcus oralis in two-species biofilms (28) and does not co-aggregate with Streptococcus mutans (29). We have reported earlier that S. cristatus represses expression of fimA in P. gingivalis and attenuates bacterial colonization in vitro (22, 30). The present study was designed to test our hypothesis that S. cristatus ArcA interferes with P. gingivalis colonization in the murine oral cavity and attenuates the subsequent P. gingivalis-induced alveolar bone loss, an indicator of P. gingivalis pathogenesis in periodontitis.
P. gingivalis-induced alveolar bone loss in rodent models has been widely accepted as a valid model for P. gingivalis pathogenicity critical for periodontitis. Furthermore, studies utilizing these rodent models have indicated that P. gingivalis – induced alveolar bone loss depends on the expression of FimA protein in the bacterium (17, 18, 31). These studies have shown that isogenic mutants lacking FimA or its associated minor proteins are less virulent and induce significantly less alveolar bone loss relative to the wild-type bacterium. Since we have previously demonstrated that ArcA of S. cristatus represses fimA gene expression in P. gingivalis in vitro (20, 22, 30), in this study we intended to determine if biofilms of S. cristatus CC5A, relative to those of a isogenic mutant lacking ArcA would interfere with the pathogenesis of P. gingivalis in the murine oral cavity.
S. cristatus is a component of earlier colonizers in human dental plaque and has been detected in the 8-h biofilms on freshly cleaned enamel chips (26). In this study, we demonstrated that S. cristatus, like some other oral streptococci (32-34) could colonize the rodent oral cavity. The presence of S. cristatus in the oral cavity could be detected as monobiofilms (Fig. 1) and also as heterotypic biofilms with P. gingivalis (Fig. 3B).
The objective of this study is to determine if S. cristatus ArcA attenuates P. gingivalis colonization and subsequent P. gingivalis-induced alveolar bone loss in vivo. Therefore, P. gingivalis infection was carried out in the oral cavity with existing biofilms of wild-type S. cristatus CC5A or its arcA knockout mutant. This experimental design allows us to test our hypothesis, since the only difference between the two groups is the presence or absence of S. cristatus ArcA. As demonstrated in Fig. 4, the wild-type CC5A strain expressing ArcA is more potent than the ArcA deficient mutant in blocking P. gingivalis-induced alveolar bone loss. Since P. gingivalis was not recovered from P. gingivalis alone infected group two-weeks post infection, in contrast to the S. cristatus colonized groups where P. gingivalis was recovered persistently, the P. gingivalis alone infected group was considered an inappropriate control for the study and was excluded. However, in light of our findings, it would be interesting to compare the P. gingivalis alone infected group with the S. cristatus precolonized groups in the bone loss experiments in the future.
Inter-species interactions play important roles in the initiation and development of oral infectious diseases, since it can affect the pathogenecity of a particular pathogen. Our results provide direct evidence that S. cristatus ArcA attenuates P. gingivalis biofilm formation (Fig. 3A) and subsequent pathogenesis (as indicated by P. gingivalis-induced alveolar bone loss) in vivo (Fig. 4). The presence of S. cristatus CC5A did not completely eliminate P. gingivalis colonization and P. gingivalis-induced alveolar bone loss in mice. However, both the quantity of colonized P. gingivalis and P. gingivalis-induced bone loss were significantly less in the S. cristatus CC5A-infected group than in the corresponding S. cristatus ArcAE-infected animals. Further in vivo studies are warranted to determine if purified S. cristatus ArcA or a synthetic ArcA functional domain attenuates P. gingivalis pathogenesis, as they did in P. gingivalis biofilm formation in vitro (20, 30
ACKNOWLEDGEMENTS
This study was supported in part by grants NIH NIDCR DE017708 to BYW and NCRR 1U54RR026140 and UL1RR024975 to HX.
Footnotes
The authors declare no potential conflicts of interest with respect to the authorship and/or publication of this article.
