SUMMARY
Antibodies developed during HIV-1 infection lose efficacy as the viral spike mutates. We postulated that anti-HIV-1 antibodies primarily bind monovalently because HIV’s low spike density impedes bivalent binding through inter-spike crosslinking, and the spike structure prohibits bivalent binding through intra-spike crosslinking. Monovalent binding reduces avidity and potency, thus expanding the range of mutations permitting antibody evasion. To test this idea, we engineered antibody-based molecules capable of bivalent binding through intra-spike crosslinking. We used DNA as a “molecular ruler” to measure intra-epitope distances on virion-bound spikes and construct intra-spike crosslinking molecules. Optimal bivalent reagents exhibited up to 2.5 orders-of-magnitude increased potency (>100-fold average increases across virus panels) and identified conformational states of virion-bound spikes. The demonstration that intra-spike crosslinking lowers the concentration of antibodies required for neutralization supports the hypothesis that low spike densities facilitate antibody evasion and the use of molecules capable of intra-spike crosslinking for therapy or passive protection.
INTRODUCTION
The HIV-1 envelope (Env) spike trimer, a trimer of gp120 and gp41 subunits, is the only target of neutralizing antibodies. The spike utilizes antibody-evasion strategies including mutation, glycan shielding, and conformational masking (West et al., 2014). While important, these features are not unique to HIV-1: other viruses employing these strategies elicit IgG antibody responses that provide sterilizing immunity or viral clearance. A potentially unique antibody-evasion strategy for HIV-1 involves hindering IgGs from using both antigen-binding fragments (Fabs) to bind bivalently to spikes (Klein and Bjorkman, 2010; Mouquet et al., 2010). This is accomplished by the small number and low density of Env spikes (Chertova et al., 2002; Liu et al., 2008; Zhu et al., 2006), which prevent most IgGs from inter-spike crosslinking (bivalent binding between spikes), and the architecture of the Env trimer, which impedes intra-spike crosslinking (bivalent binding within a spike trimer) (Klein et al., 2009; Luftig et al., 2006).
On a typical virus with closely-spaced envelope spikes, an IgG antibody can bind using both Fabs to crosslink neighboring spikes, leading to a nearly irreversible antibody-antigen interaction (Mattes, 2005). Avidity effects from bivalent binding of IgG antibodies have been shown to be critical for neutralization of many viruses, including polio and influenza (Icenogle et al., 1983; Schofield et al., 1997). By contrast, the small number of spikes (~14) present on the surface of HIV-1 (Chertova et al., 2002; Liu et al., 2008; Zhu et al., 2006) impedes simultaneous engagement of both antibody combining sites (Klein and Bjorkman, 2010; Mouquet et al., 2010): most spikes are separated by distances that far exceed the ~15 nm reach of the two Fab arms of an IgG (Liu et al., 2008; Zhu et al., 2006) (Figure 1A). Inter-spike crosslinking might still be possible if spikes could freely diffuse within the viral membrane. However, cryo-electron tomography of HIV-1 (Zhu et al., 2006) and evidence for interactions between the cytoplasmic tail of gp41 and the matrix protein of HIV (Bhatia et al., 2009; Crooks et al., 2008; Yu et al., 1992) suggest that a virion’s spike distribution is likely to be relatively static over time scales relevant to neutralization. Taken together, the mechanisms to hinder inter- and intra-spike crosslinking imply that most anti-HIV-1 IgGs bind monovalently to virions.
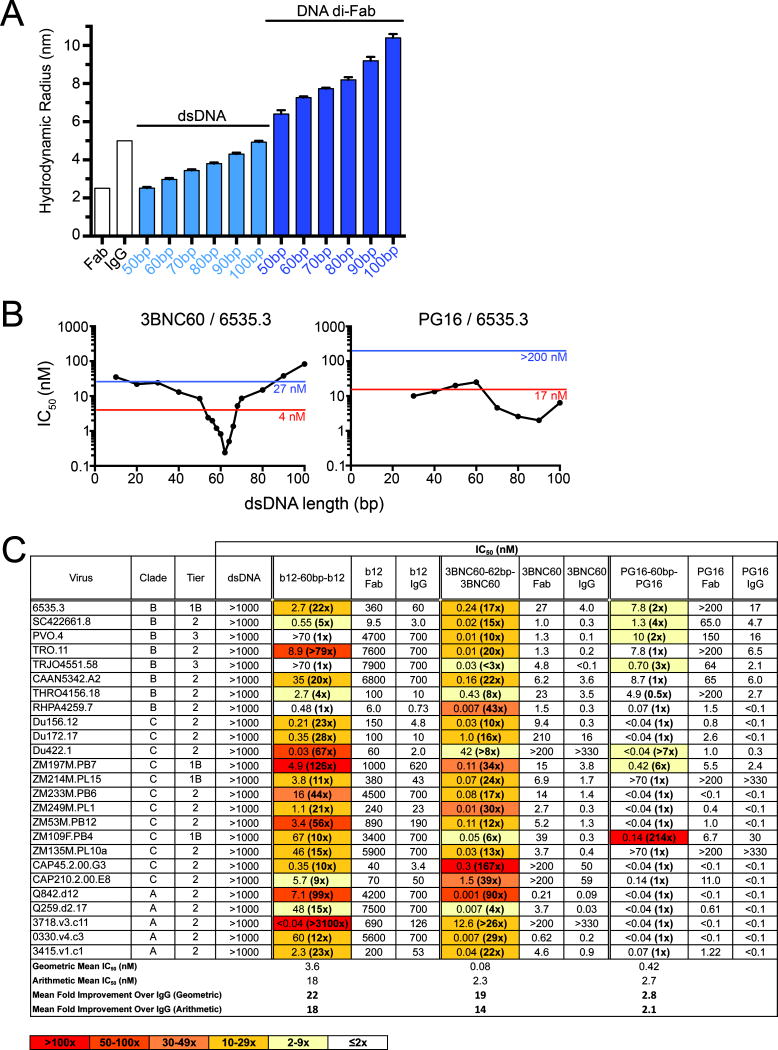
Figure 1. IgG and diFab reagents binding to viral spikes.
(A) Top: IgG binding monovalently to spikes on HIV-1 surfaces, which include a small number (~14) and low density of Env (Chertova et al., 2002; Liu et al., 2008; Zhu et al., 2006). Bottom: Homo-diFab reagent binding bivalently to HIV-1 Env by intra-spike crosslinking. Schematic representations of Env adapted from figures in (Liu et al., 2008). (B) Schematic of method used to produce homo- and hetero-diFabs. See also Figure S1.
It seems an unlikely coincidence that HIV-1, among the most adept of viruses at evading antibody-mediated neutralization, has an unusually low density of surface envelope spikes with restricted mobility as well as an unusually high mutation rate. We speculated that HIV-1 evolved a low spike density to hinder bivalent binding by antibodies (Klein and Bjorkman, 2010) and postulated that the combination of predominantly monovalent IgG binding and HIV-1’s rapid mutation rate creates an additional effective antibody evasion strategy (Klein and Bjorkman, 2010). If the affinity between an IgG Fab and a viral spike is high enough, monovalent IgG binding to a virion should not, in and of itself, hinder or prevent viral neutralization. Thus affinity-matured anti-Env IgGs raised against a particular strain of virus can effectively neutralize autologous virus (Klein et al., 2013; West et al., 2014). However, upon mutation of an antibody epitope on Env, the low affinity of the monovalent Fab-antigen interaction would result in either complete loss of neutralization or neutralization only at very high concentrations. These concepts are illustrated by comparisons of binding and neutralization for variants of IgG and Fab forms of palivizumab, a neutralizing IgG against respiratory syncytial virus (RSV) (Wu et al., 2005), a virus with a high density of Env spikes (Liljeroos et al., 2013). Palivizumab Fabs with fast off-rates/low affinities exhibited 2–3 log improvements in neutralization potencies when converted to bivalent IgGs and the potencies of the IgGs were not affected by mutations that increased the off-rates of their corresponding monovalent Fabs by >100-fold (Wu et al., 2005), illustrating the importance of avidity for IgGs with weak or moderate affinity Fabs. However, high affinity/slow off-rate palivizumab Fabs were equally as potent as their IgG counterparts, which could bind bivalently to RSV through inter-spike crosslinking. In the palivizumab example, binding and neutralization potencies were evaluated for a single strain of virus and antibodies. In the case of HIV-1, we are interested in the effects of mutations in the virus on binding of the same antibody, but the effects of mutation are expected to be similar. Thus we postulate that avidity effects through bivalent binding can serve as a buffer to dampen the effects of viral mutations on neutralization potencies of IgGs.
