Abstract
Infectious agents develop intricate mechanisms to interact with host cell pathways and hijack the genetic and epigenetic machinery to change phenotypic states. Amongst the Apicomplexa phylum of obligate intracellular parasites which cause veterinary and human diseases, Theileria is the only genus which transforms its mammalian host cells1. Theileria infection of bovine leukocytes induces proliferative and invasive phenotypes associated with activated signalling pathways, notably JNK and AP-12. The transformed phenotypes are reversed by treatment with the theilericidal drug Buparvaquone3. We used comparative genomics to identify a homologue of the Peptidyl Prolyl Isomerase Pin1 (designated TaPin1) in T. annulata which is secreted into the host cell and modulates oncogenic signalling pathways. Here we show that TaPin1 is a bona fide prolyl isomerase and that it interacts with the host ubiquitin ligase FBW7 leading to its degradation and subsequent stabilization of c-Jun which promotes transformation. We performed in vitro analysis and in vivo zebrafish xenograft experiments to demonstrate that TaPin1 is directly inhibited by the anti-parasite drug Buparvaquone (and other known Pin1 inhibitors) and is mutated in a drug-resistant strain. Prolyl isomerisation is thus a conserved mechanism which is important in cancer and is used by Theileria parasites to manipulate host oncogenic signaling.
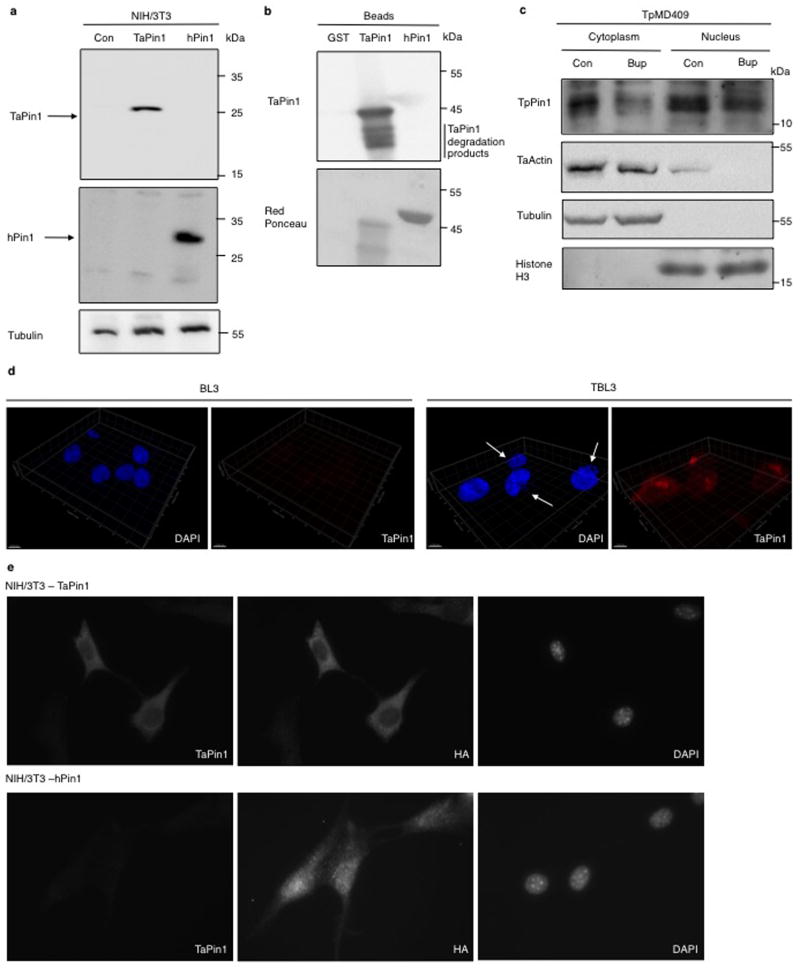
To identify proteins secreted by Theileria into the host cell which could contribute to transformation4–6, we conducted an in silico screen of parasite genomes; we identified 689 proteins in the T. annulata genome with a predicted signal peptide. Comparison with T. gondii (a non-transforming apicomplexan parasite) proteome, narrowed the candidate list to 33 proteins with a Theileria-specific signal peptide (Extended Data Fig. 1a). We focused on the TA18945 gene encoding a homologue of the human parvulin Pin1 (hPin1) Peptidyl Prolyl Isomerase (PPIase) as mammalian Pin1 regulates cell proliferation, pluripotency and survival7,8 and contributes to tumorigenesis9,10. hPin1 catalyzes the cis/trans isomerization of peptidyl-prolyl bonds in phosphorylated Ser/Thr-Pro motifs inducing conformational changes that affect substrate stability and activity11,12 and there are several small-molecule inhibitors of hPin113–15. The TA18945-encoded protein has a signal peptide and a highly conserved PPIase domain (Extended Data Fig. 1b–c), but lacks the WW domain important for substrate recognition of mammalian Pin111. A gene in the T. parva genome, also associated with transformation, encodes a conserved TpPin1 predicted protein, whereas the signal peptide is not conserved in the related T. orientalis genome which does not transform host cells16 (Extended Data Fig. 2a–b). We detected Theileria Pin1 transcripts in B cells infected with T. annulata or T. parva and they decreased upon Buparvaquone treatment (Fig. 1a). The levels of host bovine BtPin1 transcripts were unaffected by Theileria infection or Buparvaquone treatment (Extended Data Fig. 3). An antibody generated against a TaPin1-specific peptide (NPVNRNTGMAVTR) recognized parasite Pin1 protein or transfected TaPin1 in mouse fibroblasts, but not mammalian Pin1 (Fig. 1b, Extended Data Fig. 4a–e). Confocal microscopy and immunoblot analysis located the parasite Pin1 protein to both the host cell cytoplasm and nucleus (Fig. 1b–c, Extended Data Fig. 4c–d). The host nuclear signal in the confocal images was 10-fold over background in parasitized cells (205.0 ± 15.48 nuclear fluorescence intensity/pixel compared to 21.45 ± 8.50 in controls p<0.0001, n=31). Thus, comparative parasite genomics identified TaPin1 which is secreted into the host cytoplasm and nucleus.
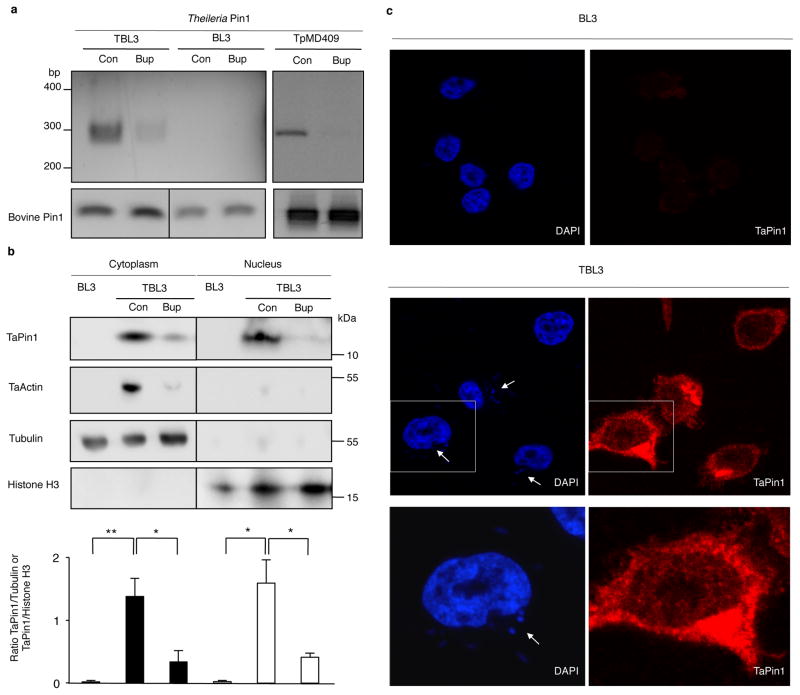
Fig. 1. Theileria parasites secrete a conserved Pin1 PPIase protein.
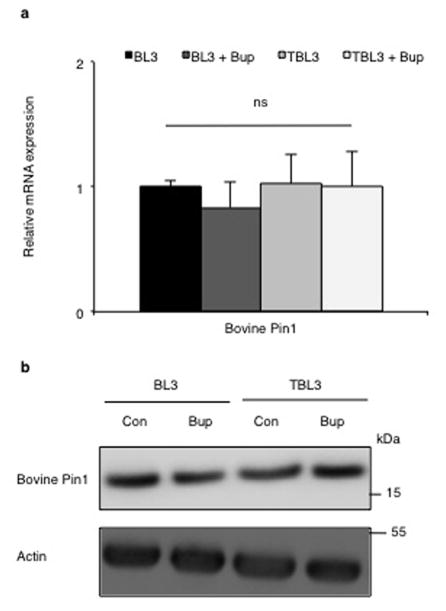
a. Expression of Pin1 RNA in T. annulata-infected TBL3 cells, uninfected BL3 cells or T. parva-infected TpMD409 cells, treated with Buparvaquone (Bup) or control (Con). Bovine Pin1 expression was used as loading control.
b. TaPin1 protein was detected in the host cytoplasm and nucleus, in contrast Apicomplexan actin (TaActin). Bovine Histone H3 (nuclear) and Tubulin (cytoplasmic) proteins were controls. Relative quantification showing TaPin1/Tubulin or TaPin1/Histone H3 ratios calculated with Image J software (average ± sd, n=3). The p-values were corrected for the multiple comparisons using the Bonferroni correction based on the total overall number of pairwise comparisons. *p<0.05, **p<0.01.
c. TaPin1 was detected in the cytoplasm and nucleus of infected cells by confocal microscopy using an affinity-purified antibody specific for TaPin1, counterstaining with DAPI (white arrows indicate parasites).
Results are representative of 3 independent experiments.
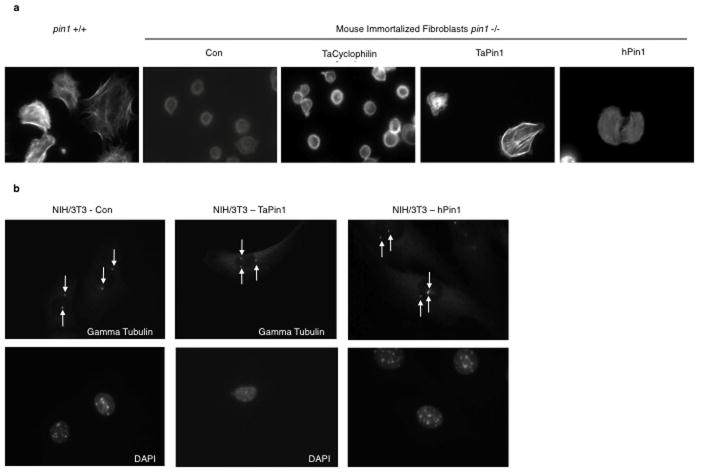
To explore the functional PPIase activity of the secreted TaPin1 protein, we developed a chymotrypsin-coupled in vitro assay and found that TaPin1 and hPin1 catalytic activities were comparable (Fig. 2a). TaPin1 and hPin1 were also equivalent in activation of the cyclinD1-Luciferase reporter in bovine B cells (Fig. 2b), an established readout for Pin1 activity9. We mutated key C92 and K38 residues in TaPin1 and showed loss of the PPIase activity (Fig. 2c). Furthermore, TaPin1 rescued cyclinD1 promoter activity and cell spreading defects in pin1-/- mouse fibroblasts (Fig. 2d, Extended Data Fig. 5a). Mammalian Pin1 overexpression disrupts cell cycle regulation causing centrosome amplification and cell transformation17. TaPin1 also induced centrosome duplication when overexpressed in mouse fibroblasts (Fig. 2e, Extended Data Fig. 5b). Furthermore, TaPin1 functionally replaced mammalian Pin1 and rescued colony formation as effectively as hPin1 in human breast cancer cells with knocked-down Pin1 (Fig. 2f). These combined results show that Theileria secretes a bona fide phosphorylation-dependent PPIase which could contribute to host cell transformation.
