Abstract
The Cbl proteins are a family of ubiquitin ligases (E3s) that regulate signaling through many tyrosine kinase dependent pathways. A predominant function is to negatively regulate receptor tyrosine kinase (RTK) signaling by ubiquitination of active RTKs, targeting them for trafficking to the lysosome for degradation. Also, Cbl-mediated ubiquitination can regulate signaling protein function by altered cellular localization of proteins without degradation. In addition to their role as E3s, Cbl proteins play a positive role in signaling by acting as adaptor proteins which can recruit signaling molecules to the active RTKs. Cbl-b, a second family member, negatively regulates the costimulatory pathway of CD8 T-cells and also negatively regulates Natural Killer (NK) cell function. The different functions of Cbl proteins, and their roles both in the development of cancer and the regulation of immune responses provide multiple therapeutic opportunities. Mutations in Cbl which inactivate the negative E3 function while maintaining the positive adaptor function have been described in approximately 5% of myeloid neoplasms. Understanding how the signaling pathways (e.g. Fms-like tyrosine kinase 3 (Flt3), PI-3 kinase, and signal transducer and activator of transcription (Stat)) are dysregulated by these mutations in Cbl has identified potential targets for therapy of myeloid neoplasms. Conversely, the loss of Cbl-b leads to increased adaptive and innate antitumor immunity suggesting that inhibiting Cbl-b may be a means to increase antitumor immunity across a wide variety of tumors. Thus, targeting the pathways regulated by Cbl proteins may provide attractive opportunities for treating cancer.
Background
Cbl proteins are a highly conserved family of ubiquitin ligases (E3s) primarily found in metazoans that negatively regulate signal transduction through many tyrosine kinase (TK) dependent pathways (comprehensively reviewed in (1)). Mutations in Cbl proteins contribute to the pathogenesis of cancer by dysregulating RTK signaling pathways. Further, Cbl-b, the second mammalian Cbl protein, negatively regulates T-cell and NK cell anti-tumor function. Together, the data emerging about how Cbl proteins contribute to the pathogenesis of cancer and how they regulate anti-tumor immunity may provide a number of attractive approaches to cancer treatment.
The Cbl proteins as regulators of signaling
First identified as the cellular homologues of the v-Cbl transforming gene of the Casitas B lymphoma murine retrovirus, Cbl proteins have been found throughout metazoans (2). There are three mammalian Cbl proteins: Cbl (a.k.a., c-Cbl; CBL2; RNF55), Cbl-b (a.k.a., RNF56), and Cbl-c (a.k.a., Cbl-3, Cbl-SL, RNF57) (2). Cbl proteins are characterized by a highly conserved N-terminal tyrosine kinase binding (TKB) domain and a C3HC4 RING finger (RF) which is the catalytic domain for the E3 activity (2). These two domains are separated by a highly conserved alpha-helical linker region that is critical to the regulation of Cbl E3 function. The Cbl proteins vary more in the C-terminus which contains motifs (e.g., proline rich domains, tyrosines which become phosphorylated, and an ubiquitin associated domain) which mediate a broad array of protein interactions with signaling molecules (3). The Cbl proteins are tyrosine phosphorylated upon activation of a variety of growth factor receptors, and they associate with many proteins containing SH2 and SH3 domains (reviewed in (4–6)). These diverse interactions modulate signaling both negatively and positively through many pathways (1, 4–6).
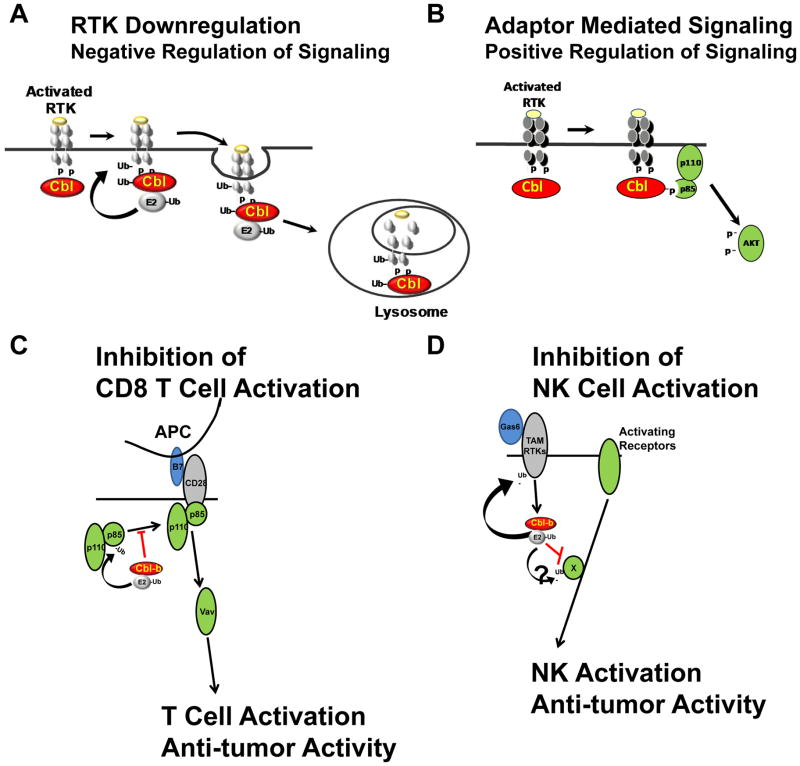
The E3 activity of Cbl proteins is critical to the negative regulation of signaling by activated RTKs (Fig. 1A). The covalent modification of proteins by ubiquitin occurs via the sequential activation and conjugation of ubiquitin to target proteins by a ubiquitin activating enzyme (E1), a ubiquitin-conjugating enzyme (E2), and an E3 (7). The majority of E3s contain a RF and mediate the transfer of ubiquitin directly from the associated E2 to one or more lysines of the specific target protein. Thus the E3 confers specificity to the process. The Cbl proteins normally exist in the cytosol in an inactive state where the catalytic RF is masked by the N-terminal TKB domain (8–11). Upon activation of the kinases, the Cbl proteins bind directly (via the TKB)(12–14) or indirectly via adaptor dependent mechanisms (15, 16) to phosphotyrosines on the RTK (Fig. 1A). Phosphorylation of a conserved tyrosine