This line of reasoning suggests that bivalent HIV-1 binders would be optimal for passive prevention or immunotherapy, but because inter-spike distances are not constant even on a single virion, it is not possible to engineer reagents that could consistently accomplish inter-spike crosslinking. In contrast, reagents that can bind bivalently to a single trimeric spike would function independently of both spike density and distribution. (Pace et al., 2013) To test the idea that intra-spike crosslinking results in increased neutralization potency, we used molecular rulers to map epitopes on virion-bound HIV-1 spikes and created new molecules designed to synergize through bivalent interactions within single Env trimers (Figure 1A). We developed methods to produce multiple combinations of Env-binders separated by different distances by attaching broadly neutralizing antibody (bNAb) Fabs to variable-length double-stranded DNA (dsDNA) (Figure 1B; Figure S1). We chose dsDNA as a linker because its long persistence length (460–500 Å (Bednar et al., 1995) compared with ~30 Å for peptides (Zhou, 2004)) permits its use as a molecular ruler with 3.4 Å/basepair (bp) increments. Here we show that homo- and hetero-diFabs joined by optimal-length dsDNA bridges can achieve neutralization potency increases of 2–3 orders of magnitude and provide evidence that the synergy results from intraspike crosslinking. Upon determining the optimal distances between Env trimer-bound Fabs, we show that it is possible to convert the dsDNA bridge to a protein linker to create a protein-based reagent with similar synergistic properties. These results illustrate the importance of avidity in antibody-pathogen interactions, elucidate mechanisms by which HIV-1 evades the host immune system, and are relevant to the choice of potential protein therapeutics to be delivered to prevent or treat HIV-1 infections.
RESULTS
Homo-diFabs exhibit length-dependent avidity effects consistent with intra-spike crosslinking
Fabs were modified to contain a free thiol and then conjugated to maleimide-activated single-stranded DNA (ssDNA) (Figure 1B). Different lengths of dsDNA (designed to lack secondary structures (Zadeh et al., 2011) (Supplementary Experimental Procedures) were annealed with and ligated to the ssDNA-Fab conjugates to create homo- or hetero-diFabs, in which the two Fabs were the same or different, respectively. Dynamic light scattering confirmed that conjugates with longer DNA bridges were more extended (Figure 2A), supporting the use of dsDNA as a ruler. Inter-Fab distances calculated from dsDNA lengths were regarded as approximate because the DNA linkers included short regions of ssDNA (persistence length 22 Å) (Chi et al., 2013) to permit orientational flexibility.
Figure 2. Characterization of homo-diFabs.
(A) Dynamic light scattering measurements of hydrodynamic radii for IgG and Fab proteins, different lengths of dsDNA alone, and di-Fabs with different dsDNA linkers. (B) Effects of dsDNA bridge length on neutralization potencies of 3BNC60 and PG16 homo-diFabs against the Tier 1B HIV-1 strain 6535.3. Neutralization IC50s are plotted against the length of the dsDNA linker. IC50s for the parent IgG and Fab are indicated as red and blue lines, respectively. (C) Neutralization of primary HIV-1 strains by b12 and PG16 homo-diFabs, each constructed with a 60bp dsDNA bridge. IC50s are reported for the homo-diFabs, the parental Fabs and IgGs, and dsDNA alone. As a measure of potential synergy, the molar ratio of the IC50 values for the IgG and the homo-diFab is listed for each strain in parentheses beside the IC50 for the homo-diFab. See also Figure S2.
We first determined the optimal dsDNA linker for a homo-diFab constructed from 3BNC60, a bNAb against the CD4 binding site (CD4bs) on the gp120 subunit of Env (Scheid et al., 2011), by evaluating homo-diFabs with different dsDNA lengths using in vitro neutralization assays. The 50% inhibitory concentrations (IC50s) against HIV-1 strain 6535.3 depended on the dsDNA length, with the most potent homo-diFab containing a bridge of 62bp (211 Å) (Figure 2B; Figure S2). This length is close to the predicted distance (~198 Å) between the C-termini of adjacent 3BNC60 Fabs bound to the open structure of an HIV-1 trimer (Merk and Subramaniam, 2013) (Figure 3, Figure S3). Bridge lengths of ~60bp also exhibited the best potencies for 3BNC60 homo-diFabs against DU172.17 HIV-1 and for homo-diFabs constructed from VRC01 (Wu et al., 2010), a related CD4bs bNAb (Figure S2). The ~100-fold increased potency of 3BNC60-62bp-3BNC60 compared with 3BNC60 IgG against HIV-1 6535.3 (Figure 2B) suggested synergy resulting from avidity effects due to bivalent binding. The bivalent interaction likely resulted from intra-spike crosslinking rather than inter-spike crosslinking since the latter should not manifest with a sharp length-dependence because inter-spike distances are variable within and between virions (Liu et al., 2008; Zhu et al., 2006).
Figure 3. Comparison of intra-spike distances for three conformations found for virion-associated HIV-1 Env spike trimers.
(A) Three conformations of Env trimers shown as surface representations (top row: gp120 coordinates only) and schematically (bottom two rows). Schematic representations of Env trimers adapted from figures in (Liu et al., 2008). Env spikes are shown as seen from above (top and middle rows) and the side (bottom row). V1V2 loops are cyan, V3 loops are purple, the CD4 binding site is yellow, the remainder of gp120 is maroon, gp41 is green, and the membrane bilayer is gray. The closed structure (PDB code 4NCO) was observed for unliganded trimers (Liu et al., 2008) and trimers associated with Fabs from potent VRC01-like (PVL) antibodies (Lyumkis et al., 2013; Merk and Subramaniam, 2013). The open structure was observed for trimers associated with CD4 or the Fab from the CD4-induced antibody 17b (Merk and Subramaniam, 2013; Tran et al., 2012) (coordinates obtained from S. Subramaniam). The partially-open structure was observed for trimers associated with the Fab from b12 (Liu et al., 2008; Merk and Subramaniam, 2013) (PDB code 3DNL). (B) Measured distances between homo-diFabs bound to HIV-1 trimer structures. Fabs from the indicated bNAbs shown bound to the gp120 portions of Env in the three conformation shown in panel A. Fabs are shown as ribbons; gp120 subunits are shown as surface representations with V1V2 loops in cyan, V3 in purple, the CD4 binding site in yellow, and the remainder of gp120 in maroon. The distance between the Cys233heavy chain carbon-α atoms of adjacent bound Fabs is indicated by a gray line as an approximation of an optimal length for a dsDNA bridge attached to Cys233heavy chain. Assuming three-fold symmetry of trimers, only one distance is possible for bound 3BNC60, b12 and 10-1074 homo-diFabs. See also Figure S3.
To formally assess the extent to which inter-spike crosslinking could contribute to synergy, we evaluated homo-diFabs constructed from the V1V2 loop-specific bNAb PG16 (Walker et al., 2009), which cannot crosslink within a single spike because only one anti-V1V2 Fab binds per Env trimer (Julien et al., 2013b). PG16 homo-diFabs with different dsDNA bridges did not exhibit length-dependent neutralization profiles against strain 6535.3 (Figure 2B) and other viral strains (Figure S2D). However, increased potencies were observed for PG16 homo-diFabs with ≥ 70bp or 80bp (≥248 Å or 272 Å) bridges, perhaps reflecting increased inter-spike crosslinking with longer separation distances (Figure 2B; Figure S2D).
Comparison of homo-diFabs that can or cannot exhibit intra-spike crosslinking
To evaluate the potential for intra-spike crosslinking across different viral strains, we compared homo-diFabs designed to be capable (b12 and 3BNC60) or incapable (PG16) of intra-spike crosslinking (Figure 2C). To minimize inter-spike crosslinking, the homo-diFabs were constructed with 60–62bp bridges. The b12-60bp-b12 homo-diFab exhibited increased potency compared with b12 IgG in 21 of 25 strains in a cross-clade panel of primary HIV-1, with potency increases ≥ 10-fold for 16 strains and a geometric mean potency increase of 22-fold. 3BNC60-62bp-3BNC60 showed even more consistent synergy, being more potent than 3BNC60 IgG against all 25 strains tested, with ≥10-fold increases for 20 strains and a mean increase of 19-fold. By contrast, the PG16-60bp-PG16 homo-diFab showed potency increases compared with PG16 IgG against only six strains, with relatively small (2- to 7-fold) increases in five strains and an overall 2.8-fold mean potency change.