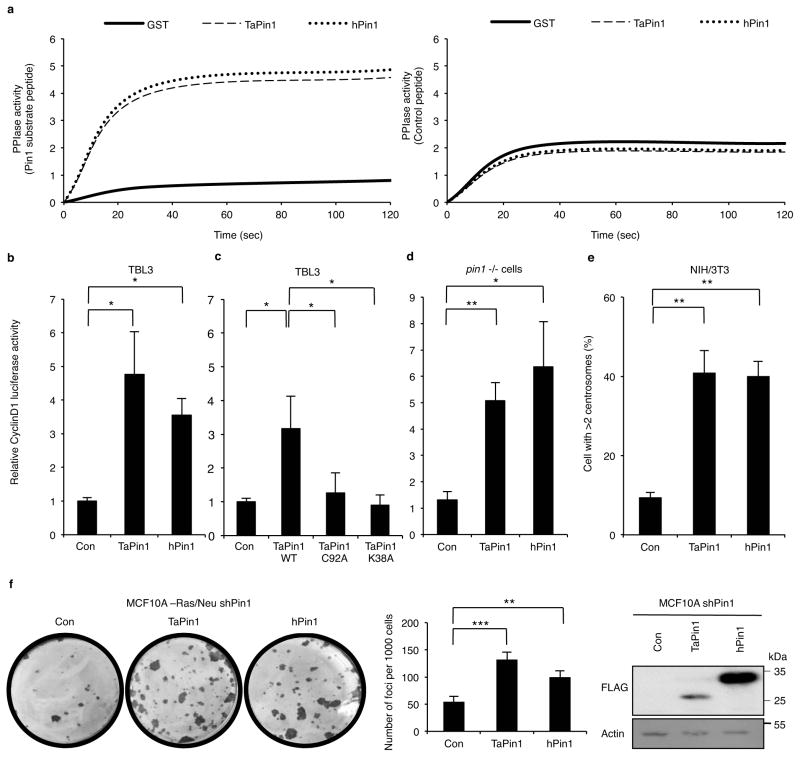
Fig. 2. TaPin1 is a functional homologue of hPin1 involved in transformation.
a. hPin1 and TaPin1 catalytic PPIase activities measured by in vitro chymotrypsin-coupled using a Pin1 substrate peptide (Suc-Ala-Glu-Pro-Phe-pNA). No activity was detected for GST alone or control substrate peptide (Suc-Ala-Ala-Pro-Phe-pNA).
b. TaPin1 and hPin1 increased CyclinD1-Luciferase promoter activity when transfected in TBL3 cells.
c. C92A and K38A TaPin1 mutants showed reduced activation of cyclinD1 promoter when transfected in TBL3 cells.
d. TaPin1 or hPin1 induced CyclinD1-Luciferase promoter activity in pin1-/- immortalized fibroblasts.
e. TaPin1 causes centrosome amplification. NIH3T3 fibroblasts stably expressing TaPin1 or hPin1 were arrested at the G1/S transition by aphidicolin, stained with anti-γ-tubulin antibody. 300 cells were scored.
f. TaPin1 or hPin1 transfection increased colony foci formation in Pin1 knockdown MCF10A-Ras/Neu cells. Expression of transfected TaPin1 and hPin1 in MCF10A-Ras/Neu-shPin1 detected with an anti-FLAG antibody. Actin was a loading control.
Note: “Con” indicates Control empty vector transfection.
Data represent 3 independent experiments (average ± sd). The p-values were calculated using the Dunnett method for multiple comparisons with the TaPin1WT for Figure 2c. For Figures 2b / 2d / 2e / 2f : the p-values were corrected using Dunnett multiple comparisons with the control. *p<0.05, **p<0.01, ***p<0.001.
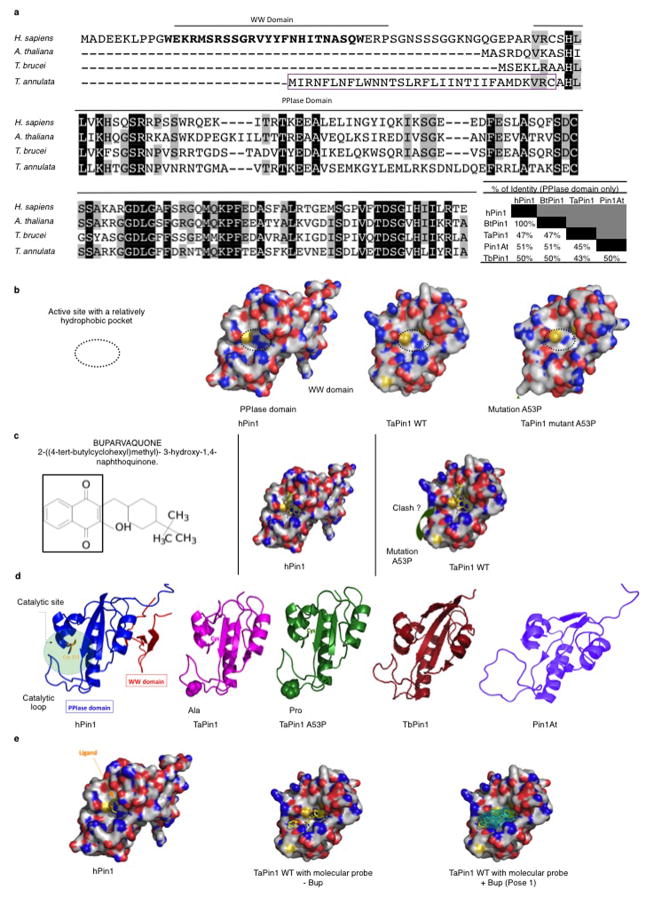
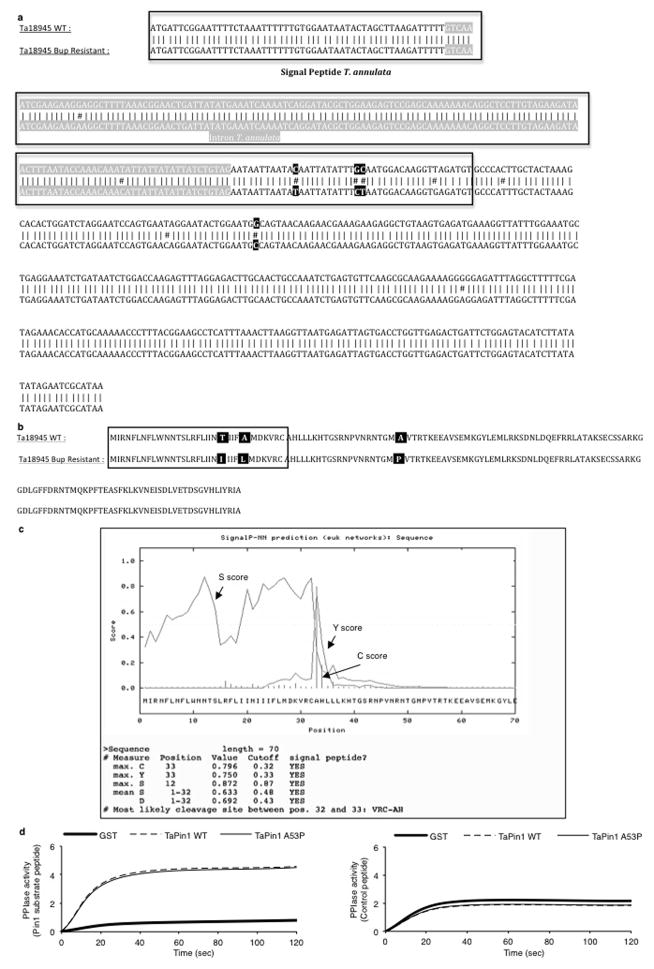
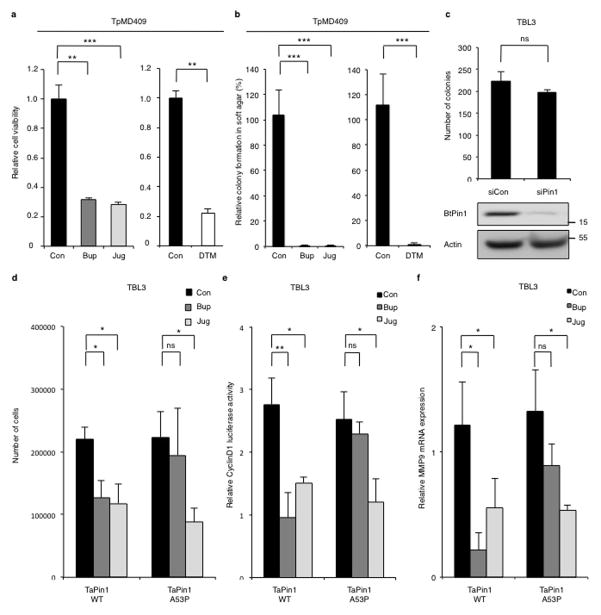
In a search for potential inhibitors, we noted that the chemical structure of Buparvaquone is similar to Juglone, a well-characterized inhibitor of mammalian Pin113. The TaPin1 sequence exhibits over 47% identity with hPin1 in the PPIase domain (Extended Data Fig. 6a). Our homology models of TaPin1 protein based on published hPin1 experimental data suggest a similar structure with a conserved catalytic pocket (Fig. 3a, Extended Data Fig. 6b). Notably, several Pin1 homologues also lack the WW domain, including Arabidopsis thaliana Pin1At18–20, MdPin1 in Malus domestica and the parasite Trypanosoma brucei TbPin1 homologue20–22, and the predicted TaPin1 model closely resembles these structures (Extended Data Fig. 6d). We investigated the hPin1 experimental structure and the TaPin1 predicted model with the binding pocket and hot-spot detection algorithm FTMap, using the server FTFlex. Notably, we found key hot-spot regions in the catalytic site area, matching the substrate binding region of hPin1 (Extended Data Fig. 6). Juglone and Buparvaquone molecules could be docked into the active site of both TaPin1 and hPin1 by in silico analysis (Fig. 3a, Extended Data Fig. 6c). We predicted that Buparvaquone might target TaPin1 directly and that Juglone (or other Pin1 inhibitors) could functionally replace Buparvaquone to block parasite transformation. Both Buparvaquone and Juglone inhibited TaPin1 PPIase activity in vitro, as did the unrelated non-quinone inhibitor DTM14, albeit to a lesser degree (Fig. 3b). Buparvaquone-resistant Theileria strains are an emerging clinical concern for cattle in infected areas23 and mutations in the cytochrome B gene were recently reported24. But mitochondrial and non-mitochondrial pathways might cooperate in transformation and participate in drug resistance. We sequenced the TaPin1 gene in genomic DNA from a drug-resistant isolate and identified a mutation (A53>P substitution) in the catalytic loop of TaPin1 (Extended Data Fig. 7). Structural modeling suggested that this mutation could affect the nearby catalytic region and disturb ligand binding; computational docking indicated that the small Juglone molecule could react with the thiolate group of C113 or C92 in hPin1, TaPin1 or mutant TaPin1-A53P. However, the A53P mutation might impede interaction with the bulky, hydrophobic moiety of the larger Buparvaquone compound (MW=326, cf. Juglone MW=176), creating steric clashes between the inhibitor and residues in the modified structure (Fig. 3a, Extended Data Fig. 6c). Mutant TaPin1-A53P was catalytically active on the Pin1 substrate and was inhibited by Juglone and DTM, but not by Buparvaquone (Fig. 3b). The Pin1 inhibitors (Buparvaquone, Juglone and DTM) all reduced parasite load and viability of host cells infected with T. annulata or T. parva (Fig. 3c–d, Extended Data Fig. 8a) and blocked colony growth of parasitized cells in soft-agar assays in vitro (Fig. 3e, Extended Data Fig. 8b). In contrast, knocking down the endogenous bovine BtPin1 did not affect colony formation (Extended Data Fig. 8c). Transfection with mutant TaPin1-A53P rendered TBL3 cells resistant to Buparvaquone, but not Juglone, treatment (Extended Data Fig. 8d). Similarly, Juglone inhibited both WT and mutant TaPin1 activity in the cyclinD1-luciferase assay, but only the mutant was resistant to Buparvaquone (Extended Data Fig. 8e). Fish xenograft models are effective for monitoring in vivo tumor formation and for drug testing25 and are emerging as important experimental models to study cancer26. We used a zebrafish xenograft experimental system to test drug effects on tumor growth in vivo and observed a two-fold increase in tumor growth of infected cells which was efficiently inhibited by the anti-Pin1 drugs (Fig. 3f–g). Thus, our modelling predictions, biochemical analysis in vitro, transformation assays and tumor growth in vivo all support the targeting of TaPin1 by Buparvaquone and the role of TaPin1 in Theileria-induced cell transformation.