in the linker region that separates the TKB from the RF of the Cbl proteins by the activated RTK (or other TKs) results in a dramatic structural rearrangement of the Cbl protein in which the N-terminal TKB rotates ~180 degrees (8, 10, 17). This exposes the RF allowing increased E2 binding to Cbl proteins and markedly increased E3 activity of the Cbl proteins. This structural rearrangement also positions the E2 in closer proximity to the RTK facilitating the transfer of ubiquitin from the E2 to lysines on the RTK (8, 10, 17). Thus activation of the RTK serves both to create a phosphotyrosine based docking site on the RTK for the Cbl proteins and to phosphorylate and stimulate the E3 activity of the Cbl proteins. The ubiquitinated RTK trafficks through the endocytic compartments to the lysosome where it is degraded (Fig.1A) (reviewed in (18)).
Figure 1.
Cbl pathways. A. All Cbl proteins are recruited to activated RTKs where they mediate ubiquitination and downregulation of the RTKs. The ubiquitinated RTKs are degraded by the lysosome. Thus loss of the E3 function of Cbl results in sustained signaling by RTKs. B. Cbl proteins can serve as adaptor proteins which recruit signaling molecules such as PI3 Kinase to the activated RTK. The mutant proteins that have lost E3 function frequently retain the ability to activate PI3K by this mechanism and so function as oncogenes. C. Cbl-b is a negative regulator of the CD28 costimulatory pathway in T-Cells. CD28 is activated by B7 molecules on the surface of antigen presenting cells (APC). Cbl-b ubiquitinates the p85 subunit of PI3K, preventing its recruitment to the activated CD28. The loss of Cbl-b results in hyperactive immunity, including anti-tumor immunity. D. Cbl-b is a negative regulator of NK cell anti-tumor acitivity. Growth arrest specific-6 (Gas6) is an activating ligand for the TAM receptors. Cbl-b is activated downstream of the TAM RTKs and inhibits NK cell activation – presumably by ubiquitinating an unknown substrate (X) that is required for activation. Cbl-b also can ubiquitinate the TAM receptors. The loss of Cbl-b results in increased NK cell anti-tumor activity.
While the E3 activity of the Cbl proteins has been most extensively studied, the Cbl proteins can also function as adaptor molecules that recruit signaling molecules to activated RTKs (4–6, 18). This results in a positive role in signaling. For example, several studies have shown that the Cbl protein serves as an adaptor to recruit phosphatidylinositol-4,5-bisphosphate 3-kinase (PI3K) to activated RTKs with subsequent activation of the PI3K/AKT pathway (Fig. 1B) (19, 20).
Clinical-Translational Advances
Cbl proteins as drivers of cancer
v-Cbl was originally identified as an oncogene, causing leukemia in mice and transforming NIH3T3 cells (21). The v-Cbl protein contains only the TKB domain of Cbl and, when expressed in cells, prevents RTK ubiquitination and downregulation most likely by acting as a dominant negative protein preventing the recruitment of endogenous Cbl proteins to the RTK (22, 23). Other transforming mutants of the murine Cbl protein have been identified from chemically induced lymphomas (70Z Cbl and p95 Cbl)(24, 25). These mutant proteins have in frame deletions of part or all of the linker and RF domains thus losing E3 activity. Interestingly, these deletions result from point mutations which lead to mis-splicing of the Cbl mRNA.
Mice deficient in Cbl, Cbl-b, or Cbl-c do not develop leukemia (26). In contrast, mice that have a knockin of a RF mutant Cbl develop myeloid leukemia (26). The absence of leukemia in Cbl knockout mice and the development of leukemia in mice with a Cbl RING finger mutant knockin can be explained by a dominant negative function of the mutant protein, whereby the mutant Cbl protein binds to activated RTKs and prevents recruitment of the wild type Cbl or Cbl-b proteins to the RTK. Consistent with this, mice deficient in both Cbl and Cbl-b in hematopoietic stem cells develop early onset of myeloid leukemia (27). However, the positive functions of Cbl proteins in signaling based on the adaptor function of Cbl suggest that the mutant proteins may have both loss of tumor suppressor function (i.e., the loss of the negative regulatory E3 function) and gain of oncogene function (e.g., coupling the RTK to downstream signaling pathways such as PI3K). Consistent with this, the transforming 70Z form of Cbl activates the EGFR in the absence of ligand and enhances activity of the EGFR and downstream signaling upon ligand stimulation (28).