Hetero-diFabs exhibit dramatic potency increases consistent with intra-spike crosslinking
To determine whether heterotypic bivalent binding can produce synergy and to measure distances between epitopes, we used dsDNA to link Fabs recognizing different epitopes on gp120. We first evaluated hetero-diFabs constructed with Fabs from V1V2 (PG16 or PG9) (Walker et al., 2009) and CD4bs (b12 or 3BNC60) (Roben et al., 1994; Scheid et al., 2011) bNAbs linked with 60bp dsDNA bridges. PG16-60bp-b12 hetero-diFabs were evaluated in neutralization assays against HIV-1 strains SC4226618 (more sensitive to b12 than PG16) and CAP210 (more sensitive to PG16 than b12). According to the model being tested, in the absence of synergistic binding; i.e., when only one Fab can bind to a spike at a time, a hetero-diFab would be no more potent than a non-covalent mixture of the dsDNA and the two Fabs against each viral strain, whereas synergistic binding would result in avidity effects exhibited by increased potency of the hetero-diFab. For both viral strains, the PG16-60bp-b12 hetero-diFab was ~10-fold more potent than the mixture of Fabs plus dsDNA or the more potent of the two Fabs alone (Figure 4; Figure S4). To more systematically explore potential synergy, we evaluated PG16-60bp-b12 against a 25-member panel of HIV-1 strains, finding synergistic effects (between 2- and 145-fold more potent than the corresponding non-covalent mixture for most strains; geometric mean improvement of 4.7-fold) (Table S1). When Fabs from PG16 or PG9 were combined with a more potent CD4bs-recognizing bNAb (3BNC60), the resulting hetero-diFabs exhibited greater synergy – several examples of >150-fold improvement for PG16-60bp-3BNC60 and PG9-60bp-3BNC60 and geometric mean potency improvements of 29- and 68-fold, respectively (Figure 4, Tables S2,S3). Other hetero-diFabs, constructed with combinations of Fabs recognizing the CD4bs (3BNC60 (Scheid et al., 2011)), the gp120 V3 loop (10-1074 (Mouquet et al., 2012)), and a gp41 epitope (10E8 (Huang et al., 2012)), also showed synergistic effects (Figure 4, Table S4), and a 3BNC60-60bp-b12 hetero-diFab exhibited up to 660-fold synergy and a geometric mean potency increase of 90-fold (Figure 4, Table S5). In contrast, analogous IgG heterodimers, constructed with two different Fabs linked to a single Fc (Schaefer et al., 2011), did not show synergy when evaluated against the same viruses, demonstrating that synergistic effects required optimal separation distances that permitted each Fab to achieve its specific binding orientation (Figure S4; Tables S1–S5). We conclude that hetero-diFabs can achieve synergy through simultaneous recognition of two different epitopes on the same HIV-1 Env trimer.
Figure 4. Synergistic dsDNA-based hetero-diFabs.
Neutralization of primary HIV-1 strains by hetero-diFabs. IC50s are reported for the hetero-diFabs. See Tables S1–S5 for IC50s of parental Fabs and IgGs, dsDNA alone, and the non-covalent mixtures of Fabs and dsDNA. As a measure of potential synergy of each hetero-diFab, the molar ratio of the IC50 values for the non-covalent mixture and the hetero-diFab is listed for each strain in parentheses beside the IC50 for the hetero-diFab. NT = not tested. See also Figure S4.
To more precisely define optimal intra-epitope separation distances, we evaluated hetero-diFabs with different bridge lengths, finding length-dependent synergy effects. For example, PG16-3BNC60 hetero-diFabs with 40bp and 50bp dsDNA bridges showed improved neutralization potencies when compared to the 60bp (204 Å) version, achieving ≥100-fold potency increases against over half of the tested strains and geometric mean improvements of 98- and 107-fold respectively (Figure 4, Table S4). The 40bp and 50bp bridges (136 Å and 170 Å, respectively) corresponded to the approximate separation distances between PG16 and 3BNC60 Fabs when bound to the same gp120 within a trimer (147 Å) or to neighboring protomers within open or partially-open trimers (167 Å) (Figure S3). In a second length dependency example, 10-1074-40bp-3BNC60 was more potent than 10-1074-60bp-3BNC60 (Figure 4, Table S4). The ~136 Å distance between the two Fabs in 10-1074-40bp-3BNC60 corresponded to the approximate separation between these Fabs bound to the same gp120 (141 Å), while 60bp more closely approximated Fabs bound to neighboring protomers on an open trimer (193 Å) (Figure S3). The 40bp and 50bp versions of 10E8-3BNC60 showed consistent synergy (Figure 4, Table S4); however, the lack of structural information concerning 10E8 binding to Env trimer hindered interpretation of 10E8-containing hetero-diFabs.
A hetero-diFab constructed with a protein linker exhibits synergistic potency increases
Bivalent molecules involving dsDNA linkers were effective for demonstrating synergistic neutralization, but a protein reagent would be preferable as an anti-HIV-1 therapeutic. We recently described a series of protein linkers of various lengths and rigidities (Klein et al., 2014) that can mimic the properties of different lengths of dsDNA. Thus we can substitute a comparable protein linker for an optimal dsDNA bridge to create a protein reagent capable of simultaneous binding to two different epitopes on a single HIV-1 spike trimer. As a proof-of-principle example, we used sortase-catalyzed protein ligation and click chemistry (Witte et al., 2013) to construct a bivalent reagent analogous to PG16-40bp-3BNC60 by substituting the dsDNA linker with 12 domains of a designed tetratricopeptide-repeat (TPR) protein (Kajander et al., 2007) (Figure 5A; Figure S5). We chose a TPR linker because tandem repeats of TPR domains form a rigid rod-like structure whose length corresponds predictably with the number of repeats, with each domain contributing ~10 Å (Kajander et al., 2007). PG16 Fab was expressed with a C-terminal sortase signal, and the C-terminus of the 3BNC60 Fab was modified to include twelve TPR repeats and a sortase signal. The tagged Fabs were covalently attached to peptides containing click handles using sortase-catalyzed ligation, and then incubated to allow the click reaction to form PG16 Fab linked to 3BNC60 Fab by twelve TPR repeats (PG16-TPR12-3BNC60). Together with the remnants of the click handles, the linker would occupy ~131 Å, approximately the same length as the dsDNA linker in PG16-40bp-3BNC60 reagent (Figure 5A; Figure S5). The protein-based molecule, PG16-TPR12-3BNC60, exhibited between 11- and >200-fold synergy against 12 primary HIV-1 strains (Figure 5B; 33-fold geometric mean increased potency).
Figure 5. Synergistic protein-based hetero-diFab.
(A) Schematic representation of PG16-TPR12-3BNC60 (not to scale). Approximate lengths are indicated (120 Å for the TRP12 linker plus ~11 Å for the fused click handles). (B) Neutralization of primary HIV-1 strains. IC50s are reported for PG16-TPR12-3BNC60, the parental components of the reagent (PG16 Fab and 3BNC60 Fab-TPR12), and TPR12 alone. As a measure of potential synergy of PG16-TPR12-3BNC60, the molar ratio of the IC50 values for the most potent component and PG16-TPR12-3BNC60 is listed for each strain in parentheses beside the IC50 for PG16-TPR12-3BNC60. See also Figure S5.
Simulations of the effects of avidity on IgG binding to tethered antigens
To better understand the effects of avidity arising from bivalent binding of IgGs to antigens tethered to a surface such as a viral membrane, we used modeling software to simulate the saturation of surface-bound antigens by monovalent Fabs and bivalent IgGs. We chose a 1 hour incubation time based upon conditions under which in vitro neutralization assays are conducted (Montefiori, 2005). We varied the density of the tethered antigens, the concentrations of Fab or IgG, and investigated a range of intrinsic association and dissociation rate constants for the binding interaction. The fraction of antigen bound by a Fab or IgG was calculated as a function of on- and off-rates (ka and kd), whose ratio (kd/ka) is equal to the affinity (KD, or equilibrium dissociation constant). We compared saturation by Fabs (top row), IgGs in which only monovalent binding was permitted (center row), and IgGs that bound bivalently through crosslinking of neighboring antigens (bottom row) (Figure 6A). As expected, saturation by Fabs and IgGs was nearly identical for monovalent binding conditions (Figure 6A, first two rows). By contrast, across a range of input concentrations, there were ka and kd combinations for IgGs binding bivalently that exhibited saturation binding under conditions in which monovalent Fabs and IgGs binding monovalently did not (Figure 6A, bottom row). Thus, consistent with experimental results in the palivizumab/RSV system (Wu et al., 2005), the simulations suggested that bivalency through crosslinking can rescue binding of IgGs whose Fabs exhibit weak binding affinities as a result of fast dissociation rate constants, whereas IgGs whose Fabs exhibit high affinities because of slow dissociation rates did not display strong avidity enhancement.