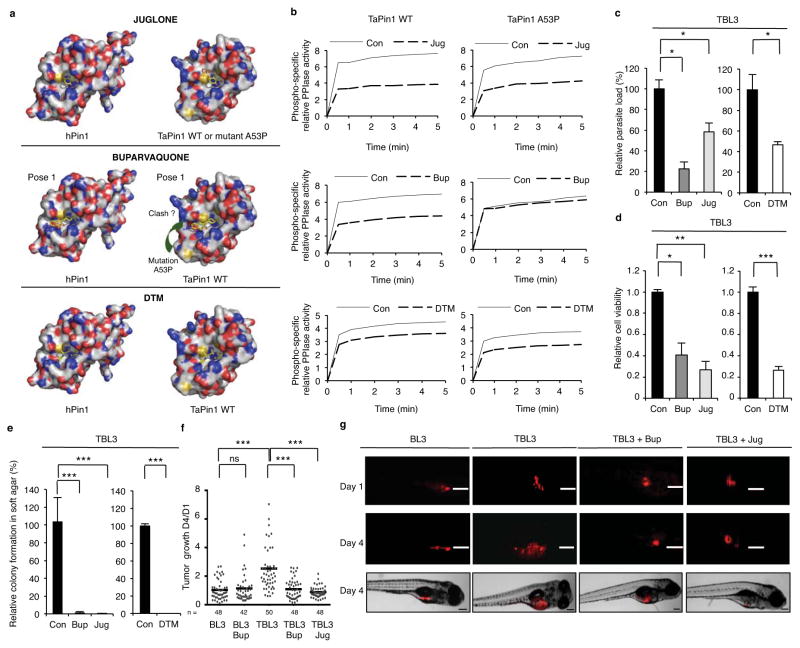
Fig. 3. Inhibition of TaPin1 activity blocks transformation in vitro and in vivo.
a. Homology prediction models for TaPin1 and TaPin1-A53P Mutant based on similarity with hPin1. The TaPin1-A53P mutation induces a conformational change near the catalytic loop (green arrow). Computational analysis predicted docking of Juglone, Buparvaquone or DTM molecules in the Pin1 active sites (see Extended Data Fig. 6c for alternative). Bup/Jug: red=polar and negatively-charged residues; blue=polar and positively charged residues; yellow= S atoms; white=remaining residues. DTM: blue = N atoms; amber = S atoms. Colours in small molecule: O atoms in red; other atoms in yellow.
b. TaPin1 and TaPin1 A53P catalytic PPIase activity, measured in vitro chymotrypsin-coupled assay, upon treatment with Buparvaquone (Bup), Juglone (Jug) or DTM. “Con” indicates Control solutions.
c. Drug treatment (72h) eliminated Theileria parasites in infected cells (parasite nuclei counted following DAPI staining).
d. Pin1 inhibitors decreased the viability of infected TBL3 cells (XTT assay 72h).
e. Drug treatment decreased colony formation of parasitized cells in soft-agar (72h treatment with Buparvaquone (Bup), Juglone (Jug) or DTM): macroscopic colonies/plate after 10 days.
f. TaPin1 inhibition reduced Xenograft tumor growth in zebrafish embryos (with a Zeiss AxioZoom V16 Macroscope at day 1 and day 4 post-injection). The median tumor development Day4:Day1 is shown. “n” = number of embryos.
g. Representative images from individual zebrafish embryos photographed with a Zeiss AxioZoom V16 Macroscope. Scalebar = 200 μm.
“Con” = vector solutions alone.
Data represent 3 independent experiments (average ± sd, n=3). For Figures 3c / 3d / 3e: the p-values were corrected using the Dunnett test multiple comparisons with the control. An unpaired Mann-Whitney test was performed for the zebrafish experiments to analyze the significant difference between the control and treatment groups. *p<0.05, **p<0.01, ***p<0.001.
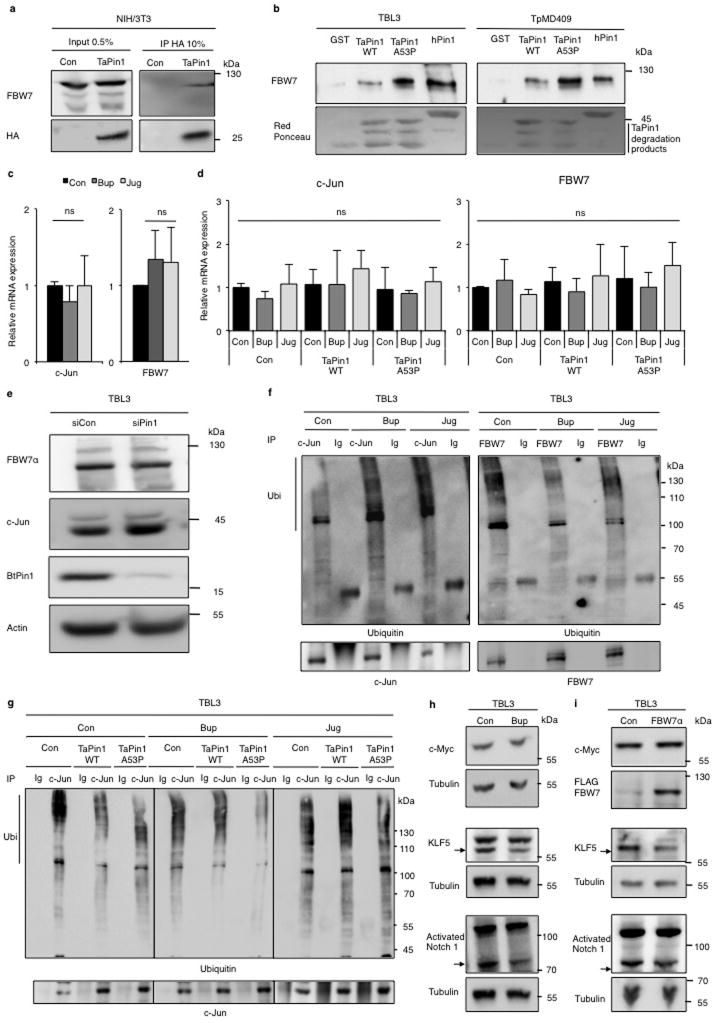
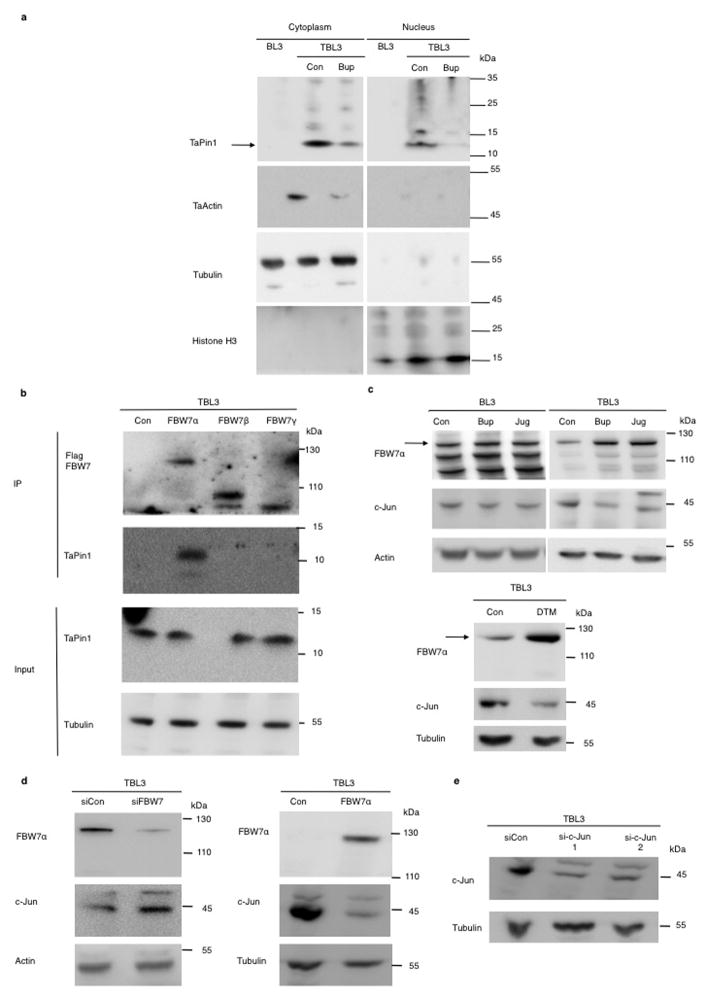
To investigate how TaPin1 affects host signaling pathways, we studied relevant substrates targeted by TaPin1. hPin1 targets many proteins, including the ubiquitin ligase FBW727, which exerts anti-tumor function by degrading oncoproteins required for cellular proliferation, such as c-Jun28,29. Since c-Jun is induced (and critical) during Theileria-induced transformation2, we examined whether TaPin1 targets the conserved bovine FBW7 protein. We found that TaPin1 interacts with host FBW7 in Theileria-infected cells or murine fibroblasts (Extended Data Fig. 9a–b). Conversely, FBW7α in particular, but not other isoforms, co-immunoprecipitated TaPin1 from parasitized cells (Fig. 4a). FBW7α protein levels were reduced in parasitized cells compared with uninfected cells and correlated with elevated c-Jun levels (Fig. 4b). Pharmacological TaPin1 inhibition restored FBW7 protein expression and reduced c-Jun levels, without affecting mRNA expression (Fig. 4b, Extended Data Fig. 9c–d). But knocking-down bovine Pin1 did not affect FBW7 or c-Jun protein levels (Extended Data Fig. 9e). siRNA FBW7 knockdown caused accumulation of c-Jun protein, whereas exogenous FBW7α transfection decreased c-Jun protein levels (Fig. 4c). Furthermore, knocking-down FBW7 increased AP-1 activity, measured by a Luciferase reporter assay, and rescued the inhibition of AP-1 by Buparvaquone or Juglone (Fig. 4d–e). TaPin1 inhibition caused increased c-Jun ubiquitination in TBL3 cells and decreased FBW7 (auto)-ubiquitination (Extended Data Fig. 9f). c-Jun ubiquitination was FBW7-dependent, as this effect was abolished by siRNA targeting bovine FBW7 (Fig. 4f). Half-life analysis using cycloheximide showed that siFBW7 increased c-Jun stability and rescued c-Jun levels following TaPin1 inhibition (Fig. 4g). In addition, the TaPin1-A53P mutant rescued the effect of Buparvaquone, but not Juglone, on c-Jun ubiquitination and transcriptional activity (reflected by the MMP-9 AP-1 target gene) (Extended Data Fig. 8f, Fig. 9g). As mammalian FBW7 targets many protein substrates, we examined the effects of Buparvaquone treatment or FBW7α transfection, but noted no changes in levels of endogenous c-Myc or activated Notch 1 proteins, while there was a modest effect on the KLF5 transcription factor (Extended Data Fig. 9h, i). These combined results suggest that c-Jun is the major target of TaPin1-FBW7α in our cells. Finally, FBW7α overexpression or c-Jun knockdown both caused significant reduction of colony growth in soft-agar proliferation assays of parasitized TBL3 cells (Fig. 4h).