Cbl mutations have been found in ~5% of a wide variety of myeloid neoplasms including myelodysplastic syndrome, myelofibrosis, refractory anemia with excess blasts, de novo and secondary acute myeloid leukemia (AML and sAML, respectively), atypical chronic myelogenous leukemia (aCML), CML in blast crisis, chronic myelomonocytic leukemia (CMML), and juvenile myelomonocytic leukemia (JMML) (reviewed in (29)). The frequency of Cbl mutations appears to be highest in JMML (~15%), CMML (~13%), sAML (~10%), and aCML (8%) (29). The majority of these mutations are missense mutations that cluster within the linker region and within the RF domain leading to disruption of E3 activity (reviewed in (29)). The linker tyrosine (Y371 in Cbl), whose phosphorylation is required for E3 activity (as described above), is frequently mutated in myeloid neoplasms accounting for ~15% of all missense mutations (29, 30). These Y371 mutations occur mostly in patients with JMML and CMML (30–34). Deletions of all or portions of the Cbl exon containing the distal portion of the linker region and the proximal portion of the RF have been described (29, 30). As seen in the murine Cbl deletion mutants, these deletions result from mis-splicing due to mutations, insertions, or deletions in the splice donor and acceptor sites surrounding exon 8. Nonsense mutations, frame shift mutations, and insertions within the linker and RF regions have been found as well (29). The missense mutations of Cbl are usually homozygous mutations (resulting from copy neutral loss of heterozygosity – also known as uniparental disomy) while the deletions that arise from splicing mutations are more commonly heterozygous (31–41). Transformation assays in NIH 3T3 cells found that deletions of the linker domain were transforming while point mutations in the linker or RF were not (42). In addition, one group found that 70Z Cbl induces greater ligand independent proliferation and survival than the R420Q mutation (43). However, others found no difference in transformation efficiency between 70Z Cbl and a variety of point mutants found in patients (34). Thus it is unclear why most missense mutations are homozygous and the deletion mutations are heterozygous.
Mutations of Cbl-b and Cbl-c are uncommon in myeloid neoplasms, and the mutations found have not been functionally characterized (37, 39). A total of five mutations of Cbl-b (out of ~2000 patients evaluated) that are either frame shift or missense mutations within the RF domain have been reported in myeloid neoplasms (31, 37, 39–41). Several cases of frame shift or polymorphisms in the RF domain of Cbl-c have been reported, but Cbl-c expression is restricted to epithelial cells, so the significance of these abnormalities is unclear (39, 44–46).
v-Cbl also caused B-cell lymphomas in mice, but mutation in human lymphoid malignancies is rare. Sequencing of Cbl in more than 500 lymphoid malignancies found five somatic mutations, three of which represent splice site mutations resulting in the loss of a portion of the RF (30, 33, 47, 48). The Cancer Genome Atlas (TCGA) sequencing programs have identified copy number variations and mutations in solid tumors of the three Cbl genes in 0.3–19.6 % of the tumors but the significance of these aberrations is unknown (the results included here are in whole or part based upon data generated by the TCGA Research Network: http://cancergenome.nih.gov [49]). Somatic mutations of Cbl have been found in 10 non-small cell lung tumors out of 452 samples (30, 50). All but one of the mutations described are outside the linker and RING finger, and all are heterozygous. For those mutants analyzed, E3 activity was maintained, but overexpression of these mutants in lung cancer cells resulted in increased viability and motility (50). This suggests that they may impair the association of Cbl with a critical substrate, but the mechanism by which these mutants affected viability or motility is unknown.
Therapeutic approaches to myeloid neoplasms containing mutant Cbl proteins will require targeting the activated pathways since the mutations in Cbl are at least partly loss of function mutations. Activating mutations in Flt3 are found in ~30% of patients with AML, and inhibitors of Flt3 are being tested for treatment of Flt3 mutant AML (51). Cbl ubiquitinates and mediates lysosomal degradation of activated Flt3 (41). The development of leukemia in Cbl RF mutant knockin mice is dependent on Flt3 activity as crossing these mice to Flt3 deficient mice abrogates the development of leukemia (26). Treatment of the Cbl RF mutant knockin mice which had developed a myeloproliferative disorder with the high potency Flt3 inhibitor quizartinib (AC220) significantly reduced the white blood count (73% reduction, P<0.01), the spleen weight (69% reduction, p=0.037), and infiltration of the liver and lungs by myeloid cells (52). Treatment with AC220 induced quiescence in Flt3 dependent multipotent progenitor cells (52). This suggests that myeloid neoplasms containing a Cbl mutation are driven by Flt3 RTK and that inhibiting this pathway may have efficacy for the treatment of these neoplasms (26, 41). However, in the animal studies, the effect of Flt3 inhibition by AC220 was not maintained once the drug was discontinued so that long term treatment or combination therapy may be required (52). Alternatively, studies of the effects of mutant Cbl proteins on signaling have found enhanced activation of the PI3K/AKT and STAT5 pathways in the absence and presence of ligand (Fig. 1B) (33, 34, 41). Importantly, the ligand independent growth was inhibited ~70–80% by PI3K and mTOR inhibitors (33). Thus targeting the PI3K pathway also is worth exploring in Cbl mutant myeloid neoplasms.