REFERENCES
- 1.Beck JD, Eke P, Heiss G, et al. Periodontal disease and coronary heart disease: a reappraisal of the exposure. Circulation. 2005;112:19–24. doi: 10.1161/CIRCULATIONAHA.104.511998. [DOI] [PubMed] [Google Scholar]
- 2.Scannapieco FA. Periodontal inflammation: from gingivitis to systemic disease? Compend Contin Educ Dent. 2004;25:16–25. [PubMed] [Google Scholar]
- 3.Offenbacher S. Maternal periodontal infections, prematurity, and growth restriction. Clin Obstet Gynecol. 2004;47:808–821. doi: 10.1097/01.grf.0000141894.85221.f7. discussion 881-802. [DOI] [PubMed] [Google Scholar]
- 4.Socransky SS, Haffajee AD. Periodontal microbial ecology. Periodontol. 2000;38:135–187. doi: 10.1111/j.1600-0757.2005.00107.x. 2005. [DOI] [PubMed] [Google Scholar]
- 5.Socransky SS, Haffajee AD, Cugini MA, Smith C, Kent RL., Jr Microbial complexes in subgingival plaque. J Clin Periodontol. 1998;25:134–144. doi: 10.1111/j.1600-051x.1998.tb02419.x. [DOI] [PubMed] [Google Scholar]
- 6.Suzuki Y, Yoshimura F, Takahashi K, Tani H, Suzuki T. Detection of fimbriae and fimbrial antigens on the oral anaerobe Bacteroides gingivalis by negative staining and serological methods. J Gen Microbiol. 1988;134:2713–2720. doi: 10.1099/00221287-134-10-2713. [DOI] [PubMed] [Google Scholar]
- 7.Suzuki Y, Yoshimura F, Tani H, Suzuki T. Fimbriae from the oral anaerobe Bacteroides gingivalis: a screening of clinical isolates from various places. Adv Dent Res. 1988;2:301–303. doi: 10.1177/08959374880020021601. [DOI] [PubMed] [Google Scholar]
- 8.Griffen AL, Lyons SR, Becker MR, Moeschberger ML, Leys EJ. Porphyromonas gingivalis strain variability and periodontitis. J Clin Microbiol. 1999;37:4028–4033. doi: 10.1128/jcm.37.12.4028-4033.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Perez-Chaparro PJ, Lafaurie GI, Gracieux P, et al. Distribution of Porphyromonas gingivalis fimA genotypes in isolates from subgingival plaque and blood sample during bacteremia. Biomedica. 2009;29:298–306. [PubMed] [Google Scholar]
- 10.Beikler T, Peters U, Prajaneh S, Prior K, Ehmke B, Flemmig TF. Prevalence of Porphyromonas gingivalis fimA genotypes in Caucasians. Eur J Oral Sci. 2003;111:390–394. doi: 10.1034/j.1600-0722.2003.00065.x. [DOI] [PubMed] [Google Scholar]
- 11.Amano A, Fujiwara T, Nagata H, et al. Prophyromonas gingivalis fimbriae mediate coaggregation with Streptococcus oralis through specific domains. J Dent Res. 1997;76:852–857. doi: 10.1177/00220345970760040601. [DOI] [PubMed] [Google Scholar]
- 12.Goulbourne PA, Ellen RP. Evidence that Porphyromonas (Bacteroides) gingivalis fimbriae function in adhesion to Actinomyces viscosus. J Bacteriol. 1991;173:5266–5274. doi: 10.1128/jb.173.17.5266-5274.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hajishengallis G, Shakhatreh MA, Wang M, Liang S. Complement receptor 3 blockade promotes IL-12-mediated clearance of Porphyromonas gingivalis and negates its virulence in vivo. J Immunol. 2007;179:2359–2367. doi: 10.4049/jimmunol.179.4.2359. [DOI] [PubMed] [Google Scholar]
- 14.Lamont RJ, Bevan CA, Gil S, Persson RE, Rosan B. Involvement of Porphyromonas gingivalis fimbriae in adherence to Streptococcus gordonii. Oral Microbiol Immunol. 1993;8:272–276. doi: 10.1111/j.1399-302x.1993.tb00573.x. [DOI] [PubMed] [Google Scholar]
- 15.Lamont RJ, Jenkinson HF. Life below the gum line: pathogenic mechanisms of Porphyromonas gingivalis. Microbiol Mol Biol Rev. 1998;62:1244–1263. doi: 10.1128/mmbr.62.4.1244-1263.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lin X, Wu J, Xie H. Porphyromonas gingivalis minor fimbriae are required for cell-cell interactions. Infect Immun. 2006;74:6011–6015. doi: 10.1128/IAI.00797-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Malek R, Fisher JG, Caleca A, et al. Inactivation of the Porphyromonas gingivalis fimA gene blocks periodontal damage in gnotobiotic rats. J Bacteriol. 1994;176:1052–1059. doi: 10.1128/jb.176.4.1052-1059.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang M, Shakhatreh MA, James D, et al. Fimbrial proteins of porphyromonas gingivalis mediate in vivo virulence and exploit TLR2 and complement receptor 3 to persist in macrophages. J Immunol. 2007;179:2349–2358. doi: 10.4049/jimmunol.179.4.2349. [DOI] [PubMed] [Google Scholar]