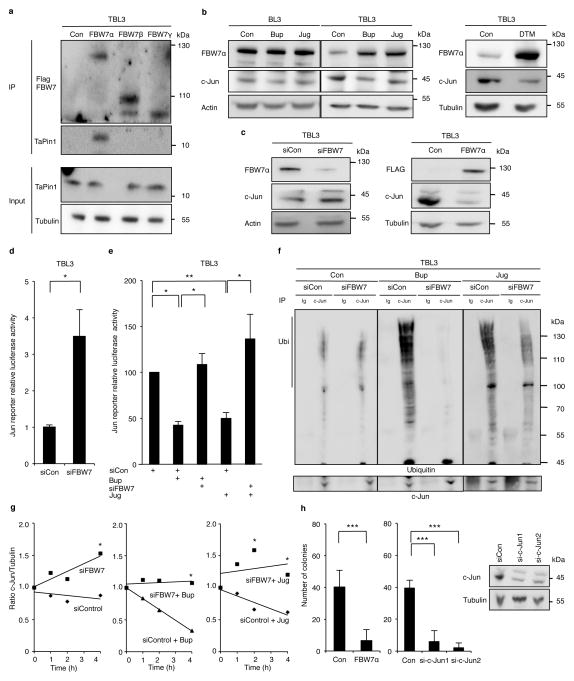
Fig. 4. TaPin1 activates the oncogenic c-Jun pathway via FBW7 ubiquitination.
a. Endogenous TaPin1 interacts with FBW7α isoform. Protein extracts from TBL3 expressing Flag-hFBW7 isoforms or Flag-Control “Con” were immunoprecipitated and immunoblotted with TaPin1 or Flag antibodies.
b. Inhibition of TaPin1 by Bup, Jug or DTM increased FBW7α protein levels and decreased c-Jun expression in TBL3 cells. Actin/Tubulin were loading controls.
c. Inhibition of FBW7 increases c-Jun protein levels in TBL3 cells, whereas ectopic FBW7α expression reduced c-Jun protein levels. Bovine actin/Tubulin were loading controls. Con = empty vector.
d. Inhibition of Jun reporter activity (BIC promoter-Luciferase, an AP-1 target gene) in TBL3 cells transfected with siRNA against FBW7 or siControl (siCon).
e. Jun reporter activity (BIC promoter-Luciferase) in TBL3 cells treated with Buparvaquone (Bup) or Juglone (Jug) was rescued by siRNA against FBW7 but not siControl.
f. siFBW7 inhibition rescued drug effects on c-Jun ubiquitination in parasitized TBL3 cells, incubated with MG132, followed by immunoprecipitation of endogenous c-Jun and immunoblotting with c-Jun and ubiquitin antibodies. “Ig” = non-specific control immunoglobulin.
g. siFBW7 depletion increased the half-life of endogenous c-Jun protein. TBL3 cells treated with Buparvaquone (Bup) or Juglone (Jug) were incubated with cycloheximide, followed by immunoblotting with c-Jun or Tubulin antibodies. Relative c-Jun protein levels at T0 were set at 1.
h. Colony formation of TBL3 cells was markedly reduced by ectopic expression of FBW7α isoform or c-Jun siRNA. Efficiency of two independent si-c-Jun is shown. Bovine Tubulin loading control.
Data are representative of 3 independent experiments (average ± sd, n=3). The p-values with the Bonferroni method based on the number of pairwise comparisons were calculated for Figure 4e. The statistics in Figure 4h used the Dunnett procedure. *p<0.05, **p<0.01, ***p<0.001.
c-Jun is critical for Theileria transformation and host cell proliferation2, but it was previously unknown how the parasite initiated this effect. In TBL3 cells, c-Jun seems to be activated by reduced FBW7 degradation rather than phosphorylation by JNK signaling2. Subsequent activation of a feedback loop, involving c-Jun control of the miR-155 oncomiR, could create a epigenetic switch to maintain transformation and proliferation30. We studied B lymphocytes infected naturally with T. parva (TpMD409) or artificially with T. annulata (TBL3) and are currently investigating Theileria-transformed T lymphocytes and macrophages. Our discovery of mutation in the TaPin1 gene, suggests that new anti-Pin1 compounds might be effective clinical reagents to treat drug-resistant theileriosis. Finally, the evolution of a Pin1 homolog with an acquired signal peptide only in T. annulata and T. parva genomes, provides fascinating insight into how apicomplexan species have hijacked oncogenic pathways to maintain host cell transformation.
Materials and Methods
Cell lines and culture conditions
All infected bovine cell lines used in this study were previously described: TBL3 cells were derived from in vitro infection of the spontaneous bovine-B lymphosarcoma cell line, BL3, with Hissar stock of T.annulata. The TpMD409 lymphocyte cell line is infected with T. parva. The culture conditions of these cell lines were described previously31. All parasite-infected cell lines were provided by the Langsley laboratory. Cells were cultured in RPMI 1640 (Gibco-BRL), supplemented with 10% heat-inactivated Fetal calf serum, 4mM L-Glutamine, 25mM HEPES, 10μM β-mercaptoethanol and 100μg/ml penicillin/streptomycin in a humidified 5% CO2 atmosphere at 37°C. NIH3T3 and Murine Immortalized Fibroblasts cells were kindly provided by C. Francastel (UMR7216 Epigenetics and Cell Fates, Paris, France) and G. Del Sal (LNCIB - Laboratorio Nazionale CIB, Trieste, Italy), respectively. MCF10A Ras/Neu – shPin1 were previously described 17. Murine and human cell lines were cultured in DMEM (Gibco-BRL, Paisley, UK), supplemented with 10% heat-inactivated Fetal calf serum, 4mM L-Glutamine and 100 μg/ml penicillin/streptomycin in a humidified 5% CO2 atmosphere at 37°C. Cell numbers, as judged by Trypan Blue exclusion test, were determined by counting cells using a Countess automated cell counter (Invitrogen). All cell lines were mycoplasma negatives. The anti-parasite drug Buparvaquone (BW720c)3 was used at 200ng/ml for 72 hours (Chemos GmbH, Ref: 88426-33-9). BW720c has no effect on growth of uninfected cells (Hudson, 1985). Cells were treated with Juglone at 5μM resuspended in Ethanol (Sigma, Ref: H47003), DTM at 1μM resuspended in DMSO (DTM was kindly provided by T. Uchida, Tokyo University, Japan).
Plasmids
Plasmids p3xFLAG-myc-CMV-24: FBW7alpha, beta or gamma were kindly provided by BE. Clurman (Fred Hutchinson Cancer Research Center, Seattle, USA). Human gene hPin1 and parasite genes TaPin1 WT (TA18945) or TaCyclophilin (TA19600) were cloned between restriction sites XhoI and NotI in pREV-HA-FLAG-RIL2 using oligonucleotides: hPin1 (Fwd - CCGCTCGAGGCGGACGAGGAGAAGCTG, Rev – AAGGAAAAAAGCGGCCGCTCACTCAGTGCGGAGGATGA), TaPin1 WT (Fwd – CCGCTCGAGGCCCACTTGCTACTAAAG, Rev – ATAAGAATGCGGCCGCTTATGCGATTCTATATATAAGATG), and TaCyclophilin (Fwd – CCGCTCGAGTTCTACAATCAACCCAAGCAT, Rev - AAGGAAAAAAGCGGCCGCTCACAATAATTCTCCACAGTCC). Point mutations TaPin1 K38A, A53P and C92A were created from pRev-HA-FLAG-TaPin1 WT-RIL2 using a set of primers following a 3-step PCR protocol: TaPin1 K38A (Fwd - GCCCACTTGCTACTAGCGCACACTGGATCTAGG, Rev – CCTAGATCCAGTGTGCGTAGTAGCAAGTGGGC), TaPin1 A53P (Fwd – GGAATACTGGAATGCCAGTAACAAGAAC, Rev – GTTCTTGTTACTGGCATTCCAGTATTCC) and TaPin1 C92A (Fwd – GCAACTGCCAAATCTGAGGCTTCAAGCGCAAGAAAAGG, Rev – CCTTTTCTTGCGCTTGAAGCCTCAGATTTGGCAGTTGC).
siRNA
BL3 and TBL3 cells were transfected using Neon Transfection kit (Invitrogen). Cells were double transfected with 400nM of the indicated siRNA: FBW7 (CATCATTAGTGGATCCACGG), Pin1 (GCCATTTGAAGACGCCTCC), c-Jun-1 (CCACGGCCAAUAUGCUCAGG), c-Jun-2 (AUGACUGCAAAGAUGGAAA).