As described above, while the E3 function of Cbl is lost due to mutations in cancers, the positive adaptor function is frequently maintained. Data suggest that the positive function contributes to the transforming potential of the Cbl mutants ((34) and reviewed in (29)). A novel approach to targeting Cbl in myeloid neoplasms is to block the adaptor function of the Cbl protein to prevent activation of the downstream signaling pathway. The Cbl proteins bind to the activated RTKs via their TKB domain, and this allows recruitment of signaling proteins bound to other domains of Cbl to the RTK (e.g., PI3 kinase (19, 20)). Thus inhibition of the interaction of the Cbl TKB with the activated RTK would prevent the recruitment of signaling proteins to the activated RTK. Based on this, Kumar et al. are developing strategies to identify small molecules or peptides that bind the Cbl TKB and block interaction with the RTK (53, 54). The efficacy of such an approach remains to be tested.
Cbl-b and adaptive and innate immune system
The loss of Cbl-b is associated with hyperactive T-cell immunity resulting in spontaneous and induced autoimmunity (55, 56). T-cells from mice lacking Cbl-b have excessive proliferation and production of the cytokine interleukin 2 that is uncoupled from the requirement for activation of the CD28 costimulatory pathway (Fig. 1C). This function is specific to Cbl-b as loss of the other Cbl proteins (Cbl or Cbl-c) does not result in increased autoimmunity or activation of the costimulatory pathway (44, 57, 58). Cbl-b has been shown to ubiquitinate the regulatory p85 subunit of PI3K (Fig. 1C) (59, 60). Rather than leading to degradation, this prevents the recruitment of PI3K to CD28 upon activation of the costimulatory pathway (59, 60). The loss of Cbl-b results in increased PI3K activity and activation of Vav (Fig. 1C). Most intriguingly, mice lacking Cbl-b have enhanced CD8 T-cell mediated killing of transplanted and spontaneous tumors, including lymphomas, lung carcinomas, and ultraviolet irradiation B induced skin carcinomas (61, 62). Importantly, reconstitution experiments of wild type or RF mutant Cbl-b into Cbl-b null mice have demonstrated that the catalytic activity of Cbl-b is essential for inhibition of the costimulatory pathway (63). Recently, Paolino et al. observed that Cbl-b null mice lacking functional T and B cells had delayed growth of breast and melanoma tumors and further that Cbl-b null mice developed fewer metastases (64). This was due to enhanced NK cell anti-tumor function. The exact mechanism by which Cbl-b inhibits NK function is not yet defined. The Tyro3, Axl and Mer (TAM) RTKs inhibit NK antitumor function, and Cbl-b acts downstream of the kinases and is required for this inhibitory function (Fig. 1D) (64). As with the role of Cbl-b in the costimulatory pathway, the E3 activity of Cbl-b is essential to this function. Thus, the loss of the Cbl-b E3 activity leads to increased NK cell antitumor activity. Cbl and Cbl-b can both ubiquitinate the TAM receptors and lead to their downregulation (64, 65). However, this does not explain the positive role that Cbl-b plays in TAM receptor signaling that leads to inhibition of NK cell function. The data suggest that Cbl-b ubiquitinates and inhibits a protein required for NK activity. Specific inhibitors of the TAM receptors also lead to increased NK cell mediated anti-tumor immunity (64). Interestingly, warfarin, which has been known to inhibit metastases was found to work by inhibition of the TAM RTKs (64). While mechanistic details are not completely worked out, the results described above demonstrate that the loss of Cbl-b promotes both adaptive and innate anti-tumor immunity.
The activation of costimulatory T-cell pathways is already used clinically for the treatment of cancer. The inhibition of Cbl-b function (either genetically or by small molecular inhibitors) would be another approach to activate the costimulatory pathway for antitumor benefit. Indeed, Stromness et al. demonstrated that RNAi mediated knockdown of Cbl-b in effector CD8+ T-cells improved the anti-leukemia efficacy of these cells in a mouse model of adoptive transfer of T-cells (66). The benefit of developing Cbl-b inhibitors would be the combined effects of increasing both adaptive (T-cell) and innate (NK cell) antitumor activity.
Conclusions
The Cbl proteins have diverse roles as regulators of signal transduction. The consequences of defects in Cbl proteins can lead to malignancy and/or to immune dysfunction. As we gain more knowledge of the signaling pathways affected in each case, novel therapeutic opportunities are arising for the treatment of cancer.
Acknowledgments
The authors thank Marion Nau for critical review of this manuscript.
Grant Support
This research was supported by the Intramural Research Program of the NIH, NCI, Center for Cancer Research.