Figure 6. Simulations of avidity effects due to bivalent binding of IgG to a tethered antigen.
(A) The fraction of tethered antigen bound by different concentrations of IgG or Fab after 1 hour shown as a heat map (cooler colors representing a lower percentage bound and warmer colors representing a higher percentage bound) as a function of kinetic constants for the IgG-antigen or Fab-antigen interaction. The fraction of antigen bound by a Fab or IgG was calculated as a function of ka and kd. The intrinsic affinities are strongest in the lower right corner (1 pM) and weakest in the upper left corner (100 mM) of each graph. For IgG, binding was forced to 100% monovalent binding (middle row) or 100% bivalent binding (bottom row). Saturation by Fabs and IgGs was nearly identical for monovalent binding conditions because the binding kinetics of IgGs would be enhanced by at most 2-fold. Comparisons of the simulations for bivalent binding (bottom row) and monovalent binding (top two panels) showed regions of saturation binding resulting from avidity effects. (B) The fraction of antigen bound as a function of time for IgGs binding to surface-tethered antigens at an input concentration of 10 nM. When the dissociation rate constant of the Fab portion of the IgG is slow (top panel) and the input concentration is approximately 100-fold higher than the affinity of the Fab, IgGs can reach saturation binding after an hour whether binding monovalently or bivalently to the surface – hence avidity effects are not apparent after an hour. However, weakening the affinity of the Fab by making the dissociation rate 1000-fold faster (bottom panel) prevents saturation when binding monovalently, but has no affect on saturation when binding bivalently – hence avidity effects are apparent throughout the incubation.
The simulations also demonstrate that the effects of avidity on binding are a complicated mixture of kinetics, input concentration, and incubation time. At any particular concentration, the threshold at which avidity is observed is controlled by kinetics rather than affinity because different combinations of kinetic constants yield the same KD. The kinetic threshold at which avidity effects are observed varies depending on the difference between the input concentration and the KD. For concentrations near or below the KD, there is a kinetic threshold such that for on- and off-rates slower than ~103 M−1s−1 and ~10−5 s−1, respectively, avidity enhancement is not observed (Figure 6A). The binding reactions are also affected by the length of incubation, such that the lower the input concentration, the longer it takes to reach saturation (Figure 6B).
We note that the simulations model binding interactions only, whereas our homo- and hetero-diFabs were evaluated for their ability to enhance neutralization of viral infectivity, a process more complicated than binding. For example, neutralization mechanisms may involve conformational changes in Env that were not accounted for in our binding simulation. In addition, kinetics constants for antibody-mediated neutralization of HIV-1 are not known; nor is the fraction of Env spikes on a virion that are required for neutralization or for fusion. In any case, it appears that the kinetic properties of the bNAb Fab components in our reagents were appropriate to realize avidity-enhanced neutralization since hetero-diFab reagents displayed ~100-fold mean improved neutralization potencies. The data therefore support the hypothesis that intra-spike crosslinking by anti-HIV-1 binding molecules represents a valid strategy for increasing potency and resistance to HIV-1 Env mutations.
DISCUSSION
We engineered new HIV-1 spike-binding molecules designed to bind bivalently to demonstrate the importance of avidity effects in antibody efficacy in HIV-1 neutralization and to establish that lack of bivalent binding by physiologic IgGs is an additional antibody evasion strategy utilized by HIV-1.
The importance for HIV-1 in maintaining a low spike density to avoid inter-spike crosslinking by IgGs was suggested by the relatively small improvements in neutralization potencies of intact anti-HIV-1 IgGs compared with their Fab counterparts (Klein and Bjorkman, 2010) and by the discovery that polyreactivity increased the apparent affinity of anti-HIV-1 antibodies through a mechanism of heteroligation (Mouquet et al., 2010). Comparison of the neutralization potencies of IgGs versus Fabs in the current study provides further support for the observation that anti-HIV-1 IgGs generally exhibit relatively small increased potencies compared to Fabs. To quantify potential avidity effects, we previously defined the molar neutralization ratio (MNR) for IgG versus Fab forms of an antibody as IC50 Fab (nM)/IC50 IgG (nM). In the absence of avidity or other advantages of the IgG compared with the Fab (e.g., increased size), the ratio would be 2.0 (Klein and Bjorkman, 2010). In the current study, the mean MNR for PG16, an IgG that cannot exhibit intra-spike crosslinking (Julien et al., 2013b), was 8.0 (data from Figure 2C), similar to the 10.5 mean MNR in a previous study (West et al., 2012). These values are lower than MNRs observed for IgGs against densely-packed viruses, which can be over 1000, but are consistent with a limited amount of inter-spike crosslinking by anti-HIV-1 IgGs whose epitopes on neighboring spikes are accessible to simultaneous engagement of the combining sites of the two Fabs of an IgG, which are separated by ~150 Å (Klein and Bjorkman, 2010). Our current results suggested that inter-spike crosslinking can be increased by creating homo-diFabs with 70bp–100bp dsDNA linkers (Figure 2B, Figure S2D). These linkers would separate the combining sites of the Fabs by ≥240–340 Å, distances that should enhance inter-spike crosslinking.
Indirect evidence for the hypothesis that HIV-1 evolved a low spike density to avoid interspike crosslinking IgGs comes from studies of a cytoplasmic tail deletion in the simian immunodeficiency virus (SIV) spike trimer. Cytoplasmic tail deletion has been suggested to increase the number of spikes per virion (Zingler and Littman, 1993) and/or the spike mobility in the virion bilayer (Crooks et al., 2008), both of which could enhance inter-spike crosslinking. Although tail-deleted mutant viruses can be produced in vitro, propagation of the virus in macaques favors viruses containing the full-length envelope spike (Zingler and Littman, 1993). These findings are consistent with the idea that an intact host immune system selects against those viruses that facilitate the ability of host IgGs to bind bivalently through inter-spike crosslinking.
Here we present a method to create potential intra-spike crosslinking antibody-based molecules using dsDNA- and protein-based linkers, and demonstrate that these reagents can exhibit up to three orders of magnitude increases in neutralization potency. We argue that the optimized versions of our new molecules achieve potency increases through intra-spike, rather than inter-spike, crosslinking because (i) distances measured between epitopes on virion-bound spike trimers corresponded to approximate intra-epitope distances on HIV-1 spike trimer structures, and (ii) increases in inter-spike crosslinking by homotypic and heterotypic reagents should not exhibit sharp linker length-dependent neutralization potencies since distances between spikes vary within a single virion and between virions. The latter point is valid even if HIV-1 spikes are clustered on mature virions, as suggested by fluorescence nanoscopy (Chojnacki et al., 2012), but not cryoelectron microscopy (Liu et al., 2008; Zhu et al., 2006). Whether HIV-1 spikes cluster upon encountering a target cell to form an entry claw (Sougrat et al., 2007) is not relevant to the mechanism of action of our reagents, since neutralization assays are conducted by incubating potential inhibitors with virions prior to addition of target cells (Montefiori, 2005), a mechanism that is also presumably relevant for most in vivo interactions of antibody and antibody-like inhibitors. Because avidity effects require recognition of two or more antigens tethered to the same surface, another potential action of our reagents, inter-virion crosslinking, would not result in avidity effects by analogy to the lack of avidity enhancement for an IgG binding two soluble antigens, one per Fab. In this respect, we note that although IgAs are capable of inter-virion crosslinking (Stieh et al., 2014), conversion of IgG bNAbs to IgAs did not result in potency increases (Kunert et al., 2004; Wolbank et al., 2003).