Parasite genomic DNA extraction and Sequencing
Buparvaquone-resistant infected cells were cultured in RPMI 1640 and parasite DNA was extracted using the kit Promega (Wizard Genomic DNA Purification Kit, Ref: A1125) following the manufacturer’s instructions. To sequence the TaPin1 gene, we first performed a PCR with specific oligonucleotides (Fwd – GTCTGTCAAATAGGTAGAAATC, Rev - GAGAGGAAGTTGAATCAAACAT) using High Fidelity Platinium Taq Polymerase (Invitrogen, Ref: 11304) and sequencing was performed using the same oligonucleotides.
RNA extraction and Reverse Transcription-qPCR
Total cellular RNAs were extracted using a NucleoSpin RNA Kit (Macherey Nagel, Ref: 740955) and parasite RNAs were extracted using the classical Trizol Protocol. cDNA synthesis was performed with the Reverse Transcriptase Superscript III (Invitrogen, Ref: 18080051). Quantitative PCR amplification was performed using the Sybr Green reagent (Applied Biosystems, Ref: 4309155). c-Jun (Fwd – ACGTTTTGAGGCGAGACTGT, Rev – TCTGTTTCCCTCTCGCAACT), FBW7 (Fwd – AGCTGGAGTGGACCAGAGAAATTG, Rev – GAATGAGAGCACGTAAAGTGC), Pin1 (Fwd – GGCCGGGTGTACTACTTCAA, Rev – TTGGTTCGGGTGATCTTCTC), MMP9 (Fwd - CCCATTAGCACGCACGACAT, Rev – TCACGTAGCCCACATAGTCCA), H2A (Fwd – GTCGTGGCAAGCAAGGAG, Rev – GATCCGGCCGTTAGGTACTC) and β-ACTIN (Fwd – GGCATCCTGACCCTCAAGTA, Rev - CACACGGAGCTCGTTGTAGA). The detection of a single product was verified by dissociation curve analysis. Relative quantities of mRNA were analyzed using the delta Ct method. The β-actin and H2A qPCR were used for normalization.
Nucleus/Cytoplasmic protein extraction
Cells were lysed in the following buffer: 5mM Tris HCl pH7.5, 40mM KCl, 2mM MgCl2, 0.5mM EDTA, 0.05mM Spermidin and Spermin, 0.1% NP-40, H2O. Lysates were incubated for 5min on ice and centrifuged for 10min at 5000rpm. The supernatant constitutes the cytoplasmic fraction and the pellet constitutes the nuclear fraction.
Anti-TaPin1 antibody purification
Anti-Pin1 rabbit polyclonal antibody raised against TaPin1 peptide “NPVNRNTGMAVTR” was prepared and purified by ProteoGenix SAS (Schiltigheim, France). Antiserum was obtained by immunizing rabbits with keyhole limpet hemocyanin (KLH)-conjugated peptide. The resulting IgG fraction was purified from antiserum by affinity chromatography against the TaPin1 peptide.
Immunoblot analysis and immunostaining
Total proteins were extracted with Laemmli lysis buffer, sonicated: 30 secs ON/30 secs OFF for 5min, resolved on 10.5% acrylamide/bis-acrylamide SDS-PAGE gels and transferred to nitrocellulose membranes (Thermo Fisher Scientific, MA, USA) in transfer buffer. Protein transfer was assessed by Ponceau-red staining. Membranes were blocked in Tris-buffered saline pH7.4 containing 0.1% Tween-20 and 5% milk for 1 h at room temperature. Incubations with primary antibodies were carried out at 4°C overnight using antibody dilutions as manufacturer recommendations in Tris-buffered saline pH7.4, 0.05% Tween-20 and 5% milk. Following 1 h incubation with an anti-rabbit or anti-mouse peroxidase-conjugated antibody (Jackson ImmunoResearch, Ref: 111-035-003 or Ref: 115-035-003) at room temperature, proteins were detected by chemiluminescence (Thermo Fisher Scientific) following the manufacturer’s instructions. We used these antibodies: Rabbit anti-TaPin1 (Home-made antibody, Proteogenix, see previous section), rabbit anti-Pin1 (Cell Signalling, Ref: 3722), rabbit anti-TaActin (kindly provided by J. Baum, Walter and Eliza Hall Institute of Medical Research, Australia), rabbit anti-c-Jun (Santa Cruz, Ref: sc1694), mouse anti-αTubulin (Sigma, Ref: T9026), mouse anti-Ubiquitin (P4D1) (Santa Cruz, Ref: sc-8017), mouse anti-c-Myc (Santa Cruz, Ref: sc-40), mouse anti-GST (Pierce Biotechnology, Ref: MA4-004), rabbit anti-KLF5 (Abcam, Ref: ab24331), rabbit anti-activated Notch1 (Abcam, Ref: ab8925), mouse anti-HA (Roche, Ref: 11583816001), rabbit anti-FBW7 (Bethyl Laboratories, Ref: A301-721A), rabbit anti-Histone H3 (Abcam, Ref: ab1791), mouse anti-Actin (Sigma, Ref: A1978) and monoclonal anti-FLAG M2-Peroxidase (Sigma, Ref: A8592).
Parasite Quantification
After indicated treatments, parasite-infected cells were plated on slides using CytoSpin centrifugation at 2000rpm for 10min. Cells were fixed in PBS 3.7% Formaldehyde for 15min at room temperature. Slides were mounted and coverslipped with ProLong Gold Antifade Reagent with DAPI (Invitrogen, Ref: P-36931). Images of immunofluoresence staining were photographed with a fluorescent microscope (Leica Inverted 6000) and the number of parasites per cells was counted. Staining was repeated for three independent biological replicates.
PPIase Assay
The PPIase activity of GST constructs: GST-Control, GST-hPin1, GST-TaPin1 WT and GST-TaPin1 A53P were determined using the protease-free PPIase activity assay11. The sample buffer was 35 mM Hepes (pH 7.5). We prepared stock solutions of the substrates (3mg/ml), Suc-Ala-Glu-Pro-Phe-pNA (Pin1 substrate peptide), or a control peptide Suc-Ala-Ala-Pro-Phe-pNA (Bachem, Ref: L-1635 or Ref: L-1400) in 0.47 M LiCl/TFE (anhydrous). Stock solution of chymotrypsin (100 mg/mL, Sigma, Ref: C4129) was prepared in 35 mM Hepes (pH 7.8). We measured the PPIase activity with the substrate (0.03mg/mL – 50 μM) in presence of chymotrypsin (0.2mg/mL) and GST-PPIases (25nM) (pre-incubated or not with Bup, Jug or DTM for 4h at 4°C) at 390 and 510 nm using a Flexstation III spectrophotometer.
Implantation of cells in zebrafish embryos
This study was approuved by the Institutional Comitee for animal Welfare of the University of Burgundy. Zebrafish and embryos were raised, staged and maintained according to standard procedures in compliance with the local animal welfare regulations (University of Burgundy). Dechorionized 2 old zebrafish embryos were anaesthetized with 0.003% tricain (Sigma) and positioned on a 10 cm Petridish before inplantation, TBL3 or BL3 cells were treated for 24h with DTM, Buparvaquone or Juglone, rinsed with PBS, labeled with the fluorescent cell tracker CM-DiI (Invitrogen) according to the manufacturer’s instructions and resuspended in PBS. The cell suspensions were loaded into borosilicate glass capillary needles and the injections were performed using a microinjector (Femtojet, Eppendorf). 20–100 cells, manually counted in injection droplets, were injected in the yolk within 3-4 hours after labelling. Around 30-100 embryos were implanted per cell line. After implantation, zebrafish embryos (including non-implanted controls) were maintained at 34 °C in egg water containing 0.003% Phenylthiourea (PTU). For individual tumor development analysis, each xenografted embryo was grown in separate well in 12 well plates. Tumor growth was monitored at day1 and and day 4 after injection by imaging the zebrafish embryos with a Zeiss AxioZoom V16 Macroscope. Images were acquired using 2.3x objective and analyzed with Zen software. For the estimation of tumor foci size, red fluorescent aerea was mesured with Zen software and data were transfered to excel for further calculations. No method of randomization was used to determine how animals were allocated to experimental groups.
Immunofluorescence
BL3 and TBL3 cells were plated on Fibronectin coated slides (Sigma; Ref: F1141). NIH/3T3 cells and Mouse Immortalized Fibroblasts cells-transfected by indicated constructs were plated on slides. All cells were then fixed in PBS 3.7% Formaldehyde for 15min at room temperature. Slides with bovines cells or NIH/3T3 cells were rinsed in PBS and permeabilized with PBS 0.2% Triton X-100 for 5min and then blocked for 30min with PBS 1% SVF and 1% BSA to prevent non-specific staining. These slides were incubated with rabbit anti-TaPin1 (1/250) and/or mouse anti-HA (1/1000, Roche, Ref: 11583816001) in PBS 1% SVF and 1% BSA at room temperature for 40min. After washing in PBS 0.2% Tween, the slides were incubated with Texas Red dye-conjugated AffinyPure Donkey anti-rabbit IgG and/or Cy2 AffinyPure Donkey anti-mouse IgG (1/5000, Jackson Immunology, Ref: 711-075-152 or Ref: 715-225-150) for 30min. Slides with Murine Immortalized Fibroblasts cells were incubated for 15min with Phalloïdin-TRITC (Life Technologies, Ref: R415). All slides were subsequently washed in PBS 0.2% Tween, mounted on slides and covered with ProLong Gold Antifade Reagent with DAPI (Invitrogen, Ref: P-36931). Images of immunofluorescence staining (Mouse Immortalized Fibroblasts cells) were photographed with a fluorescent microscope (Leica Inverted 6000). Staining was repeated for three independent biological replicates.
Confocal microscopy analysis
Acquisitions were made on a ZEISS LSM710 laser scanning confocal. Texas Red was acquired using a 561nm DPSS laser diode, emission captured between 587 and 690 nm. DAPI was acquired using a 405nm laser diode, emission captured between 410 and 506nm. Images were taken with a 60x/NA 1.4 objective, with a 2.5 zoom factor so that image pixel size was about 100nm. Optical sections were acquired every 320nm. Image analysis and nucleus fluorescence intensity/pixel quantifications were performed using the software Imaris 6.7.5 (Bitplane). Quantifications were done on the whole nucleus of n=31 bovine infected or non-infected cells after 3D construction and normalized to the background signal obtained after staining with the anti-Rabbit secondary antibody alone.
Analysis of centrosome duplication during S-phase
Centrosome duplication assays in NIH3T3 cells were conducted, as described previously17. Cells were arrested in G1/S phase by adding Aphidicolin (Sigma, Ref: A0781) at final concentration of 10μg/ml for 24h. Cells were fixed with cold-Methanol for 10min at −20°C, then stained for centrosome with anti-Gamma-Tubulin antibody (Sigma, Ref: Clone GTU-88, T5326), and analyzed by fluorescent microscopy, as described in the “ Immunofluorescence” section.