Footnotes
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
References
- 1.Tsygankov A, editor. Cbl proteins. New York: Nova Science Publishers; 2008. [Google Scholar]
- 2.Nau MM, Lipkowitz S. Welcome to the family: Cbl-family gene organization, overview of structure and functions of Cbl-related proteins in various taxonomical groups. In: Tsygankov AY, editor. Cbl proteins. New York: Nova Science Publishers; 2008. pp. 3–25. [Google Scholar]
- 3.Schmidt MH, Dikic I. The Cbl interactome and its functions. Nat Rev Mol Cell Biol. 2005;6:907–19. doi: 10.1038/nrm1762. [DOI] [PubMed] [Google Scholar]
- 4.Rao N, Dodge I, Band H. The Cbl family of ubiquitin ligases: critical negative regulators of tyrosine kinase signaling in the immune system. J Leukoc Biol. 2002;71:753–63. [PubMed] [Google Scholar]
- 5.Thien CB, Langdon WY. Cbl: many adaptations to regulate protein tyrosine kinases. Nat Rev Mol Cell Biol. 2001;2:294–307. doi: 10.1038/35067100. [DOI] [PubMed] [Google Scholar]
- 6.Tsygankov AY, Teckchandani AM, Feshchenko EA, Swaminathan G. Beyond the RING: CBL proteins as multivalent adapters. Oncogene. 2001;20:6382–402. doi: 10.1038/sj.onc.1204781. [DOI] [PubMed] [Google Scholar]
- 7.Weissman AM. Themes and variations on ubiquitylation. Nat Rev Mol Cell Biol. 2001;2:169–78. doi: 10.1038/35056563. [DOI] [PubMed] [Google Scholar]
- 8.Dou H, Buetow L, Hock A, Sibbet GJ, Vousden KH, Huang DT. Structural basis for autoinhibition of and phosphorylation-dependent activation of c-Cbl. Nat Struct Mol Biol. 2012;19:184–92. doi: 10.1038/nsmb.2231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kassenbrock CK, Anderson SM. Regulation of ubiquitin protein ligase activity in c-Cbl by phosphorylation-induced conformational change and constitutive activation by tyrosine to glutamate point mutations. J Biol Chem. 2004;279:28017–27. doi: 10.1074/jbc.M404114200. [DOI] [PubMed] [Google Scholar]
- 10.Kobashigawa Y, Tomitaka A, Kumeta H, Noda NN, Yamaguchi M, Inagaki F. Autoinhibition and phosphorylation-induced activation mechanisms of human cancer and autoimmune disease-related E3 protein Cbl-b. Proc Natl Acad Sci U S A. 2011;108:20579–84. doi: 10.1073/pnas.1110712108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ryan PE, Sivadasan-Nair N, Nau MM, Nicholas S, Lipkowitz S. The N-terminus of Cbl-c regulates ubiquitin ligase activity by modulating affinity for the ubiquitin conjugating enzyme. J Biol Chem. 2010;285:23687–98. doi: 10.1074/jbc.M109.091157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Levkowitz G, Waterman H, Ettenberg SA, Katz M, Tsygankov AY, Alroy I, et al. Ubiquitin ligase activity and tyrosine phosphorylation underlie suppression of growth factor signaling by c-Cbl/Sli-1. Mol Cell. 1999;4:1029–40. doi: 10.1016/s1097-2765(00)80231-2. [DOI] [PubMed] [Google Scholar]
- 13.Lill NL, Douillard P, Awwad RA, Ota S, Lupher ML, Jr, Miyake S, et al. The evolutionarily conserved N-terminal region of Cbl is sufficient to enhance down-regulation of the epidermal growth factor receptor. J Biol Chem. 2000;275:367–77. doi: 10.1074/jbc.275.1.367. [DOI] [PubMed] [Google Scholar]
- 14.Lupher ML, Jr, Reedquist KA, Miyake S, Langdon WY, Band H. A novel phosphotyrosine-binding domain in the N-terminal transforming region of Cbl interacts directly and selectively with ZAP-70 in T cells. J Biol Chem. 1996;271:24063–8. doi: 10.1074/jbc.271.39.24063. [DOI] [PubMed] [Google Scholar]
- 15.Scott RP, Eketjall S, Aineskog H, Ibanez CF. Distinct turnover of alternatively spliced isoforms of the RET kinase receptor mediated by differential recruitment of the Cbl ubiquitin ligase. J Biol Chem. 2005;280:13442–9. doi: 10.1074/jbc.M500507200. [DOI] [PubMed] [Google Scholar]
- 16.Waterman H, Katz M, Rubin C, Shtiegman K, Lavi S, Elson A, et al. A mutant EGF-receptor defective in ubiquitylation and endocytosis unveils a role for Grb2 in negative signaling. Embo J. 2002;21:303–13. doi: 10.1093/emboj/21.3.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kales SC, Ryan PE, Lipkowitz S. Cbl exposes its RING finger. Nat Struct Mol Biol. 2012;19:131–3. doi: 10.1038/nsmb.2241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ma K, Kales SC, Nau MM, Lipkowitz S. Cbl as a master regulator of receptor tyrosine kinase trafficking. In: Yarden Y, Tarcic G, editors. Vesicle trafficking in cancer. New York: Springer; 2013. pp. 219–44. [Google Scholar]