The use of dsDNA- and protein-based molecular rules to measure inter-epitope distances presented here can be used to probe conformations of virion-bound Env trimers. By contrast, EM and X-ray structures (Bartesaghi et al., 2013; Julien et al., 2013a; Lyumkis et al., 2013; Pancera et al., 2014) cannot capture dynamic information concerning Env conformations during neutralization. Single-molecule fluorescence resonance energy transfer (smFRET) measurements suggested that Env trimers on the surface of HIV-1 virions transition between different conformations (Munro et al., 2014), and spike trimers have been visualized by EM in different conformations: the closed structure of unliganded trimers and trimers associated with VRC01-like bNAbs (Bartesaghi et al., 2013; Liu et al., 2008; Lyumkis et al., 2013) (also observed in Fab-bound crystal structures (Julien et al., 2013a; Pancera et al., 2014)), a CD4-and/or 17b-bound open structure (Liu et al., 2008; Tran et al., 2012), and a partially-open b12-bound structure (Liu et al., 2008) (Figure 3A). Homo- and hetero-diFabs joined by different lengths of dsDNA bridges offer a new methodology to probe Env trimer conformational states on virions and potentially to address strain-specific conformational differences.
Homo-diFabs constructed from VRC01-like bNAbs showed greatest potency when binding to epitopes separated by distances most closely approximating the open structure (Liu et al., 2008; Tran et al., 2012), rather than the closed structure observed for soluble and virion-associated spike trimers bound to VRC01-like Fabs (Bartesaghi et al., 2013; Liu et al., 2008; Lyumkis et al., 2013) (Figure 2B, 3; Figure S3–S4). These results suggest that optimal intraspike crosslinking molecules can inhibit a different state than recognized by monovalent Fabs binding to spike trimers in static EM and X-ray structures (Bartesaghi et al., 2013; Liu et al., 2008; Lyumkis et al., 2013; Merk and Subramaniam, 2013; Tran et al., 2012). If so, one Fab of a homo-diFab could first bind to its epitope on a closed trimer, allowing the second Fab to latch on to a transiently-populated open form of that trimer. Alternatively, binding of the first Fab may trap the trimer into a conformation allowing increased accessibility of the second Fab, or both Fabs could bind simultaneously to a transiently-appearing open trimer. Interestingly, the distance dependence of two CD4bs antibodies, 3BNC60 and b12, was more strongly pronounced for a Tier 1B HIV-1 strain, 6535.3, than for Tier 2 or 3 strains against which the homo-diFabs were tested (Figure 2B, Figure S2). Tier categorization of HIV-1 strains refers to the sensitivity of a strain to antibody neutralization, with Tier 1 strains being more sensitive in general to antibodies than Tier 2 or 3 strains (Seaman et al., 2010). The differences in length dependence for CD4bs homo-diFabs may reflect differences in conformational variability within Env trimers from different tiers, with Tier 1 Env perhaps more easily able to adopt the open conformations likely recognized by the CD4bs antibodies with optimal bridge lengths.
For the PG16-3BNC60 hetero-diFabs, the optimal 40bp and 50bp bridge lengths (136 Å and 170 Å, respectively) corresponded to the approximate separation distances between PG16 and 3BNC60 Fabs when bound to the same gp120 within a trimer (147 Å) or to neighboring protomers within open or partially-open trimers (167 Å) (Figure S3). In a second hetero-diFab bridge length dependency example, 10-1074-40bp-3BNC60 was more potent than 10-1074-60bp-3BNC60 (Figure 4, Table S4). The ~136 Å distance between the two Fabs in 10-1074-40bp-3BNC60 corresponded to the approximate separation between these Fabs bound to the same gp120 (141 Å), while 60bp more closely approximated Fabs bound to neighboring protomers on an open trimer (193 Å) (Figure S3). In general, it is more difficult to deduce information about Env trimer conformations recognized by hetero-diFabs because the intra-epitope distance is the same in the three conformations for Fabs binding to the same gp120 subunit within an Env trimer (Figure S3), and length-dependence data for some of the hetero-diFabs, e.g., 10-1074-40bp-3BNC60, was consistent with binding to a single gp120 within an Env trimer as well as to adjacent gp120s (Figure S2). However, whether binding to the same or to adjacent protomers within the spike trimer, the increased synergy of optimal hetero-diFabs suggested a mechanism in which the more potent/tighter-binding Fab of the hetero-diFab initially bound to the viral spike, thereby allowing the second Fab, even when only weakly neutralizing on its own, to attach.
In summary, our results demonstrated that optimal length homo- and hetero-diFabs are capable of synergistic effects that increased neutralization potencies, and in some cases, allowed neutralization of viral strains resistant to conventional IgGs. These results are consistent with the hypothesis that most anti-HIV-1 IgGs bind monovalently to single Env spikes, which leaves them vulnerable to Env mutations that weaken monovalent interactions but would still permit bivalent interactions (Klein and Bjorkman, 2010). The demonstration that anti-HIV-1 reagents designed to be capable of intra-spike binding with avidity can more potently and broadly neutralize HIV-1 than conventional anti-spike IgGs is relevant to the choice of anti-HIV-1 proteins or genes to be delivered passively to prevent infection or suppress active infections. Bispecific antibodies that simultaneously bind to HIV-1 Env and to CD4 or CCR5 host receptors on the target cell represent a conceptually distinct method to increase the potency and breadth of anti-HIV-1 reagents (Pace et al., 2013). In contrast to these reagents, antibodies that achieve synergy via bivalent binding to Env by intra-spike crosslinking offer significant advantages for passive delivery; for example, neutralizing antibodies against HIV-1 Env protect more effectively in vivo than antibodies against CD4 (Pegu et al., 2014), and anti-self antibodies such as anti-CD4 IgGs have short half-lives in vivo (Bruno and Jacobson, 2010). We propose that the ideal therapeutic molecule would utilize avidity achieved by intra-spike crosslinking to reduce the concentration required for sterilizing immunity and render the low spike density of HIV-1 irrelevant to its efficacy. Moreover, analogous to using several drugs or antibodies during antiretroviral therapy, simultaneous binding to different HIV-1 epitopes should reduce or abrogate sensitivity to Env mutations.
EXPERIMENTAL PROCEDURES
Expression and purification of Fabs
Genes encoding IgG light chain genes were modified by site-directed mutagenesis to replace Cys263Light Chain, the C-terminal cysteine that forms a disulfide bond with Cys233Heavy Chain, with a serine. Modified light chain genes and genes encoding 6x-His- or StrepII-tagged Fab heavy chains (VH–CH1–tag) were subcloned separately into the pTT5 mammalian expression vector (NRC Biotechnology Research Institute). Fabs were expressed by transient transfection in HEK 293-6E (NRC Biotechnology Research Institute) cells as described (Diskin et al., 2011) and purified from supernatants by Ni-NTA or StrepII affinity chromatography followed by size exclusion chromatography in PBS pH 7.4 using a Superdex 200 10/300 or Superdex 200 16/600 column (Amersham Biosciences).
IgG heterodimers were expressed and purified as described in the Supplementary Experimental Procedures.
DNA conjugation to Fabs
DNA was conjugated to free thiol-containing Fabs using a modified version of a previously-described protocol (Hendrickson et al., 1995). Briefly, Fabs were reduced in a buffer containing 10mM TCEP-HCl pH 7–8 for two hours, and then buffer exchanged three times over Zeba desalting columns (Thermo Scientific). The percentage of reduced Fab was determined using Invitrogen’s Measure-IT Thiol Assay. Concurrently, a 5–20 base ssDNA containing a 5′ amino group (Integrated DNA Technologies, IDT-DNA) was incubated with a 100-fold molar excess of an amine-to-sulfhydryl crosslinker (Sulfo-SMCC; Thermo Scientific) for 30 minutes to form a maleimide-activated DNA strand, which was buffer exchanged as described above. The reduced Fab and activated ssDNA were incubated overnight, and the Fab-ssDNA conjugate was purified by Ni-NTA or StrepII affinity chromatography (GE Biosciences) to remove unreacted Fab and ssDNA.
ssDNA was synthesized, phosphorylated, and PAGE purified by Integrated DNA Technologies. For di-Fabs containing dsDNA bridges longer than 40bp, complementary ssDNAs were annealed by heating (95°C) and cooling (room temperature) to create dsDNA containing overhangs complementary to the Fab-ssDNA conjugates. dsDNA was purified by size exclusion chromatography (Superdex 200 10/300) and incubated overnight with the corresponding tagged Fab-ssDNA conjugates. Homo- and hetero-diFab reagents were purified by Ni-NTA and StrepII affinity chromatography when appropriate to remove free DNA and excess Fab-ssDNA conjugates, treated with T4 DNA ligase (New England Biolabs), and purified again by size exclusion chromatography (Figure S1B). To make di-Fabs containing dsDNA bridge lengths less than 40bp, two complementary ssDNA-conjugated Fabs were incubated at 37°C without a dsDNA bridge and then purified as described above. Protein-DNA reagents were stable at 4°C for >6 months as assessed by SDS-PAGE.