Luciferase Assay
Non-treated or treated bovine cells were transfected with the cyclinD1 or BIC Luciferase reporters, using electroporation (Neon kit, Invitrogen, Ref: MPK1096). Mouse cells were transfected using Lipofectamine 2000 (Invitrogen, Ref: 11668019). Transfection efficiencies were normalized to Renilla activity by co-transfection of a pRL-TK Renilla reporter plasmid (Promega, Ref: E6241). Luciferase assays were performed 36h post-transfection using the Dual-Luciferase Reporter Assay System (Promega, Ref: E1980) in a microplate luminometer. Relative luminescence was represented as the ratio Firefly/Renilla luminescence, compared with the corresponding empty vector control.
Colony Forming Assay
MCF10A cells were transfected by the indicated plasmids using Fugene HD transfection system (Promega, Ref: E2311) following manufacturer’s instructions. After 36h, 1000 cells were plated in 6-well plates. Cultures were incubated in humidified 37°C incubators with an atmosphere of 5% CO2 in air, and control plates were monitored for growth using a microscope. At the time of maximum foci formation (8-10 days in culture), final foci numbers were counted manually after fixation a staining with 0.5% Crystal Violet (Sigma, Ref: C3886).
Soft Agar Colony Forming Assay
A two-layer soft agar culture system was used. A total of 20,000 bovine cells (treated with Bup or Jug) or 40,000 bovine cells (treated with DTM or transfected with indicated plasmids/siRNA) were plated in a volume of 1.5 ml (0.7% SeaKem ME Agarose: Lonza, Ref: 50011) + 2x DMEM 20% Fetal calf Serum over 1.5-ml base layer (1% SeaKem ME Agarose + 2x DMEM 20% Fetal calf Serum) in 6-well plates. Cultures were incubated in humidified 37°C incubators with an atmosphere of 5% CO2 in air, and control plates were monitored for growth using a microscope. At the time of maximum colony formation (10-15 days in culture), final colony numbers were counted manually after fixation a staining with 0.005% Crystal Violet (Sigma, Ref: C3886).
GST pull-down
hPin1, TaPin1 WT and TaPin1 A53P were cloned between restriction sites BamHI and EcoRI in pGEX-2T plasmid, which was kindly provided by G. Del Sal (LNCIB - Laboratorio Nazionale CIB, Trieste, Italy). TaPin1 WT or A53P (Fwd – CGCGGATCCGCCCACTTGCTACTAAAG, Rev - CCGGAATTCTTATGCGATTCTATATATAAGATG). Plasmid constructs were expressed in E. coli strain BL21 and purified using glutathione-sepharose beads. Concentration of purified protein was estimated by Coomassie staining. Beads coated with 1μg of GST fusion proteins were incubated with 250μl of cell lysate (see Immunoprecipitation – HA) in 50mM Tris pH7.6, 150mM NaCl, 0.1% Triton, for 2h at 4°C. Beads were washed 5 times with 50mM Tris pH7.6, 300mM NaCl, 0.5 % Triton. Proteins were revealed by Western Blot analysis using specific antibodies.
Immunoprecipitation - HA
NIH3T3 cells stably expressing TaPin1 or TBL3 transiently expressing the FBW7 constructs were lysed in the following buffer: 20mM Tris HCL pH8, 150mM NaCl, 0.6% NP-40 and 2mM EDTA. Protein complexes were affinity-purified on anti-HA antibody-conjugated agarose (Sigma, Ref: A2095) for NIH/3T3 lysates or on anti-Flag antibody-conjugated agarose (Sigma, Ref: A2220) for bovine lysates and eluted with the HA peptide or Flag peptide respectively. After 5 washes, immunopurified complexes were resolved on 4-12% SDS-PAGE bis-Tris acrylamide gradient gel in MOPS buffer (Invitrogen, Ref: NP 0322 BOX, NP0001-02, respectively).
Immunoprecipitation – Ubiquitin
Cells were treated for 3h at 37°C with 20μM MG132 and lysed 10min on ice in the following buffer: 150mM NaCl, 1% Nonidet P-40, 0.5% Deoxycholate, 0.1% SDS, 50mM Tris HCl pH 7.5, 20mM NEM, 5mM Iodoacetamide, 100uM MG132, 2mg/ml Pefabloc SC (Roche) and 5μg/mL each Aprotinin, Leupeptin, Pepstatin. Equal amounts of total cellular proteins were immunoprecipitated with rabbit Anti-c-Jun (E254) (Abcam, Ref: ab32137) or rabbit Anti-FBW7 (Bethyl Laboratories, Ref: A301-721A), coupled to protein G sepharose beads (Sigma, Ref: P3296) for 90min at 4°C. After three washes, immunoprecipitated proteins were eluted in Laemmli sample buffer at 95°C for 5min, resolved by SDS-PAGE and analysed by Western blot using the indicated antibodies. Immunoprecipitation was repeated for three independent biological replicates.
Cycloheximide chase assay
Infected bovine cells (TBL3) were treated for 72h with Buparvaquone, Juglone or DTM and transiently transfected with indicated siRNA. Then cells were treated 30, 60 or 120 min with 100 mg/mL Cycloheximide. Cells were lysed in Laemmli sample buffer, resolved by SDS-PAGE and analyzed by western blot using the indicated antibodies. Relative quantification indicates the c-Jun/Tubulin ratios calculated with Image J software (NIH) and c-Jun levels at time 0 was set as 1. Cycloheximide chase experiments were repeated for four independent biological replicates.
Viability assays
1×104 cells were plated in 96-well plates in triplicate and Buparvaquone, Juglone or DTM was added. Cell viability was measured after 72h using the Cell proliferation Kit II–XTT (Roche) and the GloMax-Multi Detection System (Promega).
Data and statistical analysis
The GraphPad PRISM 6 program (GraphPad Software Inc. La Jolla, CA, USA) was used for statistics. The results presented in all the figures represent the average ± sd of at least three independent experiments. Statistical analysis was performed using the One-way ANOVA test – multiple comparisons test. The p-values were corrected for the multiple comparisons using the Bonferroni correction based on the total overall number of pairwise comparisons for the Figure 1b. The p-values were calculated using the approach of Dunnett for multiple comparisons with the TaPin1WT for Figure 2c. For Figures 2b / 2d / 2e / 2f / 3c / 3d / 3e: the p-values were corrected using the Dunnett multiple comparisons with the control. The p-values with the Bonferroni method based on the number of pairwise comparisons were calculated for Figure 4e. The statistics in Figure 4h used the Dunnett procedure. Finally an unpaired Mann-Whitney test was performed for the zebrafish experiments to analyze the significant difference between the control and treatment groups. The SPSS 19.0 program (SPSS Inc. Chicago, IL, USA) was used for statistics in all Extended Data Figures. The results presented in these Extended Data Figures represent the average ± sd of at least three independent experiments. p values of <0.05 were considered statistically significant and are indicated with asterisks *,p<0.05; **,p<0.01; ***,p<0.001.
Bioinformatic screen
On 9-10-2011 search strategies at EuPathDB32 were used to search for all T. annulata protein-encoding genes with predicted signal peptides (SignalP 2.0). 689 genes were returned. 138 of these were found to have a predicted SP only in T. annulata and not in their T. gondii orthologs. Among these proteins, we excluded (i) hypothetical proteins, (ii) proteins which are not expressed at the macroschizont stage, and (iii) proteins which are predicted to be targeted to the apicoplast of the parasite. We obtained 33 proteins in Extended Data Table 1. This search strategy was repeated on 4/22/2013 to ensure that results were consistent with any EuPathDB updates. All 33 proteins from Extended Data Table 1 were returned in the updated search.
Structures analysis
Homology models of TaPin1 WT and Mutant A53P were built with the online server EsyPred33. The experimental structure of human Pin134 co-crystallized with a dipeptide Ala-Pro (resolution 1.35 Å) was used as a template. In order to check the protonation state of the protein titratable groups, we used our online server PCE35. The most likely binding pocket areas for TaPin1 WT were predicted with FTMap and investigation of side chain flexibility (if any) in the area of the predicted binding cavities was carried out with the server FTFlex36. The 2D structures of Juglone, Buparvaquone and DTM were obtained from PubChem and the 3D structures were generated with our package DG-AMMOS37. Three docking tools, Surflex38, Molegro Virtual Docker39 and our tool MS-DOCK40 were used to search for possible poses of these compounds in hPin1, for TaPin1 WT and Mutant A53P. Calibration of our docking tools was performed on the thiol-stress sensing regulator co-crystallized with a quinone molecule, which in this structure is covalently attached to a Cys residue (PDB file 4HQM)41. Visualization and figures were prepared with PyMol.
Extended Data
Extended Data Fig. 1. Bioinformatic screen and in silico predicted signal peptide for TaPin1.
a. On 9-10-2011 search strategies at EuPathDB were used to search for all T. annulata protein-encoding genes with predicted signal peptides (SignalP 2.0 HMM). 689 genes were returned. 138 of these were found to have a predicted SP only in T. annulata and not in their T. gondii orthologs. Among these proteins, we excluded (i) hypothetical proteins, (ii) proteins which are not expressed at the macroschizont stage, (iii) proteins which are predicted to be targeted to the apicoplast of the parasite. We obtained 33 proteins (right panel).
b. Sequence alignment of Pin1 genes in H. sapiens (Human), B. taurus (Cow), M. musculus (Mouse) and T. annulata revealed a predicted signal peptide (underlined) and conserved PPIase domain in TaPin1. Stars indicate TaPin1 residues mutated in the study.
c. Example signal peptide predicted with SignaIP 3.0 Server.
Extended Data Fig. 2. TaPin1 is conserved between Theileria annulata and Theileria parva.
a. As the T.parva TpPin1 sequence did not have an annotated signal peptide, we re-analyzed sequence alignment of pin1 genes in T. annulata (TaPin1) and T. parva (TpPin1) (blast-NCBI). The annotated T. annulata intron is indicated in grey and the annotated Methionine codon (ATG) in T. parva in black (underlined). TaPin1 and TpPin1 are well conserved including in the intronic sequence. We conclude that the T.parva TpPin1 annotation should be modified in light of our PCR and Western blot analysis (see Figures 1b and Extended Data 5c).
a. Sequence alignment of pin1 genes in T. annulata (TaPin1) and T. orientalis (ToPin1) (blast-NCBI). The annotated T. annulata intron is indicated in grey and the annotated Methionine codon in T. orientalis in black (underlined). TaPin1 and ToPin1 are less conserved than TaPin1 and TpPin1.
Extended Data Fig. 3. Bovine Pin1 is not regulated by the parasite.
Buparvaquone treatment had no effect on the mRNA levels (a) of bovine Pin1 in infected (TBL3) or non-infected (BL3) cells as assessed by qPCR analysis and on the protein level (b) as assessed by immunoblot analysis. “Con” indicates Control and “Bup” indicates Buparvaquone treatment. (average ± sd, n=3). The result in 3b is representative of 3 independent experiments.