- 19.Dombrosky-Ferlan PM, Corey SJ. Yeast two-hybrid in vivo association of the Src kinase Lyn with the proto-oncogene product Cbl but not with the p85 subunit of PI 3-kinase. Oncogene. 1997;14:2019–24. doi: 10.1038/sj.onc.1201031. [DOI] [PubMed] [Google Scholar]
- 20.Ueno H, Sasaki K, Honda H, Nakamoto T, Yamagata T, Miyagawa K, et al. c-Cbl is tyrosine-phosphorylated by interleukin-4 and enhances mitogenic and survival signals of interleukin-4 receptor by linking with the phosphatidylinositol 3'-kinase pathway. Blood. 1998;91:46–53. [PubMed] [Google Scholar]
- 21.Langdon WY, Hartley JW, Klinken SP, Ruscetti SK, Morse HC., III v-cbl, an oncogene from a dual-recombinant murine retrovirus that induces early B-lineage lymphomas. Proc Natl Acad Sci U S A. 1989;86:1168–72. doi: 10.1073/pnas.86.4.1168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bonita DP, Miyake S, Lupher MLJ, Langdon WY, Band H. Phosphotyrosine binding domain-dependent upregulation of the platelet-derived growth factor receptor alpha signaling cascade by transforming mutants of Cbl: implications for Cbl's function and oncogenicity. Mol Cell Biol. 1997;17:4597–610. doi: 10.1128/mcb.17.8.4597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Levkowitz G, Waterman H, Zamir E, Kam Z, Oved S, Langdon WY, et al. c-Cbl/Sli-1 regulates endocytic sorting and ubiquitination of the epidermal growth factor receptor. Genes Dev. 1998;12:3663–74. doi: 10.1101/gad.12.23.3663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bisson SA, Ujack EE, Robbins SM. Isolation and characterization of a novel, transforming allele of the c-Cbl proto-oncogene from a murine macrophage cell line. Oncogene. 2002;21:3677–87. doi: 10.1038/sj.onc.1205510. [DOI] [PubMed] [Google Scholar]
- 25.Blake TJ, Shapiro M, Morse HC, III, Langdon WY. The sequences of the human and mouse c-cbl proto-oncogenes show v-cbl was generated by a large truncation encompassing a proline-rich domain and a leucine zipper-like motif. Oncogene. 1991;6:653–7. [PubMed] [Google Scholar]
- 26.Rathinam C, Thien CB, Flavell RA, Langdon WY. Myeloid leukemia development in c-Cbl RING finger mutant mice is dependent on FLT3 signaling. Cancer Cell. 2010;18:341–52. doi: 10.1016/j.ccr.2010.09.008. [DOI] [PubMed] [Google Scholar]
- 27.Naramura M, Nandwani N, Gu H, Band V, Band H. Rapidly fatal myeloproliferative disorders in mice with deletion of Casitas B-cell lymphoma (Cbl) and Cbl-b in hematopoietic stem cells. Proc Natl Acad Sci U S A. 2010;107:16274–9. doi: 10.1073/pnas.1007575107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Thien CB, Langdon WY. Tyrosine kinase activity of the EGF receptor is enhanced by the expression of oncogenic 70Z-Cbl. Oncogene. 1997;15:2909–19. doi: 10.1038/sj.onc.1201468. [DOI] [PubMed] [Google Scholar]
- 29.Kales SC, Ryan PE, Nau MM, Lipkowitz S. Cbl and human myeloid neoplasms: the Cbl oncogene comes of age. Cancer Res. 2010;70:4789–94. doi: 10.1158/0008-5472.CAN-10-0610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.COSMIC: Catalogue of Somatic Mutations in Cancer [database on the Internet] Cambridge (UK): Wellcome Trust Sanger Institute, Genome Research Limited; c2004. [cited 2014 Oct 19]. Available from: http://www.sanger.ac.uk/genetics/CGP/cosmic/ [Google Scholar]
- 31.Grand FH, Hidalgo-Curtis CE, Ernst T, Zoi K, Zoi C, McGuire C, et al. Frequent CBL mutations associated with 11q acquired uniparental disomy in myeloproliferative neoplasms. Blood. 2009;113:6182–92. doi: 10.1182/blood-2008-12-194548. [DOI] [PubMed] [Google Scholar]
- 32.Loh ML, Sakai DS, Flotho C, Kang M, Fliegauf M, Archambeault S, et al. Mutations in CBL occur frequently in juvenile myelomonocytic leukemia. Blood. 2009;114:1859–63. doi: 10.1182/blood-2009-01-198416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Reindl C, Quentmeier H, Petropoulos K, Greif PA, Benthaus T, Argiropoulos B, et al. CBL exon 8/9 mutants activate the FLT3 pathway and cluster in core binding factor/11q deletion acute myeloid leukemia/myelodysplastic syndrome subtypes. Clin Cancer Res. 2009;15:2238–47. doi: 10.1158/1078-0432.CCR-08-1325. [DOI] [PubMed] [Google Scholar]