Bridge and linker sequences are listed in Supplementary Experimental Procedures.
Characterization of DNA-Fab reagents
Fractions from the center of an SEC elution peak were concentrated using Amicon Ultra-15 Centrifugal Filter Units (Millipore) (MW cutoff = 10 kDa) to a volume of 500 μL, and DLS measurements were performed on a DynaPro® NanoStar™ (Wyatt Technology) using the manufacturer’s suggested settings. Hydrodynamic radii were determined as described (Dev and Surolia, 2006). Briefly, a nonlinear least squares fitting algorithm was used to fit the measured correlation function to obtain a decay rate. The decay rate was converted to the diffusion constant that can be interpreted as the hydrodynamic radius via the Stokes-Einstein equation.
Hetero-diFab with TPR linker
PG16-TPR12-3BNC60, a C-to-C linked hetero-diFab containing 12 consensus tetratricopeptide-repeat (TPR) domains (Kajander et al., 2007) as a protein linker (Klein et al., 2014), was prepared from modified PG16 and 3BNC60 Fabs using a combination of sortase-catalyzed peptide ligation and click chemistry (Witte et al., 2013). The C-terminus of the PG16 Fab heavy chain was modified to include the amino acid sequence GGGGASLPETGGLNDIFEAQKIEWHEHHHHHH, comprising a flexible linker, the recognition sequence for S. aureus Sortase A (underlined), a BirA tag, and a 6x-His tag. The C-terminus of The 3BNC60 Fab heavy chain C-terminus was modified to include a (Gly4Ser)3 linker followed by 12 tandem TPR domains and the amino acid sequence ASGGGGSGGGGSGGGGSLPETGGHHHHHH, comprising a second (Gly4Ser)3 linker, the Sortase A recognition sequence (underlined), and a 6x-His tag. The Fabs were expressed in HEK-6E cells and purified with Ni-NTA and gel filtration chromatography as described above. Peptides (GGGK with C-terminal azide and cyclooctyne click handles) were synthesized by GenScript, and sortase-catalyzed peptide ligation was used to attach the azide-containing peptide to PG16 Fab and the cyclooctyne-containing peptide to the 3BNC60-TPR12 fusion protein as described (Guimaraes et al., 2013). Approximate yields after each sortase reaction were ~30%. Peptide-ligated PG16 and 3BNC60 Fabs were passed over a Ni-NTA column to remove His-tagged enzyme and Fabs that did not lose their His tags during the reaction, mixed at equimolar ratios, and the click reaction was accomplished by incubating overnight at 25°C. The approximate yield for the click reaction was ~65%. The resulting PG16-TPR12-3BNC60 hetero-diFab was purified by size exclusion chromatography to remove unreacted Fabs for an overall yield of ~22%.
Measurements of intra-spike distances
To derive predicted distances between two adjacent Fab bound to HIV-1 Env, we superimposed Fabs bound to their epitopes on the structures of Env trimers in three different conformations: closed (a 4.7 Å crystal structure of a gp140 SOSIP trimer; PDB code 4NCO), open (a 9 Å EM structure of a SOSIP trimer–17b Fab complex (Tran et al., 2012); coordinates obtained from S. Subramaniam), partially-open (an ~20 Å EM structure of a viral spike bound to b12 Fab; PDB code 3DNL). The positions of the CH1 and CL domains in Fab structures used for docking were adjusted to create Fabs with the average elbow bend angle found in a survey of human Fab structures (Stanfield et al., 2006). The VH-VL domains of the adjusted Fabs were then superimposed on crystal structures of Fab-gp120 or Fab-gp140 complexes (PDB codes 3NGB, 2NY7 and 4CNO for complexes with VRC01, b12 and PGT122 Fabs, respectively) or a PG16-epitope scaffold complex (PDB code 4DQO). The position on Env trimer of 10-1074, a clonal variant of the PGT121–PGT123 family (Mouquet et al., 2012), was approximated using the 4CNO gp140-PGT122 structure. In other cases, related antibodies, e.g., PG9/PG16 and VRC01/3BNC117/3BNC60, were also assumed to bind similarly. The complex structures were superimposed on the Env trimer structures by aligning the common portions. The distance between the Cys233heavy chain carbon-α atoms of adjacent Fabs was then measured using PyMol (Schrödinger, 2011) to approximate the length of dsDNA bridges attached to Cys233heavy chain. Measurements derived using other EM structures for the closed and open trimers (PDB codes 3DNN, 3J5M and 3DNO) or using a recent 3.5 Å Env trimer crystal structure (Pancera et al., 2014) resulted in differences of ≤10 Å for analogous distance measurements.
In vitro neutralization assays
Neutralization of pseudoviruses derived from primary HIV-1 isolates was monitored by the reduction of HIV-1 Tat-induced luciferase reporter gene expression in the presence of a single round of pseudovirus infection in TZM-bl cells as described previously (Montefiori, 2005) and in the Supplementary Experimental Procedures.
Simulation of Fab and IgG saturation of surface-bound antigens
Numerical analysis (Mathematica, v. 10) was used to simulate saturation of surface-bound antigens by monovalent Fabs (Equation 1), bivalent IgGs to unpaired antigen (Ag) (Equation 2), and paired antigen (pAg) (Equations 3,4), where “paired antigen” was defined as antigens that are spaced such that an IgG can bind two epitopes simultaneously (e.g., intra-spike crosslinking of two epitopes on the same viral spike or inter-spike crosslinking between two viral spikes). In the bivalent model (Equations 3,4), the surface concentrations of antigen and IgG-antigen complexes were approximated by the inverse of the volume of a sphere (Vs) with radius equal to the hydrodynamic radius of the molecule multiplied by Avogadro’s number (Na) as described previously (Müller et al., 1998).
Fab binding to antigen:
| [1] |
IgG binding to unpaired antigen:
| [2] |
IgG binding to paired antigen:
| [3] |
| [4] |
Supplementary Material
Acknowledgments
We thank Martin Witte, Jessica Ingram, Chris Theile, and Hidde Ploegh for advice and help with sortase/click chemistry experiments, Sriram Subramaniam for helpful discussions, coordinates, and files to make schematic figures, David Baltimore, Bil Clemons, Ron Diskin, Jennifer Keeffe, Sarkis Mazmanian, Stuart Sievers, Louise Scharf, and Kai Zinn for suggestions, Jost Vielmetter and the Caltech Protein Expression Center for assistance with protein production and neutralization assay development, Priyanthi Gnanapragasam for doing in-house neutralization assays, the NIH AIDS Reagent Program, the Fraunhofer Institut IBMT, and Rene Mares for pseudoviruses, Erin Isaza, Siduo Jiang, and Jennifer Keeffe for reagents, Sonal Patel and Marta Murphy for help with figures. This work was supported by the Director’s Pioneer Award [1DP1OD006961-01 to P.J.B.], National Institutes of Health HIVRAD [P01 Al100148 to P.J.B.], and Collaboration for AIDS Vaccine Discovery (CAVD) grants with support from the Bill and Melinda Gates Foundation [grant 38660 (P.J.B.) and grant 1032144 (M.S.S.). P.J.B. and M.C.N. are HHMI Investigators.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Author Contributions
R.P.G. and P.J.B. conceived the study, J.S.K. performed simulations to assess avidity effects; R.P.G. and M.S.P. prepared dsDNA-Fab reagents, R.P.G., M.S.P., J.S.K., S.B., and A.P.W. perfected methods to attach dsDNA to Fabs, R.P.G. and M.S.S. performed neutralization assays, R.P.G., A.P.W., and P.J.B. analyzed the data, and R.P.G., M.C.N, and P.J.B. wrote the paper with contributions from all coauthors.