Extended Data Fig. 4. Characterization of the TaPin1 antibody and secretion of TpPin1 in Theileria parva.
a The antibody raised against TaPin1 specifically recognizes Theileria version in TaPin1-stably transfected NIH3T3 cells but not the empty vector in Control-stably transfected NIH3T3 cells or the human version in hPin1-stably transfected NIH3T3 cells. Both Theileria and Human versions of Pin1 are present respectively in TaPin1-stably transfected NIH3T3 and hPin1-stably transfected NIH3T3 cells.
b. The antibody raised against TaPin1 specifically recognizes GST-TaPin1 but not GST and GST-hPin1 beads.
c. TpPin1 protein was detected in the nuclear and cytoplasmic fractions of T.parva-infected TpMD409 cells and decreased upon Buparvaquone (Bup) treatment. “Con” indicates Control. Antibodies recognizing apicomplexan actin, bovine Tubulin or bovine Histone H3 were used as controls.
d. TaPin1 expression in BL3 or parasite-infected TBL3 cells was examined by confocal microscopy analysis using an antibody raised specifically against Theileria TaPin1, counterstaining with DAPI. 3D reconstruction in BL3 and TBL3 cells are shown.
e. Immunostaining of TaPin1 and HA in NIH-3T3 stably expressing TaPin1 and NIH-3T3 stably expressing hPin1 counterstaining with DAPI.
All the results are representatives of 3 independent experiments.
Extended Data Fig. 5. TaPin1 functionally replaces hPin1.
a. The HA-tagged version of TaPin1 and hPin1 can rescue cell spreading defects in knockout pin1-/- murine immortalized fibroblats. Cell spreading was assessed by Phalloïdin-TRITC staining. “Con” indicates Control = transfection with the appropriate empty vector.
b. TaPin1 causes centrosome amplification. NIH3T3 fibroblasts stably expressing TaPin1 or hPin1 were arrested at the G1/S transition by aphidicolin, stained with anti-γ-tubulin antibody (arrow) and counterstained with DAPI.
These photos are representative of cell in 3 independent experiments
Extended Data Figure 6. The PPIase domain of Pin1 is well conserved.
a. Sequence alignment of pin1 genes in H. sapiens, A. thaliana, T. brucei and T. annulata revealed the presence or the absence of a conserved WW domain. Percentages of identity are indicated. The magenta box indicates the predicted signal peptide of TaPin1.
b. Homology models for TaPin1 WT and Mutant A53P based on sequence identity/similarity with hPin1. The mutation Ala>Pro in TaPin1 appears to induce a conformational change within and nearby the catalytic loop.
c. The Buparvaquone molecule can be docked in the active site of hPin1 or in the active site of WT TaPin1 predicted structure. Here, the second lowest docked energy pose is shown (the best predicted energy pose is shown Fig. 3a). These two predicted binding poses are fully consistent with the different binding modes of some inhibitors co-crystallized with hPin1. Yet, considering computations shown below in Extended Data Fig. 6e, we suggest that the most likely pose for Buparvaquone corresponds to the pose 1 reported in Fig 3a. However, independently of the selected poses, we expect that Buparvaquone would not fit well in the catalytic pocket due to structural changes induced by the A53P mutation.
d. 3D structure of the experimental hPin1 structure and the predicted TaPin1 WT, A53P mutant, alongside Trypanosome TbPin1, Arabidopsis Pin1At models (ribbon diagram). The 3D structures are well conserved among these proteins with some differences, for instance, in the catalytic loops. The TaPin1, TbPin1 and PinAt enzymes lack the WW domain present in hPin1 yet the overall fold in the catalytic area is well-conserved suggesting that accurate homology models for TaPin1 can be built using the approach used in the present study.
e. The experimental structure of hPin1 is represented as a solid surface with a view down the active site, showing a small co-crystallized ligand next to the catalytic site residue Cys113. In the same orientation, small chemical fragments are predicted to bind in the catalytic site region with FTmap. Using this information, we propose that the most likely binding pose for Buparvaquone is the one shown in Fig. 3.
Extended Data Fig. 7. Buparvaquone-resistant parasites are mutated in the TaPin1 PPIase domain.
a. Sequence alignment of the TaPin1 gene in T. annulata WT or Buparvaquone-resistant genomes (Blast-NCBI). The annotated T. annulata intron is indicated in grey. Buparvaquone-resistant parasites have 3 mutations indicated in black.
b. Sequence alignment of the TaPin1 protein in T. annulata WT or Buparvaquone-resistant strains. Buparvaquone-resistant parasites have a mutation in the PPIase domain (residue N°53): Ala to Pro mutation.
c. Signal peptide predicted of TaPin1 Mutant A53P with SignaIP 3.0 Server.
d. The measure of TaPin1-GST and TaPin1 A53P-GST catalytic activity were determined using PPIase assay (chymotrypsin-coupled assay) (average ± sd, n=4).
Extended Data Fig. 8. Inhibition of bovine Pin1 does not affect Theileria-associated cell transformation.
a. Buparvaquone (Bup), Juglone (Jug) and DTM decreased the viability of cells infected with T. parva (TpMD409). Host cell viability was assessed using the XTT assay after 72h.
b. Colony formation of parasite-infected TpMD409 cells in soft-agar was lost after 72h treatment with Buparvaquone (Bup), Juglone (Jug) or DTM. Number of macroscopic colonies per plate were counted after 10 days.
c. siPin1 has no effect on TBL3 transformed phenotypes. TBL3 were transiently transfected with siControl or siPin1. The average number of colonies per plate is shown. (average ± sd, n=3). Indicated protein levels were detected by Western blot analysis using specific antibodies. Actin was used as loading control (Results representative of 3 independent experiments).
d/f. Overexpression of the TaPin1 Mutant A53P rescued the Buparvaquone but not Juglone effects on Theileria infected cells. All data represent 3 independent experiments (average ± sd, n=3). The SPSS 19.0 program (SPSS Inc. Chicago, IL, USA) was used for statistics. *p<0.05, **p<0.01.
Extended Data Fig. 9. TaPin1 changes c-Jun stability through the regulation of FBW7.
a. TaPin1 interacts with endogenous mouse FBW7. Protein extracts from NIH3T3 cells stably expressing HA-Flag-TaPin1 or HA-Flag-Control “Con” were used for HA immunoprecipitation, followed by immunoblot analysis using FBW7 and HA antibodies.
b. TaPin1 interacts with endogenous bovine FBW7 protein. Parasite-infected TBL3 and TpMD409 cell lysates were incubated with indicated GST-coupled beads, followed by immunoblot analysis using a FBW7 antibody.
c/d Analysis of indicated bovine gene expression by qPCR in TBL3 cells following Buparvaquone (Bup) or Juglone (Jug) treatment with or without TaPin1 WT or Mutant A53P. β-actin and H2A mRNAs were used for normalization (data represent 3 independent experiments - average ± sd, n=3).
e. siPin1 does not affect FBW7a or c-Jun protein levels. TBL3 were transiently transfected by siControl “siCon” or si-bovine-Pin1. Indicated protein levels were detected by Western blot analysis using specific antibodies. Actin was used as loading control.
f. Inhibition of TaPin1 by Buparvaquone/Juglone increased c-Jun ubiquitination and decreased FBW7 ubiquitination. Parasite-infected TBL3 cells (+/- Buparvaquone (Bup) or Juglone (Jug) - “Con” indicates Control) were incubated with MG132, followed by immunoprecipitation of endogenous c-Jun or FBW7 and immunoblot analysis with the indicated antibodies.
g. The effect of Buparvaquone on c-Jun ubiquitination was rescued by overexpression of TaPin1 Mutant A53P. Parasite-infected TBL3 cells (+/- Buparvaquone (Bup) or Juglone (Jug) - “Con” indicates Control) were transfected with TaPin1 WT, TaPin1 Mutant A53P or empty vector “Con” and then treated with MG132, followed by immunoprecipitation of endogenous c-Jun and immunoblot analysis with the indicated antibodies.
h. Measure of c-Myc, KLF5 and activated Notch 1 protein levels in TBL3 upon Buparvaquone (Bup) treatment. “Con” indicates Control. Tubulin was used as loading control.
i. Measure of c-Myc, KLF5 and activated Notch 1 protein levels in TBL3 upon FBW7α ectopic expression. “Con” indicates Control = transfection with the appropriate empty vector. Tubulin was used as loading control.
All the results in a, b, e, f, g, h and i are representatives of 3 independent experiments.
Extended Data Fig. 10. Original Blot.
a. From Figure 1b : TaPin1 protein was detected in the host cytoplasm and nucleus, in contrast Apicomplexan actin (TaActin). Bovine Histone H3 (nuclear) and Tubulin (cytoplasmic) proteins were controls.
b. From Figure 4a: Endogenous TaPin1 interacts with FBW7α isoform. Protein extracts from TBL3 expressing Flag-hFBW7 isoforms or Flag-Control “Con” were immunoprecipitated and immunoblotted with TaPin1 or Flag antibodies.
c. From Figure 4b: Inhibition of TaPin1 by Bup, Jug or DTM increased FBW7α protein levels and decreased c-Jun expression in TBL3 cells. Actin/Tubulin were loading controls.
d. From Figure 4c: Inhibition of FBW7 increases c-Jun protein levels in TBL3 cells, whereas ectopic FBW7α expression reduced c-Jun protein levels. Bovine actin/Tubulin were loading controls. Con = empty vector.
e. From Figure 4h: Efficiency of two independent si-c-Jun. Bovine Tubulin loading control.
Acknowledgments
We thank Gordon Langsley, Mohamed Aziz Darghouth and Matthew Weitzman for critical reading of the manuscript and invaluable advice on this study. We thank members of the UMR7216 for helpful discussions. JW thanks Claire Gawer for advice and support. We thank the following for providing reagents: G. Langsley (France) for TBL3, BL3 and TpMD409 infected cells and for a Theileria cDNA library; C. Francastel (France) for NIH3T3 cells; G. Del Sal (Italy) for pin1-/- murine immortalized fibroblasts; BE. Clurman (USA) for FBW7 plasmids; J. Baum (Australia) for the rabbit anti-Apicomplexa Actin antibody, T. Uchida (Japan) for DTM. Measurements of the PPIase activities were performed at the Flexstation III facility of the Biologie Fonctionnelle et Adaptative laboratory (Paris, France). Confocal analysis was performed at the microscopy facility of the ImagoSeine platform (Jacques Monod Institute, Paris, France). This work was supported by grants NIH grant R01CA167677 to K.P.L., the Association for International Cancer Research (AICR, #08-0111), the French National Research Agency (ANR) (Blanc SVSE 3 #090601), and the “Who Am I?” Laboratory of Excellence #ANR-11-LABX-0071 funded by the French Government through its “Investments for the Future” program operated by the ANR under grant #ANR-11-IDEX-0005-01.