- 34.Sanada M, Suzuki T, Shih LY, Otsu M, Kato M, Yamazaki S, et al. Gain-of- function of mutated C-CBL tumour suppressor in myeloid neoplasms. Nature. 2009;460:904–8. doi: 10.1038/nature08240. [DOI] [PubMed] [Google Scholar]
- 35.Abbas S, Rotmans G, Lowenberg B, Valk PJ. Exon 8 splice site mutations in the gene encoding the E3-ligase CBL are associated with core binding factor acute myeloid leukemias. Haematologica. 2008;93:1595–7. doi: 10.3324/haematol.13187. [DOI] [PubMed] [Google Scholar]
- 36.Beer PA, Delhommeau F, Lecouedic JP, Dawson MA, Chen E, Bareford D, et al. Two routes to leukemic transformation following a JAK2 mutation-positive myeloproliferative neoplasm. Blood. 2010;115:2891–900. doi: 10.1182/blood-2009-08-236596. [DOI] [PubMed] [Google Scholar]
- 37.Caligiuri MA, Briesewitz R, Yu J, Wang L, Wei M, Arnoczky KJ, et al. Novel c-CBL and CBL-B ubiquitin ligase mutations in human acute myeloid leukemia. Blood. 2007;110:1022–4. doi: 10.1182/blood-2006-12-061176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dunbar AJ, Gondek LP, O'Keefe CL, Makishima H, Rataul MS, Szpurka H, et al. 250K single nucleotide polymorphism array karyotyping identifies acquired uniparental disomy and homozygous mutations, including novel missense substitutions of c-Cbl, in myeloid malignancies. Cancer Res. 2008;68:10349–57. doi: 10.1158/0008-5472.CAN-08-2754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Makishima H, Cazzolli H, Szpurka H, Dunbar A, Tiu R, Huh J, et al. Mutations of e3 ubiquitin ligase cbl family members constitute a novel common pathogenic lesion in myeloid malignancies. J Clin Oncol. 2009;27:6109–16. doi: 10.1200/JCO.2009.23.7503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Muramatsu H, Makishima H, Jankowska AM, Cazzolli H, O'Keefe C, Yoshida N, et al. Mutations of E3 ubiquitin ligase Cbl family members but not TET2 mutations are pathogenic in juvenile myelomonocytic leukemia. Blood. 2010;115:1969–75. doi: 10.1182/blood-2009-06-226340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sargin B, Choudhary C, Crosetto N, Schmidt MH, Grundler R, Rensinghoff M, et al. Flt3-dependent transformation by inactivating c-Cbl mutations in AML. Blood. 2007;110:1004–12. doi: 10.1182/blood-2007-01-066076. [DOI] [PubMed] [Google Scholar]
- 42.Thien CB, Walker F, Langdon WY. RING finger mutations that abolish c-Cbl-directed polyubiquitination and downregulation of the EGF receptor are insufficient for cell transformation. Mol Cell. 2001;7:355–65. doi: 10.1016/s1097-2765(01)00183-6. [DOI] [PubMed] [Google Scholar]
- 43.Bandi SR, Brandts C, Rensinghoff M, Grundler R, Tickenbrock L, Kohler G, et al. E3 ligase-defective Cbl mutants lead to a generalized mastocytosis and myeloproliferative disease. Blood. 2009;114:4197–208. doi: 10.1182/blood-2008-12-190934. [DOI] [PubMed] [Google Scholar]
- 44.Griffiths EK, Sanchez O, Mill P, Krawczyk C, Hojilla CV, Rubin E, et al. Cbl-3-deficient mice exhibit normal epithelial development. Mol Cell Biol. 2003;23:7708–18. doi: 10.1128/MCB.23.21.7708-7718.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Keane MM, Ettenberg SA, Nau MM, Banerjee P, Cuello M, Penninger J, et al. cbl-3: a new mammalian cbl family protein. Oncogene. 1999;18:3365–75. doi: 10.1038/sj.onc.1202753. [DOI] [PubMed] [Google Scholar]
- 46.Kim M, Tezuka T, Suziki Y, Sugano S, Hirai M, Yamamoto T. Molecular cloning and characterization of a novel cbl-family gene, cbl-c. Gene. 1999;239:145–54. doi: 10.1016/s0378-1119(99)00356-x. [DOI] [PubMed] [Google Scholar]
- 47.Nicholson L, Knight T, Matheson E, Minto L, Case M, Sanichar M, et al. Casitas B lymphoma mutations in childhood acute lymphoblastic leukemia. Genes Chromosomes Cancer. 2012;51:250–6. doi: 10.1002/gcc.20949. [DOI] [PubMed] [Google Scholar]
- 48.McKeller MR, Robetorye RS, Dahia PL, Aguiar RC. Integrity of the CBL gene in mature B-cell malignancies. Blood. 2009;114:4321–2. doi: 10.1182/blood-2009-08-239988. [DOI] [PubMed] [Google Scholar]
- 49.cBio Portal for Cancer Genomics [database on the Internet] Bethesda (MD): The Cancer Genome Atlas Program Office; c2010. cited 2014 Oct 19]. Available from: http://www.cbioportal.org/public-portal/index.do. [Google Scholar]