References
- Bartesaghi A, Merk A, Borgnia MJ, Milne JL, Subramaniam S. Prefusion structure of trimeric HIV-1 envelope glycoprotein determined by cryo-electron microscopy. Nat Struct Mol Biol. 2013;20:1352–1357. doi: 10.1038/nsmb.2711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bednar J, Furrer P, Katritch V, Stasiak AZ, Dubochet J, Stasiak A. Determination of DNA persistence length by cryo-electron microscopy. Separation of the static and dynamic contributions to the apparent persistence length of DNA. J Mol Biol. 1995;254:579–594. doi: 10.1006/jmbi.1995.0640. [DOI] [PubMed] [Google Scholar]
- Bhatia AK, Kaushik R, Campbell NA, Pontow SE, Ratner L. Mutation of critical serine residues in HIV-1 matrix result in an envelope incorporation defect which can be rescued by truncation of the gp41 cytoplasmic tail. Virology. 2009;384:233–241. doi: 10.1016/j.virol.2008.10.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruno CJ, Jacobson JM. Ibalizumab: an anti-CD4 monoclonal antibody for the treatment of HIV-1 infection. The Journal of antimicrobial chemotherapy. 2010;65:1839–1841. doi: 10.1093/jac/dkq261. [DOI] [PubMed] [Google Scholar]
- Chertova E, Bess JW, Jr, Crise BJ, Sowder IR, Schaden TM, Hilburn JM, Hoxie JA, Benveniste RE, Lifson JD, Henderson LE, et al. Envelope glycoprotein incorporation, not shedding of surface envelope glycoprotein (gp120/SU), Is the primary determinant of SU content of purified human immunodeficiency virus type 1 and simian immunodeficiency virus. J Virol. 2002;76:5315–5325. doi: 10.1128/JVI.76.11.5315-5325.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chi Q, Wang G, Jiang J. The persistence length and length per base of single-stranded DNA obtained from fluorescence correlation spectroscopy measurements using mean field theory. Physica A: Statistical Mechanics and its Applications. 2013;392:1072–1079. [Google Scholar]
- Chojnacki J, Staudt T, Glass B, Bingen P, Engelhardt J, Anders M, Schneider J, Muller B, Hell SW, Krausslich HG. Maturation-dependent HIV-1 surface protein redistribution revealed by fluorescence nanoscopy. Science. 2012;338:524–528. doi: 10.1126/science.1226359. [DOI] [PubMed] [Google Scholar]
- Crooks ET, Jiang P, Franti M, Wong S, Zwick MB, Hoxie JA, Robinson JE, Moore PL, Binley JM. Relationship of HIV-1 and SIV envelope glycoprotein trimer occupation and neutralization. Virology. 2008;377:364–378. doi: 10.1016/j.virol.2008.04.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dev S, Surolia A. Dynamic light scattering study of peanut agglutinin: size, shape and urea denaturation. Journal of biosciences. 2006;31:551–556. doi: 10.1007/BF02708406. [DOI] [PubMed] [Google Scholar]
- Diskin R, Scheid JF, Marcovecchio PM, West AP, Jr, Klein F, Gao H, Gnanapragasam PN, Abadir A, Seaman MS, Nussenzweig MC, et al. Increasing the potency and breadth of an HIV antibody by using structure-based rational design. Science. 2011;334:1289–1293. doi: 10.1126/science.1213782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guimaraes CP, Witte MD, Theile CS, Bozkurt G, Kundrat L, Blom AE, Ploegh HL. Site-specific C-terminal and internal loop labeling of proteins using sortase-mediated reactions. Nat Protoc. 2013;8:1787–1799. doi: 10.1038/nprot.2013.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendrickson ER, Truby TM, Joerger RD, Majarian WR, Ebersole RC. High sensitivity multianalyte immunoassay using covalent DNA-labeled antibodies and polymerase chain reaction. Nucleic Acids Res. 1995;23:522–529. doi: 10.1093/nar/23.3.522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J, Ofek G, Laub L, Louder MK, Doria-Rose NA, Longo NS, Imamichi H, Bailer RT, Chakrabarti B, Sharma SK, et al. Broad and potent neutralization of HIV-1 by a gp41-specific human antibody. Nature. 2012;491:406–412. doi: 10.1038/nature11544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Icenogle J, Shiwen H, Duke G, Gilbert S, Rueckert R, Anderegg J. Neutralization of poliovirus by a monoclonal antibody: kinetics and stoichiometry. Virology. 1983;127:412–425. doi: 10.1016/0042-6822(83)90154-x. [DOI] [PubMed] [Google Scholar]
- Julien JP, Cupo A, Sok D, Stanfield RL, Lyumkis D, Deller MC, Klasse PJ, Burton DR, Sanders RW, Moore JP, et al. Crystal structure of a soluble cleaved HIV-1 envelope trimer. Science. 2013a;342:1477–1483. doi: 10.1126/science.1245625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Julien JP, Lee JH, Cupo A, Murin CD, Derking R, Hoffenberg S, Caulfield MJ, King CR, Marozsan AJ, Klasse PJ, et al. Asymmetric recognition of the HIV-1 trimer by broadly neutralizing antibody PG9. Proc Natl Acad Sci U S A. 2013b;110:4351–4356. doi: 10.1073/pnas.1217537110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kajander T, Cortajarena AL, Mochrie S, Regan L. Structure and stability of designed TPR protein superhelices: unusual crystal packing and implications for natural TPR proteins. Acta Crystallographica Section D-Biological Crystallography. 2007;63:800–811. doi: 10.1107/S0907444907024353. [DOI] [PubMed] [Google Scholar]
- Klein F, Mouquet H, Dosenovic P, Scheid JF, Scharf L, Nussenzweig MC. Antibodies in HIV-1 vaccine development and therapy. Science. 2013;341:1199–1204. doi: 10.1126/science.1241144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein JS, Bjorkman PJ. Few and far between: how HIV may be evading antibody avidity. PLoS Pathog. 2010;6:e1000908. doi: 10.1371/journal.ppat.1000908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein JS, Gnanapragasam PN, Galimidi RP, Foglesong CP, West AP, Jr, Bjorkman PJ. Examination of the contributions of size and avidity to the neutralization mechanisms of the anti-HIV antibodies b12 and 4E10. Proc Natl Acad Sci USA. 2009;106:7385–7390. doi: 10.1073/pnas.0811427106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein JS, Jiang S, Galimidi RP, Keeffe JR, Bjorkman PJ. Design and characterization of structured protein linkers with differing flexibilities. Protein Engineering, Design, and Selection. 2014;27:325–330. doi: 10.1093/protein/gzu043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunert R, Wolbank S, Stiegler G, Weik R, Katinger H. Characterization of molecular features, antigen-binding, and in vitro properties of IgG and IgM variants of 4E10, an anti-HIV type 1 neutralizing monoclonal antibody. AIDS Res Hum Retroviruses. 2004;20:755–762. doi: 10.1089/0889222041524571. [DOI] [PubMed] [Google Scholar]
- Liljeroos L, Krzyzaniak MA, Helenius A, Butcher SJ. Architecture of respiratory syncytial virus revealed by electron cryotomography. Proc Natl Acad Sci U S A. 2013;110:11133–11138. doi: 10.1073/pnas.1309070110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Bartesaghi A, Borgnia MJ, Sapiro G, Subramaniam S. Molecular architecture of native HIV-1 gp120 trimers. Nature. 2008;455:109–113. doi: 10.1038/nature07159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luftig MA, Mattu M, Di Giovine P, Geleziunas R, Hrin R, Barbato G, Bianchi E, Miller MD, Pessi A, Carfi A. Structural basis for HIV-1 neutralization by a gp41 fusion intermediate-directed antibody. Nat Struct Mol Biol. 2006;13:740–747. doi: 10.1038/nsmb1127. [DOI] [PubMed] [Google Scholar]
- Lyumkis D, Julien JP, de Val N, Cupo A, Potter CS, Klasse PJ, Burton DR, Sanders RW, Moore JP, Carragher B, et al. Cryo-EM structure of a fully glycosylated soluble cleaved HIV-1 envelope trimer. Science. 2013;342:1484–1490. doi: 10.1126/science.1245627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattes MJ. Binding parameters of antibodies: pseudo-affinity and other misconceptions. Cancer Immunol Immunother. 2005;54:513–516. doi: 10.1007/s00262-004-0644-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merk A, Subramaniam S. HIV-1 envelope glycoprotein structure. Curr Opin Struct Biol. 2013;23:268–276. doi: 10.1016/j.sbi.2013.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montefiori DC. Evaluating neutralizing antibodies against HIV, SIV, and SHIV in luciferase reporter gene assays. In: Coligan John E, et al., editors. Current protocols in immunology. Unit 12. Chapter 12. 2005. p. 11. [DOI] [PubMed] [Google Scholar]