Footnotes
Author information
JM, MP: performed the experiments
JDD: performed the comparative genomics bioinformatics screen
BOV: performed the structural 3D modeling
JC, TL, CG: designed and executed the zebrafish experiments
XZZ, KPL : provided critical reagents and advice MM: provided genomic DNA from Buparvaquone-resistant parasites
JM, SM, and JBW : conceived the study and designed experiments, analysed the data and wrote the paper.
References
- 1.Dobbelaere D, Heussler V. Transformation of leukocytes by Theileria parva and T. annulata. Annu Rev Microbiol. 1999;53:1–42. doi: 10.1146/annurev.micro.53.1.1. [DOI] [PubMed] [Google Scholar]
- 2.Chaussepied M, et al. Upregulation of Jun and Fos family members and permanent JNK activity lead to constitutive AP-1 activation in Theileria-transformed leukocytes. Mol Biochem Parasitol. 1998;94:215–226. doi: 10.1016/s0166-6851(98)00070-x. [DOI] [PubMed] [Google Scholar]
- 3.McHardy N, Wekesa LS, Hudson AT, Randall AW. Antitheilerial activity of BW720C (buparvaquone): a comparison with parvaquone. Res Vet Sci. 1985;39:29–33. [PubMed] [Google Scholar]
- 4.Shiels BR, et al. A Theileria annulata DNA Binding Protein Localized to the Host Cell Nucleus Alters the Phenotype of a Bovine Macrophage Cell Line. Eukaryot Cell. 2004;3:495–505. doi: 10.1128/EC.3.2.495-505.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pain A, et al. Genome of the host-cell transforming parasite Theileria annulata compared with T. parva. Science. 2005;309:131–133. doi: 10.1126/science.1110418. [DOI] [PubMed] [Google Scholar]
- 6.Witschi M, et al. Proteomic analysis of the Theileria annulata schizont. Int J Parasitol. 2013;43:173–180. doi: 10.1016/j.ijpara.2012.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lu KP, Hanes SD, Hunter T. A human peptidyl-prolyl isomerase essential for regulation of mitosis. Nature. 1996;380:544–547. doi: 10.1038/380544a0. [DOI] [PubMed] [Google Scholar]
- 8.Winkler KE, Swenson KI, Kornbluth S, Means AR. Requirement of the Prolyl Isomerase Pin1 for the Replication Checkpoint. Science. 2000;287:1644–1647. doi: 10.1126/science.287.5458.1644. [DOI] [PubMed] [Google Scholar]
- 9.Wulf GM, et al. Pin1 is overexpressed in breast cancer and cooperates with Ras signaling in increasing the transcriptional activity of c-Jun towards cyclin D1. EMBO J. 2001;20:3459–3472. doi: 10.1093/emboj/20.13.3459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ryo A, et al. PIN1 Is an E2F Target Gene Essential for Neu/Ras-Induced Transformation of Mammary Epithelial Cells. Mol Cell Biol. 2002;22:5281–5295. doi: 10.1128/MCB.22.15.5281-5295.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yaffe MB, et al. Sequence-Specific and Phosphorylation-Dependent Proline Isomerization: A Potential Mitotic Regulatory Mechanism. Science. 1997;278:1957–1960. doi: 10.1126/science.278.5345.1957. [DOI] [PubMed] [Google Scholar]
- 12.Lu KP, Finn G, Lee TH, Nicholson LK. Prolyl cis-trans isomerization as a molecular timer. Nat Chem Biol. 2007;3:619–629. doi: 10.1038/nchembio.2007.35. [DOI] [PubMed] [Google Scholar]
- 13.Hennig L, et al. Selective Inactivation of Parvulin-Like Peptidyl-Prolyl cis/trans Isomerases by Juglone†. Biochemistry (Mosc ) 1998;37:5953–5960. doi: 10.1021/bi973162p. [DOI] [PubMed] [Google Scholar]
- 14.Tatara Y, Lin YC, Bamba Y, Mori T, Uchida T. Dipentamethylene thiuram monosulfide is a novel inhibitor of Pin1. Biochem Biophys Res Commun. 2009;384:394–398. doi: 10.1016/j.bbrc.2009.04.144. [DOI] [PubMed] [Google Scholar]
- 15.Moore JD, Potter A. Pin1 inhibitors: Pitfalls, progress and cellular pharmacology. Bioorg Med Chem Lett. 2013;23:4283–4291. doi: 10.1016/j.bmcl.2013.05.088. [DOI] [PubMed] [Google Scholar]
- 16.Hayashida K, et al. Comparative Genome Analysis of Three Eukaryotic Parasites with Differing Abilities To Transform Leukocytes Reveals Key Mediators of Theileria-Induced Leukocyte Transformation. mBio. 2012;3 doi: 10.1128/mBio.00204-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chen C-H, et al. SENP1 desumoylates and regulates Pin1 protein activity and cellular function. Cancer Res. 2013 doi: 10.1158/0008-5472.CAN-12-4360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Landrieu I, et al. The Arabidopsis thaliana PIN1At Gene Encodes a Single-domain Phosphorylation-dependent Peptidyl Prolylcis/trans Isomerase. J Biol Chem. 2000;275:10577–10581. doi: 10.1074/jbc.275.14.10577. [DOI] [PubMed] [Google Scholar]
- 19.Landrieu I, Wieruszeski JM, Wintjens R, Inzé D, Lippens G. Solution Structure of the Single-domain Prolyl Cis/Trans Isomerase PIN1At from Arabidopsis thaliana. J Mol Biol. 2002;320:321–332. doi: 10.1016/S0022-2836(02)00429-1. [DOI] [PubMed] [Google Scholar]
- 20.Yao JL, Kops O, Lu PJ, Lu KP. Functional Conservation of Phosphorylation-specific Prolyl Isomerases in Plants. J Biol Chem. 2001;276:13517–13523. doi: 10.1074/jbc.M007006200. [DOI] [PubMed] [Google Scholar]
- 21.Goh JY, et al. Functional characterization of two novel parvulins in Trypanosoma brucei. FEBS Lett. 2010;584:2901–2908. doi: 10.1016/j.febslet.2010.04.077. [DOI] [PubMed] [Google Scholar]
- 22.Sun L, et al. Solution structural analysis of the single-domain parvulin TbPin1. PloS One. 2012;7:e43017. doi: 10.1371/journal.pone.0043017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mhadhbi M, et al. In vivo evidence for the resistance of Theileria annulata to buparvaquone. Vet Parasitol. 2010;169:241–247. doi: 10.1016/j.vetpar.2010.01.013. [DOI] [PubMed] [Google Scholar]
- 24.Sharifiyazdi H, Namazi F, Oryan A, Shahriari R, Razavi M. Point mutations in the Theileria annulata cytochrome b gene is associated with buparvaquone treatment failure. Vet Parasitol. 2012;187:431–435. doi: 10.1016/j.vetpar.2012.01.016. [DOI] [PubMed] [Google Scholar]
- 25.Konantz M, et al. Zebrafish xenografts as a tool for in vivo studies on human cancer. Ann N Y Acad Sci. 2012;1266:124–137. doi: 10.1111/j.1749-6632.2012.06575.x. [DOI] [PubMed] [Google Scholar]
- 26.White R, Rose K, Zon L. Zebrafish cancer: the state of the art and the path forward. Nat Rev Cancer. 2013;13:624–636. doi: 10.1038/nrc3589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Min SH, et al. Negative Regulation of the Stability and Tumor Suppressor Function of Fbw7 by the Pin1 Prolyl Isomerase. Mol Cell. 2012;46:771–783. doi: 10.1016/j.molcel.2012.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nateri AS, Riera-Sans L, Costa CD, Behrens A. The Ubiquitin Ligase SCFFbw7 Antagonizes Apoptotic JNK Signaling. Science. 2004;303:1374–1378. doi: 10.1126/science.1092880. [DOI] [PubMed] [Google Scholar]
- 29.Wei W, Jin J, Schlisio S, Harper JW, Kaelin WG. The v-Jun point mutation allows c-Jun to escape GSK3-dependent recognition and destruction by the Fbw7 ubiquitin ligase. Cancer Cell. 2005;8:25–33. doi: 10.1016/j.ccr.2005.06.005. [DOI] [PubMed] [Google Scholar]
- 30.Marsolier J, et al. OncomiR Addiction Is Generated by a miR-155 Feedback Loop in Theileria-Transformed Leukocytes. PLoS Pathog. 2013;9:e1003222. doi: 10.1371/journal.ppat.1003222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Moreau MF, et al. Theileria annulata in CD5+Macrophages and B1 B Cells. Infect Immun. 1999;67:6678–6682. doi: 10.1128/iai.67.12.6678-6682.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Aurrecoechea C, et al. ApiDB: integrated resources for the apicomplexan bioinformatics resource center. Nucleic Acids Res. 2007;35:D427–D430. doi: 10.1093/nar/gkl880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lambert C, Léonard N, Bolle XD, Depiereux E. ESyPred3D: Prediction of proteins 3D structures. Bioinformatics. 2002;18:1250–1256. doi: 10.1093/bioinformatics/18.9.1250. [DOI] [PubMed] [Google Scholar]
- 34.Ranganathan R, Lu KP, Hunter T, Noel JP. Structural and Functional Analysis of the Mitotic Rotamase Pin1 Suggests Substrate Recognition Is Phosphorylation Dependent. Cell. 1997;89:875–886. doi: 10.1016/s0092-8674(00)80273-1. [DOI] [PubMed] [Google Scholar]
- 35.Miteva MA, Tufféry P, Villoutreix BO. PCE: web tools to compute protein continuum electrostatics. Nucleic Acids Res. 2005;33:W372–W375. doi: 10.1093/nar/gki365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Grove LE, Hall DR, Beglov D, Vajda S, Kozakov D. FTFlex: accounting for binding site flexibility to improve fragment-based identification of druggable hot spots. Bioinformatics. 2013;29:1218–1219. doi: 10.1093/bioinformatics/btt102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lagorce D, Pencheva T, Villoutreix BO, Miteva MA. DG-AMMOS: A New tool to generate 3D conformation of small molecules using Distance Geometry and Automated Molecular Mechanics Optimization for in silico Screening. BMC Chem Biol. 2009;9:6. doi: 10.1186/1472-6769-9-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Spitzer R, Jain AN. Surflex-Dock: Docking benchmarks and real-world application. J Comput Aided Mol Des. 2012;26:687–699. doi: 10.1007/s10822-011-9533-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Thomsen R, Christensen MH. MolDock:_ A New Technique for High-Accuracy Molecular Docking. J Med Chem. 2006;49:3315–3321. doi: 10.1021/jm051197e. [DOI] [PubMed] [Google Scholar]
- 40.Sauton N, Lagorce D, Villoutreix BO, Miteva MA. MS-DOCK: Accurate multiple conformation generator and rigid docking protocol for multi-step virtual ligand screening. BMC Bioinformatics. 2008;9:184. doi: 10.1186/1471-2105-9-184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ji Q, et al. Molecular mechanism of quinone signaling mediated through S-quinonization of a YodB family repressor QsrR. Proc Natl Acad Sci. 2013;110:5010–5015. doi: 10.1073/pnas.1219446110. [DOI] [PMC free article] [PubMed] [Google Scholar]