- 50.Tan YH, Krishnaswamy S, Nandi S, Kanteti R, Vora S, Onel K, et al. CBL is frequently altered in lung cancers: its relationship to mutations in MET and EGFR tyrosine kinases. PLoS One. 2010;5:e8972. doi: 10.1371/journal.pone.0008972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Grunwald MR, Levis MJ. FLT3 inhibitors for acute myeloid leukemia: a review of their efficacy and mechanisms of resistance. Int J Hematol. 2013;97:683–94. doi: 10.1007/s12185-013-1334-8. [DOI] [PubMed] [Google Scholar]
- 52.Taylor SJ, Dagger SA, Thien CB, Wikstrom ME, Langdon WY. Flt3 inhibitor AC220 is a potent therapy in a mouse model of myeloproliferative disease driven by enhanced wild-type Flt3 signaling. Blood. 2012;120:4049–57. doi: 10.1182/blood-2012-06-436675. [DOI] [PubMed] [Google Scholar]
- 53.Kumar EA, Charvet CD, Lokesh GL, Natarajan A. High-throughput fluorescence polarization assay to identify inhibitors of Cbl(TKB)-protein tyrosine kinase interactions. Anal Biochem. 2011;411:254–60. doi: 10.1016/j.ab.2010.11.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kumar EA, Yuan Z, Palermo NY, Dong L, Ahmad G, Lokesh GL, et al. Peptide truncation leads to a twist and an unusual increase in affinity for casitas B-lineage lymphoma tyrosine kinase binding domain. J Med Chem. 2012;55:3583–7. doi: 10.1021/jm300078z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bachmaier K, Krawczyk C, Kozieradzki I, Kong YY, Sasaki T, Oliveira-dos-Santos A, et al. Negative regulation of lymphocyte activation and autoimmunity by the molecular adaptor Cbl-b. Nature. 2000;403:211–6. doi: 10.1038/35003228. [DOI] [PubMed] [Google Scholar]
- 56.Chiang YJ, Kole HK, Brown K, Naramura M, Fukuhara S, Hu RJ, et al. Cbl-b regulates the CD28 dependence of T-cell activation. Nature. 2000;403:216–20. doi: 10.1038/35003235. [DOI] [PubMed] [Google Scholar]
- 57.Murphy MA, Schnall RG, Venter DJ, Barnett L, Bertoncello I, Thien CBF, et al. Tissue hyperplasia and enhanced T-cell signalling via ZAP-70 in c-cbl- deficient mice. Mol Cell Biol. 1998;18:4872–82. doi: 10.1128/mcb.18.8.4872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Naramura M, Kole HK, Hu RJ, Gu H. Altered thymic positive selection and intracellular signals in Cbl-deficient mice. Proc Natl Acad Sci U S A. 1998;95:15547–52. doi: 10.1073/pnas.95.26.15547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Fang D, Liu YC. Proteolysis-independent regulation of PI3K by Cbl-b-mediated ubiquitination in T cells. Nat Immunol. 2001;2:870–5. doi: 10.1038/ni0901-870. [DOI] [PubMed] [Google Scholar]
- 60.Fang D, Wang HY, Fang N, Altman Y, Elly C, Liu YC. Cbl-b, a RING-type E3 ubiquitin ligase, targets phosphatidylinositol 3-kinase for ubiquitination in T cells. J Biol Chem. 2001;276:4872–8. doi: 10.1074/jbc.M008901200. [DOI] [PubMed] [Google Scholar]
- 61.Chiang JY, Jang IK, Hodes R, Gu H. Ablation of Cbl-b provides protection against transplanted and spontaneous tumors. J Clinical Invest. 2007;117:1029–36. doi: 10.1172/JCI29472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Loeser S, Loser K, Bijker MS, Rangachari M, van der Burg SH, Wada T, et al. Spontaneous tumor rejection by cbl-b-deficient CD8+ T cells. J Exp Med. 2007;204:879–91. doi: 10.1084/jem.20061699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Paolino M, Thien CB, Gruber T, Hinterleitner R, Baier G, Langdon WY, et al. Essential role of E3 ubiquitin ligase activity in Cbl-b-regulated T cell functions. J Immunol. 2011;186:2138–47. doi: 10.4049/jimmunol.1003390. [DOI] [PubMed] [Google Scholar]
- 64.Paolino M, Choidas A, Wallner S, Pranjic B, Uribesalgo I, Loeser S, et al. The E3 ligase Cbl-b and TAM receptors regulate cancer metastasis via natural killer cells. Nature. 2014;507:508–12. doi: 10.1038/nature12998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Valverde P. Effects of Gas6 and hydrogen peroxide in Axl ubiquitination and downregulation. Biochem Biophys Res Commun. 2005;333:180–5. doi: 10.1016/j.bbrc.2005.05.086. [DOI] [PubMed] [Google Scholar]
- 66.Stromnes IM, Blattman JN, Tan X, Jeevanjee S, Gu H, Greenberg PD. Abrogating Cbl-b in effector CD8(+) T cells improves the efficacy of adoptive therapy of leukemia in mice. J Clin Invest. 2010;120:3722–34. doi: 10.1172/JCI41991. [DOI] [PMC free article] [PubMed] [Google Scholar]