- 19.Weinberg A, Belton CM, Park Y, Lamont RJ. Role of fimbriae in Porphyromonas gingivalis invasion of gingival epithelial cells. Infect Immun. 1997;65:313–316. doi: 10.1128/iai.65.1.313-316.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Xie H, Cook GS, Costerton JW, Bruce G, Rose TM, Lamont RJ. Intergeneric communication in dental plaque biofilms. J Bacteriol. 2000;182:7067–7069. doi: 10.1128/jb.182.24.7067-7069.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang BY, Wu J, Lamont RJ, Lin X, Xie H. Negative correlation of distributions of Streptococcus cristatus and Porphyromonas gingivalis in subgingival plaque. J Clin Microbiol. 2009;47:3902–3906. doi: 10.1128/JCM.00072-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Xie H, Lin X, Wang BY, Wu J, Lamont RJ. Identification of a signalling molecule involved in bacterial intergeneric communication. Microbiology. 2007;153:3228–3234. doi: 10.1099/mic.0.2007/009050-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pathirana RD, O'Brien-Simpson NM, Brammar GC, Slakeski N, Reynolds EC. Kgp and RgpB, but not RgpA, are important for Porphyromonas gingivalis virulence in the murine periodontitis model. Infect Immun. 2007;75:1436–1442. doi: 10.1128/IAI.01627-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tran SD, Rudney JD. Improved multiplex PCR using conserved and species-specific 16S rRNA gene primers for simultaneous detection of Actinobacillus actinomycetemcomitans, Bacteroides forsythus, and Porphyromonas gingivalis. J Clin Microbiol. 1999;37:3504–3508. doi: 10.1128/jcm.37.11.3504-3508.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sharma A, Inagaki S, Honma K, Sfintescu C, Baker PJ, Evans RT. Tannerella forsythia-induced alveolar bone loss in mice involves leucine-rich-repeat BspA protein. J Dent Res. 2005;84:462–467. doi: 10.1177/154405910508400512. [DOI] [PubMed] [Google Scholar]
- 26.Diaz PI, Chalmers NI, Rickard AH, et al. Molecular characterization of subject-specific oral microflora during initial colonization of enamel. Appl Environ Microbiol. 2006;72:2837–2848. doi: 10.1128/AEM.72.4.2837-2848.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nyvad B, Kilian M. Microbiology of the early colonization of human enamel and root surfaces in vivo. Scand J Dent Res. 1987;95:369–380. doi: 10.1111/j.1600-0722.1987.tb01627.x. [DOI] [PubMed] [Google Scholar]
- 28.Periasamy S, Kolenbrander PE. Central role of the early colonizer Veillonella sp. in establishing multispecies biofilm communities with initial, middle, and late colonizers of enamel. J Bacteriol. 2010;192:2965–2972. doi: 10.1128/JB.01631-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lamont RJ, El-Sabaeny A, Park Y, Cook GS, Costerton JW, Demuth DR. Role of the Streptococcus gordonii SspB protein in the development of Porphyromonas gingivalis biofilms on streptococcal substrates. Microbiology. 2002;148:1627–1636. doi: 10.1099/00221287-148-6-1627. [DOI] [PubMed] [Google Scholar]
- 30.Wu J, Xie H. Role of arginine deiminase of Streptococcus cristatus in Porphyromonas gingivalis colonization. Antimicrob Agents Chemother. 2010;54:4694–4698. doi: 10.1128/AAC.00284-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Umemoto T, Hamada N. Characterization of biologically active cell surface components of a periodontal pathogen. The roles of major and minor fimbriae of Porphyromonas gingivalis. J Periodontol. 2003;74:119–122. doi: 10.1902/jop.2003.74.1.119. [DOI] [PubMed] [Google Scholar]
- 32.Ooshima T, Sumi N, Izumitani A, Sobue S. Effect of inoculum size and frequency on the establishment of Streptococcus mutans in the oral cavities of experimental animals. J Dent Res. 1988;67:964–968. doi: 10.1177/00220345880670061501. [DOI] [PubMed] [Google Scholar]
- 33.Loach DM, Jenkinson HF, Tannock GW. Colonization of the murine oral cavity by Streptococcus gordonii. Infect Immun. 1994;62:2129–2131. doi: 10.1128/iai.62.5.2129-2131.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sharma A, Honma K, Evans RT, Hruby DE, Genco RJ. Oral immunization with recombinant Streptococcus gordonii expressing porphyromonas gingivalis FimA domains. Infect Immun. 2001;69:2928–2934. doi: 10.1128/IAI.69.5.2928-2934.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]