- Mouquet H, Scharf L, Euler Z, Liu Y, Eden C, Scheid JF, Halper-Stromberg A, Gnanapragasam PN, Spencer DI, Seaman MS, et al. Complex-type N-glycan recognition by potent broadly neutralizing HIV antibodies. Proc Natl Acad Sci U S A. 2012;109:E3268–3277. doi: 10.1073/pnas.1217207109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mouquet H, Scheid JF, Zoller MJ, Krogsgaard M, Ott RG, Shukair S, Artyomov MN, Pietzsch J, Connors M, Pereyra F, et al. Polyreactivity increases the apparent affinity of anti-HIV antibodies by heteroligation. Nature. 2010;467:591–595. doi: 10.1038/nature09385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munro JB, Gormann J, Ma X, Zhou Z, Arthos J, Burton DR, Koff WC, Courtner JR, Smith AB, 3rd, Kwong PD, et al. Conformational dynamics of single HIV-1 envelope trimers on the surface of native virions. Science Xpress. 2014 doi: 10.1126/science.1254426. published online 8 October 2014 [DOI:10.1126/science.1254426] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pace CS, Song R, Ochsenbauer C, Andrews CD, Franco D, Yu J, Oren DA, Seaman MS, Ho DD. Bispecific antibodies directed to CD4 domain 2 and HIV envelope exhibit exceptional breadth and picomolar potency against HIV-1. Proc Natl Acad Sci U S A. 2013;110:13540–13545. doi: 10.1073/pnas.1304985110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pancera M, Zhou T, Druz A, Georgiev IS, Soto C, Gorman J, Huang J, Acharya P, Chuang GY, Ofek G, et al. Structure and immune recognition of trimeric pre-fusion HIV-1 Env. Nature. 2014 doi: 10.1038/nature13808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pegu A, Yang ZY, Boyington JC, Wu L, Ko SY, Schmidt SD, McKee K, Kong WP, Shi W, Chen X, et al. Neutralizing antibodies to HIV-1 envelope protect more effectively in vivo than those to the CD4 receptor. Science translational medicine. 2014;6:243ra288. doi: 10.1126/scitranslmed.3008992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roben P, Moore JP, Thali M, Sodroski J, Barbas CF, 3rd, Burton DR. Recognition properties of a panel of human recombinant Fab fragments to the CD4 binding site of gp120 that show differing abilities to neutralize human immunodeficiency virus type 1. J Virol. 1994;68:4821–4828. doi: 10.1128/jvi.68.8.4821-4828.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaefer W, Regula JT, Bahner M, Schanzer J, Croasdale R, Durr H, Gassner C, Georges G, Kettenberger H, Imhof-Jung S, et al. Immunoglobulin domain crossover as a generic approach for the production of bispecific IgG antibodies. Proc Natl Acad Sci U S A. 2011;108:11187–11192. doi: 10.1073/pnas.1019002108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheid JF, Mouquet H, Ueberheide B, Diskin R, Klein F, Olivera TY, Pietzsch J, Fenyo D, Abadir A, Velinzon K, et al. Sequence and Structural Convergence of Broad and Potent HIV Antibodies That Mimic CD4 Binding. Science. 2011;333:1633–1637. doi: 10.1126/science.1207227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schofield DJ, Stephenson JR, Dimmock NJ. Variations in the neutralizing and haemagglutination-inhibiting activities of five influenza A virus-specific IgGs and their antibody fragments. J Gen Virol. 1997;78(Pt 10):2431–2439. doi: 10.1099/0022-1317-78-10-2431. [DOI] [PubMed] [Google Scholar]
- Schrödinger L. The PyMOL Molecular Graphics System (The PyMOL Molecular Graphics System) 2011 [Google Scholar]
- Seaman MS, Janes H, Hawkins N, Grandpre LE, Devoy C, Giri A, Coffey RT, Harris L, Wood B, Daniels MG, et al. Tiered categorization of a diverse panel of HIV-1 Env pseudoviruses for assessment of neutralizing antibodies. J Virol. 2010;84:1439–1452. doi: 10.1128/JVI.02108-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sougrat R, Bartesaghi A, Lifson JD, Bennett AE, Bess JW, Zabransky DJ, Subramaniam S. Electron tomography of the contact between T cells and SIV/HIV-1: implications for viral entry. PLoS Pathog. 2007;3:e63. doi: 10.1371/journal.ppat.0030063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanfield RL, Zemla A, Wilson IA, Rupp B. Antibody elbow angles are influenced by their light chain class. J Mol Biol. 2006;357:1566–1574. doi: 10.1016/j.jmb.2006.01.023. [DOI] [PubMed] [Google Scholar]
- Stieh DJ, King DF, Klein K, Liu P, Shen X, Hwang K, Ferrari G, Montefiori DC, Haynes B, Pitisuttithum P, et al. Aggregate complexes of HIV-1 induced by multimeric antibodies. Retrovirology. 2014;11:78. doi: 10.1186/s12977-014-0078-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran EE, Borgnia MJ, Kuybeda O, Schauder DM, Bartesaghi A, Frank GA, Sapiro G, Milne JL, Subramaniam S. Structural mechanism of trimeric HIV-1 envelope glycoprotein activation. PLoS Pathog. 2012;8:e1002797. doi: 10.1371/journal.ppat.1002797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker LM, Phogat SK, Chan-Hui PY, Wagner D, Phung P, Goss JL, Wrin T, Simek MD, Fling S, Mitcham JL, et al. Broad and Potent Neutralizing Antibodies from an African Donor Reveal a New HIV-1 Vaccine Target. Science. 2009;326:285–289. doi: 10.1126/science.1178746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- West AP, Jr, Galimidi RP, Gnanapragasam PN, Bjorkman PJ. Single-Chain Fv-Based Anti-HIV Proteins: Potential and Limitations. J Virol. 2012;86:195–202. doi: 10.1128/JVI.05848-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- West AP, Jr, Scharf L, Scheid JF, Klein F, Bjorkman PJ, Nussenzweig MC. Structural Insights on the Role of Antibodies in HIV-1 Vaccine and Therapy. Cell. 2014;156:633–648. doi: 10.1016/j.cell.2014.01.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witte MD, Theile CS, Wu T, Guimaraes CP, Blom AE, Ploegh HL. Production of unnaturally linked chimeric proteins using a combination of sortase-catalyzed transpeptidation and click chemistry. Nat Protoc. 2013;8:1808–1819. doi: 10.1038/nprot.2013.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolbank S, Kunert R, Stiegler G, Katinger H. Characterization of human class-switched polymeric (immunoglobulin M [IgM] and IgA) anti-human immunodeficiency virus type 1 antibodies 2F5 and 2G12. J Virol. 2003;77:4095–4103. doi: 10.1128/JVI.77.7.4095-4103.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H, Pfarr DS, Tang Y, An LL, Patel NK, Watkins JD, Huse WD, Kiener PA, Young JF. Ultra-potent antibodies against respiratory syncytial virus: effects of binding kinetics and binding valence on viral neutralization. J Mol Biol. 2005;350:126–144. doi: 10.1016/j.jmb.2005.04.049. [DOI] [PubMed] [Google Scholar]
- Wu X, Yang ZY, Li Y, Hogerkorp CM, Schief WR, Seaman MS, Zhou T, Schmidt SD, Wu L, Xu L, et al. Rational design of envelope identifies broadly neutralizing human monoclonal antibodies to HIV-1. Science. 2010;329:856–861. doi: 10.1126/science.1187659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu X, Yuan X, Matsuda Z, Lee TH, Essex M. The matrix protein of human immunodeficiency virus type 1 is required for incorporation of viral envelope protein into mature virions. J Virol. 1992;66:4966–4971. doi: 10.1128/jvi.66.8.4966-4971.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zadeh JN, Steenberg CD, Bois JS, Wolfe BR, Pierce MB, Khan AR, Dirks RM, Pierce NA. NUPACK: Analysis and design of nucleic acid systems. J Comput Chem. 2011;32:170–173. doi: 10.1002/jcc.21596. [DOI] [PubMed] [Google Scholar]
- Zhou HX. Polymer models of protein stability, folding, and interactions. Biochemistry. 2004;43:2141–2154. doi: 10.1021/bi036269n. [DOI] [PubMed] [Google Scholar]
- Zhu P, Liu J, Bess J, Jr, Chertova E, Lifson JD, Grise H, Ofek GA, Taylor KA, Roux KH. Distribution and three-dimensional structure of AIDS virus envelope spikes. Nature. 2006;441:847–852. doi: 10.1038/nature04817. [DOI] [PubMed] [Google Scholar]
- Zingler K, Littman DR. Truncation of the cytoplasmic domain of the simian immunodeficiency virus envelope glycoprotein increases env incorporation into particles and fusogenicity and infectivity. J Virol. 1993;67:2824–2831. doi: 10.1128/jvi.67.5.2824-2831.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.